Abstract
Introduction
Dopamine (DA) is a neurotransmitter that regulates the rewarding and motivational processes underlying food intake and eating behaviors. This study hypothesized associations of DNA methylation signatures at genes modulating DA signaling with obesity features, metabolic profiles, and dietary intake.
Methods
An adult population within the Methyl Epigenome Network Association project was included (n = 473). DNA methylation levels in white blood cells were measured by microarray (450K). Differentially methylated genes were mapped within the dopaminergic synapse pathway using the KEGG reference database (map04728). Subsequently, network enrichment analyses were run in the pathDIP portal. Associations of methylation patterns with anthropometric markers of general (BMI) and abdominal obesity (waist circumference), the blood metabolic profile, and daily dietary intakes were screened.
Results
After applying a correction for multiple comparisons, 12 CpG sites were strongly associated (p < 0.0001) with BMI: cg03489495 (ITPR3), cg22851378 (PPP2R2D), cg04021127 (PPP2R2D), cg22441882 (SLC18A1), cg03045635 (DRD5), cg23341970 (ITPR2), cg13051970 (DDC), cg08943004 (SLC6A3), cg20557710 (CACNA1C), cg24085522 (GNAL), cg16846691 (ITPR2), and cg09691393 (SLC6A3). Moreover, average methylation levels of these genes differed according to the presence or absence of abdominal obesity. Pathway analyses revealed a statistically significant contribution of the aforementioned genes to dopaminergic synapse transmission (p = 4.78E−08). Furthermore, SLC18A1 and SLC6A3 gene methylation signatures correlated with total energy (p < 0.001) and carbohydrate (p < 0.001) intakes.
Conclusions
The results of this investigation reveal that methylation status on DA signaling genes may underlie epigenetic mechanisms contributing to carbohydrate and calorie consumption and fat deposition.
Keywords: diet, dopamine, epigenetics, obesity, SLC18A1, SLC6A3
1. INTRODUCTION
Besides homeostatic processes concerning energy and nutrient metabolic control, eating behavior is also regulated by hedonic (nonhomeostatic) mechanisms (Hernández Ruiz de Eguilaz et al., 2018), which are thought to be driven by the rewarding properties of foods and specific nutritional and behavioral afferent signals (Ziauddeen, Alonso‐Alonso, Hill, Kelley, & Khan, 2015). In this context, it has been reported that similar to alcohol and other drugs of abuse, highly palatable foods (rich in sugars and fat) can trigger neuroadaptive responses in brain reward circuitries (Alonso‐Alonso et al., 2015). These effects can stimulate feeding behavior and related attitudes independent of energy status or overcome other signals of satiety and hunger, contributing to overeating and weight gain (Kenny, 2011). Because of the rising prevalence of obesity and the widespread availability of calorie‐dense foods, understanding the hedonic processes underlying food consumption and behavioral cues beyond metabolic needs has become a priority in obesity research (Stice, Figlewicz, Gosnell, Levine, & Pratt, 2013).
Reward and gratification associated with palatable food consumption are partially mediated by abrupt dopamine (DA) increases in the nucleus accumbens and the ventral tegmental area (Singh, 2014). Moreover, the amount of DA released after consuming a preferred meal eventually correlates with the degree of experienced pleasure (Small, Jones‐Gotman, & Dagher, 2003). Thus, disruption of DA activity can lead to loss of control over intake and continued consumption despite negative consequences, being both behaviors commonly seen in addiction and obesity (Volkow, Wang, Tomasi, & Baler, 2013). Consistently, deficits in mesolimbic DA neurotransmission have been linked to diet‐induced obesity in rats (Geiger et al., 2009). In humans, imaging studies suggest that obese subjects may suffer impairments in dopaminergic pathways involved in reward sensitivity, incentive motivation, conditioning, and control (Volkow, Wang, Fowler, Tomasi, & Baler, 2012). Therefore, some novel strategies in the prevention and treatment of obesity target to manage DA functions (Blum et al., 2018).
Emerging evidences suggest that several genetic and epigenetic factors modulate the relationships between DA signaling, overconsumption, and obesity (Blum, Thanos, & Gold, 2014; Stice, Yokum, Zald, & Dagher, 2011). For instance, polymorphisms near or within key genes regulating dopaminergic synapse, including catechol‐o‐methyltransferase (COMT), D2 receptor (DRD2), and DA active transporter (DAT, SLC6A3) have been associated with altered reward circuitry responsivity related to a spectrum of addictive behaviors (Stice et al., 2011). Moreover, differential DNA methylation patterns at DAT and tyrosine hydroxylase (TH) were linked to altered DA‐related gene expression in response to chronic intake of high‐fat diet in mice (Vucetic, Carlin, Totoki, & Reyes, 2012). Furthermore, a set of transcriptional and epigenetic changes in the hypothalamus of prenatally stressed female rats were implicated in an increased susceptibility to a high‐fat‐sucrose diet (Paternain et al., 2012). This study hypothesized associations of DNA methylation signatures at genes modulating DA signaling with obesity features and accompanying metabolic profiles as well as an epigenetic influence on macronutrient intake.
2. MATERIALS AND METHODS
2.1. Subjects
A transversal nutriepigenomic analysis was conducted in a general adult population within the Methyl Epigenome Network Association (MENA) project (n = 473). The MENA cohort is constituted by previous clinical trials analyzing genome‐environmental interactions concerning weight management and associated metabolic outcomes (Abete et al., 2015; Huerta, Navas‐Carretero, Prieto‐Hontoria, Martínez, & Moreno‐Aliaga, 2015; Larsen et al., 2010; Martínez‐González et al., 2014; Petersen et al., 2006; San‐Cristobal et al., 2015; Santos et al., 2016; Zulet et al., 2011). Each study received ethical approval from appropriate local Human Research Ethics Committees. In addition, all procedures carried out throughout this investigation were in agreement with the ethical principles of the 2013 Helsinki Declaration (World Medical Association, 2013). Also, subject's information was coded to insure full anonymity. All participants gave their informed consent before inclusion in the study.
2.2. Anthropometric measurements and blood pressure
Anthropometric measurements including weight, height, and waist circumference (WC) were collected by trained health personnel using conventional methods (de la Iglesia et al., 2014; Mansego, Milagro, Zulet, & Martinez, 2015). Body mass index (BMI) was calculated dividing weight (kg) by squared height (m2). The World Health Organization (2017) classification of BMI in adults was used to characterize normal weight (BMI 18.5–24.9 kg/m2) and overweight/obese individuals (BMI ≥25 kg/m2). Abdominal obesity (AO) was defined based on established WC cutoffs for men (>102 cm) and women (>88 cm) as reported by the National Cholesterol Education Program (2002). Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were measured from the right arm of each participant with a sphygmomanometer after a 15‐min rest. The average of two successful readings was recorded following the World Health Organization criteria (2004) (Whitworth, & Chalmers, 2004).
2.3. Biochemical tests
Venous blood samples were drawn from each participant by venipuncture after a 12‐hr overnight fast. Glucose, total cholesterol (TC), high‐density lipoprotein cholesterol (HDL‐c), and triglycerides were determined in the automatic analyzer Pentra C200 (HORIBA Medical, Madrid, Spain) with appropriate commercial kits provided by this company. Low‐density lipoprotein cholesterol was calculated using the Friedewald equation: LDL‐c = TC − HDL‐c − triglycerides/5 as described elsewhere (Ramos‐Lopez et al., 2018b). Plasma concentrations of insulin (Mercodia, Uppsala, Sweden) were measured using specific enzyme‐linked immunosorbent assays and assessed by means of an automated analyzer system (Triturus, Grifols, Barcelona, Spain). Insulin resistance was estimated by the homeostatic model assessment‐insulin resistance (HOMA‐IR) index according to the following formula: (fasting insulin (mU/L) × plasma glucose (mmol/L)/22.5) as previously reported (Crujeiras et al., 2014). Triglyceride‐glucose (TyG) index was calculated as: (ln [fasting triglycerides (mg/dl) × fasting plasma glucose (mg/dl)/2]) as described elsewhere (Navarro‐González, Sánchez‐Íñigo, Pastrana‐Delgado, Fernández‐Montero, & Martinez, 2016).
2.4. Dietary assessment
Dietary data were additionally obtained from 247 subjects of the MENA cohort, which presented similar characteristics regarding the whole population. The habitual consumption of 137 food items during the previous year was evaluated with a validated, semiquantitative food frequency questionnaire (de la Fuente‐Arrillaga, Ruiz, Bes‐Rastrollo, Sampson, & Martinez‐González, 2010). Food frequencies (daily, weekly, monthly or never), portions, and serving sizes were computed and further converted to daily energy (kcal) and macronutrient intakes (g) using recognized Spanish food composition tables, as described elsewhere (Goni, Aray, Martínez, & Cuervo, 2016). Nutrients from the diet (carbohydrates, protein, and fat) were adjusted by total energy intake using the residual method, as previously reported (Carraro et al., 2016).
2.5. DNA methylation analyses
Blood samples were centrifuged (2,000 g, at 4°C for 15 min) to isolate white blood cells (WBCs) from whole blood. WBCs were immediately frozen at −80°C in buffy coat until use as described elsewhere (Arpón et al., 2016). Genomic DNA was extracted from WBC using the Master Pure DNA purification kit (Epicentre Biotechnologies, Madison, WI, USA) following instructions provided by the supplier. DNA quality was assessed with the PicoGreen® dsDNA Quantitation Reagent (Invitrogen, Carlsbad, CA, USA). A total of 500 ng of purified DNA was treated with sodium‐bisulfite using the EZ‐96 DNA Methylation Kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer's protocol. Modified DNA samples were whole‐genome amplified and hybridized to Infinium Human Methylation 450K BeadChips (Illumina, San Diego, CA, USA) as detailed elsewhere (Mansego, Garcia‐Lacarte, Milagro, Marti, & Martinez, 2017). The scanning of the samples was carried out with the Illumina HiScanSQ system, and the image intensities were extracted with the GenomeStudio Methylation Software Module, v1.9 (Illumina).
DNA methylation data preprocessing has been recently described (Ramos‐Lopez, Riezu‐Boj, Milagro, & Martinez, 2018c; Ramos‐Lopez et al., 2018a). Briefly, CpG methylation levels were expressed as β values, which are calculated as the ratio between the Illumina methylated probe intensities and the overall probe intensities (sum of methylated and unmethylated probe intensities). β values ranging from 0 (unmethylated) to 1 (completely methylated) were used, as previously reported (Weinhold, Wahl, Pechlivanis, Hoffmann, & Schmid, 2016). Methylation data were peak‐based corrected for type I and type II bias and subsequently normalized using a categorical Subset Quantile Normalization method (Touleimat & Tost, 2012). Probes containing single nucleotide polymorphisms, those hybridizing to multiple genomic locations, or associated with X and Y chromosomes, were removed (Naeem et al., 2014; Nordlund et al., 2013). The ComBat normalization method was applied to adjusting for nonbiological experimental variation (Johnson, Li, & Rabinovic, 2007). Moreover, an additional analysis to estimate the variation explained due to different cell subtypes (granulocytes, monocytes, B cells, T cells‐CD8+, T cells‐CD4+, and natural killer cells) was performed according to the Houseman criteria (Houseman et al., 2012).
2.6. Pathway analyses
To test the hypothesis of this study, differentially methylated genes were mapped to the dopaminergic synapse pathway (map04728) using the online Kyoto Encyclopedia of Genes and Genomes (KEGG) reference database (http://www.genome.jp/kegg/pathway.html). The Pathway Data Integration Portal (pathDIP) platform (http://ophid.utoronto.ca/pathdip/) was used to perform pathway enrichment analyses, with a confidence level of 99%. p value corresponding to KEGG source was then reported.
2.7. Statistical analyses
The Kolmogorov–Smirnov test was used to determine data distribution. All study variables were normally distributed (p > 0.05). Results are expressed as means ± standard deviations (SD), meanwhile, men and women are presented as number of cases. Statistical differences between AO groups were analyzed by student t test (continuous variables) and chi‐square test (dichotomous variables). A linear regression model concerning BMI outcomes was computed using the LIMMA package for R software, which was adjusted by covariates such as age, sex, study cohorts, and DNA methylation chips. The Benjamini–Hochberg correction for multiple comparisons was applied. Statistically significant thresholds were based on False Discovery Rate (FDR) cutoffs (p < 0.05) and B‐statistic values from LIMMA (B > 0). The LIMMA B‐statistic is the log‐odds that a determined gene is differentially methylated. The cutoff B value above 0 implies that a CpG is more likely to be differentially methylated than to not be differentially methylated, giving a reasonable balance of false positives and false negatives (Yang et al., 2011). Best BMI‐associated CpGs were selected according to stricter FDR values (p < 0.0001). Further linear regression analyses adjusted by age and sex were performed to evaluate associations of methylation values at DA signaling genes with anthropometric measurements, the metabolic profile, and dietary intakes. p < 0.05 was considered statistically significant. Statistical analyses were performed in the IBM SPSS software version 20 for Windows (IBM Inc., Armonk, NY, USA). GraphPad Prism® program version 6.0C (La Jolla, CA, USA) was used to graphically illustrate significant correlations.
3. RESULTS
Demographic, anthropometric, and metabolic characteristics as well as dietary intake of the study population categorized by the presence or absence of AO are reported (Table 1). About 82% of the study population presented excessive body weight according to the BMI classification of the World Health Organization (BMI ≥25 kg/m2). Moreover, 57% of the whole sample presented AO based on WC values. No differences between AO groups concerning age and sex were found. Subjects with AO had statistically significant higher levels of blood pressure, insulin, HOMA‐IR, TyG index, and worse lipid profile as well as greater daily dietary consumption of calories, carbohydrates, protein, and fat compared to non‐AO individuals.
Table 1.
Demographic, anthropometric, and metabolic characteristics as well as dietary intake of the study population categorized by the presence or absence of abdominal obesity
| Variable | Non‐AO | AO | p value |
|---|---|---|---|
| n | 205 | 268 | — |
| Age (years) | 46.0 ± 17.7 | 47.8 ± 11.0 | 0.182 |
| Men/women | 83/122 | 87/181 | 0.082 |
| Anthropometric and clinical data | |||
| Weight (kg) | 68.2 ± 10.7 | 91.9 ± 17.8 | <0.001 |
| BMI (kg/m2) | 25.5 ± 3.2 | 33.5 ± 4.6 | <0.001 |
| WC (cm) | 83.1 ± 11.2 | 105.5 ± 11.9 | <0.001 |
| SBP (mmHg) | 121.5 ± 36.4 | 106.8 ± 42.2 | <0.001 |
| DBP (mmHg) | 75.1 ± 22.1 | 89.3 ± 38.2 | <0.001 |
| Metabolic profile | |||
| Glucose (mg/dl) | 99.4 ± 33.4 | 104.4 ± 26.6 | 0.080 |
| Insulin (mIU/L) | 6.8 ± 3.7 | 11.1 ± 7.8 | <0.001 |
| HOMA‐IR index | 1.38 ± 0.84 | 2.99 ± 2.59 | <0.001 |
| TC (mg/dl) | 197.7 ± 40.3 | 210.0 ± 39.6 | 0.001 |
| HDL‐c (mg/dl) | 57.4 ± 13.5 | 50.7 ± 12.8 | <0.001 |
| LDL‐c (mg/dl) | 118.6 ± 38.2 | 134.4 ± 34.2 | <0.001 |
| TG (mg/dl) | 111.4 ± 62.7 | 125.4 ± 77.3 | 0.041 |
| TyG index | 4.57 ± 0.31 | 4.65 ± 0.33 | 0.015 |
| Dietary intake | |||
| Energy (Kcal/day) | 2,373 ± 508 | 2,679 ± 844 | 0.003 |
| Carbohydrates (g/day) | 239.6 ± 68.1 | 272.1 ± 107.3 | 0.013 |
| Protein (g/day) | 93.2 ± 18.6 | 109.5 ± 30.7 | <0.001 |
| Fat (g/day) | 107.3 ± 22.6 | 119.5 ± 41.2 | 0.012 |
Continuous variables are represented as means ± standard deviations. Men and women are number of cases. AO: abdominal obesity; BMI: body mass index; DBP: diastolic blood pressure; HDL‐c: high‐density lipoprotein cholesterol; HOMA‐IR index: homeostatic model assessment‐insulin resistance index; LDL‐c: low‐density lipoprotein cholesterol; SBP: systolic blood pressure; TC: total cholesterol; TG: triglycerides; TyG index: triglyceride‐glucose index; WC: waist circumference. Dietary intake was available from 247 subjects.
Overall, 119 CpG sites at genes integrating the dopaminergic synapse pathway correlated with BMI (kg/m2). Of these, 44 CpGs showed best associations (p < 0.0001). After adjusting by age plus sex and the appropriate correction for multiple comparisons, 12 CpGs at 9 genes remained statistically significant: cg03489495 (ITPR3), cg22851378 (PPP2R2D), cg04021127 (PPP2R2D), cg22441882 (SLC18A1), cg03045635 (DRD5), cg23341970 (ITPR2), cg13051970 (DDC), cg08943004 (SLC6A3), cg20557710 (CACNA1C), cg24085522 (GNAL), cg16846691 (ITPR2), and cg09691393 (SLC6A3). Genomic and statistical data of these CpG sites sorted by FDR values are presented (Table 2). Most of them are located in coding (n = 5) and promoter (n = 4) regions, meanwhile, the rest is mapped within untranslated trailers (n = 3).
Table 2.
Genomic and statistical data of CpG sites at dopamine pathway genes statistically associated with BMI
| CpG_IDa | Illumina_ID | Gene name | Gene symbol | CHR positionb | Genomic region | p value | FDR | B | r 2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | cg03489495 | Inositol 1,4,5‐trisphosphate receptor type 3 | ITPR3 | 6:33,588,875 | Body | 2.5E−14 | 9.2E−11 | 20.82 | 0.105 |
| 2 | cg22851378 | Protein phosphatase 2 regulatory subunit Bdelta | PPP2R2D | 10:133,747,932 | Body | 2.4E−09 | 3.9E−07 | 9.55 | 0.072 |
| 3 | cg04021127 | Protein phosphatase 2 regulatory subunit Bdelta | PPP2R2D | 10:133,747,926 | TSS1500 | 1.6E−08 | 1.6E−06 | 7.67 | 0.059 |
| 4 | cg22441882 | Solute carrier family 18 member A1 | SLC18A1 | 8:20,040,654 | 3′UTR | 3.7E−08 | 2.9E−06 | 6.87 | 0.054 |
| 5 | cg03045635 | Dopamine receptor D5 | DRD5 | 4:9,783,198 | 5′UTR | 2.6E−07 | 1.2E−05 | 4.98 | 0.079 |
| 6 | cg23341970 | Inositol 1,4,5‐trisphosphate receptor type 2 | ITPR2 | 12:26,782,390 | TSS1500 | 4.6E−07 | 1.8E−05 | 4.42 | 0.055 |
| 7 | cg13051970 | Dopa decarboxylase | DDC | 7:50,628,968 | Body | 4.7E−07 | 1.8E−05 | 4.42 | 0.045 |
| 8 | cg08943004 | Solute carrier family 6 member 3 | SLC6A3 | 5:1,416,873 | 1stExon | 4.8E−07 | 1.9E−05 | 4.38 | 0.050 |
| 9 | cg20557710 | Calcium voltage‐gated channel subunit alpha1 C | CACNA1C | 12:2,788,782 | Body | 5.8E−07 | 2.1E−05 | 4.20 | 0.052 |
| 10 | cg24085522 | G protein subunit alpha L | GNAL | 18:11,849,055 | 3′UTR | 7.4E−07 | 2.5E−05 | 3.97 | 0.042 |
| 11 | cg16846691 | Inositol 1,4,5‐trisphosphate receptor type 2 | ITPR2 | 12:26,986,520 | TSS1500 | 2.3E−06 | 5.8E−05 | 2.87 | 0.067 |
| 12 | cg09691393 | Solute carrier family 6 member 3 | SLC6A3 | 5:1,417,003 | TSS1500 | 2.5E−14 | 9.8E−05 | 2.16 | 0.034 |
Data are sorted by FDR values.
B: LIMMA B‐statistic from LIMMA; BMI: body mass index; CHR: chromosome; FDR: False Discovery Rate.
aStudied CpG identifier. bCpG locations were mapped using GRCh37 version of the genome from Ensembl platform.
In a multiple regression model, methylation signatures of the aforementioned 12 CpG sites accounted for about 21% of the variability in BMI (adj. r 2 = 0.207, p < 0.001). Statistically relevant associations between methylation status and BMI are plotted (Figure 1). Of note, seven CpG sites positively correlated with BMI values, whereas in the remaining analyzed CpGs, negative correlations were found (n = 5). Moreover, average methylation levels of each CpG differed according to the presence or absence of AO (Figure 2), with a robust level of significance in most cases (p < 0.0001). No statistically significant relationships between methylation patterns at DA signaling genes with serum levels of glucose, insulin, lipid profile, or blood pressure were detected.
Figure 1.
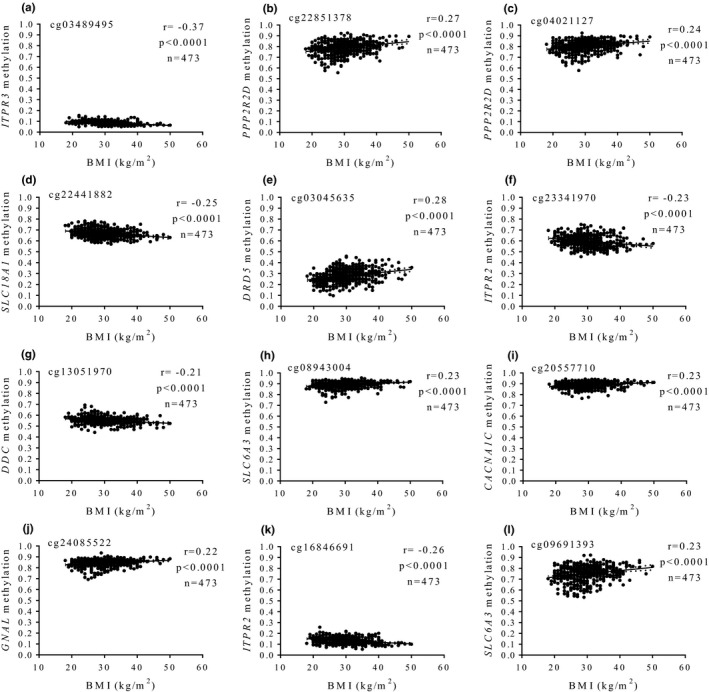
Associations between methylation levels (beta values) at dopamine pathway genes and BMI values. (a) cg03489495, ITPR3, (b) cg22851378, PPP2R2D, (c) cg04021127, PPP2R2D, (d) cg22441882, SLC18A1, (e) cg03045635, DRD5, (f) cg23341970, ITPR2, (g) cg13051970, DDC, (h) cg08943004, SLC6A3, (i) cg20557710, CACNA1C, (j) cg24085522, GNAL, (k) cg16846691, ITPR2, (l) cg09691393, SLC6A3
Figure 2.
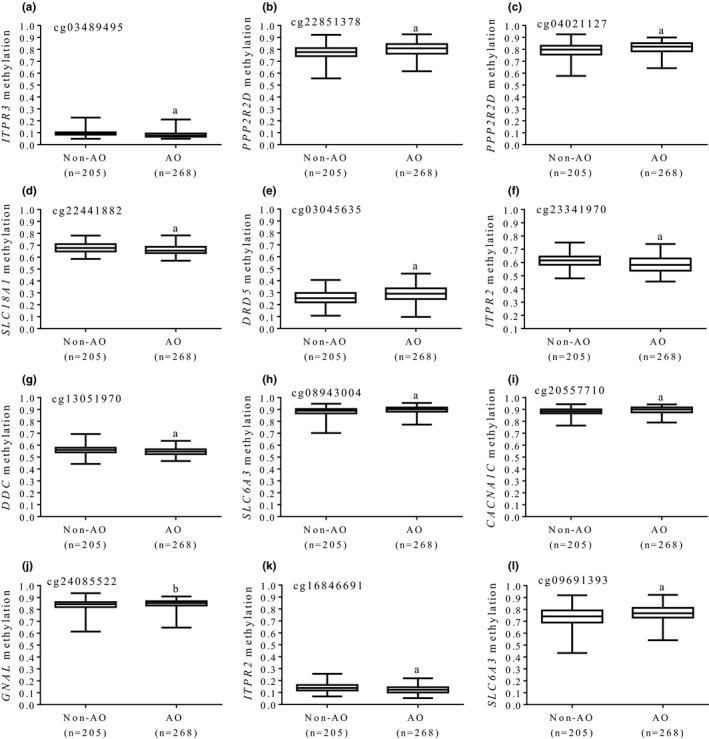
Average methylation levels (beta values) at dopamine pathway genes according to the presence or absence of abdominal obesity. (a) cg03489495, ITPR3, (b) cg22851378, PPP2R2D, (c) cg04021127, PPP2R2D, (d) cg22441882, SLC18A1, (e) cg03045635, DRD5, (f) cg23341970, ITPR2, (g) cg13051970, DDC, (h) cg08943004, SLC6A3, (i) cg20557710, CACNA1C, (j) cg24085522, GNAL, (k) cg16846691, ITPR2, (l) cg09691393, SLC6A3. AO: abdominal obesity. a p < 0.0001; b p < 0.001
Pathway mapping of the BMI‐associated genes within the DA signaling cascade is shown (Figure 3). Interestingly, pathway enrichment analyses revealed a significant contribution of BMI‐associated genes to dopaminergic synapse transmission (p = 4.78E−08), involving complex interactions between presynaptic and postsynaptic cells (Figure 3). These genes modulated key processes involving physiological DA actions such as transport, uptake/reuptake, covalent modifications, and appropriate downstream signal flow.
Figure 3.
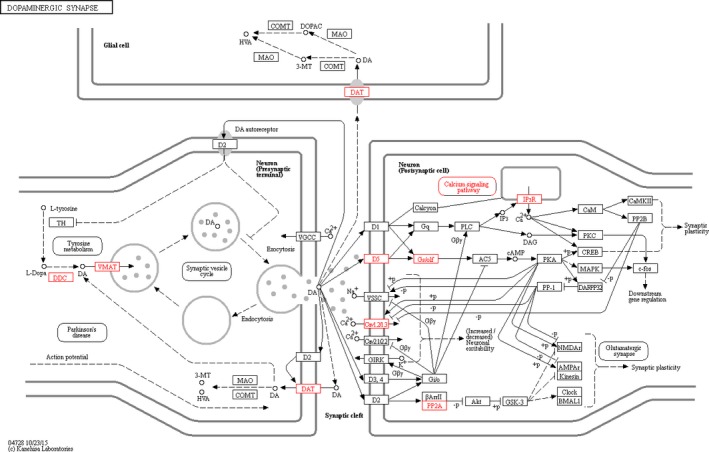
Mapping of BMI‐associated genes within the dopaminergic synapse pathway (red boxes). The following genes were computed: ITPR3, PPP2R2D, SLC18A1, DRD5, ITPR2, DDC, SLC6A3, CACNA1C, GNAL. Figure taken from KEGG reference database (map04728). Pathway enrichment analyses, based on pathDIP (p = 4.78E−08)
Furthermore, potential associations between DA gene methylation profiles and available daily dietary intakes were evaluated in 247 subjects of the MENA cohort (Figure 4). Thus, methylation at cg22441882 (SLC18A1), cg08943004 (SLC6A3), and cg09691393 (SLC6A3) consistently correlated with total energy consumption (p < 0.001) and carbohydrate intake (p < 0.001). However, no relationships between methylation patterns of these CpG sites and protein or fat intakes were found.
Figure 4.
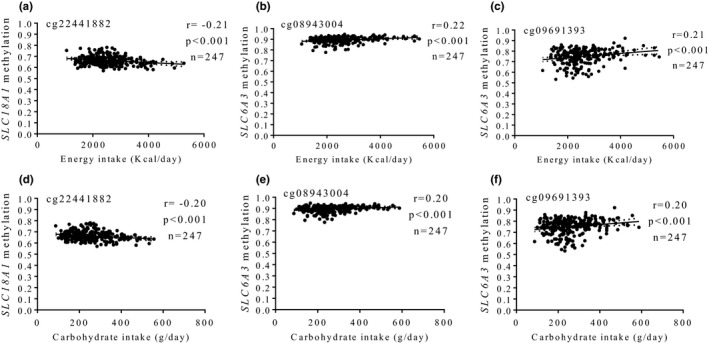
Associations between methylation levels (beta values) at dopamine signaling genes and energy (a–c) and carbohydrate (d–f) intakes. (a, d) cg22441882, SLC18A1 (b, e) cg08943004, SLC6A3 (c, f) cg09691393, SLC6A3. AO: abdominal obesity
4. DISCUSSION
DA is a major (nonhomeostatic) regulator of food intake behaviors (Alonso‐Alonso et al., 2015). In agreement with our hypothesis, the present investigation evidenced associations of DA gene methylation patterns with BMI, AO, and carbohydrate intake, which might serve as epigenetic biomarkers of feeding behavior attitudes, excessive adiposity, and fat deposition. These results are consistent with the fact that disruptions in dopaminergic synapse may lead to overconsumption by altering the rewarding effects elicited by palatable foods (Ziauddeen et al., 2015). In this sense, it has been reported that high‐carbohydrate diets can trigger addictive‐like neurochemical and behavioral responses in vulnerable individuals, contributing to weight gain (Lennerz & Lennerz, 2018). The link between body weight regulation and fat storage and dopaminergic signaling may also rely on the endocrine effects of DA in peripheral tissues such as insulin secretion and specific actions on adipocytes (Rubí & Maechler, 2010). Furthermore, human adipose cells express DA receptors during adipogenesis, suggesting a controlling role of DA in adipose tissue processes (Borcherding et al., 2011).
DA is synthesized through DOPA decarboxylase (DDC) activity and subsequently packed into synaptic vesicles via the SLC18 family of transporter proteins including VMAT1 (SLC18A1) (Lawal & Krantz, 2013). In this study, both DDC and SLC18A1 gene methylation levels negatively correlated with BMI and were downregulated under AO conditions. In addition, a negative correlation between SLC18A1 methylation and carbohydrate intake was found. Interestingly, decreased AADC activity has been reported in obese mice fed a high‐fat high‐simple‐carbohydrate diet (Moreira‐Rodrigues et al., 2012). Moreover, genome wide and candidate gene studies identified SLC18A1 as one potential pleiotropic gene overlapped between mood disorders and cardiometabolic diseases (Amare, Schubert, Klingler‐Hoffmann, Cohen‐Woods, & Baune, 2017). Also, a genetic variation in SLC18A1 made statistically significant contributions to BMI in Chinese subjects (Chen et al., 2013).
Once released from presynaptic axonal terminals, DA interacts with at least five distinct, but closely related G protein‐coupled receptor subtypes (D1 to D5) in the postsynaptic cells, which regulate the physiological actions of DA (Beaulieu, Espinoza, & Gainetdinov, 2015). In particular, the DA receptor D5 (DRD5) belongs to the D1‐class receptors, whose activation stimulates cAMP production by adenylyl cyclase on DA‐receptive cells (Beaulieu et al., 2015). Here, DRD5 methylation levels positively correlated with BMI and differed according to AO. In a previous work, it was shown that peripheral blood mononuclear cells from individuals presenting AO expressed lower DRD5 levels compared to subjects without AO (Leite, Lima, Marino, Cosentino, & Ribeiro, 2016). Furthermore, DRD5 expression negatively correlated with weight, BMI, and WC values, suggesting that AO is associated with downregulation of dopaminergic pathways in blood cells.
The regulation of synaptic and extrasynaptic DA concentrations is an important process that contributes to efficient DA neurotransmission and compartmentalization (Lohr, Masoud, Salahpour, & Miller, 2017). This function is driven by the DA transporter (DAT, SLC6A3), a membrane protein located perisynaptically, where it rapidly recaptures and transports DA from the extracellular space into the cytosol of the presynaptic neuron (Sotnikova, Beaulieu, Gainetdinov, & Caron, 2006). In this study, two CpG sites at SLC6A3 gene correlated with BMI and carbohydrate intake with a positive trend. Consistently, it was reported that hypothalamic SLC6A3 was hypermethylated in the promoter region in response to high‐fat‐sucrose diet in prenatally stressed female adult rats (Paternain et al., 2012). Similarly, a significant increase in DNA methylation within the promoter region of SLC6A3 was found in the ventral tegmental area of mice fed a high‐fat diet, which associated with repressed expression (Vucetic et al., 2012). In humans, methylation changes at the SLC6A3 gene have been related to prematurity, a known risk factor for obesity (Arpón et al., 2018). Also, SLC6A3 gene polymorphisms were associated with palatable food intake and WC in children in early stages of development (Fontana et al., 2015). Additionally, genetic variants in SLC6A3 have been associated with obesity risk in some populations (Bieliński et al., 2017; González‐Giraldo, Trujillo, & Forero, 2017).
Regarding DA‐evoked downstream transducers, different methylation patterns at ITPR3, PPP2R2D, ITPR2, CACNA1C, and GNAL genes were found to be associated with BMI and AO in this research. According to our results, it has been proposed that a mutation in Itpr3 gene could influences food choice by impairing the detection of nutrients in mice (Tordoff, Jaji, Marks, & Ellis, 2012). Likewise, a genetic variant in ITPR3 gene was related to the linking for particular foods in a Silk Road population (Pirastu et al., 2012). Meanwhile, ITPR2 and CACNA1C have been identified as candidate genes associated with addictive tendencies toward food (Pedram, Zhai, Gulliver, Zhang, & Sun, 2017). Of note, CACNA1C methylation levels (a concomitant taste signaling molecule) were previously associated with BMI in an adult population (Ramos‐Lopez et al., 2018a, 2018b). Also, a linkage between ITPR2 locus and central adiposity was reported (Graff et al., 2013; Liu et al., 2014). Until now, there is no evidence showing potential relationships between PPP2R2D and GNAL genes and obesity.
The strengths of this investigation include a relatively large sample analyzed, and the analysis of DNA methylation status at all genes integrating the dopaminergic synapse pathway. In addition, several potential confounding factors were considered in the methylation‐related statistical analyses such as sex, age, study cohorts, methylation chips, cell subtypes, nonbiological experimental variation, as well as multiple comparison correction. On the other hand, a limitation of this investigation was the lack of expression assays, but RNA samples were not available. This drawback makes it difficult to predict the effects on gene expression of methylation signatures and phenotypic impact, especially CpGs located in nonpromoter regions or those with small changes when comparing AO groups. For example, although the mean methylation levels at cg03489495 (ITPR3) statistically differed between non‐AO and AO individuals, it represented approximately a 2% difference in methylation status. Additionally, type I and type II bias cannot be completely ruled out despite of appropriate statistical settings. Of note, some obtained relevant data could have also been lost because of using robust FDR values to select best BMI‐associated CpG sites in the regression analyses.
Another point to comment is the measurement of DNA methylation signatures in peripheral WBC as surrogate of brain cells methylome profiles. Although previous studies support tissue‐specific DNA methylation patterns (Lokk et al., 2014), there is growing evidence in humans suggesting that some methylation marks detected in leukocytes can be reflected in other target tissues, including oral mucosa (San‐Cristobal et al., 2016) and subcutaneous adipose tissue (Crujeiras et al., 2017). Also, homologies between genomic signatures (including DNA methylation patterns) from blood and brain were reported in a rodent model of concussive injury (Meng et al., 2017). Moreover, it has been shown that in addition to human brain, main DA signaling genes are also expressed in circulating human blood cells, including DDC (Kokkinou, Nikolouzou, Hatzimanolis, Fragoulis, & Vassilacopoulou, 2009), SLC18A1 (Amenta et al., 2001), SLC6A3 (Mill, Asherson, Browes, D'Souza, & Craig, 2002), and DRD5 (Leite et al., 2016).
Indeed, epigenetic phenomena are important regulators of genome expression and function, which have an impact on diverse physiological and behavioral processes related to food intake, and energy homeostasis (Milagro, Mansego, De Miguel, & Martínez, 2013). Not surprisingly, many epigenetic mechanisms can be implicated in the development of excessive adiposity and associated metabolic risk, including those affecting DA function (Martínez, Milagro, Claycombe, & Schalinske, 2014). In this context, epigenetic modifications at genes involved in DA signaling transmission may help to explain putative relationships between brain reward circuitries, eating behaviors, and body weight status. This knowledge may also be useful for individual disease risk prediction, the search for therapeutic targets, and the design/implementation of nutriepigenomic strategies aimed to prevention, prognosis, and integral management of obesity and accompanying metabolic complications (Ramos‐Lopez et al., 2017).
In conclusion, the results of this investigation reveal that methylation status of DA signaling genes may be one epigenetic regulator contributing to carbohydrate and calorie consumption and obesity development.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest concerning this research.
ACKNOWLEDGMENTS
Authors thank all the participants of this study and the personnel of the primary health centers. Technical and laboratory support from Asuncion Redin, Laura Olazaran, Iosune Zubieta, Maria Hernandez, Salome Perez, Blanca Martinez de Morentin, Ana Lorente, Veronica Ciaurriz and Maria Zabala is gratefully acknowledged. The MENA Project is constituted by the cohorts PREDIMED, OBEPALIP, GEDYMET, DiOGenes, RESMENA, NUGENOB, Food4Me, and ICTUS, where each of the following researchers in alphabetical order is involved: Alonso A, Arancibia C, Arós F, Astrup A, Babio N, Blázquez V, Bondia‐Pons I, Brennan L, Buil‐Cosiales P, Campión J, Cataldo LR, Celis‐Morales C, Corella D, Covas MI, Dalskov S, Daniel H, De Arce A, de la Iglesia R, Estruch R, Fernández‐Crehuet J, Fiol M, Fitó M, Flores M, Forga L, Galgani J, Gibney ER, Gibney MJ, Gómez‐Úriz AM, González‐Muniesa P, Goyenechea E, Guy‐Grand B, Handjieva‐Darlenska T, Holst C, Huerta AE, Jebb S, Kafatos A, Kunesová M, Lamuela‐Raventós RM, Langin D, Lapetra J, Larsen TM, López De Munain A, López‐Legarrea P, Lovegrove JA, Macdonald I, Manios Y, Martínez‐Zabaleta MT, Mathers JC, Morales M, Muñoz MA, Olmos P, Pedersen O, Petersen M, Pfeiffer A, Pintó X, Pollak F, PREDIMED Investigators, Prieto‐Hontoria PL, Ros E, Rössner S, Ruiz‐Gutierrez V, Salas‐Salvadó J, Saris WH, Serra‐Majem L, Sørensen TI, Sorlí JV, Stich V, Taylor MA, Toledo E, Toubro S, Traczyk I, Valderas JP, van Baak M, Vega J, Verdich C, Walsh M, Yévenes I. Conceptual support from IUNS Task Force ‐ Gene Nutrients Interactions: Knowledge to Action (2017) is also credited.
APPENDIX 1.
1.1.
Other members of the MENA project in alphabetical order are: I Abete, AB Crujeiras, M Cuervo, L Goni, A Marti, MA Martinez‐Gonzalez, MJ Moreno‐Aliaga, S Navas‐Carretero, R San‐Cristobal, JL Santos, and MA Zulet.
Ramos‐Lopez O, Riezu‐Boj JI, Milagro FI, Martinez JA; MENA Project . Dopamine gene methylation patterns are associated with obesity markers and carbohydrate intake. Brain Behav. 2018;8:e01017 10.1002/brb3.1017
Funding information
This investigation was supported by the grants from the Government of Navarra (PT024), CIBERobn (CB12/03/30002), MINECO (AGL2013‐45554‐R), and NUTRIGENIO (AGL2013‐45554‐R). O.R.L. was supported by a 2‐year postdoctoral grant from National Council of Science and Technology, Mexico (CONACyT, Num. CVU. 444175) in collaboration with the PhD Program in Molecular Biology in Medicine, University of Guadalajara, Mexico (CONACyT, PNPC 000091) and the University of Navarra, Spain (LE/97).
Contributor Information
J. Alfredo Martinez, Email: jalfmtz@unav.es.
MENA Project:
I Abete, AB Crujeiras, M Cuervo, L Goni, A Marti, MA Martinez‐Gonzalez, MJ Moreno‐Aliaga, S Navas‐Carretero, R San‐Cristobal, JL Santos, and MA Zulet
REFERENCES
- Abete, I. , Gómez‐Úriz, A. M. , Mansego, M. L. , De Arce, A. , Goyenechea, E. , Blázquez, V. , … Milagro, F. I. (2015). Epigenetic changes in the methylation patterns of KCNQ1 and WT1 after a weight loss intervention program in obese stroke patients. Current Neurovascular Research, 12, 321–333. 10.2174/1567202612666150731110247 [DOI] [PubMed] [Google Scholar]
- Alonso‐Alonso, M. , Woods, S. C. , Pelchat, M. , Grigson, P. S. , Stice, E. , Farooqi, S. , … Beauchamp, G. K. (2015). Food reward system: Current perspectives and future research needs. Nutrition Reviews, 73, 296–307. 10.1093/nutrit/nuv002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amare, A. T. , Schubert, K. O. , Klingler‐Hoffmann, M. , Cohen‐Woods, S. , & Baune, B. T. (2017). The genetic overlap between mood disorders and cardiometabolic diseases: A systematic review of genome wide and candidate gene studies. Translational Psychiatry, 7, e1007 10.1038/tp.2016.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenta, F. , Bronzetti, E. , Cantalamessa, F. , El‐Assouad, D. , Felici, L. , Ricci, A. , & Tayebati, S. K. (2001). Identification of dopamine plasma membrane and vesicular transporters in human peripheral blood lymphocytes. Journal of Neuroimmunology, 117, 133–142. 10.1016/S0165-5728(01)00317-4 [DOI] [PubMed] [Google Scholar]
- Arpón, A. , Milagro, F. I. , Laja, A. , Segura, V. , de Pipaón, M. S. , Riezu‐Boj, J. I. , & Alfredo, Martínez. J. (2018). Methylation changes and pathways affected in preterm birth: A role for SLC6A3 in neurodevelopment. Epigenomics, 10, 91–103. 10.2217/epi-2017-0082 [DOI] [PubMed] [Google Scholar]
- Arpón, A. , Riezu‐Boj, J. I. , Milagro, F. I. , Marti, A. , Razquin, C. , Martínez‐González, M. A. , … Martínez, J. A. (2016). Adherence to Mediterranean diet is associated with methylation changes in inflammation‐related genes in peripheral blood cells. Journal of Physiology and Biochemistry, 73, 445–455. 10.1007/s13105-017-0552-6 [DOI] [PubMed] [Google Scholar]
- Beaulieu, J. M. , Espinoza, S. , & Gainetdinov, R. R. (2015). Dopamine receptors ‐ IUPHAR review 13. British Journal of Pharmacology, 172, 1–23. 10.1111/bph.12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieliński, M. , Jaracz, M. , Lesiewska, N. , Tomaszewska, M. , Sikora, M. , Junik, R. , … Borkowska, A. (2017). Association between COMT Val158Met and DAT1 polymorphisms and depressive symptoms in the obese population. Neuropsychiatric Disease and Treatment, 13, 2221–2229. 10.2147/NDT [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, K. , Thanos, P. K. , & Gold, M. S. (2014). Dopamine and glucose, obesity, and reward deficiency syndrome. Frontiers in Psychology, 5, 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, K. , Thanos, P. K. , Wang, G. J. , Febo, M. , Demetrovics, Z. , Modestino, E. J. , … Gold, M. S. (2018). The food and drug addiction epidemic: Targeting dopamine homeostasis. Current Pharmaceutical Design, 23, 6050–6061. 10.2174/1381612823666170823101713 [DOI] [PubMed] [Google Scholar]
- Borcherding, D. C. , Hugo, E. R. , Idelman, G. , De Silva, A. , Richtand, N. W. , Loftus, J. , & Ben‐Jonathan, N. (2011). Dopamine receptors in human adipocytes: Expression and functions. PLoS ONE, 6, e25537 10.1371/journal.pone.0025537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro, J. C. , Hermsdorff, H. H. , Mansego, M. L. , Zulet, M. Á. , Milagro, F. I. , Bressan, J. , & Martínez, J. A. (2016). Higher fruit intake is related to TNF‐α hypomethylation and better glucose tolerance in healthy subjects. Journal of Nutrigenetics and Nutrigenomics, 9, 95–105. 10.1159/000448101 [DOI] [PubMed] [Google Scholar]
- Chen, C. , Chen, W. , Chen, C. , Moyzis, R. , He, Q. , Lei, X. , … Dong, Q. (2013). Genetic variations in the serotoninergic system contribute to body‐mass index in Chinese adolescents. PLoS ONE, 8, e58717 10.1371/journal.pone.0058717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crujeiras, A. B. , Diaz‐Lagares, A. , Sandoval, J. , Milagro, F. I. , Navas‐Carretero, S. , Carreira, M. C. , … Martinez, J. A. (2017). DNA methylation map in circulating leukocytes mirrors subcutaneous adipose tissue methylation pattern: A genome‐wide analysis from non‐obese and obese patients. Scientific Reports, 7, 41903 10.1038/srep41903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crujeiras, A. B. , Zulet, M. A. , Lopez‐Legarrea, P. , de la Iglesia, R. , Pardo, M. , Carreira, M. C. , … Casanueva, F. F. (2014). Association between circulating irisin levels and the promotion of insulin resistance during the weight maintenance period after a dietary weight‐lowering program in obese patients. Metabolism, 63, 520–531. 10.1016/j.metabol.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Fontana, C. , Vitolo, M. R. , Campagnolo, P. D. , Mattevi, V. S. , Genro, J. P. , & Almeida, S. (2015). DRD4 and SLC6A3 gene polymorphisms are associated with food intake and nutritional status in children in early stages of development. The Journal of Nutritional Biochemistry, 26, 1607–1612. 10.1016/j.jnutbio.2015.07.030 [DOI] [PubMed] [Google Scholar]
- de la Fuente‐Arrillaga, C. , Ruiz, Z. V. , Bes‐Rastrollo, M. , Sampson, L. , & Martinez‐González, M. A. (2010). Reproducibility of an FFQ validated in Spain. Public Health Nutrition, 13, 1364–1372. 10.1017/S1368980009993065 [DOI] [PubMed] [Google Scholar]
- Geiger, B. M. , Haburcak, M. , Avena, N. M. , Moyer, M. C. , Hoebel, B. G. , & Pothos, E. N. (2009). Deficits of mesolimbic dopamine neurotransmission in rat dietary obesity. Neuroscience, 159, 1193–1199. 10.1016/j.neuroscience.2009.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goni, L. , Aray, Miranda. M. , Martínez, J. A. , & Cuervo, M. (2016). Validación de un cuestionario de frecuencia de consumo de grupos de alimentos basado en un sistema de intercambios. Nutrición Hospitalaria, 33, 1391–1399. [DOI] [PubMed] [Google Scholar]
- González‐Giraldo, Y. , Trujillo, M. L. , & Forero, D. A. (2017). Two dopaminergic genes, DRD4 and SLC6A3, are associated with body mass index in a Colombian sample of young adults. Archives of Physiology and Biochemistry, 000, 1–5. 10.1080/13813455.2017.1401643 [DOI] [PubMed] [Google Scholar]
- Graff, M. , Fernández‐Rhodes, L. , Liu, S. , Carlson, C. , Wassertheil‐Smoller, S. , Neuhouser, M. , … North, K. E. (2013). Generalization of adiposity genetic loci to US Hispanic women. Nutrition & Diabetes, 3, e85 10.1038/nutd.2013.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández Ruiz de Eguilaz, M. , Martínez de Morentin Aldabe, B. , Almiron‐Roig, E. , Pérez‐Diez, S. , San Cristóbal Blanco, R. , Navas‐Carretero, S. , & Martínez, J. A. (2018). Multisensory influence on eating behavior: Hedonic consumption. Endocrinología, Diabetes y Nutrición, 65, 114–125. [DOI] [PubMed] [Google Scholar]
- Houseman, E. A. , Accomando, W. P. , Koestler, D. C. , Christensen, B. C. , Marsit, C. J. , Nelson, H. H. , … Kelsey, K. T. (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta, A. E. , Navas‐Carretero, S. , Prieto‐Hontoria, P. L. , Martínez, J. A. , & Moreno‐Aliaga, M. J. (2015). Effects of α‐lipoic acid and eicosapentaenoic acid in overweight and obese women during weight loss. Obesity (Silver Spring), 23, 313–321. 10.1002/oby.20966 [DOI] [PubMed] [Google Scholar]
- de la Iglesia, R. , Lopez‐Legarrea, P. , Crujeiras, A. B. , Pardo, M. , Casanueva, F. F. , Zulet, M. A. , & Martinez, J. A. (2014). Plasma irisin depletion under energy restriction is associated with improvements in lipid profile in metabolic syndrome patients. Clinical Endocrinology, 81, 306–311. 10.1111/cen.12383 [DOI] [PubMed] [Google Scholar]
- Johnson, W. E. , Li, C. , & Rabinovic, A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Kenny, P. J. (2011). Reward mechanisms in obesity: New insights and future directions. Neuron, 69, 664–679. 10.1016/j.neuron.2011.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkinou, I. , Nikolouzou, E. , Hatzimanolis, A. , Fragoulis, E. G. , & Vassilacopoulou, D. (2009). Expression of enzymatically active L‐DOPA decarboxylase in human peripheral leukocytes. Blood Cells, Molecules and Diseases, 42, 92–98. 10.1016/j.bcmd.2008.10.010 [DOI] [PubMed] [Google Scholar]
- Larsen, T. M. , Dalskov, S. , van Baak, M. , Jebb, S. , Kafatos, A. , Pfeiffer, A. , … Astrup, A. (2010). The diet, obesity and genes (diogenes) dietary study in eight European countries ‐ a comprehensive design for long‐term intervention. Obesity Reviews, 11, 76–91. 10.1111/j.1467-789X.2009.00603.x [DOI] [PubMed] [Google Scholar]
- Lawal, H. O. , & Krantz, D. E. (2013). SLC18: Vesicular neurotransmitter transporters for monoamines and acetylcholine. Molecular Aspects of Medicine, 34, 360–372. 10.1016/j.mam.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, F. , Lima, M. , Marino, F. , Cosentino, M. , & Ribeiro, L. (2016). Dopaminergic receptors and tyrosine hydroxylase expression in peripheral blood mononuclear cells: A distinct pattern in central obesity. PLoS ONE, 11, e0147483 10.1371/journal.pone.0147483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennerz, B. , & Lennerz, J. K. (2018). Food addiction, high‐glycemic‐index carbohydrates, and obesity. Clinical Chemistry, 64, 64–71. 10.1373/clinchem.2017.273532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. T. , Buchkovich, M. L. , Winkler, T. W. , Heid, I. M. , African Ancestry Anthropometry Genetics Consortium , GIANT Consortium , … Adrienne, C. L. (2014). Multi‐ethnic fine‐mapping of 14 central adiposity loci. Human Molecular Genetics, 23, 4738–4744. 10.1093/hmg/ddu183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, K. M. , Masoud, S. T. , Salahpour, A. , & Miller, G. W. (2017). Membrane transporters as mediators of synaptic dopamine dynamics: Implications for disease. European Journal of Neuroscience, 45, 20–33. 10.1111/ejn.13357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokk, K. , Modhukur, V. , Rajashekar, B. , Märtens, K. , Mägi, R. , Kolde, R. , … Tõnisson, N. (2014). DNA methylome profiling of human tissues identifies global and tissue‐specific methylation patterns. Genome Biology, 15, r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansego, M. L. , Garcia‐Lacarte, M. , Milagro, F. I. , Marti, A. , Martinez, J. A. , & GENOI Members . (2017). DNA methylation of miRNA coding sequences putatively associated with childhood obesity. Pediatric Obesity, 12, 19–27. 10.1111/ijpo.12101 [DOI] [PubMed] [Google Scholar]
- Mansego, M. L. , Milagro, F. I. , Zulet, M. A. , & Martinez, J. A. (2015). SH2B1 CpG‐SNP is associated with body weight reduction in obese subjects following a dietary restriction program. Annals of Nutrition and Metabolism, 66, 1–9. 10.1159/000368425 [DOI] [PubMed] [Google Scholar]
- Martínez, J. A. , Milagro, F. I. , Claycombe, K. J. , & Schalinske, K. L. (2014). Epigenetics in adipose tissue, obesity, weight loss, and diabetes. Advances in Nutrition, 5, 71–81. 10.3945/an.113.004705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐González, M. Á. , Toledo, E. , Arós, F. , Fiol, M. , Corella, D. , Salas‐Salvadó, J. , … PREDIMED Investigators . (2014). Extravirgin olive oil consumption reduces risk of atrial fibrillation: The PREDIMED (Prevención con Dieta Mediterránea) trial. Circulation, 130, 18–26. 10.1161/CIRCULATIONAHA.113.006921 [DOI] [PubMed] [Google Scholar]
- Meng, Q. , Zhuang, Y. , Ying, Z. , Agrawal, R. , Yang, X. , & Gomez‐Pinilla, F. (2017). Traumatic brain injury induces genome‐wide transcriptomic, methylomic, and network perturbations in brain and blood predicting neurological disorders. EBioMedicine, 16, 184–194. 10.1016/j.ebiom.2017.01.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milagro, F. I. , Mansego, M. L. , De Miguel, C. , & Martínez, J. A. (2013). Dietary factors, epigenetic modifications and obesity outcomes: Progresses and perspectives. Molecular Aspects of Medicine, 34, 782–812. 10.1016/j.mam.2012.06.010 [DOI] [PubMed] [Google Scholar]
- Mill, J. , Asherson, P. , Browes, C. , D'Souza, U. , & Craig, I. (2002). Expression of the dopamine transporter gene is regulated by the 3′ UTR VNTR: Evidence from brain and lymphocytes using quantitative RT‐PCR. American Journal of Medical Genetics, 114, 975–979. 10.1002/(ISSN)1096-8628 [DOI] [PubMed] [Google Scholar]
- Moreira‐Rodrigues, M. , Quelhas‐Santos, J. , Roncon‐Albuquerque, R. , Serrão, P. , Leite‐Moreira, A. , Sampaio‐Maia, B. , & Pestana, M. (2012). Blunted renal dopaminergic system in a mouse model of diet‐induced obesity. Experimental Biology and Medicine, 237, 949–955. 10.1258/ebm.2012.012077 [DOI] [PubMed] [Google Scholar]
- Naeem, H. , Wong, N. C. , Chatterton, Z. , Hong, M. K. , Pedersen, J. S. , Corcoran, N. M. , … Macintyre, G. (2014). Reducing the risk of false discovery enabling identification of biologically significant genome‐wide methylation status using the HumanMethylation450 array. BMC Genomics, 15, 51 10.1186/1471-2164-15-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) (2002). Third Report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) final report. Circulation, 106, 3143–3421. [PubMed] [Google Scholar]
- Navarro‐González, D. , Sánchez‐Íñigo, L. , Pastrana‐Delgado, J. , Fernández‐Montero, A. , & Martinez, J. A. (2016). Triglyceride‐glucose index (TyG index) in comparison with fasting plasma glucose improved diabetes prediction in patients with normal fasting glucose: The Vascular‐Metabolic CUN cohort. Preventive Medicine, 86, 99–105. 10.1016/j.ypmed.2016.01.022 [DOI] [PubMed] [Google Scholar]
- Nordlund, J. , Bäcklin, C. L. , Wahlberg, P. , Busche, S. , Berglund, E. C. , Eloranta, M. L. , … Syvänen, A. C. (2013). Genome‐wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biology, 14, r105 10.1186/gb-2013-14-9-r105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternain, L. , Batlle, M. A. , De la Garza, A. L. , Milagro, F. I. , Martínez, J. A. , & Campión, J. (2012). Transcriptomic and epigenetic changes in the hypothalamus are involved in an increased susceptibility to a high‐fat‐sucrose diet in prenatally stressed female rats. Neuroendocrinology, 96, 249–260. 10.1159/000341684 [DOI] [PubMed] [Google Scholar]
- Pedram, P. , Zhai, G. , Gulliver, W. , Zhang, H. , & Sun, G. (2017). Two novel candidate genes identified in adults from the Newfoundland population with addictive tendencies towards food. Appetite, 115, 71–79. 10.1016/j.appet.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Petersen, M. , Taylor, M. A. , Saris, W. H. , Verdich, C. , Toubro, S. , Macdonald, I. , … Astrup, A. (2006). Randomized, multi‐center trial of two hypo‐energetic diets in obese subjects: High‐ versus low‐fat content. International Journal of Obesity, 30, 552–560. 10.1038/sj.ijo.0803186 [DOI] [PubMed] [Google Scholar]
- Pirastu, N. , Robino, A. , Lanzara, C. , Athanasakis, E. , Esposito, L. , Tepper, B. J. , & Gasparini, P. (2012). Genetics of food preferences: A first view from silk road populations. Journal of Food Science, 77, S413–S418. 10.1111/j.1750-3841.2012.02852.x [DOI] [PubMed] [Google Scholar]
- Ramos‐Lopez, O. , Arpón, A. , Riezu‐Boj, J. I. , Milagro, F. I. , Mansego, M. L. , Martinez, J. A. , & MENA project . (2018a). DNA methylation patterns at sweet taste transducing genes are associated with BMI and carbohydrate intake in an adult population. Appetite, 120, 230–239. 10.1016/j.appet.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Ramos‐Lopez, O. , Milagro, F. I. , Allayee, H. , Chmurzynska, A. , Choi, M. S. , Curi, R. , … Martínez, J. A. (2017). Guide for current nutrigenetic, nutrigenomic, and nutriepigenetic approaches for precision nutrition involving the prevention and management of chronic diseases associated with obesity. Journal of Nutrigenetics and Nutrigenomics, 10, 43–62. 10.1159/000477729 [DOI] [PubMed] [Google Scholar]
- Ramos‐Lopez, O. , Riezu‐Boj, J. I. , Milagro, F. I. , Goni, L. , Cuervo, M. , & Martinez, J. A. (2018b). Differential lipid metabolism outcomes associated with ADRB2 gene polymorphisms in response to two dietary interventions in overweight/obese subjects. Nutrition, Metabolism and Cardiovascular Diseases, 28, 165–172. 10.1016/j.numecd.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Ramos‐Lopez, O. , Riezu‐Boj, J. I. , Milagro, F. I. , Martinez, J. A. , & MENA Project . (2018c). DNA methylation signatures at endoplasmic reticulum stress genes are associated with adiposity and insulin resistance. Molecular Genetics and Metabolism, 123, 50–58. 10.1016/j.ymgme.2017.11.011 [DOI] [PubMed] [Google Scholar]
- Rubí, B. , & Maechler, P. (2010). Minireview: New roles for peripheral dopamine on metabolic control and tumor growth: Let's seek the balance. Endocrinology, 151, 5570–5581. 10.1210/en.2010-0745 [DOI] [PubMed] [Google Scholar]
- San‐Cristobal, R. , Navas‐Carretero, S. , Celis‐Morales, C. , Brennan, L. , Walsh, M. , Lovegrove, J. A. , … Martinez, J. A. (2015). Analysis of dietary pattern impact on weight status for personalised nutrition through on‐line advice: The Food4Me Spanish Cohort. Nutrients, 7, 9523–9537. 10.3390/nu7115482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San‐Cristobal, R. , Navas‐Carretero, S. , Milagro, F. I. , Riezu‐Boj, J. I. , Guruceaga, E. , Celis‐Morales, C. , … Martinez, J. A. (2016). Gene methylation parallelisms between peripheral blood cells and oral mucosa samples in relation to overweight. Journal of Physiology and Biochemistry, 73, 465–474. 10.1007/s13105-017-0560-6 [DOI] [PubMed] [Google Scholar]
- Santos, J. L. , Yévenes, I. , Cataldo, L. R. , Morales, M. , Galgani, J. , Arancibia, C. , … Pollak, F. (2016). Development and assessment of the disposition index based on the oral glucose tolerance test in subjects with different glycaemic status. Journal of Physiology and Biochemistry, 72, 121–131. 10.1007/s13105-015-0458-0 [DOI] [PubMed] [Google Scholar]
- Singh, M. (2014). Mood, food, and obesity. Frontiers in Psychology, 5, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, D. M. , Jones‐Gotman, M. , & Dagher, A. (2003). Feeding‐induced dopamine release in dorsal striatum correlates with meal pleasantness ratings in healthy human volunteers. NeuroImage, 19, 1709–1715. 10.1016/S1053-8119(03)00253-2 [DOI] [PubMed] [Google Scholar]
- Sotnikova, T. D. , Beaulieu, J. M. , Gainetdinov, R. R. , & Caron, M. G. (2006). Molecular biology, pharmacology and functional role of the plasma membrane dopamine transporter. CNS & Neurological Disorders‐Drug Targets, 5, 45–56. [DOI] [PubMed] [Google Scholar]
- Stice, E. , Figlewicz, D. P. , Gosnell, B. A. , Levine, A. S. , & Pratt, W. E. (2013). The contribution of brain reward circuits to the obesity epidemic. Neuroscience & Biobehavioral Reviews, 37, 2047–2058. 10.1016/j.neubiorev.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stice, E. , Yokum, S. , Zald, D. , & Dagher, A. (2011). Dopamine‐based reward circuitry responsivity, genetics, and overeating. Current Topics in Behavioral Neurosciences, 6, 81–93. [DOI] [PubMed] [Google Scholar]
- Tordoff, M. G. , Jaji, S. A. , Marks, J. M. , & Ellis, H. T. (2012). Macronutrient choice of BTBR.NZW mice congenic for a 21‐gene region of chromosome 17. Physiology & Behavior, 106, 556–561. 10.1016/j.physbeh.2012.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touleimat, N. , & Tost, J. (2012). Complete pipeline for Infinium(®) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics, 4, 325–341. 10.2217/epi.12.21 [DOI] [PubMed] [Google Scholar]
- Volkow, N. D. , Wang, G. J. , Fowler, J. S. , Tomasi, D. , & Baler, R. (2012). Food and drug reward: Overlapping circuits in human obesity and addiction. Current Topics in Behavioral Neurosciences, 11, 1–24. [DOI] [PubMed] [Google Scholar]
- Volkow, N. D. , Wang, G. J. , Tomasi, D. , & Baler, R. D. (2013). Obesity and addiction: Neurobiological overlaps. Obesity Reviews, 14, 2–18. 10.1111/j.1467-789X.2012.01031.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic, Z. , Carlin, J. L. , Totoki, K. , & Reyes, T. M. (2012). Epigenetic dysregulation of the dopamine system in diet‐induced obesity. Journal of Neurochemistry, 120, 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold, L. , Wahl, S. , Pechlivanis, S. , Hoffmann, P. , & Schmid, M. (2016). A statistical model for the analysis of beta values in DNA methylation studies. BMC Bioinformatics, 17, 480 10.1186/s12859-016-1347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth, J. A. , & Chalmers, J. (2004). World health organisation‐international society of hypertension (WHO/ISH) hypertension guidelines. Clinical and Experimental Hypertension, 26, 747–752. 10.1081/CEH-200032152 [DOI] [PubMed] [Google Scholar]
- World Health Organization . (2017). Fact sheet: Obesity and overweight. Available from: http://www.who.int/mediacentre/factsheets/fs311/en/
- World Medical Association . (2013). World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. Journal of the American Medical Association, 310, 2191–2194. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Zhang, K. , Dai, W. , He, X. , Zhao, Q. , Wang, J. , & Sun, Z. S. (2011). Systematic evaluation of genome‐wide methylated DNA enrichment using a CpG island array. BMC Genomics, 12, 10 10.1186/1471-2164-12-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziauddeen, H. , Alonso‐Alonso, M. , Hill, J. O. , Kelley, M. , & Khan, N. A. (2015). Obesity and the neurocognitive basis of food reward and the control of intake. Advances in Nutrition, 6, 474–486. 10.3945/an.115.008268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulet, M. A. , Bondia‐Pons, I. , Abete, I. , de la Iglesia, R. , López‐Legarrea, P. , Forga, L. , … Martínez, J. A. (2011). The reduction of the metabolyc syndrome in Navarra‐Spain (RESMENA‐S) study: A multidisciplinary strategy based on chrononutrition and nutritional education, together with dietetic and psychological control. Nutrición Hospitalaria, 26, 16–26. [PubMed] [Google Scholar]


