Abstract.
Enteric coinfections among children in low-income countries are very common, but it is not well known if specific pathogen combinations are associated or have clinical importance. In this analysis, feces samples from children in Rwanda and Zanzibar less than 5 years of age, with (N = 994) or without (N = 324) acute diarrhea, were analyzed by real-time polymerase chain reaction targeting a wide range of pathogens. Associations were investigated by comparing co-detection and mono-detection frequencies for all pairwise pathogen combinations. More than one pathogen was detected in 840 samples (65%). A negative association (coinfections being less common than expected from probability) was observed for rotavirus in combination with Shigella, Campylobacter, or norovirus genogroup II, but only in patients, which is statistically expected for agents that independently cause diarrhea. A positive correlation was observed, in both patients and controls, between Ct (threshold cycle) values for certain virulence factor genes in enteropathogenic Escherichia coli (EPEC) (eae and bfpA) and toxin genes in enterotoxigenic E. coli (eltB and estA), allowing estimation of how often these genes were present in the same bacteria. A significant positive association in patients only was observed for Shigella and EPEC-eae, suggesting that this coinfection might interact in a manner that enhances symptoms. Although interaction between pathogens that affect symptoms is rare, this work emphasizes the importance and difference in interpretation of coinfections depending on whether they are positively or negatively associated.
INTRODUCTION
Infectious diarrhea can be caused by a wide range of viruses, bacteria, and protozoa and remains a major cause of childhood morbidity and mortality in low-income countries, in particular, among children less than 5 years of age.1 Formerly, bacterial infections were mainly identified by culture, protozoa by microscopy, and some viruses (rotavirus and adenovirus) by antigen tests. Molecular methods such as polymerase chain reaction (PCR) have improved the detection of viruses and other pathogens, and the recent development of multiplex assays has allowed parallel identification of a large number of enteropathogens.2–6 Although it has been known from earlier studies using older techniques that coinfections with two or more enteric pathogens are common, the application of molecular techniques has revealed even higher rates.1,2,7–10 This raises the question whether more than one agent contributes to the symptoms and multiple infections represent a diagnostic challenge due to the difficulty in identifying the causative agent. Detection of coinfections by molecular diagnostic methods allows more sensitive studies of whether microbes are more likely or less likely to occur together,11–14 but large data sets are required to evaluate more rare pathogen combinations. In the present study, we have merged data from two recent studies in Rwanda and Zanzibar to elucidate the importance of enteric coinfections in 1,318 children with or without diarrhea.
MATERIALS AND METHODS
Samples.
The samples were collected as 1 mL of feces or as rectal swabs from children less than 5.0 years of age with or without diarrhea as part of clinical studies conducted in Rwanda (2009–2012; N = 988) or Zanzibar (2011; N = 330), the clinical data from which they have been reported previously.2,7,15,16 The inclusion criteria for patients (N = 994) were diarrhea with a duration of < 96 hours (with or without vomiting or fever), whereas the controls (N = 324) had absence of diarrhea, vomiting, or fever for > 10 days before inclusion. The median age was 13.5 months for patients, of whom 43% were girls and 24.0 months for healthy controls, of whom 51% were girls. Adenovirus was not tested in the samples from 2009 to 2010 (in 658 patients and 324 controls) and enteropathogenic Escherichia coli (EPEC)-bfpA and EPEC-eae were not tested in samples from Zanzibar (165 patients and 165 controls). The number of patients and controls from Rwanda and Zanzibar, as well as their mean ages, is summarized in Supplemental Table 1.
Nucleic acid extraction and real-time PCR.
Approximately 250 µL of feces were dissolved in 4.5 mL of saline and centrifuged for 5 minutes at 750 × g. Then, 250 µL of the dissolved feces or 250 µL of the rectal swab were mixed with 2 mL of lysis buffer, and this volume was used for extraction of total nucleic acid in an EasyMag instrument (BioMérieux, Marcy-l’Étoile, France). The nucleic acids were eluted in 110 µL and 5 µL of this volume was used in each PCR.
Real-time PCR was performed in an ABI 7900 384-well system (Applied Biosystems, Foster City, CA) in nine parallel reactions targeting adenovirus, astrovirus, norovirus genotype I (GI) or genotype II (GII), rotavirus, sapovirus, Campylobacter jejuni, Cryptosporidium parvum/Cryptosporidium hominis, enterotoxigenic E. coli (ETEC) coding for heat-labile toxin (eltB) or heat-stable toxin (estA), EPEC coding for intimin (eae) or bundle-forming pilus (bfpA), Salmonella, and Shigella. The sequences of the primers and probe have been previously described.2,17
Amplifications were performed in 20-µL reactions containing oligonucleotides and Taqman Fast Virus 1-step Master Mix (ABI, for RNA targets [Applied Biosystems]) or Universal Master Mix (ABI, for DNA targets [Applied Biosystems]). After a reverse transcription step at 46°C for 30 minutes followed by 10 minutes of denaturation at 95°C, 45 cycles of two-step PCR were performed (15 seconds at 95°C and 60 seconds at 56°C). In each run, plasmids containing the target regions for all agents were amplified in parallel with patient specimens to verify the performance of each target PCR.
The results from real-time PCR were recorded as Ct (threshold cycle) values, which is essentially the cycle in which the fluorescence from the probe reaches certain intensity. The assessment of Ct values was carried out in a standardized manner by the same person in all runs. The Ct values were used as a proxy for viral load in some of the correlation analyses.
Statistics.
Three analytical approaches were used. First, we tested whether any combination of two pathogens was more or less frequent than expected from their detection rates alone. These analyses were performed separately for patients and controls and were presented both as correlations in a heat map illustrating Pearson rho and as odds ratios (ORs) in tables, with statistical significance assessed by Fisher’s exact test. In addition, correlations between Ct values were investigated for each pathogen pair.
Second, interactions between coinfection and symptoms were evaluated by logistic regression analysis for all pairwise pathogen combinations with the presence of symptoms (i.e., patient versus control) as a dependent variable and detection (yes or no) of the two pathogens as an independent variable, and with an interaction term of the two.
Third, associations between coinfection and the presence or severity of certain symptoms were analyzed to further explore the potential pathogenic importance of coinfection: coinfection and mono-infections were compared, for each pair of pathogens, as regards the presence of vomiting or dehydration (Fisher’s exact test), as well as body temperature, frequency of diarrhea, or degree of dehydration (Mann–Whitney rank sum test).
Ethics.
The studies were conducted in accordance with the Declaration of Helsinki and approved by the ethical committee at the National University in Rwanda, by the Zanzibar Medical Research Ethics Committee (ID: ZAMREC/0001/April/010) and by the regional ethical board in Gothenburg (ID:052-08), Sweden. Written informed consent was obtained from an accompanying caretaker and was recorded on a consent form for each child included in the studies.
RESULTS
Overall detection rates and frequency of multiple infections.
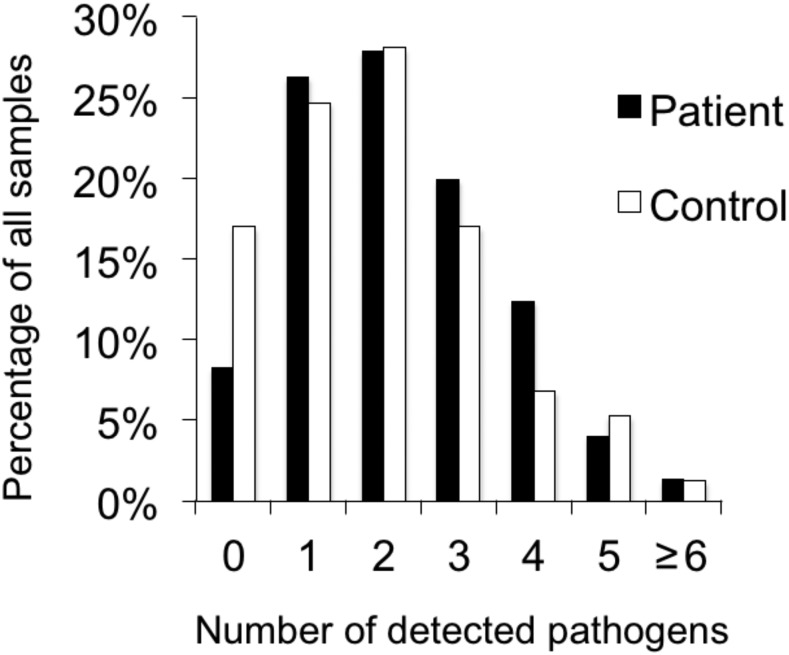
The proportions of children with none, one (N = 341), or more than one (N = 840) detected agents are shown in Figure 1. The number of patients and controls with mono- or coinfection is presented in Supplemental Table 1 and details for all pathogen combinations, in patients and controls, are presented in Supplemental Table 2.
Figure 1.
Number of pathogens detected by real-time polymerase chain reaction in patients and controls.
Pathogens associated with symptomatic infection.
The detection frequencies and Ct values for the merged data set are presented in Supplemental Table 3, showing that rotavirus, norovirus GII, astrovirus, Cryptosporidium, and EPEC-bfpA were detected more frequently in patients than in controls. The Ct values were significantly lower (i.e., microbial loads were higher) in patients than in controls for Shigella and ETEC-estA, and to a lesser extent also for ETEC-eltB, Cryptosporidium, and Campylobacter. Overall, these findings reflect what has been reported before, separately for children in Rwanda and Zanzibar.2,7,15
Positive association between detected pathogens.
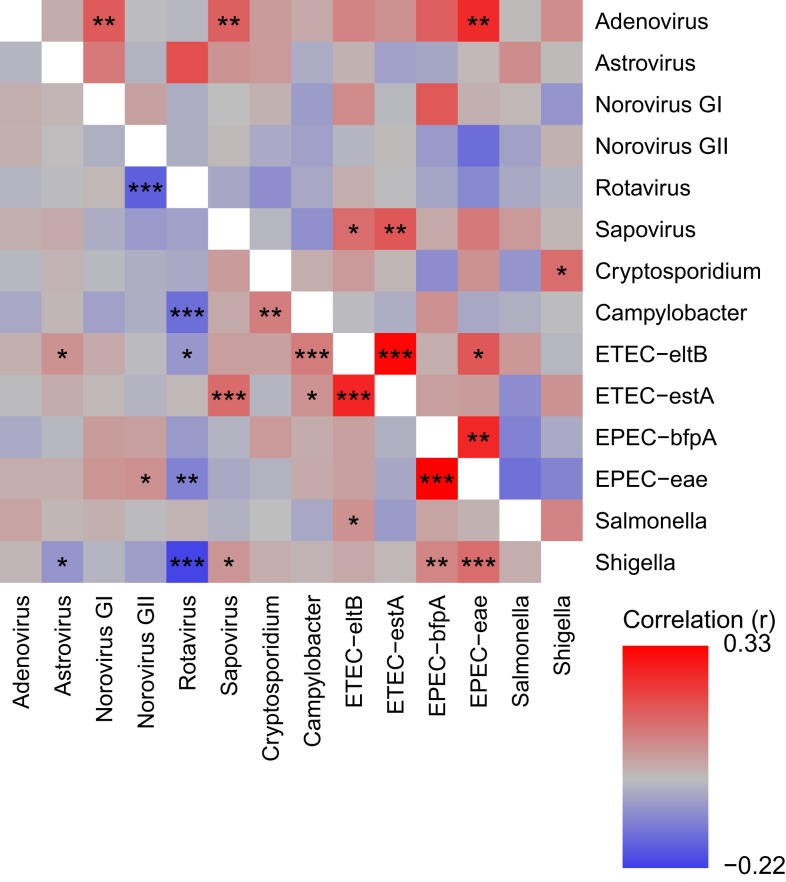
Three pathogen target pairs showed strong positive associations in both patients and controls, as indicated by a red color in the heat map in Figure 2 (with P values by Fisher’s exact test indicated by asterisks; the corresponding frequencies and OR are shown in Supplemental Table 2). In particular, the combinations between EPEC-eae and EPEC-bfpA and between ETEC-estA and ETEC-eltB were much more frequent than expected from their mono-infection rates, with 84 samples positive for both EPEC-eae and EPEC-bfpA and 153 samples being positive for both ETEC-estA and ETEC-eltB. There was also a significant association between ETEC-estA and sapovirus, with 31 samples positive for both.
Figure 2.
Heat map showing degree of correlation for all pathogen combinations, comparing their detection as coinfection vs. mono-infection. The color represents the correlation coefficient (r), as indicated in the legend in the lower right corner. Correlations in patients are shown in the lower left part and those in controls in the upper right part. Statistical significance is indicated by asterisks: *P < 0.05, **P < 0.01, and ***P < 0.001. The corresponding details (counts and P values) are described in Supplemental Table 2.
As shown in Figure 2, positive associations only in patients were observed for the following combinations: Campylobacter and ETEC-eltB, Cryptosporidium and Campylobacter, Shigella and EPEC-eae, and Shigella and EPEC-bfpA.
Positive correlation between Ct values.
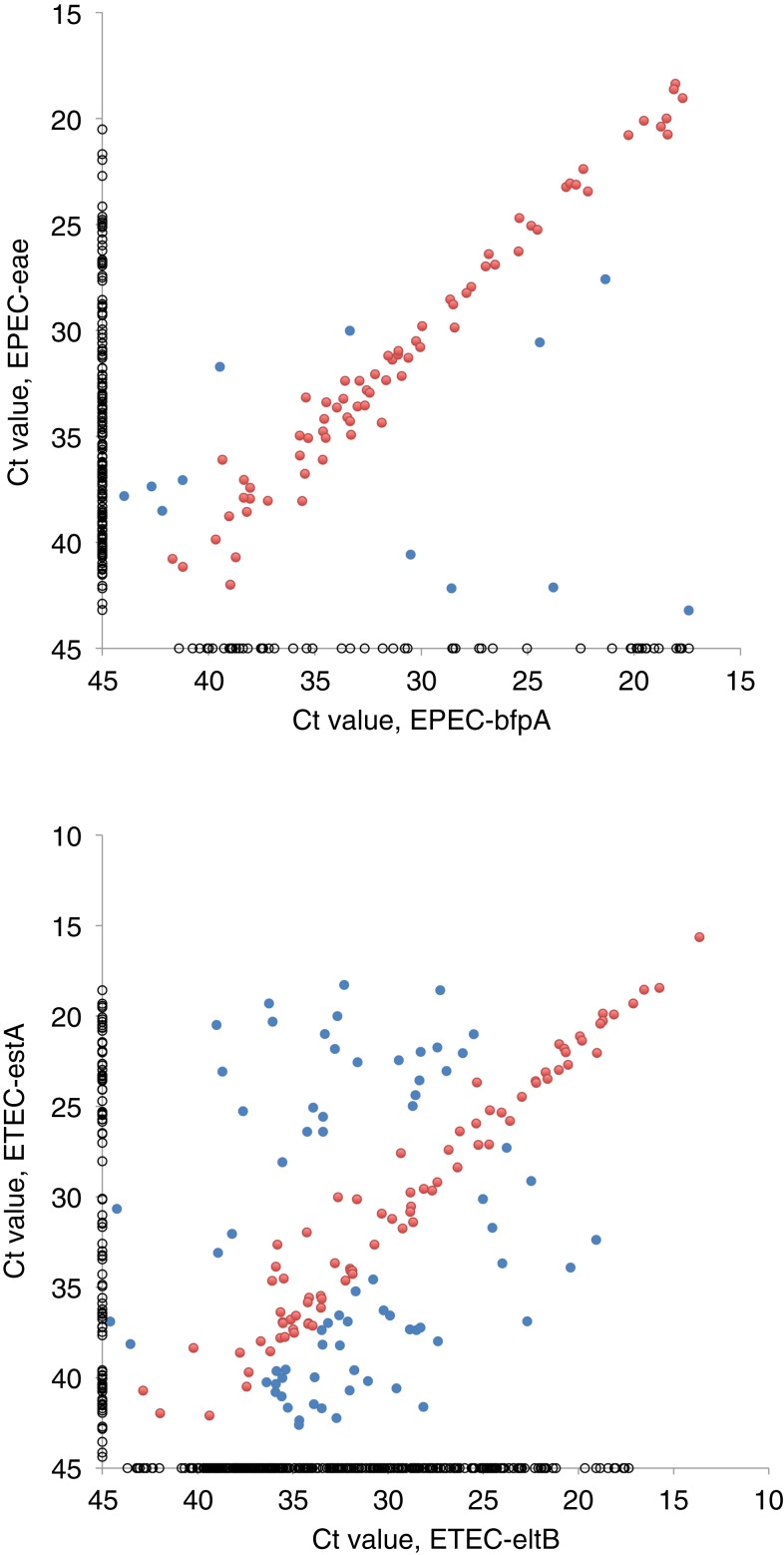
When Ct values were compared in a pairwise manner, significant correlations were only observed for two target pairs, one being EPEC-eae and EPEC-bfpA and the other ETEC-estA and ETEC-eltB. As shown in Figure 3, the Ct values were similar (< 3.3 cycles difference; corresponding to < 1 log10) for these pairs in many samples, suggesting that the target genes were likely present in the same bacterial strains: 72 samples with bfpA and eae and 82 samples with eltB and estA. In other cases (12 samples with bfpA and eae and 71 samples with eltB and estA), a greater difference in Ct indicated that the target genes were present in separate strains, each carrying only one of them. This was also the case in the relatively large number of samples that were positive for only one of the targets (303 eltB, 94 estA, 54 bfpA, and 177 eae, shown as Ct = 45 in Figure 3). Taken together, these observations indicate that 22% of EPEC were “typical,” that is, carried both eae and bfpA genes, whereas 58% had only eae and 20% only bfpA genes, and that 13% of ETEC carried both eltB and estA genes, whereas 60% had only eltB and 27% only estA genes.
Figure 3.
Threshold cycle (Ct) values for 84 samples that were positive for enteropathogenic E. coli (EPEC)-eae and EPEC-bfpA (upper) and for 153 samples that were positive for enterotoxigenic E. coli (ETEC)-eltB and ETEC-estA (lower). Pearson’s correlation coefficient (ρ) was 0.79 for eae and bfpA, and 0.61 for eltB and estA (P < 0.0001 for both). Red dots represent samples with less than 3.3 cycles difference in Ct value, suggesting that the target genes were present in the same bacterial strain (72 with bfpA and eae; 82 with eltB and estA). Samples with ≥ 3.3 cycles difference are shown as blue dots (12 bfpA and eae; 71 eltB and estA). Fifty-four samples that were positive for EPEC-bfpA alone, 177 for EPEC-eae alone, 303 for ETEC-eltB alone, and 94 for ETEC-estA alone were given a Ct value of 45 and are shown as black circles.
Negative association between detected pathogens.
Microbes with the capacity of causing diarrhea on their own should, from a statistical point of view, be negatively associated in patients, that is, in children with diarrhea. Such negative associations were indeed observed for some pathogens, as shown in Figure 2. All these negative associations included rotavirus as one of the agents, in combination with Shigella, norovirus GII, or Campylobacter.
Impact of coinfection on symptoms.
Logistic regression was performed with the presence of symptoms (i.e., patient versus control) as a dependent variable and detection of the two pathogens (yes or no) as an independent variable. This analysis indicated a positive interaction for two pathogen combinations: Shigella and EPEC-eae (OR = 4.85, P = 0.01), and norovirus GII and EPEC-eae (OR = 5.57, P = 0.006). The effects of coinfection could not be evaluated properly for rotavirus or norovirus GII because of very low counts among healthy controls.
The potential impact by coinfection on the severity of symptoms was further evaluated in 829 patients from Rwanda, for whom clinical information about symptoms had been recorded. We compared whether vomiting, frequency of diarrhea, or degree of dehydration was more frequent or more pronounced in cases with coinfections than in mono-infection for each pair of pathogens. Coinfection with Shigella and EPEC-eae was associated with an increased frequency of diarrhea (by ≈ 1/day) as compared with mono-infections of either of the two pathogens (P < 0.001, Mann–Whitney rank sum test).
DISCUSSION
It is known from numerous reports that infection with several gastroenteric agents is common among children in low-income countries, but the clinical importance of polymicrobial infection is uncertain and studies of potential associations between the agents are scarce.11,14 For infections spreading independent of each other, one would—on statistical grounds—predict that agents that alone may cause diarrhea would be negatively associated (i.e., be less common together than one would expect from their mono-infection rates) among patients but not among controls. Positive associations in patients only would be anticipated if a combined infection induced more severe symptoms. Positive associations in both patients and controls might, for example, appear for pathogens whose transmission or ability to infect was dependent on the same environmental or host factors or if the target genes were present in the same pathogen.
In the present study of associations between gastroenteric agents in feces samples from 994 children with diarrhea and from 324 healthy controls were studied. Significant negative associations were observed only among children with diarrhea and only for a few pairs of pathogens: rotavirus and Shigella, rotavirus and Campylobacter, and rotavirus and norovirus GII. These four agents were (together with ETEC-estA and Cryptosporidium) the pathogens associated with symptomatic infection in the present data set (as previously reported2,7,15), as well as in the global enteric multicenter study.12 This finding was, as mentioned, expected (from a statistical point of view) and indicates that negative associations might serve to identify pathogens that cause diarrhea even when a control group is lacking, similar to the strategy of case-only studies in genetics. Interestingly, there was no negative association between rotavirus and ETEC-estA, despite their strong association with symptomatic infection. This might be explained if coinfection with these two agents aggravated symptoms because this would result in a positive association that opposed the expected negative association between their detection. Additional studies are needed to clarify potential associations between rotavirus and ETEC-estA.
Positive associations in both patients and controls were observed for pairs of target genes in ETEC and EPEC, and correlations of Ct values allowed calculation of the proportion of ETEC and EPEC that carried both virulence genes in a manner that, to our knowledge, has not been described before. The gene coding for intimin (eae) is located to the locus of enterocyte effacement in the E. coli chromosome and the gene coding for bundle-forming pilus (bfpA) to the EPEC adherence factor plasmid.18 In the present study, 22% of all EPEC strains were deduced, on the basis of similar Ct values, to carry both genes, 20% only the bfpA gene and 58% only the eae gene. Overall, strains carrying both eae and bfpA genes (“typical EPEC”) were found in 7.7% of patients and 5.0% of controls, which is in line with previous reports.19
Likewise, there was a relatively strong correlation between Ct values for ETEC-eltB and ETEC-estA among the 153 samples that were positive for both targets. The genes encoding eltB and estA are usually located to plasmids that may or may not be present together. The proportion of ETEC that presented with similar Ct values for eltB and estA was 13%, suggesting that these genes were present in the same bacteria in a relatively small proportion of ETEC strains. Taking Ct values into account, 28% of all the 1,318 samples contained strains that were deduced to carry ETEC-eltB only, 12% ETEC-estA only, and 6.2% to carry both toxin genes. The overall detection frequency for ETEC is higher than that in most previous studies, which probably can be explained by the high sensitivity of PCR.20,21 The use of Ct values to estimate the proportion of samples with typical EPEC or with ETEC producing both toxins is, to our knowledge, novel and represents a strategy to avoid that the transition from traditional to molecular methods leads to loss of important information.9 We believe that this approach should be of value in future studies of EPEC and ETEC infections.
Positive associations in patients only were observed for Campylobacter and ETEC-eltB, Campylobacter and Cryptosporidium, Shigella and EPEC-eae, and for Shigella and EPEC-bfpA, suggesting that these combinations might potentiate symptoms. Similarly, infections with EPEC-eae in combination with Shigella or norovirus GII were more likely to be symptomatic when analyzed by a different approach, comparing the patient/control ratio for coinfection with the corresponding mono-infection. These associations should, however, be interpreted with caution because this study was not primarily designed to compare coinfection in patients and controls, as these groups differed significantly in age and were not sufficiently well matched in time and area either. This limitation in design should, however, not influence the results of analysis within patients only, for each pathogen pair, and of whether coinfections increased symptoms. The finding that severe dehydration was more frequent in patients coinfected by Shigella and EPEC-eae supports that these agents indeed might have synergistic effects on symptoms. This potential positive interaction need to be confirmed and explained in future studies, and such effects by other combinations also need to be further investigated because the number of cases was very low for many pathogen combinations.
As mentioned, few previous studies have analyzed the frequency and importance of coinfections. In a study from India that included 2,748 patients with diarrhea, there was a very strong association between rotavirus and Vibrio cholerae14 and significant negative associations were also seen between V. cholerae and Cryptosporidium, adenovirus, Shigella, ETEC, or Vibrio parahaemolyticus and between rotavirus and Shigella. These negative associations were, however, not pointed out as support for a causal role, which probably would be the correct interpretation. In a recent study of coinfections, negative associations were not described,22 whereas in another, negative associations between rotavirus and norovirus or diarrheagenic E. coli in Chinese patients with diarrhea were considered unexpected and difficult to interpret.23 We observed negative associations only in patients and only for pathogens that previously have been strongly associated with symptomatic infection. We want to point out that such negative associations are actually statistically expected and a reflection of the capacity of these agents to cause diarrhea on their own. This interpretation, which has been overlooked in previous reports, needs to be emphasized to promote correct conclusions in future studies.
Supplementary Material
Acknowledgments:
This study was supported by grants from Swedish International Development Cooperation Agency (SIDA), and by the ACT Consortium through an award from the Bill & Melinda Gates Foundation to the London School of Hygiene and Tropical Medicine.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.Kotloff KL, et al. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382: 209–222. [DOI] [PubMed] [Google Scholar]
- 2.Elfving K, Andersson M, Msellem MI, Welinder-Olsson C, Petzold M, Bjorkman A, Trollfors B, Martensson A, Lindh M, 2014. Real-time PCR threshold cycle cutoffs help to identify agents causing acute childhood diarrhea in Zanzibar. J Clin Microbiol 52: 916–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, et al. 2013. A laboratory-developed TaqMan array card for simultaneous detection of 19 enteropathogens. J Clin Microbiol 51: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, et al. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14: 716–724. [DOI] [PubMed] [Google Scholar]
- 5.Wolffs PF, Bruggeman CA, van Well GT, van Loo IH, 2011. Replacing traditional diagnostics of fecal viral pathogens by a comprehensive panel of real-time PCRs. J Clin Microbiol 49: 1926–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kharat VB, Ahmed M, Jiang Z-D, Riddle MS, DuPont HL, 2017. Colonization factors in enterotoxigenic Escherichia coli strains in travelers to Mexico, Guatemala, and India compared with children in Houston, Texas. Am J Trop Med Hyg 96: 83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabayiza JC, Andersson ME, Nilsson S, Bergstrom T, Muhirwa G, Lindh M, 2014. Real-time PCR identification of agents causing diarrhea in Rwandan children less than 5 years of age. Pediatr Infect Dis J 33: 1037–1042. [DOI] [PubMed] [Google Scholar]
- 8.Sinha A, et al. 2013. Culture-independent real-time PCR reveals extensive polymicrobial infections in hospitalized diarrhoea cases in Kolkata, India. Clin Microbiol Infect 19: 173–180. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, et al. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keddy KH, Smith AM, Page NA, 2016. GEMS extend understanding of childhood diarrhoea. Lancet 388: 1252–1254. [DOI] [PubMed] [Google Scholar]
- 11.Bhavnani D, Goldstick JE, Cevallos W, Trueba G, Eisenberg JN, 2012. Synergistic effects between rotavirus and coinfecting pathogens on diarrheal disease: evidence from a community-based study in northwestern Ecuador. Am J Epidemiol 176: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotloff KL, et al. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382: 209–222. [DOI] [PubMed] [Google Scholar]
- 13.Lindsay B, et al. 2015. Microbiota that affect risk for shigellosis in children in low-income countries. Emerg Infect Dis 21: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay B, Ramamurthy T, Sen Gupta S, Takeda Y, Rajendran K, Nair GB, Stine OC, 2011. Diarrheagenic pathogens in polymicrobial infections. Emerg Infect Dis 17: 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elfving K, et al. 2016. Acute uncomplicated febrile illness in children aged 2–59 months in Zanzibar—aetiologies, antibiotic treatment and outcome. PLoS One 11: e0146054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabayiza JC, Andersson ME, Nilsson S, Baribwira C, Muhirwa G, Bergstrom T, Lindh M, 2014. Diarrhoeagenic microbes by real-time PCR in Rwandan children under 5 years of age with acute gastroenteritis. Clin Microbiol Infect 20: O1128–O1135. [DOI] [PubMed] [Google Scholar]
- 17.Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergstrom T, Muhirwa G, Lindh M, 2013. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect Dis 13: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS, 2007. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol 189: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochoa TJ, et al. 2009. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from periurban areas in Lima, Peru. Clin Infect Dis 49: 1694–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansour A, et al. 2014. Pathogenicity and phenotypic characterization of enterotoxigenic Escherichia coli isolates from a birth cohort of children in rural Egypt. J Clin Microbiol 52: 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qadri F, Svennerholm AM, Faruque AS, Sack RB, 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18: 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li LL, Liu N, Humphries EM, Yu JM, Li S, Lindsay BR, Stine OC, Duan ZJ, 2016. Aetiology of diarrhoeal disease and evaluation of viral-bacterial coinfection in children under 5 years old in China: a matched case-control study. Clin Microbiol Infect 22: 381.e9–381.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang SX, et al. 2016. Impact of co-infections with enteric pathogens on children suffering from acute diarrhea in southwest China. Infect Dis Poverty 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.