Abstract.
Alphaviruses (Togaviridae) are arboviruses frequently associated with emerging infectious diseases. In this study, we aimed to investigate the presence of alphaviruses in Uruguay by detecting the viral genome in mosquitoes and neutralizing antibodies in equines. A total of 3,575 mosquitoes were analyzed for alphavirus genome detection. Serologic studies were performed on 425 horse sera by plaque reduction neutralization test (PRNT80) against Venezuelan equine encephalitis virus (VEEV) subtype IAB, Pixuna virus (PIXV), Rio Negro virus (RNV), western equine encephalitis virus (WEEV), and Madariaga virus (MADV). Mosquitoes belonging to six genera were captured and 82.9% were identified as Culex pipiens. Two Cx. pipiens pools collected in Fray Bentos and Las Toscas localities were alphavirus positive, and phylogenetic analyses showed that the sequences grouped into two different clusters: the lineage I of eastern equine encephalitis virus and RNV (VEEV complex), respectively. Plaque reduction neutralization test assays showed antibodies against strains of the VEEV complex, MADV, and WEEV. Rio Negro virus was the most geographically widespread virus, showing higher seroprevalences (up to 20%). Seroprevalences against VEEV IAB ranged between 4.6% and 13%; antibodies against PIXV, WEEV, and MADV were less frequent (3–4%). In conclusion, RNV exhibited the highest seroprevalence in horses, a wide geographical distribution, and viral genome was detected in Cx. pipiens mosquitoes. Madariaga virus had a low seroprevalence in equines, but an epizootic lineage typical of North America was detected in Cx. pipiens mosquitoes. Taken together, our results show that alphaviruses are present in Uruguay with variable occurrence and geographical distribution being a potential threat for human and equine health.
INTRODUCTION
Alphaviruses (Togaviridae) are arthropod-borne viruses, comprising 31 viral species and at least eight serogroups.1,2 They are enveloped, single-stranded positive RNA viruses. Their genome is 11–12 kilobases in size and encodes five structural (C, E3, E2, 6K, and E1) and four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4).
Alphaviruses are maintained in nature in enzootic or epizootic cycles between mosquitoes as vectors and vertebrates as amplifying hosts. Humans and other animals are usually dead-end hosts, where infections may lead to disease. Enzootic and epizootic cycles differ in the vectors and/or the vertebrate amplifying hosts involved; besides, enzootic strains may become epizootic, which is likely mediated through mutation and subsequent adaptation.3,4 In humans, most of the infections are asymptomatic and disease severity may range from a febrile illness to a more severe syndrome such as arthritis or encephalitis. Ecological, geographical, and climate changes, coupled with their intrinsic genetic variability and adaptability, often affect the ecological niche of alphaviruses, making them ideal emerging or reemerging viruses. Recent examples are the emergence of chikungunya and Mayaro viruses (MAYV) in Central and South America or the reemergence of Madariaga virus (MADV) in Central America.5–9
The earliest report on alphavirus circulation in Uruguay dates from 1970. In that study, a serological survey performed by complement fixation and hemagglutination inhibition tests for eastern equine encephalitis virus (EEEV), western equine encephalitis virus (WEEV), MAYV, and Mucambo virus (MUCV) showed antibodies only for WEEV and EEEV, ranging between 3.3–6% in children and 0.9–2% in adults.10 In 1972–1973, Argentina and Uruguay experienced equine epizooties caused by WEEV and a virus isolation from a sick horse was reported.11 Since then, no study was carried out until 2011 in Uruguay, when the first human case of a fatal encephalitis caused by WEEV was described.12
To gain insight into the actual prevalence of alphaviruses in Uruguay, we investigated the presence of alphavirus genome in mosquitoes and performed a serological survey in horses.
MATERIALS AND METHODS
Area of study.
Uruguay is located at 30°–35° (South latitude) and 53°–58° (West longitude) with a total surface of 176.215 km2. It is divided administratively into 19 departments (Figure 1). It has a temperate climate with four clearly delineated seasons, similar rainfall levels throughout the year, and an annual average temperature of 17.5°C.
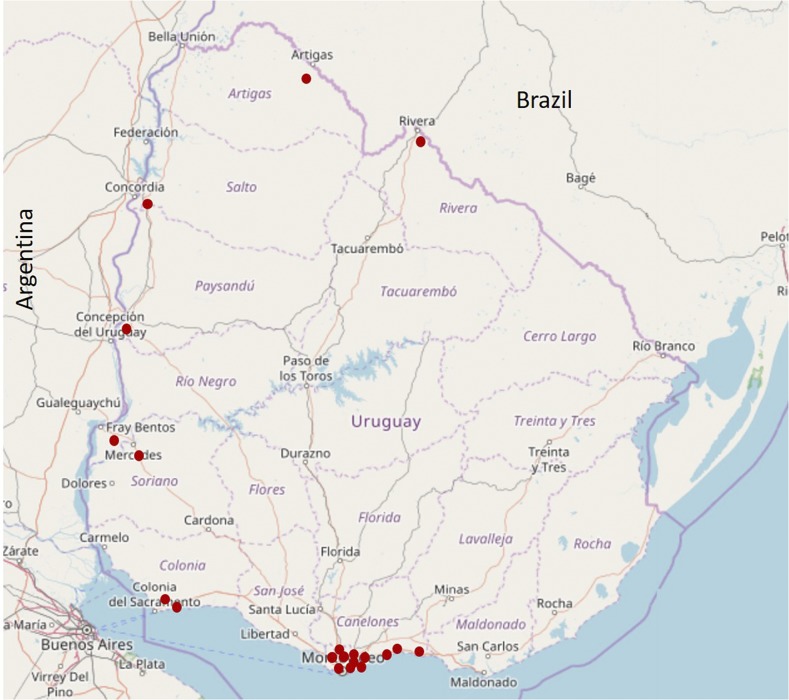
Figure 1.
Mosquito trapping sites. Mosquito traps were placed in nine of the 19 departments of Uruguay between 2006 and 2014. The map was built with the free software QMapShack v1.9.1 and edited in Microsoft PowerPoint v2016. This figure appears in color at www.ajtmh.org.
Molecular detection of alphavirus in mosquito pools.
Mosquito capture.
Mosquitoes were collected at rural, urban, and/or suburban areas at the Artigas, Canelones, Colonia, Montevideo, Paysandú, Rio Negro, Rivera, Salto, and Soriano departments (Figure 1). Captures were made mostly in public land or under permission of the owner when performed in private properties. Mosquito collections were made with Center for Disease Control (CDC) traps (models 512 and 712; The John W. Hock Company, Gainesville, Florida) supplemented with CO2 or using manual aspirator, during summer, spring, and autumn seasons. Traps were settled and remained active in the evening until the next morning. Collected mosquitoes were transported alive to the laboratory under cold conditions and classified into groups between 1 and 50 individuals by locality, species, gender, and engorged/not engorged females. Mosquito species were classified following Darsie keys.13 Pools were stored at −80°C until processing. Each pool was individually triturated using polypropylene mortars (pellet pestles; Sigma-Aldrich®, St. Louis, MI) with 1 mL of minimum essential medium (MEM-E) (Eagle Minimal Essential Medium; Gibco® or Invitrogen®, Thermo Fisher, Waltham, MA). Each homogenate was centrifuged for 15 minutes at 240 × g at 4°C for clarification and the supernatant was stored at −80°C until RNA extraction.
RNA extraction.
Total RNA of each homogenized mosquito pool was extracted using TRIZOL® (Invitrogen BRL; Life Technologies, Rockville, MD) according to the manufacturer’s instructions. Briefly, 200 µL of each homogenate was mixed with 750 µL of TRIzol®, 200 µL chloroform, and 0.5 µL glycogen (Qiagen Valencia, CA) and vortexed for 2 minutes. After a 20-minute incubation at room temperature, the mix was centrifuged for 15 minutes at 4°C and 10,600 × g. The aqueous phase was precipitated with isopropanol (overnight, −20°C). After 20 minutes of centrifugation at 4°C and 10,600 × g, the pellet obtained was washed with 70% ethanol, dried, and suspended in 35 µL of RNase-free water and stored at −80°C.
Generic nsP4 amplification by reverse transcription (RT) and nested polymerase chain reaction (PCR) (Pan-alphavirus RT-nested PCR).
A 195-base pair fragment of the nsP4 gene of alphaviruses was amplified by RT and nested polymerase chain reaction (nested PCR) assay according to Sánchez-Seco et al.14 Reverse transcription and the first round of PCR were performed using SuperScript™ One Step RT-PCR kit with Platinum® Taq polymerase (Invitrogen) in a PCR Sprint Thermal Cycler (Thermo Electron Corporation). Briefly, 5 µL of total RNA was mixed with 40 pmol of each specific primer Alpha 1+ (5′-GAYGCITAYYTIGAYATGGTIGAIGG-3′) and Alpha 1− (5′-CKYTCYTCIGTRTGYTTIGTICCIGG-3′), 2.4 mM MgSO4, 0.4 mM of each dNTP, and 1 µL of SuperScript II™/Platinum® Taq (Invitrogen) to a final volume of 50 µL. Cycle conditions were as follows: one cycle of 45°C for 30 minutes and 94°C for 2 minutes, followed by 40 cycles of 94°C/30 seconds, 52°C/1 minute, and 68°C/30 seconds. A final extension at 68°C/5 minutes was performed. The second PCR round was performed in a final volume of 50 µL using 1 µL of the first round mixed with 1.5 mM of MgCl2, 0.2 mM of each dNTPs, 5 U/µL of Taq DNA polymerase (Invitrogen), and 40 pmol of each primer: Alpha 2+ (5′-GIAAYTGYAAYGTIACICARATG-3′) and Alpha 2− (5′-GCRAAIARIGCIGCIGCYTYIGGICC-3′). Cycle conditions were as follows: 94°C for 2 minutes, followed by 40 cycles of 94°C/1 minute, 52°C/1 minute, and 72°C/30 seconds with a final extension of 72°C/5 minutes. The positive control used was total RNA extracted from an isolate of the Venezuelan equine encephalitis virus (VEEV) prototype strain TC-83 and sterile RNAse-free water was used as the negative control in every assay. Visualization of the PCR products was performed by electrophoresis in a 1.5% agarose gel and stained with 1 µg/mL ethidium bromide.
Sequencing and phylogenetic analysis.
PCR products of alphavirus positive samples were purified with the QIAquick Gel Extraction Kit (Qiagen®). Direct nucleotide sequencing (plus and minus strands) was conducted using 5 pmol of Alpha 2+ and Alpha2− primers at Macrogen, Inc., Korea. Sequences were reviewed and edited with BioEdit v7.2.5 software15 and compared with GenBank database sequences, using nucleotide Basic Local Alignment Search Tool16; nsP4 sequence alignments were constructed using ClustalW.17 Sequences of the positive samples and representative sequences of the Alphavirus genus retrieved from GenBank were included to build the alignments. To estimate the most suitable model of nucleotide substitution, Modelgenerator v0.85 software was used.18 Phylogenetic reconstruction was performed under the maximum likelihood (ML) criterion, using PhyML v3.0 software and by Bayesian analysis, using MrBayes v3.2.6 software. Statistical supports of the tree nodes were calculated by approximate likelihood ratio test (aLRT) for ML and posterior probabilities (pp) for Bayesian analysis.
Seroprevalence survey in equines.
Equine sera.
During 2007, sera from healthy horses were collected for epidemiological purposes by the Veterinary Laboratories Division (DILAVE) “Miguel C. Rubino” (Ministry of Agriculture and Fisheries of Uruguay). The animals had no record of vaccination against alphaviruses and were living in rural areas of the country. Blood samples were taken from the jugular vein and maintained at −80°C. A subsample of 425 sera from 18 of the 19 departments of Uruguay (Artigas, Canelones, Cerro Largo, Colonia, Durazno, Flores, Florida, Lavalleja, Maldonado, Paysandú, Río Negro, Rivera, Rocha, Salto, San José, Soriano, Tacuarembó, and Treinta y Tres) (Figure 1) was kindly supplied by Dr. M. A. Solari from DILAVE to perform this study. Sera were kept at −20°C while performing the serologic assays.
Antibody screening by plaque reduction neutralization test (PRNT80).
Four hundred and twenty-five (425) equine sera were tested by plaque reduction neutralization assay,19 to detect and titrate specific neutralizing antibodies (NTAbs) against alphaviruses. Sera were inactivated at 56°C for 25 minutes, then centrifuged at 11,400 × g for 30 minutes to clarify, and the supernatant was stored at −20°C until assayed. Samples were analyzed for NTAbs against VEEV subtype IAB, Pixuna virus (PIXV) (formerly subtype IV), Rio Negro virus (RNV) (formerly subtype VI), MADV, EEEV complex, and WEEV by PRNT using VeroE6 cells (ATCC® CRL-1586), as described by Earley et al.20 The serum samples were initially tested at a dilution of 1:10. Those that neutralized at least 80% of inoculated viral plaque-forming units were considered positive and titrated further using 2-fold serial dilutions, to determine the end-point titer. Viruses used in this study were as follows: 1) VEEV IAB strain TC83, 2) PIXV (VEEV subtype IV) strain BeAr35645,21 3) RNV (VEEV subtype VI) strain AG80-663,22 4) MADV strain Cba55,23 and 5) WEEV strain Cba87.23 Viral suspensions were prepared with a 10% dilution of infected suckling mice brain in MEM (Gibco™; Thermo Fisher), 10% fetal bovine serum (Invitrogen), and 1% gentamicin (Gibco™; Thermo Fisher) and then centrifuged at 11,400 × g for 30 minutes.
RESULTS
Alphavirus genome detection in mosquitoes and phylogeny.
A total of 3,575 mosquitoes grouped in 266 pools were collected at nine departments of Uruguay during March 2006 to December 2014. From the trapped mosquitoes, 3,333 (93.2%) were female and 242 (6.7%) were male. According to the morphological classification, six genera were found: Aedes, Culex, Mansonia, Psorophora, Uranotaenia, and Aedeomyia. Most of the individuals were classified at the species level as Culex pipiens (N = 2,964, 82.9%). The second most abundant species was Aedes albifasciatus (N = 202, 5.6%). Because of the damage and loss of characters, 307 mosquitoes (8.5%) were classified at the genus level as Culex spp. Other Culex species (bidens, dolosus, apicinus, and coronator) were collected, representing a minority of the total captured (N = 9, 0.25%). Regarding the other mosquitoes collected, Aedes aegypti, Mansonia spp., Psorophora spp., Uranotaenia spp., and Aedeomyia spp. accounted for 2.6% of the total captured (N = 93) (Table 1, Supplemental Table 1).
Table 1.
Mosquito captures
| Department | Habitat/locality | Genus/species | #Pool/locality | Total |
|---|---|---|---|---|
| Artigas | Rural | Cx. pipiens | 20 | 20 |
| Artigas | Rural | Culex spp. | 12 | 12 |
| Artigas | Rural | Cx. coronator | 1 | 1 |
| Artigas | Rural | Cx. bidens | 2 | 2 |
| Artigas | Rural | Ae. albifasciatus | 3 | 3 |
| Artigas | Rural | Uranotaenia spp. | 1 | 1 |
| Artigas | Rural | Aedeomyia squamipennis | 1 | 1 |
| Canelones | Las Toscas/Los Titanes/Atlántida | Cx. pipiens | 5/3/10 | 18 |
| Canelones | Los Titanes | Culex spp. | 2 | 2 |
| Canelones | Los Titanes | Cx. bidens | 1 | 1 |
| Canelones | Atlántida | Ae. albifasciatus | 2 | 2 |
| Canelones | Los Titanes | Aedes spp. | 3 | 3 |
| Canelones | Las Toscas | Ae. aegypti | 1 | 1 |
| Colonia | Colonia/Laguna de los Patos | Cx. pipiens | 4/5 | 9 |
| Colonia | Colonia capital/Laguna de los Patos | Ae. albifasciatus | 1/2 | 3 |
| Colonia | Laguna de los Patos | Mansonia titillans | 5 | 5 |
| Colonia | Colonia capital | Psorophora ferox | 1 | 1 |
| Colonia | Colonia/Laguna de los Patos | Psorophora spp. | 1/1 | 2 |
| Montevideo | Union/Cuch. Pereira/Bañados de Carrasco/Belvedere | Cx. pipiens | 7/2/1/16/2/ | 44 |
| Cerrito/V. García/Colón | 9/7 | |||
| Montevideo | Bañados de Carrasco/Belvedere/Cerrito/V. García | Culex spp. | 2/3/1/3 | 9 |
| Montevideo | Belvedere | Cx. dolosus | 1 | 1 |
| Montevideo | Buceo/Belvedere | Ae. albifasciatus | 3/5/1/3 | 12 |
| Cerrito/V. García | ||||
| Montevideo | Bañado de Carrasco | Aedes scapualris | 1 | 1 |
| Montevideo | Bañados de Carrasco | Aedes crinifer | 2 | 2 |
| Montevideo | Bañados de Carrasco | M. titillans | 1 | 1 |
| Paysandú | Paysandú capital | Cx. pipiens | 20 | 20 |
| Paysandú | Paysandú capital | Ae. albifasciatus | 4 | 4 |
| Rio Negro | Fray Bentos/Arrayanes | Cx. pipiens | 1/2 | 3 |
| Rivera | Rivera capital | Cx. pipiens | 3 | 3 |
| Rivera | Rivera capital | Culex spp. | 5 | 5 |
| Rivera | Rivera capital | Aedes fluviatilis | 1 | 1 |
| Salto | Salto capital | Cx. pipiens | 50 | 50 |
| Salto | Salto capital | Culex spp. | 3 | 3 |
| Salto | Salto capital | Cx. coronator | 1 | 1 |
| Salto | Salto capital | Cx. apicinus | 1 | 1 |
| Salto | Salto capital | Ae. albifasciatus | 3 | 3 |
| Salto | Salto capital | Ae. scapualris | 2 | 2 |
| Salto | Salto capital | Ae. aegypti | 3 | 3 |
| Salto | Salto capital | Psorophora ciliata | 1 | 1 |
| Salto | Salto capital | Psorophora spp. | 1 | 1 |
| Soriano | Mercedes | Cx. pipiens | 8 | 8 |
| 266 |
Department, habitat or locality, genus and/or species, and number of pools per locality are indicated. Every pool was analyzed for alphavirus genome detection.
To detect and identify the presence of alphavirus genomes in mosquitoes, pools were processed by a pan-alphavirus RT-nested PCR. Two pools were positive (Pool 1 and Pool 267). The first (CpFB) corresponded to a pool of 10 Cx. pipiens–engorged females, captured in 2006 in Fray Bentos city (Rio Negro department), near the Uruguay River. The other positive sample (CpLT) corresponded to 1 Cx. pipiens female, captured in 2014 at Las Toscas locality (Canelones department), in Southern Uruguay, near the Rio de la Plata River. Blast analysis of the sequences retrieved from PCR products showed that CpFB displayed a high similarity score with EEEV (96% identity, E-value 3e-57); meanwhile, CpLT was highly similar to VEEV (99% identity, E-value 1e-82).
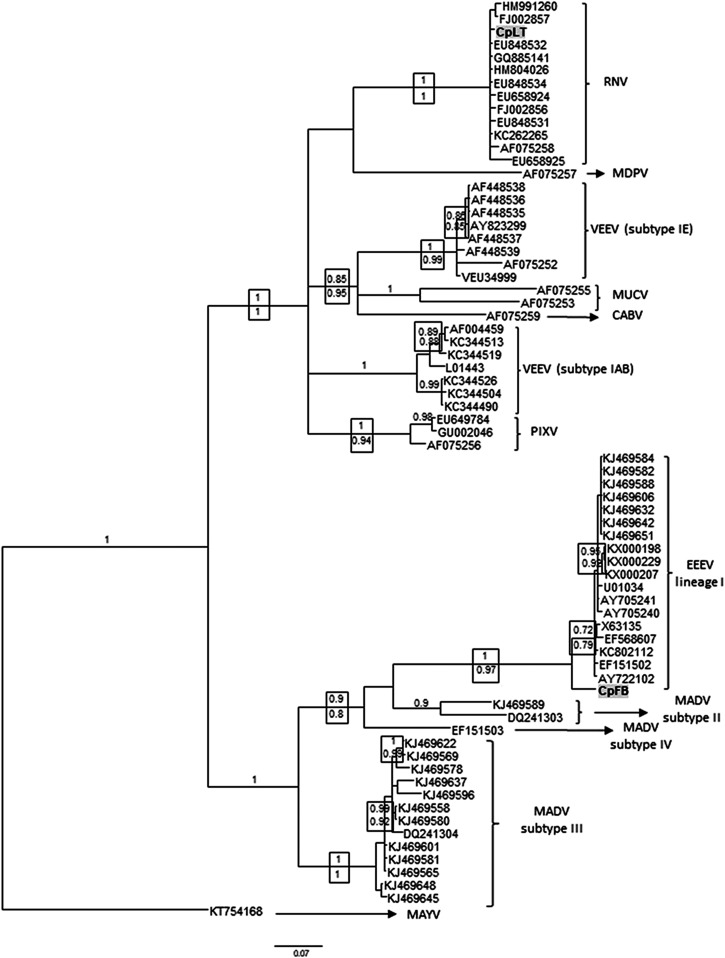
Phylogenies were inferred under Bayesian and ML criteria, using the general time reversible + gamma + proportion invariant (GTR + Γ + I) model of nucleotide substitution. The analyses included the alphavirus genomes reported herein, together with representative sequences of the VEEV antigenic complex: VEEV (subtypes IAB and IE), Mosso das Pedras virus (MDPV), MUCV, Cabassou virus (CABV), RNV, PIXV, EEEV (lineage I), and MADV (subtypes II, III, and IV). Mayaro virus was used as outgroup species.
Bayesian and ML analyses showed concordant topologies for the main phylogroups. Figure 2 shows the Bayesian tree, with pp statistical supports depicted above and aLRT below the corresponding nodes. CpFB sequence clustered together with EEEV strains from lineage I, with significant statistical support (pp = 1, aLRT = 0.97), whereas CpLT clustered with RNV with pp and aLRT both = 1. CpFB displays a basal position in the EEEV clade, with a nucleotide p-distance ranging 0.05–0.07 when compared with the EEEV group (data not shown).
Figure 2.
Bayesian phylogenetic analysis of alphavirus detected in Uruguay. Partial nsP4 genome sequences from mosquito samples were analyzed in comparison with sequences of the VEEV and EEEV complex. The analysis was conducted under the general time reversible + gamma + proportion invariant (GTR + Γ + I) model of nucleotide substitution. Two runs of four chains each (one cold and three heated, temperature 0.20) were run for three million generations; trees were sampled every 100 generations. Convergence was assessed by using the average standard deviation in partition frequency values across independent analyses with a threshold value of 0.01; burn-in was set to 25%. Supports above nodes are the posterior probabilities and below are depicted the approximate likelihood ratio test supports from maximum likelihood analysis. GenBank accession numbers: CpFB: MG009260 and CpLT: MG009261. CABV = Cabassou virus; EEEV = eastern equine encephalitis virus; MADV = Madariaga virus; MAYV = Mayaro virus; MDPV = Mosso das Pedras virus; MUCV = Mucambo virus; PIXV = Pixuna virus; RNV = Rio Negro virus; and VEEV = Venezuelan equine encephalitis virus.
Maximum likelihood analysis shows that CpLT appears more related to GQ885141, EU848532, and HM80402 sequences (Supplemental Figure 1). EU848532 and HM80402 are sequences retrieved from Cx. maxi and Cx. coronator, respectively, captured in Chaco Province (Argentina). GQ885141 was retrieved from Ae. scapularis captured in Tucumán Province (Argentina). Rio Negro virus clade is less resolved in the Bayesian tree; however, genetic p-distance showed a 100% homology between CpLT and the Argentinean strains mentioned previously, together with RNV sequence KC262265 retrieved from the rodent Akodon azarae.
Screening and titration of horse sera.
To avoid false results due to cross-reactivity between different VEEV subtypes, positive sera were simultaneously titrated against all VEEV subtypes, once determined positive. Samples with NTAbs to two or more viruses were considered positive to a given virus, if a 4-fold or greater difference in titer to one of the viruses was observed. Table 2 shows the seropositive sera found per department, the antibody titers for each virus analyzed, and the result according to the simultaneous titration.
Table 2.
Antibody titers to VEEV subtype IAB, RNV, PIXV, MADV, and WEEV found in equine sera by PRNT80
| Sera No. | Department | Antibody titer | Result | ||||
|---|---|---|---|---|---|---|---|
| VEEV IAB | RNV | PIXV | MADV | WEEV | |||
| E07/037 | Cerro Largo | – | 10 | – | – | – | RNV |
| E07/038 | Cerro Largo | – | 10 | 40 | – | – | PIXV |
| E07/102 | Cerro Largo | – | – | – | – | 80 | WEEV |
| E07/085 | Colonia | 20 | – | – | – | – | VEEV IAB |
| E07/086 | Colonia | 10 | – | – | – | – | VEEV IAB |
| E07/104 | Colonia | – | 10 | 10 | – | – | VEEV* |
| E07/187 | Colonia | – | 20 | – | – | – | RNV |
| E07/188 | Colonia | – | 160 | – | – | – | RNV |
| E07/064 | Flores | – | 10 | – | – | – | RNV |
| E07/131 | Flores | – | 10 | – | – | – | RNV |
| E07/022 | Florida | 10 | – | – | – | – | VEEV IAB |
| E07/107 | Florida | – | 10 | 20 | – | – | VEEV* |
| E07/333 | Lavalleja | – | 10 | – | – | – | RNV |
| E07/334 | Lavalleja | – | 10 | – | – | – | RNV |
| E07/335 | Lavalleja | – | 10 | – | – | – | RNV |
| E07/336 | Lavalleja | – | 10 | – | – | – | RNV |
| E07/350 | Maldonado | – | 10 | – | – | – | RNV |
| E07/353 | Maldonado | – | 10 | – | – | – | RNV |
| E07/360 | Paysandú | – | 10 | – | – | – | RNV |
| E07/362 | Paysandú | – | 10 | – | – | – | RNV |
| E07/247 | Río Negro | 80 | – | – | – | – | VEEV IAB |
| E07/368 | Rio Negro | 40 | – | – | – | – | VEEV IAB |
| E07/369 | Río Negro | 40 | 10 | – | – | – | VEEV IAB |
| E07/374 | Río Negro | – | 10 | – | – | – | RNV |
| E07/047 | Rivera | 10 | – | – | – | – | VEEV IAB |
| E07/073 | Rivera | 10 | – | – | – | – | VEEV IAB |
| E07/048 | Rivera | – | ND | ND | 10 | – | MADV |
| E07/071 | Rivera | – | – | – | 10 | – | MADV |
| E07/220 | Rivera | – | 10 | – | – | – | RNV |
| E07/052 | Rocha | – | – | – | 10 | – | MADV |
| E07/255 | San José | – | 10 | – | – | – | RNV |
| E07/403 | San José | – | – | – | 10 | – | MADV |
| E07/405 | Soriano | – | – | – | 10 | – | MADV |
| E07/009 | Tacuarembó | 10 | – | – | – | – | VEEV IAB |
| E07/258 | Tacuarembó | 80 | – | – | – | – | VEEV IAB |
| E07/081 | Tacuarembó | – | 10 | – | – | – | RNV |
| E07/230 | Tacuarembó | – | 10 | – | – | – | RNV |
MADV = Madariaga virus; PIXV = Pixuna virus; PRNT80 = plaque reduction neutralization test; RNV = Rio Negro virus; VEEV = Venezuelan equine encephalitis virus; WEEV = western equine encephalitis virus.
Subtyping inconclusive.
Titers against VEEV IAB ranged between 10 and 80, the latter being a sample from Rio Negro department. Titers against RNV were 10 for most of the sera, with a sample from Colonia neutralizing 80% of virus at a titer of 1:160. Positive sera against PIXV exhibited titers between 10 and 40 and all MADV positive samples had titers of 10. The only seropositive sample against WEEV exhibited a titer of 80.
Table 3 displays the number and percentage of seropositive sera per department. Seropositive equines against RNV were detected in 10 of the 18 departments studied. Venezuelan equine encephalitis virus subtype IAB was detected in five departments: Colonia, Florida, Rio Negro, Rivera, and Tacuarembó. Madariaga virus NTAbs were detected in samples from four departments (Rivera, Rocha, San José, and Soriano); meanwhile, PIXV and WEEV NTAbs were detected only in Cerro Largo department.
Table 3.
PRNT80 positive sera (number and percentage) per department
| Department | Total samples per department* | VEEV IAB | RNV | PIXV | MADV | WEEV |
|---|---|---|---|---|---|---|
| No. of positive (%) | No. of positive (%) | No. of positive (%) | No. of positive (%) | No. of positive (%) | ||
| Cerro Largo | 32 | – | 2 (6.2) | 1 (3.1) | – | 1 (3.1) |
| Colonia | 39 | 2 (5.1) | 2 (5.1) | – | – | – |
| Flores | 27 | – | 2 (3.7) | – | – | – |
| Florida | 21 | 1 (4.7) | – | – | – | – |
| Lavalleja | 20 | – | 4 (20) | – | – | – |
| Maldonado | 21 | – | 2 (9.5) | – | – | – |
| Paysandú | 20 | – | 2 (10) | – | – | – |
| Río Negro | 23 | 3 (13.0) | 2 (8.6) | – | – | – |
| Rivera | 43 | 2 (4.6) | 1 (2.3) | – | 2 (4.6) | – |
| Rocha | 22 | – | – | – | 1 (4.5) | – |
| San José | 24 | – | 1 (4.1) | – | 1 (4.1) | – |
| Soriano | 23 | – | – | – | 1 (4.3) | – |
| Tacuarembó | 28 | 2 (7.1) | 2 (7.1) | – | – | – |
MADV = Madariaga virus; PIXV = Pixuna virus; PRNT80 = plaque reduction neutralization test; RNV = Rio Negro virus; VEEV = Venezuelan equine encephalitis virus; WEEV = western equine encephalitis virus.
No seropositive samples found in Artigas, Salto, Canelones, Treinta y Tres, and Durazno departments. #VEEV positive samples E07/104 and E07/107 from Colonia and Florida were not included because subtyping was inconclusive.
Rio Negro virus seropositive percentages ranged from 3.5% to 20% of equines in a department. Lavalleja and Paysandú departments had the greatest number of RNV seropositive equines, with 20% and 10%, respectively. The areas with the greatest number of VEEV subtype IAB seropositive equine samples ranged from 4.6% to 13% and Rio Negro department had the highest percentage (13%). The percentage of MADV seropositive samples was similar in the four departments where detected (4.1–4.6%). Finally, PIXV and WEEV NTAbs were detected at the lowest frequency (3.1%).
DISCUSSION
Uruguay has experienced outbreaks of equine encephalitis due to alphaviruses in the past (1972–1973), concomitantly with outbreaks reported in Argentina.11 A strain of WEEV was isolated from an ill horse in 1958.10 Since then, there were few reports on the circulation of these viruses in Uruguay. After several silent decades on alphavirus outbreaks, a human case of western equine encephalitis was reported in 2011.12 Even so, no studies have been performed on the vectors potentially involved or the circulation of other alphaviruses already reported in the region. To better understand the dynamics of alphavirus circulation in the country, we carried out mosquito captures to perform viral genome detection and analyzed horse sera to look for specific NTAbs. Captures were performed from March 2006 to December 2014 and were directed to urban, suburban, and/or rural areas near the Uruguay River, all along the border with Argentina, and part of the Northeast region, near the border with Brazil and Montevideo–Canelones coast. Montevideo and Canelones departments contain more than a half of the human population of the country, are active touristic areas, and the International Carrasco Airport (in Canelones) and the Port of Montevideo are located there. Culex pipiens was the most captured mosquito species in all locations, except for rural Artigas. In this expedition, mosquitoes from four different species were captured, but the overall capture efficiency was very low. This could be explained by the date of sampling, which was autumn–winter season. Culex pipiens is a generalist mosquito adapted to live in a variety of natural or artificial habitats, which explains the high proportion found in this study.24 The second most captured species was Ae. albifasciatus, which may breed in natural or artificial water bodies and lives in semi domestic habitats.25 Other Culex species (bidens, coronator, apicinus, and dolosus) were occasionally captured, being found in Southern (Montevideo, Canelones) and Northern (Salto and Artigas) departments and associated with urban or rural areas. Mansonia spp., Psorophora spp., Uranotaenia spp., and Aedomiya spp. were also occasionally collected mostly from natural environments. Aedes aegypti accounted for five pools from Salto and Canelones, the low number of samples may be because of the placing of the CDC traps, which were not set inside the houses. Although the study was not designed to analyze abundance or habitat preferences of our mosquito populations, the species proportions and habitats of the mosquitoes captured are consistent with previous studies performed in the country.26,27
Alphavirus genomes detected in two Cx. pipiens pools from Fray Bentos (Rio Negro department) and Las Toscas (Canelones department) were further identified by phylogenetic analysis as belonging to the lineage I of the EEEV complex (CpFB) and to RNV from the VEEV complex (CpLT), respectively. Despite only analyzing a 195-bp region of the nsP4 gene, it was informative enough to identify the viruses at the species level. Moreover, as stated in Sánchez-Seco et al.,14 phylogenetic analysis of this fragment allows precise identification of the most relevant members of the genus, being a powerful tool when working with direct sequencing from mosquito homogenates. Until now, this is the first evidence of alphavirus genome detection in field caught mosquitoes in Uruguay.
Phylogenetic analysis (Figure 2) shows that sequences from the VEEV complex form a monophyletic group and are discriminated into viral species (VEEV IAB, MDPV, MUCV, CABV, RNV, and PIXV) or subtypes (IE) with significant support. The EEEV complex also appears as a monophyletic group and viruses belonging to different lineages (MADV subtypes II, III and IV, and EEEV lineage I) are consistently grouped together. This phylogeny reinforces that despite the short sequence analyzed, there is enough phylogenetic information to assign viruses to species and/or lineages. The sequence identified as RNV was closely related to Argentinean strains isolated from mosquitoes. Rio Negro virus was formerly named as VEEV subtype VI, but in 2005, the taxonomic status was changed to viral species.28 The reference strain was isolated from Cx. delpontei in Chaco Province, Argentina.22 Later, a human outbreak was recorded in Formosa Province in 1989 and soon after a RNV strain was isolated from the field mouse A. azarae.29,30 Further studies in Argentina detected this virus in different mosquito species including Ochlerotatus scapularis, Cx. maxi, Cx. coronator, and Cx. interfor in Chaco, Tucumán, and Córdoba provinces, showing a wide geographical and temporal distribution.30,31 Although RNV was detected in Argentina from mosquito species other than Cx. pipiens, sequence similarity suggests that a very similar strain is present in Uruguay. Up to now, there are no reports of human infections by RNV in Uruguay.
The virus detected in Cx. pipiens from Rio Negro department (CpFB) belongs to the EEEV lineage I, which is enzootic in the East coast of North America, Caribbean, and East Central America.32,33 Eastern equine encephalitis virus causes localized outbreaks in horses and humans, and surveillance in the USA has detected an increase in neuroinvasive human cases since 2010.34 There is only one report about the detection of EEEV lineage I in South America,35 so our finding in Cx. pipiens is intriguing. Laboratory cross contamination was ruled out because no isolates, samples, or RNA from this EEEV lineage was ever managed in our facilities. Additional sequence information could not be obtained from this sample; however, the branching position of the CpFB nsP4 fragment placed it unequivocally in lineage I. It is possible that a silent circulation or a sporadic introduction of this viral lineage occurred near May 2006 because no other positive pool was obtained along the study, with more than 3,000 samples of Culex spp. analyzed. Interestingly, the positive pool was formed by engorged females, so we cannot discard the possible infection from a viremic host.
Overall, NTAb titers were low, indicating potential past infections, and only a few sera showed titers > 80. Because we do not know the exact dynamic of the antibody secretion profile in equines, these low titers may represent recent infections as well and, therefore, the presence of local activity of these alphaviruses, which is confirmed by the molecular detection in mosquitoes. Seropositive equines were found for all the tested viruses (RNV, VEEV subtype IAB, PIXV, MADV, and WEEV), with RNV being the most prevalent with percentages ranging between 4% and 20%, in 10 of 18 departments sampled. It is important to note that in Canelones department, where the RNV positive mosquito was captured, no seropositive horses were found for RNV or any of the viruses tested. The finding of RNV genome in Cx. pipiens was not simultaneous, but 7 years later (2014), this time lapse may be explained by a recent introduction of the virus or simply by a subsampling in our horse serology. On the other hand, considering that arbovirus activity may be restricted to local foci, it is possible that two different episodes of RNV activity occurred around 2007 and later in 2014.
Venezuelan equine encephalitis virus subtype IAB was detected in four departments with percentages around 4%, except for Rio Negro department, where seroprevalence was 13%. Venezuelan equine encephalitis virus IAB may cause outbreaks in horses and humans; however, there are no previous records of epizootics or enzootic circulation in Uruguay. Seroprevalences found in this study may reveal the circulation of enzootic members of subtype I because cross-reactivity between NTAbs against TC-83 strain and other members of subtype I was described.36,37
Neutralizing antibodies against PIXV were only present at Cerro Largo department in a low percentage of equines (3%). This enzootic virus has been recovered in Argentina (Chaco, Formosa, and Tucuman provinces) from mosquitoes and detected by serology in humans.30,37
Infections by MADV in horses appear restricted to four departments (Rocha, Rivera, San José, and Soriano), with a seroprevalence of about 4.4%. There are few reports of human disease associated with MADV,38,39 the most recent coming from an encephalitis outbreak in Panama.7,8 The circulation of EEEV complex in Uruguay was reported before, in 1970. A serologic survey carried out in children (N = 481) and adults (N = 834) reported 6% and 2% prevalences for EEEV in 0–14 years old children and adults, respectively,10 thus suggesting the circulation of viruses from the EEEV complex in the human population at that time.
A single WEEV positive horse was detected in the northern department of Cerro Largo, with a titer slightly higher than most of the sera analyzed. Activity of WEEV was documented in Uruguay in 1972–1973, when an extensive epizootic affected Uruguay and Central Argentina.11 In 2009, a fatal human case of WEEV was diagnosed by RT-PCR and sequencing.12 Although we found just a single seropositive horse in our study, we may infer that there was some WEEV activity in our country in the period 2007–2009.
The results presented here suggest that several alphaviruses are circulating in Uruguay in enzootic cycles and based on previous evidence, they can also cause epizootic events. Rio Negro virus appears as the most geographically widespread. Madariaga virus, VEEV IAB, PIXV, and WEEV seem to be more restricted and showed lower prevalences, although their sporadic presence in the country was already reported, including a fatal human case occurred in 2009. To evaluate the eco-epidemiology of alphaviruses, our group has initiated a study of free-ranging birds and rodents for NTAb detection and is continuing the survey of viral genomes in mosquitoes collected in different regions and biomes of Uruguay.
Supplementary Material
Acknowledgments:
We are grateful to the entomologist Maria M. Martinez for training in mosquito classification and trapping, and to Soledad Valledor and Angélica Solari for equine sera collection. We also thank VIRORED (CYTED) for continuous support to our research.
Note: Supplemental figure and table appear at www.ajtmh.org.
REFERENCES
- 1.ICTV , 2016. Virus Taxonomy: 2016 Release Birmingham, AL: International Committee on Taxonomy of Viruses, University of Birmingham. Available at: https://talk.ictvonline.org/taxonomy/. Accessed December 1, 2017.
- 2.Fields B, 2013. Knipe DM, Howley PM, eds. Fields Virology, Vol. 2, 6th edition. Philadelphia, PA: Lippincott, Williams & Wilkins. [Google Scholar]
- 3.Weaver SC, 2005. Host range, amplification and arboviral disease emergence. Arch Virol Suppl 19: 33–44. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC, Reisen WK, 2010. Present and future arboviral threats. Antiviral Res 85: 328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes MRT, et al. 2015. Emergence and potential for spread of chikungunya virus in Brazil. BMC Med 13: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mota MT de O, Ribeiro MR, Vedovello D, 2015. Mayaro virus: a neglected arbovirus of the Americas. Future Virol 10: 1109–1122. [Google Scholar]
- 7.Carrera J-P, et al. 2013. Eastern equine encephalitis in Latin America. N Engl J Med 369: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luciani K, Abadía I, Martínez-Torres AO, Cisneros J, Guerra IGM, Estripeaut DCJ, 2015. Madariaga virus infection associated with a case of acute disseminated encephalomyelitis. Am J Trop Med Hyg 92: 1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunini S, França DDS, Silva JB, Silva LN, Silva FPA, Spadoni M, Rezza G, 2017. High frequency of mayaro virus IgM among febrile patients, central Brazil. Emerg Infect Dis 23: 1025–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somma Moreira RE, Campione-Piccardo J, Russi JC, Hortal de Giordano M, Bauzá CA, Peluffo G, Tosi HC, 1970. Arbovirus en el Uruguay. Arch Pediatr Urug 41: 359–363. [Google Scholar]
- 11.Acha PN, Szyfres B, 2003. Zoonoses and Communicable Diseases Common to Man and Animals: Chlamydioses, Rickettsioses and Viroses, 3rd edition. Washington, DC: Organización Panamericana de la Salud. [Google Scholar]
- 12.Delfraro A, Burgueño A, Morel N, González G, García A, Morelli J, Pérez W, Chiparelli H, Arbiza J, 2011. Fatal human case of western equine encephalitis, Uruguay. Emerg Infect Dis 17: 952–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darsie R, Jr., 1985. Mosquitoes of Argentina. Part I: keys for identification of adult females and fourth stage larvae in English and Spanish (Diptera, Culicidae). Mosq Syst 17: 153–253. [Google Scholar]
- 14.Sánchez-Seco MP, Rosario D, Quiroz E, Guzmán G, Tenorio A, 2001. A generic nested-RT-PCR followed by sequencing for detection and identification of members of the alphavirus genus. J Virol Methods 95: 153–161. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA, 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 16.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL, 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO, 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgueño A, Spinsanti L, Díaz LA, Rivarola ME, Arbiza J, Contigiani M, Delfraro A. 2013. Seroprevalence of St. Louis encephalitis virus and West Nile virus (Flavivirus, Flaviviridae) in horses, Uruguay. Biomed Res Int 2013: 582957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earley E, Peralta PH, Johnson KM, 1967. A plaque neutralization method for arboviruses. Exp Biol Med 125: 741–747. [DOI] [PubMed] [Google Scholar]
- 21.Shope RE, Causey OR, De Andrade AH, 1964. The Venezuelan equine encephalomyelitis complex of group A arthropod-borne viruses, including Mucambo and Pixuna from the Amazon region of Brazil. Am J Trop Med Hyg 13: 723–727. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell CJ, Monath TP, Sabattini MS, Cropp CB, Daffner JF, Calisher CH, Jakob WL, Christensen HA, 1985. Arbovirus investigations in Argentina, 1977–1980. II. Arthropod collections and virus isolations from Argentine mosquitoes. Am J Trop Med Hyg 34: 945–955. [PubMed] [Google Scholar]
- 23.Monath TP, Sabattini MS, Pauli R, Daffner JF, Mitchell CJ, Bowen GS, Cropp CB, 1985. Arbovirus investigations in Argentina, 1977–1980. IV. Serologic surveys and sentinel equine program. Arthropod collections and virus isolations from argentine mosquitoes. Am J Trop Med Hyg 34: 945–955. [PubMed] [Google Scholar]
- 24.Almirón WR, Brewer ME, 1996. Classification of immature stage habitats of Culicidae (Diptera) collected in Córdoba, Argentina. Mem Inst Oswaldo Cruz 91: 1–9. [DOI] [PubMed] [Google Scholar]
- 25.Ludueña Almeida FF, Gorla DE, 1995. The biology of Aedes (Ochlerotatus) albifasciatus Macquart, 1838 (Diptera: Culicidae) in central Argentina. Mem Inst Oswaldo Cruz 90: 463–468. [DOI] [PubMed] [Google Scholar]
- 26.Rossi GC, Martínez M, 2003. Mosquitos (Diptera: Culicidae) del Uruguay. Entomol Vect 10: 469–478. [Google Scholar]
- 27.Rossi G C, Martínez M. 2013. Lista de especies y clave ilustrada para la identificación de larvas de mosquitos (Diptera: Culicidae) halladas criando en recipientes artificiales en Uruguay. Bol Soc Zool Uruguay (2a época) 22: 49–65. [Google Scholar]
- 28.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, 2015. Virus taxonomy: VIIIth report of the international committee on taxonomy of viruses. Virus Res 2005: 221–222. [Google Scholar]
- 29.Contigiani MS, de Basualdo M, Cámara A, Ramírez A, Díaz G, González D, Medeot S, Osuna D, 1993. Presence of antibodies against Venezuelan equine encephalitis virus subtype VI in patients with acute febrile illness. Rev Argent Microbiol 25: 212–220. [PubMed] [Google Scholar]
- 30.Pisano MB, Torres C, Ré VE, Farías AA, Sánchez Seco MP, Tenorio A, Campos R, Contigiani MS, 2014. Genetic and evolutionary characterization of Venezuelan equine encephalitis virus isolates from Argentina. Infect Genet Evol 26: 72–79. [DOI] [PubMed] [Google Scholar]
- 31.Pisano MB, Spinsanti LI, Díaz LA, Farías AA, Almirón WR, Ré VE, Contigiani MS, 2012. First detection of Rio Negro virus (Venezuelan equine encephalitis complex subtype VI) in Cordoba, Argentina. Mem Inst Oswaldo Cruz 107: 125–128. [DOI] [PubMed] [Google Scholar]
- 32.Arrigo NC, Adams AP, Weaver SC, 2010. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol 84: 1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver SC, et al. 1999. Molecular epidemiological studies of veterinary arboviral encephalitides. Vet J 157: 123–138. [DOI] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention, 2005. Eastern equine encephalitis. Epidemiology and geographical distribution. Available at: https://www.cdc.gov/easternequineencephalitis/tech/epi.html. Accessed December 1, 2017.
- 35.Richard Hoyos L, Juan Suaza V, Tenorio A, Uribe S, Gallego-Gómez J, 2015. Molecular detection of eastern equine encephalitis virus in mosquitoes from La Pintada (Antioquia). Rev Mvz Cordoba 20: 4800–4806. [Google Scholar]
- 36.Pisano MB, Contigiani MS, Re V, 2016. Venezuelan equine encephalitis virus. Liu D, ed. Molecular Detection of Animal Viral Pathogens Boca Ratón, FL: CRC Press/Taylor & Francis, 269–276. [Google Scholar]
- 37.Pisano MB, Oria G, Beskow G, Aguilar J, Konigheim B, Cacace ML, Aguirre L, Stein M, Contigiani MS, 2013. Venezuelan equine encephalitis viruses (VEEV) in Argentina: serological evidence of human infection. PLoS Negl Trop Dis 7: e2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corniou B, Ardoin P, Bartholomew C, Ince W, Massiah V, 1972. First isolation of a South American strain of eastern equine virus from a case of encephalitis in Trinidad. Trop Georg Med 24: 162–167. [PubMed] [Google Scholar]
- 39.Alice F, 1956. Infeccao humana pelo virus “leste” da encefalite equine [in Brazil]. Bol Inst Biol da Bahia 3: 3–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.