Abstract.
We describe the deployment of a custom-designed molecular diagnostic TaqMan Array Card (TAC) to screen for 31 bacterial, protozoal, and viral etiologies in blood from outbreaks of acute febrile illness in Tanzania during 2015–2017. On outbreaks notified to the Tanzanian Ministry of Health, epidemiologists were dispatched and specimens were collected, transported to a central national laboratory, and tested by TAC within 2 days. This algorithm streamlined investigation, diagnosed a typhoid outbreak, and excluded dozens of other etiologies. This method is usable in-country and may be incorporated into algorithms for diagnosing outbreaks.
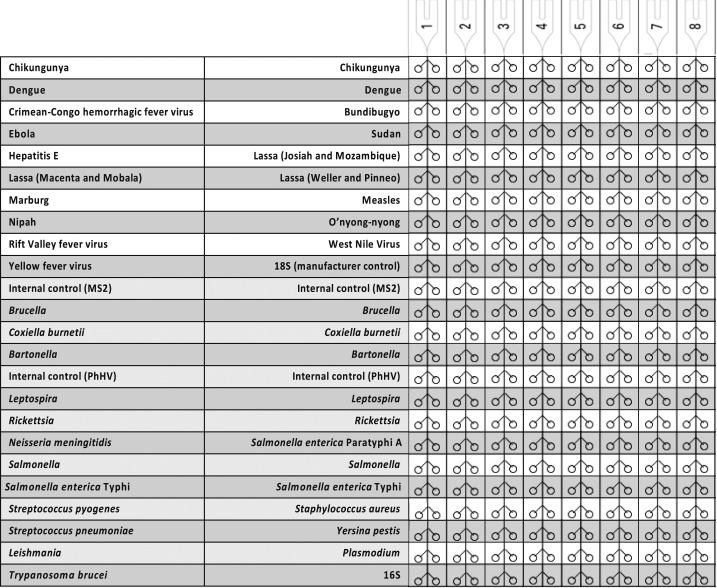
Detecting the infectious cause of acute febrile illness (AFI) is difficult, given lack of specificity in clinical signs and symptoms, myriad etiologies, different specimen types, and the multiplicity of laboratory methods required.1,2 Laboratory infrastructure is particularly sparse in many countries. Delays in laboratory confirmation contributed to the delayed detection of Ebola virus in West Africa and to its international spread. Laboratory improvements are a major initiative being undertaken by the Global Health Security Agenda, a multinational partnership, with a major goal of more rapidly detecting and controlling outbreaks at their source.3 Towards this goal, we developed a TaqMan Array Card (TAC) that detects multiple AFI organisms including viruses, parasites, and bacteria.4 The platform tests up to eight specimens with 48 customizable real-time polymerase chain reaction (PCR) assays. We previously established the technology in laboratories in several countries and have demonstrated excellent reproducibility.5 From 2015 to 2017, we deployed a customized version of this AFI TAC to Tanzania (Figure 1) to test in real-time outbreak specimens obtained by the Tanzanian Ministry of Health (MOH) and National Health Laboratory (NHL). Because the cards are modular, we included some assays previously reported from other projects (e.g., Neisseria meningitidis, Streptococcus pyogenes, Streptococcus pneumoniae, and Staphylococcus aureus).6 Cards were designed, quality-controlled, and procured at the University of Virginia and then shipped, along with other reagents, to the Kilimanjaro Clinical Research Institute in Moshi, Tanzania. When potential outbreaks were reported, Field Epidemiology and Laboratory Training Program Residents and MOH Officers would travel to conduct a rapid epidemiological study and obtain specimens. Whole blood specimens (collected in EDTA) were transported under cold chain to Dar es Salaam and then to Moshi, usually within 7 days. Nucleic acid was extracted from blood (Roche High Pure viral nucleic acid large volume kit, Indianapolis, IN), TAC testing was performed using TaqMan Fast Virus one-step master mix (Life Technologies, San Francisco, CA), and results were reviewed, double-checked, and reported back to NHL, all within 2 days of laboratory receipt. Here, we describe three outbreaks that illustrate the use of TAC for outbreak testing:
Figure 1.
TaqMan Array Card layout used for blood specimens in Tanzania, 2015–2017.
OUTBREAK 1
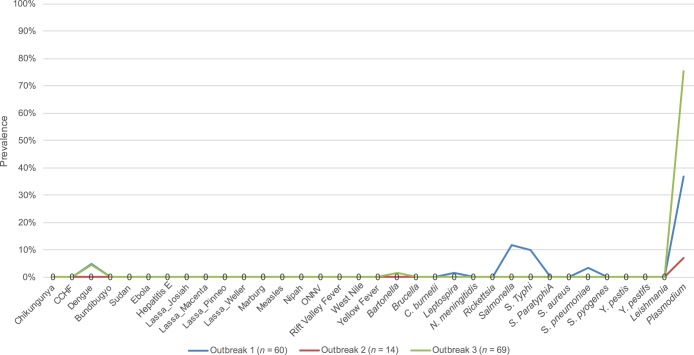
In May 2015, an outbreak of mostly fever, headache, and some diarrhea was reported from the Kigoma region in the far west of the country. Field epidemiologists were dispatched and blood and stool specimens were obtained. There had been sporadic cholera outbreaks previously and this was a leading initial diagnosis. Sixty patients were identified who met the case definition of fever and local residence and provided a blood specimen, 33 of whom also provided stool specimens. Plasmodium was detected in 22 blood specimens (37%), dengue in 3 (5%), Leptospira in 1 (2%), Salmonella or S. serotype Typhi in 10 (17%), and S. pneumoniae in 2 (3%). Stool/diarrheal specimens were tested with an enteric TAC card5 which confirmed Salmonella in eight patients (two of whom had Salmonella in blood), yielding 16 (27%) of patients with evidence of Salmonella infection. No cholera was detected.
Salmonella typhi is notoriously difficult to detect in blood by culture or PCR owing to low burden infections.7 We previously noted only an ∼71% sensitivity of TAC versus culture.4 We also find Salmonella carriage in stool to be uncommon in these settings.8 Therefore, we interpreted detection of Salmonella in 27% of patients to be strongly abnormal (Figure 2). Bacterial cultures were performed and it was confirmed that Salmonella was circulating. The epidemiologic investigation suggested that untreated water (adjusted odds ratio [aOR] versus controls, 2.9, 95% confidence interval [CI]: 1.1–7.7), bathing in the river (aOR 2.10, 95% CI: 1.2–3.7), and not washing hands after visiting toilet (aOR 2.02, 95% CI: 1.1–3.9) were associated with the illness. A chlorination program was begun and the outbreak abated.
Figure 2.
Prevalence of pathogens detected by TaqMan Array Card in whole blood from three outbreaks. Note a relative spike in Salmonella in outbreak 1, no pathogens in outbreak 2, and a spike in Plasmodium in outbreak 3. This figure appears in color at www.ajtmh.org.
OUTBREAK 2
In June 2016, in the Kondoa region of central Tanzania, an outbreak of acute liver failure with several deaths was reported. Lack of fever argued against an infectious etiology, however yellow fever and viral hepatitis were on the differential. Blood (1.5 mL) was obtained from 14 patients and all were negative for all of the 26 etiologies on the TAC except for one Plasmodium detection. These results assuaged concern for an infectious agent. Even with suboptimal sensitivity, the likelihood of all 14 specimens being falsely negative was deemed low. The epidemiologic investigation found an association with heavy rains and mold, laboratory investigation was directed to other etiologies, and the outbreak was ultimately diagnosed as aflatoxicosis.
OUTBREAK 3
In March 2017 in the Kigoma region, a new outbreak of fever developed in school children. Symptoms included headache and abdominal pain. A case control survey was performed and 104 cases had blood samples screened by malaria rapid diagnostic test (67% positive). Blood (2–2.5 mL whole blood) was obtained from 69 of the cases and 75% were positive for malaria, with few other detections. The strongest risk factors identified during epidemiologic investigation were being female, not sleeping under bednets, and recent change of water source from communal tap to river water (95% CI aOR: 1.6–4.8, 1.0–4.6, and 2.4–12, respectively). Stool specimens were obtained from 53 cases and the two most prevalent pathogens were Schistosoma mansoni (78%) and Giardia (58%). Given the dramatic prevalence of malaria by rapid diagnostic test and PCR compared with the other outbreaks (Figure 2), the higher PCR quantity for malaria versus the other outbreaks (cycle threshold 23.5 ± 6.6 versus 26.5 ± 5.6, P = 0.03), the lack of other pathogens in blood, plus the association with lack of bednet usage, we concluded this outbreak was mostly related to malaria. Schistosoma mansoni infection may also have contributed or could be an incidental finding.
CONCLUSION
Here, we show that using TAC to screen outbreak specimens for dozens of potential pathogens is feasible, can be performed in-country, and can assist epidemiologic investigation in real time. Before this project, testing in Tanzania for many of these pathogens required international shipments with weeks of delays (as happened with the Ebola outbreak in West Africa). We learned the following. We recommend testing large numbers of outbreak specimens to offset deficiencies in individual assay sensitivity because typhoid and other bacteria are frequently present at or below the limit of detection in blood. Whole blood, in large volumes (> 2 mL), is preferable over serum or plasma. We recommend the TAC laboratory have the individual PCR assays available for confirming TAC results and a plan for access to other confirmatory testing, particularly if high importance pathogens are detected such as viral hemorrhagic fevers. Finally training and a long-term financial investment is needed. The TAC real-time PCR platform is ∼100,000 USD and reagent costs per specimen are ∼50–100 USD depending on batching and volume. However, this is cheaper than the equipment and reagent costs of performing conventional microbiologic testing for all 31 pathogens. Regardless of the laboratory approach chosen, prompt epidemiologic investigation and specimen collection, transport, and submission to the country’s laboratory network remain of paramount importance and require investment as well. In conclusion, use of TAC array cards to screen specimens in AFI outbreaks rapidly assisted determining outbreak etiology and contributed to controlling the outbreaks at their source.
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Prasad N, Murdoch DR, Reyburn H, Crump JA, 2015. Etiology of severe febrile illness in low- and middle-income countries: a systematic review. PLoS One 10: e0127962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iroh Tam PY, Obaro SK, Storch G, 2016. Challenges in the etiology and diagnosis of acute febrile illness in children in low- and middle-income countries. J Pediatric Infect Dis Soc 5: 190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee SA, Arthur R, Mounts AW, 2016. Global health security: building capacities for early event detection, epidemiologic workforce, and laboratory response. Health Secur 14: 424–432. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. 2016. Development of a taqman array card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including ebola virus. J Clin Microbiol 54: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, et al. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14: 716–724. [DOI] [PubMed] [Google Scholar]
- 6.Diaz MH, et al. 2013. Optimization of multiple pathogen detection using the taqman array card: application for a population-based study of neonatal infection. PLoS One 8: e66183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nga TV, et al. 2010. The sensitivity of real-time PCR amplification targeting invasive Salmonella serovars in biological specimens. BMC Infect Dis 10: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, et al. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the gems case-control study. Lancet 388: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]