Abstract.
Plasmodium falciparum histidine-rich protein 2 (PfHRP2) forms the basis of many current malaria rapid diagnostic tests (RDTs). However, the parasites lacking part or all of the pfhrp2 gene do not express the PfHRP2 protein and are, therefore, not identifiable by PfHRP2-detecting RDTs. We evaluated the performance of the SD Bioline Malaria Ag P.f/Pan RDT together with pfhrp2 variation in Madagascar. Genomic DNA isolated from 260 patient blood samples were polymerase chain reaction (PCR)–amplified for the parasite 18S rRNA and pfhrp2 genes. Post-PCR ligation detection reaction-fluorescent microsphere assay (LDR-FMA) was performed for the identification of parasite species. Plasmodium falciparum histidine-rich protein 2 amplicons were sequenced. Polymerase chain reaction diagnosis of patient samples showed that 29% (75/260) were infected and P. falciparum was present in 95% (71/75) of these PCR-positive samples. Comparing RDT and P. falciparum detection by LDR-FMA, eight samples were RDT negative but P. falciparum positive (false negatives), all of which were pfhrp2 positive. The sensitivity and specificity of the RDT were 87% and 90%, respectively. Seventy-three samples were amplified for pfhrp2, from which nine randomly selected amplicons were sequenced, yielding 13 sequences. Amplification of pfhrp2, combined with RDT analysis and P. falciparum detection by LDR-FMA, showed that there was no indication of pfhrp2 deletion. Sequence analysis of pfhrp2 showed that the correlation between pfhrp2 sequence structure and RDT detection rates was unclear. Although the observed absence of pfhrp2 deletion from the samples screened here is encouraging, continued monitoring of the efficacy of the SD Bioline Malaria Ag P.f/Pan RDT for malaria diagnosis in Madagascar is warranted.
INTRODUCTION
Histidine-rich protein 2 (HRP2), a unique protein produced exclusively by Plasmodium falciparum, has been used as a biomarker for falciparum malaria infection1 and forms the basis of many current rapid diagnostic tests (RDTs).2,3 Over the past 10 years, HRP2-detecting RDTs have become a widely used diagnostic tool for P. falciparum, especially in endemic areas that may not have appropriate microscopy capacity available. A quality control and evaluation program was set up in 2008 between the World Health Organization (WHO) and the Foundation for Innovative New Diagnostics to evaluate malaria RDT products. Since then 202 unique products have been tested in the program; 65 of them detect P. falciparum alone, 143 detect and distinguish P. falciparum from non-P. falciparum (either pan-specific or species-specific for Plasmodium vivax or P. vivax, ovale, and malariae), and 10 detect P. falciparum and non-P. falciparum without distinguishing between them (https://eprints.qut.edu.au/111599/1/111599.pdf). Currently, 12 malaria RDTs are WHO prequalified; seven are intended to detect P. falciparum only, four detect and distinguish P. falciparum from non-P. falciparum, and one detects all species but does not distinguish between them (http://www.who.int/malaria/news/2016/rdt-procurement-criteria/en/). The SD Bioline Malaria Ag P.f/Pan RDT used in the present study is among the four WHO prequalified RDTs that can detect and distinguish P. falciparum from non-P. falciparum malaria.
Rapid diagnostic tests for malaria in Madagascar were first introduced in 2003.4,5 A policy shift toward their routine use began in 2007, and these now represent the first-line diagnostic for malaria.6 A number of RDTs detecting P. falciparum-specific HRP2 (PfHRP2) have been tested in Madagascar: MakroMED4; the Malaria Hexagon dipstick5; PALUTOP(+4)7; SD Bioline Malaria Ag P.f/Pan8; and OnSite and CareStart™.9 The study that assessed the performance of the SD Bioline Malaria Ag P.f/Pan RDT was conducted between August and October 2007 on 200 patients with suspected uncomplicated malaria, and compared the RDT results with those obtained from microscopy and real-time polymerase chain reaction (PCR) combined.8 In that study, the sensitivity and specificity of the RDT for detection of P. falciparum were 92.9% and 98.9%, respectively. The sensitivity decreased to 77.3% at parasitemia levels < 100 parasites/μL.
Several host and parasite factors may influence the accuracy and sensitivity of PfHRP2-detecting RDTs.10–13 Among these are differences in the levels of pfhrp2 transcription and PfHRP2 protein expression in different parasite strains,10 high levels of circulating antibodies against PfHRP2,11 persistence of HRP2 antigenemia after antimalarial therapy,12 and very low or high parasite densities/target antigen concentrations.13 In addition, variability in the prevalence and frequency of the epitope targeted by the monoclonal antibody may affect the performance of HRP2-based RDTs.14,15 Most importantly, parasites that lack part or all of the pfhrp2 gene do not express the PfHRP2 protein and are, therefore, not identifiable by PfHRP2-detecting RDTs. Recent studies have reported pfhrp2 gene deletions in field isolates of P. falciparum, resulting in false negative test results.13 Since the first definitive report of P. falciparum parasites lacking pfhrp2 from Peru in 2010,16 such parasites have been reported from other parts of South America,17–20 Central America,21 Asia,22–25 and Africa.26–31 The potential impact of P. falciparum parasites lacking pfhrp2 on malaria case management is highly significant. Given the emphasis on WHO Test and Treat guidelines (http://www.who.int/malaria/publications/atoz/9789241549127/en/), P. falciparum parasites lacking pfhrp2 raise concerns that antimalarial treatment could be withheld from infected patients, and this scenario could potentially undermine malaria elimination efforts. Procurement decisions regarding RDT types are also dependent upon the knowledge of their efficacy among specific populations.
Although genetic polymorphism in the pfhrp2 gene worldwide is extensive,32,33 it does not appear to affect RDT detection sensitivity at ≥ 200 parasites/μL.32 An earlier analysis based on 21 cultured lines/isolates from Africa and the Asia-Pacific tested with two falciparum malaria RDTs, ParaCheck Pf and ICT Malaria Pf, yielded a binary logistic regression model that was able to predict detection sensitivity of these two RDTs based on pfhrp2 sequence structure.33 This model predicted an isolate to be detectable at ≤ 250 parasites/μL if the number of amino acid repeats type 2 × type 7 in the pfhrp2 exon-2 was > 43, with an accuracy of 87.5%. In a later study, this analysis was repeated using the WHO product testing of malaria RDTs results from round 1 (2008).32 In this testing, 34 PfHRP2-detecting RDTs, including SD Bioline Malaria Ag P.f/Pan, were tested against 79 global isolates at 200 parasites/μL. The regression analysis this time did not show a statistically robust correlation between the pfhrp2 sequence structure and RDT detection rates.32
Two studies have previously analyzed pfhrp2 variation in samples from Madagascar.32,34 The first study was conducted on 260 P. falciparum-infected samples collected in 2006 in a countrywide survey.34 The study, conducted before the widespread rollout of RDT screening, was designed to guide the Malagasy National Malaria Control Program in its choice of malaria RDT. The study reported high pfhrp2 genetic diversity throughout Madagascar and based on the model from Baker et al.,33 predicted that 9% (range 6–14%) of Malagasy isolates with ≤ 250 parasites/μL would not be detected.34 The second study used 17 samples from Madagascar (collection date not provided), which were part of 458 isolates collected from 38 countries to assess global sequence variation in pfhrp2.32 This study also found extensive pfhrp2 variation in the Malagasy isolates. In addition, some of these 17 isolates were a part of 79 global isolates that were tested with 34 PfHRP2-detecting RDTs, including SD Bioline Malaria Ag P.f/Pan.32 No correlation was found between pfhrp2 variation and RDT detection rates.32 Finally, neither study found any evidence of pfhrp2 deletion in Madagascar.32,34
The present study was conducted on samples collected between January 2014 and August 2015, 7–8 years after RDT access was scaled-up in Madagascar and the SD Bioline Malaria Ag P.f/Pan RDT8 and pfhrp2 variation34 were first analyzed here. It is also important to mention that there has been a surge in the prevalence of pfhrp2 deletion over the past 5 years, particularly in Africa and Asia. Therefore, in this context, we reevaluated the performance of the SD Bioline Malaria Ag P.f/Pan RDT together with pfhrp2 variation as a part of our ongoing malaria epidemiological studies in Madagascar.
MATERIALS AND METHODS
Study site and subjects.
This investigation is a part of our ongoing malaria epidemiological studies being conducted in the western highlands fringe region of Madagascar, in the foothills between the central highlands and the tropical western coastal zone.6,35,36 This area is endemic for both P. falciparum and P. vivax malaria, and shows malaria seasonal trends that peak from April to May annually.6 Our studies are set up in Tsiroanomandidy district with local doctors at three health centers in the Ampasimpotsy area (GPS location: −19.24525, 46.18026).
Patients reporting to the health centers with suspected malaria were screened for an infection using the SD Bioline Malaria Ag P.f/Pan RDT (product code 05FK60; Standard Diagnostics, Inc., Republic of Korea) and light microscopy by a WHO-certified technician. The RDTs used in this study were obtained from the country’s routine point-of-care supplies. They are purchased principally by the United States Agency for International Development/President's Malaria Initiative and the Global Fund and are in line with international guidelines. Lot testing is carried out by an independent body before the products are imported to Madagascar. After approval of the batch quality, the Ministry of Health in Madagascar is sent a certificate of conformity. Malaria infections were treated by the health practitioners with an age-adjusted course of artesunate-amodiaquine, in accordance with the Madagascar Ministry of Health guidelines.37
Sample collection and processing.
All samples analyzed in this study were collected between January 2014 and August 2015. Patient samples (N = 260) were collected using finger-prick blood spotted onto Whatman 3 MM filter paper. In addition, 7–10 mL venous blood samples from five patients were collected into K+-EDTA and/or Na+-heparin vacutainers and stored at 4°C for up to 24 hours before cryopreservation for subsequent in vitro cultivation of P. falciparum parasites. Cryopreservation of the parasite-infected blood samples followed the method using Glycerolyte 57 solution as described by Normark.38 Cryopreserved samples in 2 mL tubes were shipped in a liquid nitrogen dry shipper from Madagascar to the U.S. over a 10 day period. The samples were unpacked onto dry ice to minimize thawing during transfer to longer term storage in the liquid nitrogen freezer until cultivation.
In vitro cultivation of P. falciparum patient isolates.
In vitro culture of P. falciparum isolates was initiated for the five patient venous blood samples. These were: patients Ex30704, Ex2004, 2070803, 3020205 (all collected in EDTA), and Ex2002 (collected in heparin). These P. falciparum isolates were cultured in RPMI 1640 (Corning™ cellgro™, Manassas, VA), containing 25 mM HEPES and 0.2% Sodium bicarbonate. This medium was supplemented with 200 mM l-glutamine, 200 mM Hypoxanthine, 50 mg/mL Gentamicin sulfate, and 10% Albumax. This complete malaria culture medium (CMCM)39 was stored at 4°C.
The cryopreserved parasite-infected patient blood samples were thawed using a modified, decreasing NaCl concentration gradient (12%, 1.6%, and 0.9%) method.40 After the final NaCl treatment, the blood sample pellet volume was measured (30–80 μL) and resuspended in 5 mL of CMCM in a 6-well tissue culture plate. Leukocyte-depleted blood from a local healthy North American donor was added to the CMCM containing the parasite-infected patient blood pellet to achieve 4% hematocrit. The tissue culture plate was kept in a Sterilite® box with lid, which was gassed with 5% CO2 + 5% O2 + 90% N2 mixture. Parasite cultures were maintained at 37°C in 5% CO2. The culture medium was changed daily.
Preparation of DNA template.
For the samples collected on filter papers, we used the dried blood spot protocol to extract genomic DNA using a QIAamp® 96 DNA Blood Kit (QIAGEN, Valencia, CA). For the in vitro cultured samples, DNA was extracted from 50 to 100 μL of each culture using a QIAamp® DNA Micro Kit (QIAGEN).
Molecular diagnosis of Plasmodium spp. infections.
Polymerase chain reaction–based Plasmodium spp. diagnosis used a ligase detection reaction-fluorescent microsphere assay (LDR-FMA). All methods for PCR amplification of small subunit rRNA target sequences and Plasmodium species-specific detection by LDR-FMA have been described in detail by McNamara et al.41 Genomic DNA extracted from P. falciparum, P. vivax, Plasmodium malariae, and Plasmodium ovale–infected blood samples provided by the Malaria Research and Reference Reagent Resource Center (MR4; now merged with BEI Resources) and Dr. W. E. Collins (Centers for Disease Control and Prevention) served as positive controls. Species-specific fluorescence data were collected using the Bio-Plex® Manager 3.0 software (Bio-Rad, Hercules, CA).
Amplification and sequence analysis of pfhrp2 gene.
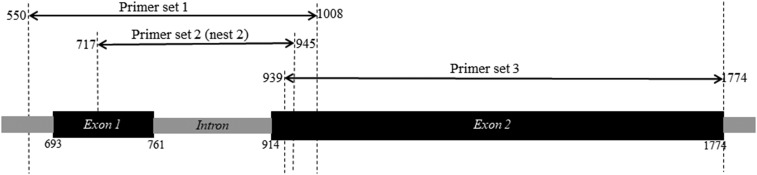
The pfhrp2 gene was amplified using three sets of primers (Primer sets #1, #2, and #3).21,34 These primer sets amplified different but overlapping regions of the pfhrp2 gene to allow for sequencing of the entire gene (Figure 1). The primer sets #1 and #3 were used to amplify the exon-1 and exon-2 regions, respectively, from the cultured isolates. The primer sets #2 and #3 were used to amplify the exon-1 and exon-2 regions, respectively, from the blood spot samples. Amplification of the exon-1 region with primer set #2 involves a nested PCR. The primer sequences, expected PCR product sizes, and amplification conditions are presented in Supplemental Table 1. Genomic DNA from the P. falciparum strains 3D7 and HB3 were used as positive controls, whereas that from the Dd2 strain, which lacks the pfhrp2 gene,42 was used as a negative control in these amplification reactions.
Figure 1.
Diagrammatic representation of the overlapping regions of the Plasmodium falciparum histidine-rich protein 2 gene amplified by using three different sets of primers. Nucleotide positions are based on the reference sequence GenBank accession number U69551.1.
Amplicons were purified using a QIAquick® PCR Purification Kit (QIAGEN). The nucleotide sequences in all purified amplicons were determined by Sanger sequencing, which was performed using a modified Applied Biosystems BigDye® Terminator v3.1 Cycle sequencing kit protocol.
CodonCode Aligner (v.6.0.2) was used for the alignment and base-calling of the raw sequences. Geneious (v.10.0.2) was used for the alignment of sequences, virtual constructing of the pfhrp2 gene, translation into protein sequences, grouping of specific amino acid repeats, identification of insertions/deletions, and for comparison with the pfhrp2 sequences from previous studies. The sequences generated in this study (N = 18; five from cultured isolates and 13 from blood spot samples, see Results) were compared with a total of 97 sequences from previous studies (GenBank accession number U69551.1,43 EU589688.1–EU589767.1,34 FJ871304.1–FJ871319.132). Among these 97 sequences, U69551.1 contains an exon-1 sequence from the ItG2 clone from a Brazilian patient. The remaining 96 sequences are exon-2 sequences from Malagasy patients from previous studies.32,34
The pfhrp2 gene sequences were translated into protein sequences and 24 amino acid repeat types (1–24) were classified as described by Baker et al.32,33 The predictive model, based on the number of type 2 × type 7 repeats being greater than 43, developed by Baker et al.33 was used to assess whether an isolate would be detected, if present at a density of ≤ 250 parasites/μL, by an RDT detecting PfHRP2. The sequences from this study (N = 18) and previous studies (N = 96) were also analyzed for the identification and distribution of 13 major epitopes, ranging 8–15 amino acids, that are recognized by 11 HRP2-specific commercially available monoclonal antibodies (MAbs).15
We calculated prevalence to determine in how many sequences from this study (N = 18) and previous studies of Malagasy samples (N = 96) a specific amino acid repeat type or epitope is present. We also calculated frequency to determine how many times a specific amino acid repeat type or epitope occurs within each sequence from this study (N = 18) and previous studies (N = 96). Frequency data was presented as average frequency, calculated as (number of each amino acid repeat type or epitope within all sequences/total number of all amino acid repeat types or epitopes within all sequences) × 100.
Ethics statement.
This study was conducted following a protocol approved by the University Hospitals of Cleveland Institutional Review Board (#09-13-01), the Division of Microbiology and Infectious Diseases/NIAID/National Institutes of Health (NIH) (#13-0067), and the Madagascar Ministry of Health Ethics Committee (#099). Patients were enrolled after informed consent by a local doctor qualified in NIH Human Subjects Training. A parent or guardian provided written informed consent on behalf of participants under the age of 18 years.
RESULTS
Molecular diagnosis of Plasmodium spp. infections.
Molecular diagnosis using LDR-FMA41 determined that all five cultured isolates were positive only for P. falciparum infection. From the filter paper blood spot samples collected during our longitudinal surveillance study, we randomly selected 260 samples for inclusion in the present analysis. Polymerase chain reaction diagnosis of these blood spot samples from patients with suspected malaria seeking treatment at the health centers showed that 29% (75/260) were infected by Plasmodium spp. The LDR-FMA analysis showed that P. falciparum was present in 95% (71/75), P. vivax in 4% (3/75), and P. ovale in 3% (2/75) of the infected samples. One of these samples showed a mixed infection with P. falciparum and P. ovale.
Amplification of the pfhrp2 gene.
Genomic DNAs from five cultured isolates were amplified using the primer sets #1 and #3. Amplicons generated with the primer set #1 were visualized on 1.5% agarose gel as band sizes of 400–500 bp. Amplification with the primer set #3 showed band sizes of 800–900 bp on 1% agarose gel.
All 260 blood spot samples were subjected to pfhrp2 amplification using primer sets #2 and #3. Of these, 73 samples produced primer set #2 nest2 amplicons of approximately 250 bp on 2% agarose gel. Amplification with primer set #3 showed band sizes of 500–900 on 1% agarose gels for the same 73 samples that were positive with primer set #2.
A comparison between the two molecular assays, pfhrp2 PCR and P. falciparum detection by LDR-FMA, is presented in Table 1. This analysis showed that among the 73 samples that were pfhrp2 PCR positive, 68 samples were P. falciparum positive and five samples were P. falciparum negative. Among the 187 pfhrp2 PCR negative samples, 180 were not infected, three were infected with P. vivax, two were infected with P. ovale, and three were P. falciparum positive. The overall concordance between these two molecular assays was 97%, suggesting variation in amplification of two different target sequences.
Table 1.
Comparison of pfhrp2 PCR and Plasmodium falciparum LDR-FMA
| P. falciparum+ | P. falciparum− | Total | |
|---|---|---|---|
| Pfhrp2+ | 68 | 5 | 73 |
| Pfhrp2− | 3 | 184 | 187 |
| Total | 71 | 189 | 260 |
LDR-FMA = ligase detection reaction-fluorescent microsphere assay; PCR = polymerase chain reaction; pfhrp2 = Plasmodium falciparum histidine-rich protein 2.
Comparison with RDT analysis.
All five cultured isolates were RDT positive for P. falciparum. From our 260 blood spot samples, RDT data was available for 226 samples, of which microscopy data was available for 136 samples. In this analysis, only P. falciparum mono- and mixed-infection samples were included; three P. vivax mono-infection samples were removed. A comparison between RDT and microscopy data (N = 133) showed that the overall concordance between these two diagnoses was 91%. The sensitivity and specificity of the RDT were 96% and 88%, respectively.
A comparison between RDT data and P. falciparum detection by using LDR-FMA (N = 223) is presented in Table 2. In this analysis, 54 samples were positive for both RDT and P. falciparum, and these samples included those three that were pfhrp2 PCR negative but P. falciparum positive (Table 1). There were eight RDT negative but P. falciparum positive samples (RDT− P. falciparum+, = false negatives), all of which were pfhrp2 positive. This analysis also showed that 16 samples were RDT positive but P. falciparum negative by molecular diagnosis (RDT+ P. falciparum−, = false positives); 15 of them were pfhrp2 negative by PCR. The overall concordance between these two diagnoses was 89%. The sensitivity and specificity of the RDT were 87% and 90%, respectively.
Table 2.
Comparison of RDT and P. falciparum LDR-FMA
LDR-FMA = ligase detection reaction-fluorescent microsphere assay; RDT = rapid diagnostic test.
2, microscopy+; 6, microscopy−
7, microscopy+; 8, microscopy−; 1, microscopy not available
Among the eight samples that were RDT− P. falciparum+, six samples were microscopy negative, whereas two samples were microscopy positive; thus, these two samples were “true” false negatives. Among the 16 samples that were RDT+ P. falciparum−, a microscopy result was not available for one sample, seven samples were microscopy positive, whereas eight samples were microscopy negative; thus, these eight samples were “true” false positives.
Amplification of the pfhrp2 gene, combined with RDT analysis and P. falciparum detection by using LDR-FMA, showed that no sample was pfhrp2− RDT− P. falciparum+, meaning that there was no indication of pfhrp2 deletion.
Sequence analysis of pfhrp2 gene from cultured isolates.
Sequence analysis of the primer set #1 amplicons revealed that their actual sizes were: Ex30704, 442 bp; Ex2004, 451 bp; 2070803, 459 bp; Ex2002, 450 bp; and 3020205, 459 bp. Exon-1 was 69 bp (23 amino acids) in length in all five isolates. The nucleotide sequence of the intronic region ranged 147–165 bp in length. Sequence analysis also showed that exon-1 nucleotide/amino acid sequence was identical in all five isolates and was identical to the exon-1 sequence used as reference (U69551.1) (Figure 2).
Figure 2.
Alignment of amino acids in the exon-1 from the reference sequence and the five Plasmodium falciparum culture isolates from Madagascar.
Sequence analysis of the primer set #3 amplicons revealed that their actual sizes were: Ex30704, 859 bp (286 amino acids); Ex2004, 844 bp (281 amino acids); 2070803, 873 bp (291 amino acids); Ex2002, 844 bp (281 amino acids); and 3020205, 866 bp (289 amino acids). Each exon-2 sequence was unique, i.e., no sequence was identical to another. Furthermore, all these five sequences were different from the 96 sequences reported previously from Madagascar.32,34 Complete gene sequences, generated with primer sets #1 and #3, from this analysis were submitted to GenBank (accession number KX886207.1–KX8862011.1).
Sequence analysis of pfhrp2 gene from filter paper blood spots.
As mentioned earlier, we did not observe any sequence variation in exon-1 among the five cultured isolates. Therefore, we did not sequence any of the exon-1 nest2 amplicons generated from 73 blood spot samples using primer set #2.
From the 73 amplicons generated using primer set #3, we randomly selected nine amplicons. From these nine amplicons, a total of 13 exon-2 sequences were generated, ranging 528–877 bp (175–291 amino acids). These sequence sizes were similar to those reported previously from Madagascar34 and globally.32,33 A comparison among these 13 sequences revealed multiple-strain infections, inferred by the presence of two different sequences, in four of the nine samples. It also showed that 11 of them were unique, and one sequence was present in two different samples. All these 12 sequences (11 unique and one common) were different from the five exon-2 sequences generated from the cultured isolates. Finally, a comparison between these 12 sequences and the 96 sequences from the previous studies conducted in Madagascar32,34 showed that no sequence was shared between these two groups, i.e., each sequence was unique. Exon-2 sequences from this analysis were submitted to GenBank (accession number MF554693–MF554705).
To summarize the results from sequence analysis of the pfhrp2 gene, we found no variation in exon-1 (N = 5, all identical sequences) and large variation in exon-2 (N = 18, with 16 unique sequences) in our study samples.
Distribution of amino acid repeat types in pfhrp2 exon-2.
Translation of nucleotide sequences into protein sequences enabled classification into amino acid repeat types (called Baker repeats).32 Of the 24 repeats (1–24), 20 repeats (1–14, 19–24) are present in pfhrp2.32 Of these 20 repeats, we identified 12 repeats in our sequences (N = 18) (Table 3). The same 12 repeats were also present in the previous sequences (N = 96) from Madagascar (Table 3).32,34 In addition, two repeats were present only in the previous sequences. Despite differences in length, almost all exon-2 sequences started with type 1 repeat (AHHAHHVAD) and ended with type 12 repeat (AHHAAAHHEAATH). The other repeat types were dispersed throughout the protein sequence, except for type 10 repeat (AHHAAAHHATD), which mostly occurred near the terminal end before the end repeat, type 12.
Table 3.
Prevalence of Baker repeat types in pfhrp2 sequences
| Repeat type | Sequence | Prevalence | |
|---|---|---|---|
| This study (N = 18) | Previous studies (N = 96) | ||
| % (n) | % (n) | ||
| 1 | AHHAHHVAD | 94 (17) | 99 (95) |
| 2 | AHHAHHAAD | 100 (18) | 100 (96) |
| 3 | AHHAHHAAY | 94 (17) | 93 (89) |
| 4 | AHH | 22 (4) | 26 (25) |
| 5 | AHHAHHASD | 83 (15) | 80 (77) |
| 6 | AHHATD | 94 (17) | 100 (96) |
| 7 | AHHAAD | 94 (17) | 100 (96) |
| 8 | AHHAAY | 89 (16) | 98 (94) |
| 9 | AAY | 0 (0) | 0 (0) |
| 10 | AHHAAAHHATD | 83 (15) | 79 (76) |
| 11 | AHN | 0 (0) | 0 (0) |
| 12 | AHHAAAHHEAATH | 100 (18) | 100 (96) |
| 13 | AHHASD | 11 (2) | 7 (7) |
| 14 | AHHAHHATD | 0 (0) | 8 (8) |
| 15* | AHHAHHAAN | 0 (0) | 0 (0) |
| 16* | AHHAAN | 0 (0) | 0 (0) |
| 17* | AHHDG | 0 (0) | 0 (0) |
| 18* | AHHDD | 0 (0) | 0 (0) |
| 19 | AHHAA | 17 (3) | 3 (3) |
| 20 | SHHDD | 0 (0) | 0 (0) |
| 21 | AHHAHHATY | 0 (0) | 0 (0) |
| 22 | AHHAHHAGD | 0 (0) | 0 (0) |
| 23 | ARHAAD | 0 (0) | 1 (1) |
| 24 | AHHTHHAAD | 0 (0) | 0 (0) |
pfhrp2 = Plasmodium falciparum histidine-rich protein 2.
Not present in pfhrp2, present in pfhrp3 (Baker et al.32).
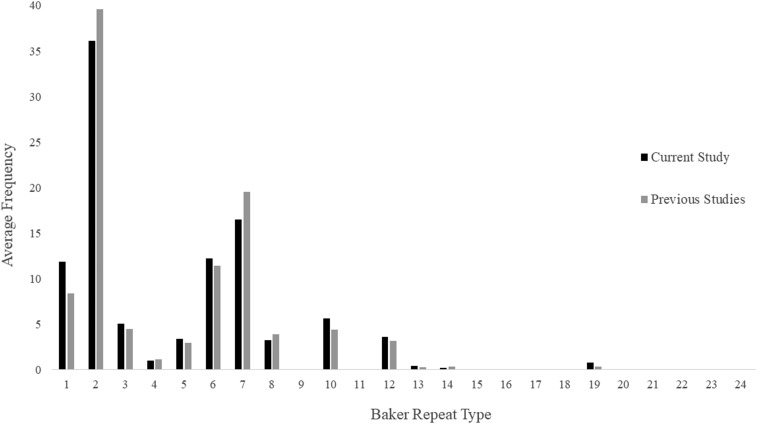
The prevalence of each repeat type in our and previous sequences is presented in Table 3. Repeat types 1, 2, 6, 7, and 12 were present in almost all sequences (94–100%). Repeat types 3, 5, 8, and 10 were also highly prevalent in all sequences (79–98%). Prevalence of repeat types 4, 13, and 19 was low to moderate in all samples (3–26%). The average frequency of each repeat within our and previous sequences is presented in Figure 3. Repeat types 2 and 7 were the most frequent within our present (39%, range = 4–14, mean = 11; 16%, range = 0–7, mean = 4, respectively) and previous (40%, range = 4–16, mean = 12; 20%, range = 2–13, mean = 6, respectively) sequences.
Figure 3.
Average frequency of amino acid repeat types (Baker repeat types) in the sequences from the current study (N = 18) and previous studies (N = 96) from Madagascar.
Baker repeat types and RDT performance.
A comparison of Baker repeat type 2 × type 7 numbers and all three diagnostic analyses, namely RDT, microscopy, and P. falciparum detection by LDR-FMA, for the 20 samples that were sequenced is presented in Table 4. Among the samples that were RDT positive, 12 had > 43 repeats, whereas five had < 43 repeats. Among the samples that were RDT negative, one sample had > 43 repeats, whereas two had < 43 repeats.
Table 4.
Baker repeat types and malaria infection diagnosis
| Sample | Repeat type number (2 × 7) | RDT | Microscopy | LDR-FMA |
|---|---|---|---|---|
| Ex2002* | 48 (12 × 4) | + | + | + |
| Ex2004* | 55 (11 × 5) | + | + | + |
| Ex30704* | 55 (11 × 5) | + | + | + |
| 2070803* | 44 (11 × 4) | + | + | + |
| 3020205* | 48 (12 × 4) | + | + | + |
| 1021506† | 10 (10 × 1) | + | + | + |
| Ext2246A† | 0 (12 × 0) | + | + | + |
| Ext2246B† | 20 (4 × 5) | + | + | + |
| Ext2263† | 28 (7 × 4) | + | + | + |
| Ex30507A† | 44 (11 × 4) | + | + | + |
| Ex30507B† | 44 (11 × 4) | + | + | + |
| Ex30508† | 91 (13 × 7) | + | + | + |
| Ext2229A† | 70 (14 × 5) | + | + | + |
| Ext2229B† | 78 (13 × 6) | + | + | + |
| Ext2223A† | 44 (11 × 4) | + | + | + |
| Ext2223B† | 50 (10 × 5) | + | + | + |
| 2100306† | 78 (13 × 6) | − | − | − |
| Ext2230† | 28 (7 × 4) | + | + | + |
| 3091101‡ | 0 (14 × 0) | − | + | + |
| 2011303‡ | 32 (8 × 4) | − | + | + |
LDR-FMA = ligase detection reaction-fluorescent microsphere assay; RDT = rapid diagnostic test.
GenBank accession number KX886207.1–KX8862011.1.
GenBank accession number MF554693–MF554705.
“true” false negatives (Table 2).
Major epitopes in pfhrp2 exon-2 targeted by MAbs in RDTs.
Epitope identification analysis on all sequences revealed that seven of the 13 major epitopes were present in almost all sequences (94–100%) (Table 5). Among these, three major epitopes, DAHHAHHA, AHHAADAHHA, and AHHAADAHH, were present within those sequences at much higher average frequencies (17–26%) than the other four major epitopes (3–9%). These three major epitopes have been shown to be recognized by 3A4 and PTL-3 (DAHHAHHA), C1-13 (AHHAADAHHA), and S2-5 and C2-3 (AHHAADAHH) MAbs.15
Table 5.
Distribution of major epitopes in pfhrp2 sequences
| This study (n = 18) | Previous studies (n = 96) | ||||
|---|---|---|---|---|---|
| Prevalence | Average frequency | Prevalence | Average frequency | ||
| Major epitope | % (n) | % | % (n) | % | MAb* |
| DAHHAHHA | 100 (18) | 19 | 100 (96) | 17 | 3A4, PTL-3 |
| DAHHAADAHH | 94 (17) | 8 | 100 (96) | 9 | 2G12-1C12 |
| DAHHVADAHH | 11 (2) | 0 | 0 (0) | 0 | 2G12-1C12 |
| YAHHAHHA | 100 (18) | 4 | 100 (96) | 4 | 1E1-A9, PTL-3 |
| DAHHAHHV | 94 (17) | 5 | 98 (94) | 3 | 1E1-A9 |
| HATDAHHAAD | 67 (12) | 1 | 83 (80) | 2 | A6-4 |
| HATDAHHAAA | 78 (14) | 2 | 88 (84) | 2 | A6-4 |
| AHHAADAHHA | 100 (18) | 23 | 100 (96) | 26 | C1-13 |
| DAHHAADAHHA | 94 (17) | 8 | 100 (96) | 9 | N7 |
| AHHAADAHH | 100 (18) | 27 | 100 (96) | 26 | S2-5, C2-3 |
| AHHASDAHH | 94 (17) | 2 | 79 (76) | 1 | S2-5 |
| TDAHHAADAHHAADA | 67 (12) | 1 | 76 (73) | 1 | TC-10 |
| AAYAHHAHHAAY | 0 (0) | 0 | 0 (0) | 0 | Genway |
MAb = monoclonal antibodies; pfhrp2 = Plasmodium falciparum histidine-rich protein 2.
Lee et al.15
DISCUSSION
Based on the amplification of both exon-1 and exon-2, together with the use of the SD Bioline Malaria Ag P.f/Pan RDT and molecular diagnosis of P. falciparum infection, we found no indication of pfhrp2 deletion in our study samples from central Madagascar. This result is consistent with those from the previous studies conducted 7–8 years ago in Madagascar,32,34 one of which was conducted countrywide.34 Given that pfhrp2 deletion has been reported from the continent of Africa within the past 5 years,26–31 resulting from the strong selective pressure on the P. falciparum population, no indication of deletion in our samples is encouraging. However, the observation that eight of 223 (3.6%) samples were RDT− P. falciparum+ (false negatives), two of which were microscopy positive, warrants continued monitoring.
The detection limit of the SD Bioline Malaria Ag P.f/Pan RDT, used in this present study, is 50 parasites/μL for malaria antigen HRP2 and 100 parasites/μL for malaria antigen pLDH (manufacturer’s note). Among our 223 study samples for which an RDT result was available, one sample had P. falciparum parasites 30/μL (an expert microscopist can detect ≥ 5–10 parasites/μL44). This sample was found to be RDT positive but pfhrp2 and Plasmodium spp. Polymerase chain reaction negative. Comparison of RDT and P. falciparum LDR-FMA results (N = 223) showed that the RDT sensitivity was 87%. Among the eight RDT− P. falciparum+ samples, all of which were pfhrp2 PCR positive, six were microscopy negative, indicating submicroscopic infections. Ironically, two samples that had 4,830 and 6,310 P. falciparum parasites/μL were also RDT negative (“true” false negatives). Although anti-PfHRP2 antibody levels in plasma were not measured in the present study, reduced sensitivity of PfHRP2-detecting RDTs has been seen among people with high anti-PfHRP2 antibody levels.11 The RDT specificity was 90%. Among the 16 RDT+ P. falciparum− samples, 15 of which were pfhrp2 negative, seven were microscopy positive, whereas eight were microscopy negative (“true” false positives). Regarding the seven microscopy-positive samples, one possibility that was not explored in this study is that in the absence of pfhrp2, the RDT positivity could be because of HRP3 detection16 (see Limitations). Regarding the eight microscopy negative samples, a reason for their RDT positivity could be the persistence of HRP2 antigenemia after antimalarial therapy,12 which needs to be further examined. Regardless of the reasons, the sensitivity and specificity results of this study, when compared with those of the previous study,8 substantiate the need to continually monitor the performance of the SD Bioline Malaria Ag P.f/Pan RDT in Madagascar.
In the present study, 89% of pfhrp2 exon-2 sequences were found to be unique. This observation is in agreement with previous observations from Madagascar: in a countrywide assessment of polymorphism in exon-2, 96% unique sequences were identified.34 In a global analysis of exon-2 sequence variation, 100% variability was observed in Malagasy samples.32 In this global analysis, the pfhrp2 sequence diversity was higher in countries with high transmission intensity such as in continental Africa (87–100%).32 A strong correlation between malaria transmission intensity and pfhrp2 diversity may be expected, as malaria infections in high transmission settings, including those in Madagascar, often involve multiple-strain infections.45 In our study, two different exon-2 sequences were present in four of the nine blood spot samples, implying multiple-strain infections. On the other hand, all five pfhrp2 exon-1 sequences were identical. They were identical to the exon-1 sequence from a Brazilian isolate,43 and those from 29 isolates from French Guiana.46 This low-level variation in exon-1 may be because of the fact that exon-1 codes for a signal peptide.47 The signal sequences of several processed and translocated P. falciparum proteins are known, and there is evidence that these parts of the protein are less variable among different strains.48
Although comparative analysis of the exon-2 sequences showed that no two sequences were the same between our study (N = 18) and previous studies (N = 96), the prevalence (Table 3) and frequency (Figure 3) of each of the 12 Baker repeat types, common between these two groups, were comparable. This was also true for the repeat types 2 and 7, which were used in a predictive model to assess whether an isolate would be detected, if present at a density of ≤ 250 parasites/μL, by an RDT detecting PfHRP2.33,34 Previously, this analysis predicted that 9% (range 6–14%) of Malagasy isolates with parasite densities ≤ 250 parasites/μL would not be detected.34 In the present study, using the SD Bioline Malaria Ag P.f/Pan RDT, we found that correlation between the predictive model and the performance of this RDT was unclear.
Our 17 of the 18 exon-2 sequences were generated from samples that were positive by all three diagnostic analyses (RDT, P. falciparum LDR-FMA, and microscopy) (Table 4). Among these 17 sequences, we identified five sequences where the number of type 2 × type 7 repeats were < 43, ranging from 0 (12 × 0, respectively) to 28 (7 × 4, respectively). The parasitemia in these samples ranged 400–15,450/μL. On the other hand, one sequence was generated from a sample that was negative by all three diagnostic analyses; a very low-level parasitemia and variation in amplification between two different PCRs may explain this result. In this sequence, the number of type 2 × type 7 repeats was 78 (13 × 6, respectively). In addition, when we sequenced the two “true” false negative samples that had 4,830 and 6,310 P. falciparum parasites/μL, the number of type 2 × type 7 repeats were 0 (14 × 0, respectively) and 32 (8 × 4, respectively), respectively. These results generally agree with those reported by Baker et al.32 in a later study, which did not find a correlation between PfHRP2 structure and the overall RDT detection rates. Although limited, these results also suggest that the diversity of pfhrp2 may not be a primary factor influencing the performance of the SD Bioline Malaria Ag P.f/Pan RDT in Madagascar. This is also supported by the finding that there was no correlation between pfhrp2 sequence length or repeat type and PfHRP2 plasma concentration in African children.49
Studies that evaluated the effect of pfhrp2 sequence variation on the binding of MAbs1 have led to the characterization of 13 major epitopes recognized by 11 MAbs (Table 5).14,15 Using the amino acid sequences encoded by a total of 448 pfhrp2 genes originating from P. falciparum isolates collected from a range of geographical regions,32 these 13 major epitopes were found to vary in their prevalence and average frequencies.15 Because durability to heat is critical to the usefulness of RDTs in the tropical field setting, thermostability of those MAbs was determined, and was also found to vary considerably.15 Results of these experiments enabled the identification of MAbs with the most desirable characteristics for inclusion in RDTs. To our knowledge, the information regarding PfHRP2-specific MAbs being used in the SD Bioline Malaria Ag P.f/Pan RDT is not disclosed by the manufacturer. When we performed epitope identification in all Madagascar sequences, we observed that epitopes DAHHAHHA, AHHAADAHHA, and AHHAADAHH were present in high prevalence and frequencies. Epitopes AHHAADAHHA and DAHHAHHA are recognized by MAbs C1-13 and PTL-3, respectively, both of which have shown the best potential (based on the prevalence and frequency of epitopes together with the heat durability profile) for use in an RDT.15 Our epitope identification results should be helpful for studies comparing performances of different RDTs in Madagascar, and for those evaluating new antibodies for the development of improved RDTs.50,51
LIMITATIONS
In our study, we did not attempt to amplify the pfhrp2 paralog gene pfhrp3. HRP3 has a sequence homology of more than 75% in the tandem repeat region to HRP2 and is recognized by most anti-HRP2 MAbs including C1-13.14,15 Normally, this cross-reactivity may not have a major impact on the sensitivity of the RDTs because of the lower abundance of HRP3.10 However, in situations of low-level parasitemia or when parasites have pfhrp2 deletions,16 it could enhance the sensitivity of the RDTs. In our study, three samples, positive by P. falciparum LDR-FMA, were pfhrp2 PCR negative but RDT positive (false negatives in Table 1). In addition, seven samples, negative by LDR-FMA but positive by microscopy, were pfhrp2 PCR negative but RDT positive (false positives in Table 2). This raises the possibility that we may have missed pfhrp2 deletion because the RDT positive results for all these samples were because of HRP3 detection.
The other limitation might be that our samples had limited geographic origin, all were from a single location in Madagascar. Two previous studies analyzed pfhrp2 sequence variation in Madagascar and did not find pfhrp2 deletion.32,34 One of these studies was conducted countrywide but did not use any RDT.34 Further studies should evaluate the performance of the PfHRP2-detecting RDT together with pfhrp2 variation elsewhere in Madagascar.
Three major conclusions can be drawn from this study. First, pfhrp2 gene deletion, assessed by analyzing both exons, was not found in the samples screened here. Second, the overall performance of the SD Bioline Malaria Ag P.f/Pan RDT (sensitivity and specificity 87% and 90%, respectively) includes the observation that the RDT did not detect eight P. falciparum PCR-positive samples (two microscopy-positive samples). Third, the RDT performance did not seem to be correlated with pfhrp2 gene variation, as described by Baker et al.32 Primary, community-based healthcare facilities in Madagascar do not have the capacity for microscopy-based diagnosis, this is limited to referral hospitals.37 A heavy reliance is therefore placed on RDTs for point-of-care malaria diagnosis in Madagascar. The observed absence of pfhrp2 deletion from the samples screened here supports this continued dependency on this method of diagnosis. However, because pfhrp2 gene deletions yielding false negative RDTs have been confirmed in Africa and Asia, and RDT negative samples without this gene deletion were found in this study, it is essential from a public health perspective to perform routine screening so that continued efficacy of RDT-based diagnosis is ensured in Madagascar.
Supplementary Material
Acknowledgments:
We thank all study participants, local health officials, field doctors, and project technicians (Michel Abrahim Marolahy, Claude Louis de Gonzague Rajaona, Stéphane Rabearimanana, Brunette Razanadrazanina, Fanomezantsoa Ralinoro, Noeline Rasoarilalao, and Yvon Ralaiseheno) for their participation and support. R. K. M. thanks D’Arbra Blankenship for her excellent technical assistance during P. falciparum in vitro cultivation. We are thankful to Simone Edelheit and Milena Rajak (Genomics Core Facility, C.W.R.U.) for performing the pfhrp2 sequencing. Study permission from Association ASA (Ankohonana Sahirana Arenina, www.asa-madagascar.org), Madagascar Ministry of Health, and logistical support of Pact Madagascar (www.pact-madagascar.org) are also acknowledged.
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1.Parra ME, Evans CB, Taylor DW, 1991. Identification of Plasmodium falciparum histidine-rich protein 2 in the plasma of humans with malaria. J Clin Microbiol 29: 1629–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadle C, Long GW, Weiss WR, McElroy PD, Maret SM, Oloo AJ, Hoffman SL, 1994. Diagnosis of malaria by detection of Plasmodium falciparum HRP-2 antigen with a rapid dipstick antigen-capture assay. Lancet 343: 564–568. [DOI] [PubMed] [Google Scholar]
- 3.Moody A, 2002. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev 15: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabarijaona LP, Ariey F, Matra R, Cot S, Raharimalala AL, Ranaivo LH, Le Bras J, Robert V, Randrianarivelojosia M, 2006. Low autochtonous urban malaria in Antananarivo (Madagascar). Malar J 5: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Randrianasolo L, Tafangy PB, Raharimalala LA, Ratsimbasoa AC, Randriamanantena A, Randrianarivelojosia M, 2007. Rapid diagnostic test for malaria: preliminary study in Madagascar in 2003. Sante 17: 69–73. [PubMed] [Google Scholar]
- 6.Howes RE, Mioramalala SA, Ramiranirina B, Franchard T, Rakotorahalahy AJ, Bisanzio D, Gething PW, Zimmerman PA, Ratsimbasoa A, 2016. Contemporary epidemiological overview of malaria in Madagascar: operational utility of reported routine case data for malaria control planning. Malar J 15: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakotonirina H, Barnadas C, Raherijafy R, Andrianantenaina H, Ratsimbasoa A, Randrianasolo L, Jahevitra M, Andriantsoanirina V, Menard D, 2008. Accuracy and reliability of malaria diagnostic techniques for guiding febrile outpatient treatment in malaria-endemic countries. Am J Trop Med Hyg 78: 217–221. [PubMed] [Google Scholar]
- 8.Ratsimbasoa A, Fanazava L, Radrianjafy R, Ramilijaona J, Rafanomezantsoa H, Menard D, 2008. Evaluation of two new immunochromatographic assays for diagnosis of malaria. Am J Trop Med Hyg 79: 670–672. [PubMed] [Google Scholar]
- 9.Ravaoarisoa E, Andriamiandranoro T, Raherinjafy R, Jahevitra M, Razanatsiorimalala S, Andrianaranjaka V, Randrianarivelojosia M, 2017. Evaluation of the OnSite malaria rapid test performance in Miandrivazo, Madagascar. Bull Soc Pathol Exot 110: 254–259. [DOI] [PubMed] [Google Scholar]
- 10.Baker J, Gatton ML, Peters J, Ho MF, McCarthy JS, Cheng Q, 2011. Transcription and expression of Plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One 6: e22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho MF, et al. 2014. Circulating antibodies against Plasmodium falciparum histidine-rich proteins 2 interfere with antigen detection by rapid diagnostic tests. Malar J 13: 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iqbal J, Siddique A, Jameel M, Hira PR, 2004. Persistent histidine-rich protein 2, parasite lactate dehydrogenase, and panmalarial antigen reactivity after clearance of Plasmodium falciparum monoinfection. J Clin Microbiol 42: 4237–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, Cunningham J, 2014. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J 13: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee N, Baker J, Andrews KT, Gatton ML, Bell D, Cheng Q, McCarthy J, 2006. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J Clin Microbiol 44: 2773–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee N, Gatton ML, Pelecanos A, Bubb M, Gonzalez I, Bell D, Cheng Q, McCarthy JS, 2012. Identification of optimal epitopes for Plasmodium falciparum rapid diagnostic tests that target histidine-rich proteins 2 and 3. J Clin Microbiol 50: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamboa D, et al. 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One 5: e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akinyi Okoth S, et al. 2015. Variation in Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and Plasmodium falciparum histidine-rich protein 3 (pfhrp3) gene deletions in Guyana and Suriname. PLoS One 10: e0126805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorado EJ, Okoth SA, Montenegro LM, Diaz G, Barnwell JW, Udhayakumar V, Murillo Solano C, 2016. Genetic characterisation of Plasmodium falciparum isolates with deletion of the pfhrp2 and/or pfhrp3 genes in Colombia: the Amazon region, a challenge for malaria diagnosis and control. PLoS One 11: e0163137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murillo Solano C, et al. 2015. Deletion of Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes in Colombian parasites. PLoS One 10: e0131576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rachid Viana GM, et al. 2017. Histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in Plasmodium falciparum isolates from select sites in Brazil and Bolivia. PLoS One 12: e0171150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdallah JF, et al. 2015. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar J 14: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N, 2016. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One 11: e0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas S, Tomar D, Rao DN, 2005. Investigation of the kinetics of histidine-rich protein 2 and of the antibody responses to this antigen, in a group of malaria patients from India. Ann Trop Med Parasitol 99: 553–562. [DOI] [PubMed] [Google Scholar]
- 24.Nima MK, Hougard T, Hossain ME, Kibria MG, Mohon AN, Johora FT, Rahman R, Haque R, Alam MS, 2017. Case report: a case of Plasmodium falciparum hrp2 and hrp3 gene mutation in Bangladesh. Am J Trop Med Hyg 97: 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atroosh WM, et al. 2015. Genetic variation of pfhrp2 in Plasmodium falciparum isolates from Yemen and the performance of HRP2-based malaria rapid diagnostic test. Parasit Vectors 8: 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amoah LE, Abankwa J, Oppong A, 2016. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP 2: based malaria rapid diagnostic tests in Ghana. Malar J 15: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koita OA, et al. 2012. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg 86: 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozycki CT, Umulisa N, Rulisa S, Mwikarago EI, Musabyimana JP, Habimana JP, Karema C, Krogstad DJ, 2017. False-negative malaria rapid diagnostic tests in Rwanda: impact of Plasmodium falciparum isolates lacking hrp2 and declining malaria transmission. Malar J 16: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parr JB, et al. 2017. Pfhrp2-deleted Plasmodium falciparum parasites in the Democratic Republic of the Congo: a national cross-sectional survey. J Infect Dis 216: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson OJ, Slater HC, Verity R, Parr JB, Mwandagalirwa MK, Tshefu A, Meshnick SR, Ghani AC, 2017. Modelling the drivers of the spread of Plasmodium falciparum hrp2 gene deletions in sub-Saharan Africa. Elife 6: e25008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurtz N, et al. 2013. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J 12: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker J, et al. 2010. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J 9: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker J, McCarthy J, Gatton M, Kyle DE, Belizario V, Luchavez J, Bell D, Cheng Q, 2005. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J Infect Dis 192: 870–877. [DOI] [PubMed] [Google Scholar]
- 34.Mariette N, Barnadas C, Bouchier C, Tichit M, Menard D, 2008. Country-wide assessment of the genetic polymorphism in Plasmodium falciparum and Plasmodium vivax antigens detected with rapid diagnostic tests for malaria. Malar J 7: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howes RE, Chan ER, Rakotomanga TA, Schulte S, Gibson J, Zikursh M, Franchard T, Ramiranirina B, Ratsimbasoa A, Zimmerman PA, 2017. Prevalence and genetic variants of G6PD deficiency among two Malagasy populations living in Plasmodium vivax-endemic areas. Malar J 16: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menard D, et al. 2010. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA 107: 5967–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Malaria Control Programme of Madagascar , 2015. Plan stratégique de lutte contre le paludisme Madagascar 2013–2017. Consolider les acquis en vue de l'élimination du paludisme à Madagascar. Version revisée pour 2015–2017.
- 38.Normark J, 2008. Freezing of patient isolates and strains with glycerolyte. Moll K, Ljungström I, Perlmann H, Scherf A, Wahlgren M, eds. Methods in Malaria Research. Manassas, VA: Malaria Research and Reference Reagent Resource Center, American Type Culture Collection, 14. [Google Scholar]
- 39.Grimberg BT, Erickson JJ, Sramkoski RM, Jacobberger JW, Zimmerman PA, 2008. Monitoring Plasmodium falciparum growth and development by UV flow cytometry using an optimized Hoechst-thiazole orange staining strategy. Cytometry A 73: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blomqvist K, 2008. Thawing of glycerolyte-frozen parasites with NaCl. Moll K, Ljungström I, Perlmann H, Scherf A, Wahlgren M, eds. Methods in Malaria Research. Manassas, VA: Malaria Research and Reference Reagent Resource Center, American Type Culture Collection, 15. [Google Scholar]
- 41.McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA, 2006. Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg 74: 413–421. [PMC free article] [PubMed] [Google Scholar]
- 42.Walker-Jonah A, Dolan SA, Gwadz RW, Panton LJ, Wellems TE, 1992. An RFLP map of the Plasmodium falciparum genome, recombination rates and favored linkage groups in a genetic cross. Mol Biochem Parasitol 51: 313–320. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan DJ, Jr, Ayala YM, Goldberg DE, 1996. An unexpected 5′ untranslated intron in the P. falciparum genes for histidine-rich proteins II and III. Mol Biochem Parasitol 83: 247–251. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman PA, Howes RE, 2015. Malaria diagnosis for malaria elimination. Curr Opin Infect Dis 28: 446–454. [DOI] [PubMed] [Google Scholar]
- 45.Rice BL, Golden CD, Anjaranirina EJ, Botelho CM, Volkman SK, Hartl DL, 2016. Genetic evidence that the Makira region in northeastern Madagascar is a hotspot of malaria transmission. Malar J 15: 596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trouvay M, Palazon G, Berger F, Volney B, Blanchet D, Faway E, Donato D, Legrand E, Carme B, Musset L, 2013. High performance of histidine-rich protein 2 based rapid diagnostic tests in French Guiana are explained by the absence of pfhrp2 gene deletion in P. falciparum. PLoS One 8: e74269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma YD, 1988. Genomic organization, structure and possible function of histidine-rich proteins of malaria parasites. Int J Biochem 20: 471–477. [DOI] [PubMed] [Google Scholar]
- 48.Mackay M, Goman M, Bone N, Hyde JE, Scaife J, Certa U, Stunnenberg H, Bujard H, 1985. Polymorphism of the precursor for the major surface antigens of Plasmodium falciparum merozoites: studies at the genetic level. EMBO J 4: 3823–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramutton T, et al. 2012. Sequence variation does not confound the measurement of plasma PfHRP2 concentration in African children presenting with severe malaria. Malar J 11: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leow CH, Jones M, Cheng Q, Mahler S, McCarthy J, 2014. Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2. Malar J 13: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ravaoarisoa E, Zamanka H, Fusai T, Bellalou J, Bedouelle H, Mercereau-Puijalon O, Fandeur T, 2010. Recombinant antibodies specific for the Plasmodium falciparum histidine-rich protein 2. MAbs 2: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.