Abstract
Fusarium oxysporum f. sp. cubense (Foc), the causal agent of Fusarium wilt or Panama disease on banana, is one of the major constraints in banana production worldwide. Indonesia is the centre of origin for wild and cultivated bananas, which likely co-evolved with Foc. This study explored the widest possible genetic diversity of Foc by sampling across Indonesia at 34 geographically and environmentally different locations in 15 provinces at six islands. This resulted in a comprehensive collection of ∼200 isolates from 40 different local banana varieties. Isolates were identified and assessed using sequence analysis of the translation elongation factor-1alpha (tef1), the RNA polymerase II largest subunit (rpb1), and the RNA polymerase II second largest subunit (rpb2). Phylogenetic analyses of these genes allowed the identification of 180 isolates of Fusarium oxysporum f. sp. cubense (Foc), and 20 isolates of the Fusarium fujikuroi species complex (FFSC), the Fusarium incarnatum-equiseti species complex (FIESC), and the Fusarium sambucinum species complex (FSSC). Further analyses, incorporating a worldwide collection of Foc strains, revealed nine independent genetic lineages for Foc, and one novel clade in the Fusarium oxysporum species complex (FOSC). Selected isolates from each lineage were tested on the banana varieties Gros Michel and Cavendish to characterise their pathogenicity profiles. More than 65 % of the isolates were diagnosed as Tropical Race 4 (Foc-TR4) due to their pathogenicity to Cavendish banana, which supports the hypothesis that Foc-TR4 is of Indonesian origin. Nine independent genetic lineages for Foc are formally described in this study. This biodiversity has not been studied since the initial description of Foc in 1919. This study provides a detailed overview of the complexity of Fusarium wilt on banana and its diversity and distribution across Indonesia.
Key words: Morphology, New species, Panama disease, Pathogenicity, Tropical Race 4, 11 New taxa
Taxonomic novelties: New species: Fusarium cugenangense N. Maryani, L. Lombard, Kema & Crous; F. duoseptatum N. Maryani, L. Lombard, Kema & Crous; F. grosmichelii N. Maryani, L. Lombard, Kema & Crous; F. hexaseptatum N. Maryani, L. Lombard, Kema & Crous; F. kalimantanense N. Maryani, L. Lombard, Kema & Crous; F. odoratissimum N. Maryani, L. Lombard, Kema & Crous; F. phialophorum N. Maryani, L. Lombard, Kema & Crous; F. purpurascens N. Maryani, L. Lombard, Kema & Crous; F. sangayamense N. Maryani, L. Lombard, Kema & Crous; F. tardichlamydosporum N. Maryani, L. Lombard, Kema & Crous; F. tardicrescens N. Maryani, L. Lombard, Kema & Crous
Introduction
Indonesia is one of the main centres of origin for banana in South-East Asia (Valmayor et al. 1999). Edible banana cultivars are descendants from two ancestral wild Musa species, Musa acuminata Colla (AA, 2n = 22) and Musa balbisiana Colla (BB, 2n = 22) (Simmonds 1962). These diversified into various edible varieties comprising diploids (AA, BB), triploids (AAA, AAB, ABB) and tetraploids (ABBB). Indonesia is the main contact area between species and subspecies of wild banana in sub-centres of diversity (Perrier et al. 2011) and, therefore, represents the primary gene centre for banana, resulting in a huge phenotypic and genotypic diversity. Indonesia is among the top 10 banana producing countries (FAOSTAT 2017) with over 200 varieties that are presently grown in almost every region of the Indonesian archipelago (Nasution 1993). The actual number of identified cultivated banana varieties could easily surpass 500. Banana is one of Indonesia's primary fruit commodities (BPS 2017), with most production supplying the domestic market.
Despite this great diversity and high popularity of bananas, there are some constraints on production. The most important of these is fungal diseases, including Fusarium wilt, also known as Panama disease (Stover 1962a). Fusarium wilt is caused by the soil-borne fungus Fusarium oxysporum f. sp. cubense (Foc), which first appeared in the 1900s in a banana plantation on Java (Stover 1962a) and thereafter disseminated to other banana production areas in Indonesia and beyond. This devastating agent of wilt on banana was first reported in the literature from samples collected in a Cuban banana plantation, and it subsequently gained notoriety as Fusarium cubense (Smith 1910).
The history of Fusarium wilt on banana goes back to the 20th century when this disease eliminated thousands of hectares of the favoured Gros Michel banana in Central America. The outbreak evolved into one of the worst plant epidemics of all times. The discovery of resistant Cavendish bananas eventually quenched the epidemic and the variety was so successful that it was disseminated around the world until it attained its current predominance in the global banana trade. The resistance of Cavendish bananas to the so-called Foc-Race1 strains, which caused the epidemic in Gros Michel is unique and durable. The risk of global monocultures is evident and problems surfaced again once other pathogenic Fusarium oxysporum strains appeared that were able to cause Fusarium wilt in Cavendish plantations. A harmful strain was initially reported from Taiwan, from whence it spread further into South-East Asia, and recently to the Indian subcontinent, the Middle East and Africa (Ordonez et al. 2015). The ongoing epidemic in Cavendish bananas is caused by a unique genotype, Vegetative Compatibility Group (VCG) 01213, of Foc and is called Tropical Race 4 (TR4). It has caused significant losses in commercial and subsistence production areas of Taiwan, Malaysia, and the northern territories of Australia (Su et al., 1986, Gerlach et al., 2000, Hermanto et al., 2009). In Indonesia, Nasir et al. (1999) reported that Fusarium wilt occurred from the Aceh province of Sumatra in the far west, to the far eastern Papua province. Losses in export Cavendish plantations in southern Sumatra have exceeded 70 %. In Northern Sumatra over 1 000 ha of plantations were destroyed within 3 yr after the appearance of the disease in this area (Nasir et al. 1999). Not only was Cavendish affected, but also many local popular varieties named in Bahasa Indonesia with ‘Pisang’ (=‘banana’) variety names, such as Pisang Raja Bulu, P. Raja Sereh, P. Ambon, P. Mas and P. Barangan, were damaged. The affected varieties are very important for the local markets (Hermanto et al. 2009).
To date, no control method has yet been identified or successfully implemented to effectively manage TR4. This is further complicated by the soil-borne nature of Foc and its ability to produce persistent chlamydospores that contaminate soils for decades (Booth 1971). Essentially, there are presently no control methods, except prevention by using pathogen-free tissue culture plants planted in non-infested soil (Ploetz 1994), and the adoption of quarantine strategies. However, these practices are mostly applied in large commercial plantations, but not in smallholder settings. Evidently, the development of new resistant banana cultivars would be the most effective control strategy to follow, and therefore research on the diversity of this pathogen is essential, particularly since it has been shown to be polyphyletic (O'Donnell et al., 1998, O'Donnell et al., 2009). It is therefore essential to acquire a better understanding of the differences between the genetic lineages for developing control strategies, and for effective resistance breeding.
In Fusarium systematics, Foc belongs to the Fusarium oxysporum species complex (FOSC). Four clades of FOSC have been identified using translation elongation factor 1-alpha (tef1) and mitochondrial subunit rDNA (mtssu), with Foc isolates clustering as basal lineage (O'Donnell et al. 2004). The incorporation of Foc isolates from native host populations, especially those from indigenous ecosystems, will be of great importance for diversity studies of this complex species.
Diversity studies on Foc isolates were conducted by using various physiological and molecular methods, which included VCGs (Moore et al. 1993), random amplified polymorphic DNA markers (RAPDs; Bentley et al. 1995), restriction fragment length polymorphisms (RFLPs; Koenig et al. 1997), amplified fragment length polymorphism (AFLP; Groenewald et al. 2006) and DNA sequence analyses (O'Donnell et al. 1998). These studies showed that the South-East Asian population of this fungus exhibits a high degree of variation, suggesting that Foc lineages co-evolved with their hosts in South-East Asia (Ploetz & Pegg 1997). However, these studies used Foc isolates from various disconnected geographical areas and lacked evidence on genetic diversity from the genetic centre of banana diversity, which is likely also the origin of the co-evolving Foc (Buddenhagen 2007). It has alternatively been suggested that Foc has multiple independent evolutionary origins, both within and outside the Musa genetic centre (Bentley et al. 1998). Using the phylogenetic genealogical approach, O'Donnell et al. (1998) identified five independent genetic lineages of Foc in a global population. Using a similar approach and additional data, Fourie et al. (2009) found three additional lineages. However, neither of these studies included Indonesian populations, and hence only limited information is available on the diversity of Foc at the centre of origin of banana.
Here, we explore the genetic diversity among Indonesian Foc strains that were isolated from local banana varieties in various different ecosystems across the country. This overview of the complexity of Fusarium wilt of banana enables us to greatly improve our knowledge of the taxonomic and phylogenetic position of Foc in the FOSC.
Materials and methods
Isolates
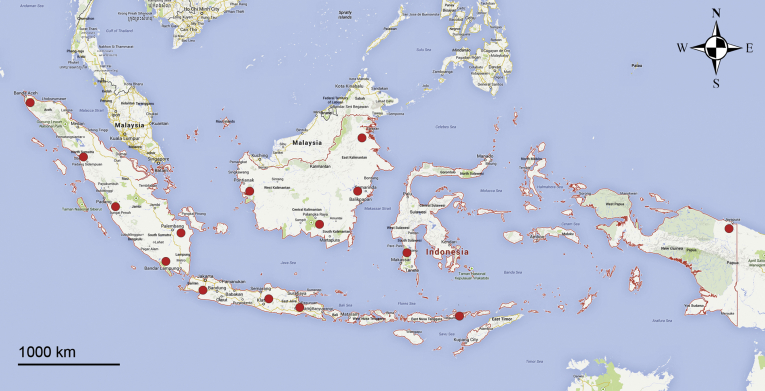
A comprehensive survey of Fusarium wilt of banana was undertaken in Indonesia. In total, 34 locations in 15 provinces were visited, representing the main banana-producing regions in Java, Sumatra, Kalimantan, Sulawesi, Papua, and Nusa Tenggara (Table 1, Fig. 1). Sampling expeditions to the former three islands were undertaken in 2014, whereas the other islands were sampled in 2015. Sampling locations were identified in two to three different regions in each province. Diagnostic specimen were collected from diseased banana plants displaying typical Fusarium wilt symptoms: yellowing of older leaf margins, collapsed leaves at the petioles, and pseudostem discolouration and splitting. The pseudostems of the diseased plants were cut and discoloured vascular strands were sampled and placed on sterile filter paper to dry, and were eventually packed in a paper envelope. Global positioning coordinates were recorded and ecological parameters, including soil pH, light intensity and vegetation of the sampling area were collected at each site. For each banana plant sampled, the youngest (cigar) leaf was taken for ploidy identification of the germplasm by flow-cytometry analyses and morphological characterisation following Valmayor et al. (1999) and Simmonds & Shepherd (1955), as well as in-situ comparisons with local banana varieties in the Musa collection at the Indonesian Institute of Sciences (LIPI) Cibinong, Bogor, Indonesia.
Table 1.
Names and geographical details of 34 sampling locations in Indonesia for establishing the Indonesian Fusarium oxysporum f. sp. cubense collection.
| Province | District | GPS |
||
|---|---|---|---|---|
| Long. | Lat. | Alt. (m) | ||
| East Kalimantan | Kutai Timur | 117.62 | 0.68 | 57 |
| Benajam | 116.77 | −1.62 | 21 | |
| Central Kalimantan | Kapuas Timur | 114.48 | −3.10 | 16 |
| Katingan | 113.42 | −1.71 | 35 | |
| Palangkaraya | 114.02 | −2.43 | 18 | |
| South Kalimantan | Kota Baru | 116.22 | −2.58 | 118 |
| Tanah Bumbu | 115.74 | −3.63 | 13 | |
| Banjar | 115.03 | −3.41 | 34 | |
| West Borneo | Kubu Raya | 109.29 | −0.06 | 8 |
| Pontianak | 109.34 | −0.04 | 17 | |
| West Java | Bogor | 107.10 | −6.68 | 657 |
| Cianjur | 107.10 | −7.02 | 875 | |
| Sukabumi | 106.79 | −7.01 | 263 | |
| Central Java | Kendal | 110.35 | −7.20 | 794 |
| Semarang | 110.59 | −7.00 | 9 | |
| Demak | 110.74 | −7.06 | 21 | |
| East Java | Lumajang | 113.11 | −8.08 | 637 |
| Bondowoso | 113.94 | −8.09 | 379 | |
| Purwodadi | 112.75 | −7.82 | 491 | |
| Jember | 113.68 | −8.24 | 39 | |
| Aceh | Jantho Aceh Besar | 95.63 | 5.35 | 133 |
| North Sumatra | Karo | 98.25 | 3 | NA |
| Brastagi | 98.51 | 3.19 | NA | |
| West Sumatra | Bukittinggi | 100.38 | −0.29 | NA |
| Padang | 100.35 | −0.94 | NA | |
| South Sumatra | Ogan Ilir | 104.70 | −3.29 | 27 |
| Palembang | 104.75 | −2.99 | NA | |
| Lampung | Way Jepara | 105.54 | −5.56 | NA |
| Papua | Sentani Jayapura | 140.83 | −2.65 | NA |
| South Sulawesi | Barru | 119.62 | −4.08 | 8 |
| Bone | 120.02 | −4.62 | 101 | |
| Maros | 119.63 | −5.10 | 48 | |
| Sidreng Rappang | 119.69 | −3.93 | 165 | |
| East Nusa Tenggara | Sikka Flores | 122.37 | −8.61 | 20 |
Fig. 1.
Map of sampling collection in 2014–2015 in the island of Java, Sumatra, Kalimantan, Sulawesi, Papua, and Flores.
Isolation
The dried pseudostem samples were cut into pieces of 2 × 3 cm and plated on Komada medium (Komada 1975). After approximately 2 d, fungal colonies resembling Fusarium were transferred to potato dextrose agar (PDA) plates (Leslie & Summerell 2006). Axenic cultures were derived by streaking a small amount of conidia, collected with the tip of an inoculation needle, on water agar (WA) plates, which allowed conidia to separate. After 24 h of incubation, plates were observed under a dissection microscope at 50× magnification and single germinating conidia were collected and transferred to PDA. Monospore isolates were either maintained on PDA or in 20 % (v/v) glycerol at −80 °C. All isolates were deposited in the Indonesian Culture Collection (InaCC) Cibinong, Indonesia. Twenty-four Foc isolates, representing the known VCG's (Ordonez et al. 2015) in the global Foc collection were included for phylogenetic analyses.
DNA isolation, amplification and analyses
Total genomic DNA was extracted from axenic isolates grown for 7 d on PDA, using the DNA Wizard Magnetic DNA Purification System for Food kit (Promega, USA) following the protocols provided by the manufacturer. Partial gene sequences were determined for the RNA polymerase largest subunit gene (rpb1) using primers RPB1-Fa & RPB1-G2R (O'Donnell et al. 2010), the RNA polymerase second largest subunit gene (rpb2) using primers RPB2-5f2 & RPB2-7cr (O'Donnell et al. 2010), and the translation elongation factor 1-alpha gene (tef1) using primers EF1 & EF2 (O'Donnell et al. 1998). Amplicons were sequenced in both directions using the same primer pairs as were used for amplification to ensure integrity of the sequences.
Consensus sequences were determined and assembled using MEGA v. 6 (Tamura et al. 2013) and compared to representative sequences from previous studies (O'Donnell et al., 1998, Fourie et al., 2009, Ordonez et al., 2015). Subsequent alignments for each individual locus were generated using MAFFT v. 7.110 (Katoh & Standley 2013) and manually corrected if necessary. The individual sequences generated in this study were compared with those maintained in the Fusarium MLST database (http://www.westerdijkinstitute.nl/fusarium/) and the NCBI's GenBank, and relevant sequences were included in the subsequent phylogenetic inference. Phylogenetic congruencies of the three loci were tested using a 70 % reciprocal bootstrap criterion (Mason-Gamer & Kellogg 1996).
Phylogenetic inference in this study was based on Maximum Likelihood (ML) and Bayesian Inference (BI). The ML analysis was performed using RAxML v. 8. (randomised accelerated (sic) maximum likelihood for high performance computing) (Stamatakis 2014) through RAxML BlackBox (http://embnet.vital-it.ch/raxml-bb/index.php). Bootstrap support (BS) was determined automatically by the software to assess the robustness of the analyses. The BI analysis was performed using MrBayes v. 3.2 (Ronquist et al. 2012). A Markov Chain Monte Carlo (MCMC) algorithm of four chains was initiated in parallel from a random tree topology with a heating parameter set at 0.3. The MCMC analyses lasted until the average standard deviations of split frequencies were below 0.01 with phylogenies saved every 1 000 generations. The first 25 % of saved phylogenies were discarded as the “burn-in” phase and posterior probabilities (PP) were determined from the remaining phylogenies. All the sequences generated in this study were deposited in the European Nucleotide Archive (ENA) and the alignments in TreeBASE.
Morphology
All Foc isolates were grown on carnation leaf agar (CLA; Fisher et al. 1982), synthetic low-nutrient agar (SNA; Nirenberg 1981) and PDA to induce sporulation under continuous light (Osram L18W/840 Cool White) for 7 d at 25 °C. Growth rates of all isolates were determined after 7 d incubation at 25 °C in the dark on PDA. Colony colours were determined using the mycological colour charts of Rayner (1970). Gross morphological characters, including microconidia, macroconidia, chlamydospores and conidiophores, were examined (50×) after mounting fungal structures in sterile water and observed using light microscopy at 1 000× magnification. For each taxonomically informative structure, the extremes are provided, but for conidia we calculated the 95 % confidence intervals and provide extremes in parentheses. All descriptions, illustrations and nomenclatural data were deposited in MycoBank (Crous et al. 2004).
Pathogenicity assays
Isolates of Foc clustering in different clades based on the MLST analyses were selected for pathogenicity assays. The Foc-TR4 reference strain FocII5-NRRL 54006 (Ordonez et al. 2015) was included as a positive control, and negative controls were treated with water. For all assays, we followed the inoculum production, inoculation and diseases assessment protocols developed by Garcia-Bastidas et al. (2018, in submission) using 2–3-mo-old Cavendish and Gros Michel plants. Prior and post-inoculation greenhouse conditions were adjusted to a constant day temperature of 25 °C (ambient light until max. 16 h), a night temperature of 23 °C, and a relative humidity of ≥ 75 %. After 7 wk, disease severities were evaluated by scoring external foliage and internal corm symptoms.
Results
Isolates
Symptoms characteristic of Fusarium wilt were observed in most of the sampling locations on a diverse suite of banana varieties in typical backyards and in a Cavendish industrial plantation (Fig. 2). In total, 40 local banana varieties showed Fusarium wilt symptoms and were sampled (Table 2, Fig. 3). However, wild banana species, including Musa acuminata var. bantamensis in West Java, M. acuminata var. rutilifes in the forest of East Java, and M. acuminata var. microcarpa and M. bornensis in Kalimantan, and the Musa-related species, Ensete glaucum in Flores, were consistently free of external Fusarium wilt symptoms. In total, 203 isolates were obtained from the symptomatic banana plants (Table 3).
Fig. 2.
Symptoms of Fusarium wilt on banana. A. External wilting symptom on leaves in a monoculture plantation in Lampung, Sumatra. B. External wilting symptom in a backyard home plantation in Cianjur, West Java. C. Splitting of the pseudostem. D. Internal symptoms, discoloration of the pseudostem. E. Discoloration of the corm.
Table 2.
List of 40 susceptible local banana varieties at six Indonesian islands from which samples were taken to isolate Fusarium oxysporum f. sp. cubense strains.
| Islands | Banana varieties |
Scientific name1 | Genome1 | ||
|---|---|---|---|---|---|
| Local name | Popular name | International name | |||
| Sumatra | Pisang Ayam | Pisang Barangan | Lakatan | Musa acuminata | AAA |
| P. Wak | P. Awak | Awak | Musa sp. | ABB | |
| P. Abe | P. Kepok | Saba | Musa sp. | ABB | |
| P. Talon | P. Raja | Raja | Musa sp. | AAB | |
| P. Barangan | P. Barangan | Lakatan | Musa acuminata | AAA | |
| P. Tanduk Bawen | P. Tanduk | Horn | Musa sp. | AAB | |
| P. Mas | P. Mas | Sucrier | Musa acuminata | AA | |
| Kalimantan | P. Sanggar/Manurun/Nipah | P. Kepok | Saba | Musa sp. | ABB |
| P. Awak/Pulau Pinang | P. Awak | Awak | Musa sp. | ABB | |
| P. Ambon | P. Ambon Hijau | Cavendish | Musa acuminata | AAA | |
| P. Susu | P. Raja Sereh | Silk | Musa sp. | AAB | |
| P. Hawa | P. Awak | Awak | Musa sp. | ABB | |
| P. Gelobok | P. Awak | Awak | Musa sp. | ABB | |
| P. Talas | P. Talas | NA | Musa acuminata | AA | |
| P. Selendang | NA | NA | Musa acuminata | AAA | |
| Dwarf Cavendish | P. Kapal | Dwarf Cavendish | Musa acuminata | AAA | |
| P. Raja | P. Raja Bulu | Raja | Musa sp. | AAB | |
| P. Kepok | P. Kepok | Saba | Musa sp. | ABB | |
| Java | P. Mas Kirana | P. Mas Kirana | Sucrier | Musa acuminata | AA |
| P. Embuk | NA | NA | Musa sp. | AAB | |
| P. Kongkong | NA | NA | Musa acuminata | AAA | |
| P. Susu | P. Raja Sereh | Silk | Musa sp. | AAB | |
| P. Glintung | NA | NA | − | NA | |
| P. Ambon | P. Ambon Kuning | Gros Michel | Musa acuminata | AAA | |
| P. Ambon Lumut | P. Ambon Hijau | Cavendish | Musa acuminata | AAA | |
| Cau Langadai | P. Siem | NA | Musa sp. | ABB | |
| Cau Apu | P. Siem | NA | Musa sp. | ABBB | |
| P. Jimbluk | P. Siem Jumbo | NA | Musa sp. | ABBB | |
| P. Uli | P. Uli | NA | Musa acuminata | AA | |
| P. Raja Nangka | P. Nangka | Laknau | Musa acuminata | AAA | |
| P. Cavendish | P. Ambon Hijau | Cavendish | Musa acuminata | AAA | |
| P. Kepok Pipik | P. Kepok Putih | NA | Musa sp. | ABB | |
| P. Raja | P. Raja Bulu | Raja | Musa sp. | AAB | |
| Papua | P. Tanduk | P. Tanduk | Horn | Musa sp. | AAB |
| P. Raja | P. Raja Bulu | Raja | Musa sp. | AAB | |
| Sulawesi | P. Kepok | P. Kepok | Saba | Musa sp. | ABB |
| P. Ambon | P. Ambon Hijau | Cavendish | Musa acuminata | AAA | |
| P. Cere | NA | NA | Musa acuminata | AAA | |
| East Nusa Tenggara | P. Kepok | P. Kepok | Saba | Musa sp. | ABB |
| P. Barangan | P. Barangan | Lakatan | Musa acuminata | AAA | |
Fig. 3.
Local Indonesian banana varieties. A. Pisang Raja Bulu (AAB). B. Pisang Awak (ABB). C. Pisang Ambon Hijau (AAA). D. Pisang Udang (ABB). E. Left, Pisang Raja Manten (AAB), right, Pisang Barangan (AAA). F. Pisang Mas Lampung (AA). G. Pisang Tanduk (AAB). H. Pisang Susu (AAB). I. Pisang Kepok (ABB). J. Pisang Jarum (AA).
Table 3.
Details of strains included in the phylogenetic analyses.
| Species name | Accession number1 | Identification2 | f. sp | Country | Host | GenBank/ENA accession3 |
||
|---|---|---|---|---|---|---|---|---|
| rpb1 | rpb2 | tef1 | ||||||
| Fusarium cugenangense | 9InaCC F983 | 7 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479559 | LS479307 | LS479756 |
| InaCC F984 | 7 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479560 | LS479308 | LS479757 | |
| NRLL 36118 | 7 | cubense | Thailand | Musa sp. var. Pisang Kepok | LS479477 | LS479221 | LS479669 | |
| NRRL 25433 | 7 | vasinvectum | Gossypium sp. | LS479462 | LS479202 | LS479648 | ||
| F. dimerum | NRRL 36140 | Citrus sp. | HM347203 | HM347218 | HM347133 | |||
| F. duoseptatum | 4,5FocMal43 | 5 | cubense | Malaysia | Musa sp. var. Pisang Rastali | – | LS479207 | LS479653 |
| InaCC F828 | 5 | cubense | Indonesia | Musa sp. var. Pisang Rastali | LS479520 | LS479266 | LS479715 | |
| InaCC F829 | 5 | cubense | Indonesia | Musa sp. var. Pisang Rastali | LS479528 | LS479274 | LS479723 | |
| InaCC F831 | 5 | cubense | Indonesia | Musa sp. var. Pisang Rastali | LS479538 | LS479285 | LS479734 | |
| InaCC F835 | 5 | cubense | Indonesia | M. acuminata var. Dwarf Cavendish | LS479567 | LS479315 | LS479764 | |
| InaCC F911 | 5 | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479234 | LS479683 | |
| InaCC F915 | 5 | cubense | Indonesia | Musa sp. Pisang Raja | LS479494 | LS479238 | LS479687 | |
| 8InaCC F916 | 5 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479495 | LS479239 | LS479688 | |
| InaCC F920 | 5 | cubense | Indonesia | Musa sp. var. Pisang Hawa | LS479499 | LS479244 | LS479693 | |
| InaCC F921 | 5 | cubense | Indonesia | Musa sp. var. Pisang Hawa | LS479500 | LS479245 | LS479694 | |
| InaCC F975 | 5 | cubense | Indonesia | Musa sp. var. Pisang Awak | LS479549 | LS479296 | LS479745 | |
| InaCC F976 | 5 | cubense | Indonesia | Musa sp. var. Pisang Awak | LS479550 | LS479297 | LS479746 | |
| InaCC F977 | 5 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479551 | LS479298 | LS479747 | |
| InaCC F978 | 5 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479552 | LS479299 | LS479748 | |
| 8InaCC F979 | 5 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479553 | LS479300 | LS479749 | |
| InaCC F980 | 5 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479554 | LS479301 | LS479750 | |
| Indo80 | 5 | cubense | Indonesia | Musa sp. var. Pisang Hawa | LS479619 | LS479387 | LS479829 | |
| NRRL 36115 | 5 | cubense | Malaysia | M. acuminata var. Pisang Ambon | LS479475 | LS479218 | LS479666 | |
| NRRL 36116 | 5 | cubense | Malaysia | Musa sp. var. Pisang Keling | – | LS479219 | LS479667 | |
| F. grosmichelii | 8InaCC F820 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479364 | LS479810 |
| InaCC F832 | 4 | cubense | Indonesia | Musa sp. var. Pisang Awak | LS479542 | LS479289 | LS479738 | |
| 8InaCC F833 | 4 | cubense | Indonesia | Musa sp. var. Pisang Awak | LS479548 | LS479295 | LS479744 | |
| 8InaCC F848 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479588 | LS479338 | LS479786 | |
| InaCC F849 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479589 | LS479339 | LS479787 | |
| InaCC F850 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479340 | LS479788 | |
| 8InaCC F851 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479341 | LS479789 | |
| 8InaCC F852 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Lumut | – | LS479342 | LS479790 | |
| InaCC F853 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Lumut | – | LS479343 | LS479791 | |
| InaCC F854 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Lumut | LS479591 | LS479345 | LS479793 | |
| InaCC F855 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Lumut | LS479592 | LS479346 | LS479794 | |
| InaCC F859 | 4 | cubense | Indonesia | M. acuminata var. Cavendish | LS479596 | LS479350 | LS479796 | |
| InaCC F861 | 4 | cubense | Indonesia | M. acuminata var. Cavendish | LS479597 | LS479351 | LS479797 | |
| InaCC F862 | 4 | cubense | Indonesia | M. acuminata var. Cavendish | LS479598 | LS479352 | LS479798 | |
| InaCC F863 | 4 | cubense | Indonesia | Musa sp. var. Pisang Siem Jumbo | LS479599 | LS479353 | LS479799 | |
| InaCC F867 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Kuning | – | LS479360 | LS479806 | |
| InaCC F868 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon Kuning | – | LS479361 | LS479807 | |
| InaCC F884 | 4 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479616 | LS479382 | LS479824 | |
| InaCC F887 | 4 | cubense | Indonesia | Musa sp. var. Pisang Siem Jumbo | LS479620 | LS479388 | LS479830 | |
| InaCC F888 | 4 | cubense | Indonesia | Musa sp. var. Pisang Siem Jumbo | LS479621 | LS479389 | LS479831 | |
| Indo83 | 4 | cubense | Indonesia | Musa sp. var. Pisang Kepok | – | LS479390 | – | |
| NRRL 36120 | 4 | cubense | Thailand | LS479478 | LS479222 | LS479670 | ||
| F. fujikuroi | CBS 221.76 | FFSC | Oryza sativa | – | – | JN695747 | ||
| F. hexaseptatum | 8InaCC F866 | 8 | cubense | Indonesia | M. acuminata var. Pisang Ambon Kuning | – | LS479359 | LS479805 |
| F. incarnatum-equiseti | NRRL 45997 | FIESC | Poaceae | – | GQ505850 | GQ505672 | ||
| F. kalimantanense | 9InaCC F917 | FOSC Clade 5 Nov. | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479497 | LS479241 | LS479690 |
| InaCC F918 | FOSC Clade 5 Nov. | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479242 | LS479691 | |
| InaCC F922 | FOSC Clade 5 Nov. | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479246 | LS479695 | |
| F. longipes | NRRL 20695 | FSSC | – | GQ915493 | GQ915509 | |||
| F. mangiferae | UMA F0924 | FFSC | Mangifera indica | KP753435 | KP753442 | KP753402 | ||
| F. odoratissimum | 7FocII5-NRRL 54006 | 1 | cubense | Indonesia | M. acuminata var. Pisang Manurung | LS479459 | LS479198 | LS479644 |
| InaCC F816 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479485 | LS479228 | LS479677 | |
| 7InaCC F817 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479556 | LS479304 | LS479753 | |
| InaCC F818 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479584 | LS479333 | LS479782 | |
| InaCC F819 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479600 | LS479354 | LS479800 | |
| InaCC F821 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479609 | LS479374 | LS479818 | |
| 7InaCC F822 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479618 | LS479386 | LS479828 | |
| 7InaCC F824 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479486 | LS479229 | LS479678 | |
| InaCC F825 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479496 | LS479240 | LS479689 | |
| 7InaCC F836 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479577 | LS479325 | LS479774 | |
| InaCC F837 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479578 | LS479326 | LS479775 | |
| InaCC F838 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479579 | LS479327 | LS479776 | |
| InaCC F839 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479580 | LS479328 | LS479777 | |
| InaCC F840 | 1 | cubense | Indonesia | Musa sp. var. Pisang Embuk | – | LS479329 | LS479778 | |
| InaCC F841 | 1 | cubense | Indonesia | Musa sp. var. Pisang Embuk | LS479581 | LS479330 | LS479779 | |
| 7InaCC F846 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | – | LS479336 | LS479785 | |
| InaCC F847 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479587 | LS479337 | – | |
| 7InaCC F856 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | LS479593 | LS479347 | – | |
| InaCC F857 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | LS479594 | LS479348 | LS479795 | |
| InaCC F858 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | LS479595 | LS479349 | – | |
| InaCC F864 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | – | LS479356 | LS479802 | |
| InaCC F865 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | – | LS479358 | LS479804 | |
| InaCC F870 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479602 | LS479363 | LS479809 | |
| InaCC F871 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | – | LS479365 | LS479811 | |
| InaCC F873 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479604 | LS479369 | LS479814 | |
| InaCC F874 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479606 | LS479371 | – | |
| InaCC F875 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479607 | LS479372 | LS479816 | |
| InaCC F876 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479608 | LS479373 | LS479817 | |
| InaCC F877 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479610 | LS479375 | LS479819 | |
| InaCC F878 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479611 | LS479376 | – | |
| InaCC F879 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479612 | LS479377 | LS479820 | |
| InaCC F880 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | – | LS479378 | LS479821 | |
| InaCC F881 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479613 | LS479379 | – | |
| InaCC F882 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479614 | LS479380 | LS479822 | |
| InaCC F883 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479615 | LS479381 | LS479823 | |
| InaCC F885 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | – | LS479384 | LS479826 | |
| InaCC F890 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479623 | LS479392 | – | |
| 7InaCC F891 | 1 | cubense | Indonesia | Musa sp. var. Pisang Glitung | – | LS479393 | LS479833 | |
| InaCC F892 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479624 | LS479394 | LS479834 | |
| InaCC F893 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479625 | LS479395 | LS479835 | |
| InaCC F894 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479626 | LS479396 | LS479836 | |
| InaCC F896 | 1 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479629 | LS479399 | LS479839 | |
| InaCC F897 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479630 | LS479400 | LS479840 | |
| InaCC F898 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479631 | LS479401 | LS479841 | |
| 7InaCC F899 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479632 | LS479402 | LS479842 | |
| InaCC F900 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479633 | LS479403 | LS479843 | |
| InaCC F901 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479634 | LS479404 | LS479844 | |
| InaCC F902 | 1 | cubense | Indonesia | Musa sp. var. Pisang Talon | LS479635 | LS479405 | LS479845 | |
| InaCC F903 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479636 | LS479406 | LS479846 | |
| InaCC F904 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479637 | LS479407 | LS479847 | |
| InaCC F905 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479638 | LS479408 | LS479848 | |
| InaCC F906 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479639 | LS479409 | LS479849 | |
| InaCC F907 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479487 | LS479230 | LS479679 | |
| 7InaCC F908 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479488 | LS479231 | LS479680 | |
| 7InaCC F909 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas | LS479489 | LS479232 | LS479681 | |
| InaCC F910 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas | LS479490 | LS479233 | LS479682 | |
| InaCC F912 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479491 | LS479235 | LS479684 | |
| InaCC F919 | 1 | cubense | Indonesia | Musa sp. var. Pisang Awak | LS479498 | LS479243 | LS479692 | |
| InaCC F923 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479501 | LS479247 | LS479696 | |
| InaCC F924 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479502 | LS479248 | LS479697 | |
| InaCC F925 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479503 | LS479249 | LS479698 | |
| InaCC F926 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479504 | LS479250 | LS479699 | |
| 7InaCC F927 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479506 | LS479252 | LS479701 | |
| InaCC F928 | 1 | cubense | Indonesia | Musa sp. var. Pisang Raja | LS479507 | LS479253 | LS479702 | |
| InaCC F929 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479508 | LS479254 | LS479703 | |
| InaCC F930 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479509 | LS479255 | LS479704 | |
| 7InaCC F931 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479510 | LS479256 | LS479705 | |
| InaCC F932 | 1 | cubense | Indonesia | Musa sp. var. Pisang Tanduk | LS479511 | LS479257 | LS479706 | |
| InaCC F933 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479512 | LS479258 | LS479707 | |
| InaCC F934 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479514 | LS479260 | LS479709 | |
| InaCC F935 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479515 | LS479261 | LS479710 | |
| 7InaCC F936 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479516 | LS479262 | LS479711 | |
| InaCC F937 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479517 | LS479263 | LS479712 | |
| InaCC F938 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479518 | LS479264 | LS479713 | |
| InaCC F939 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479519 | LS479265 | LS479714 | |
| InaCC F942 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479521 | LS479267 | LS479716 | |
| InaCC F943 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479522 | LS479268 | LS479717 | |
| InaCC F944 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479523 | LS479269 | LS479718 | |
| InaCC F945 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479524 | LS479270 | LS479719 | |
| InaCC F946 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479525 | LS479271 | LS479720 | |
| InaCC F947 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479526 | LS479272 | LS479721 | |
| InaCC F948 | 1 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479527 | LS479273 | LS479722 | |
| InaCC F953 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479529 | LS479275 | LS479724 | |
| InaCC F954 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479530 | LS479276 | LS479725 | |
| InaCC F955 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479531 | LS479277 | LS479726 | |
| InaCC F973 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479547 | LS479294 | LS479743 | |
| InaCC F985 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479562 | LS479310 | LS479759 | |
| InaCC F986 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479563 | LS479311 | LS479760 | |
| 7InaCC F988 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479565 | LS479313 | LS479762 | |
| InaCC F989 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479566 | LS479314 | LS479763 | |
| InaCC F990 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok Pipik | LS479568 | LS479316 | LS479765 | |
| InaCC F994 | 1 | cubense | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479569 | LS479317 | LS479766 | |
| 7InaCC F997 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479572 | LS479320 | LS479769 | |
| 7InaCC F998 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479573 | LS479321 | LS479770 | |
| InaCC F999 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479574 | LS479322 | LS479771 | |
| InaCC F1000 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479575 | LS479323 | LS479772 | |
| Indo4 | 1 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479590 | LS479344 | LS479792 | |
| Indo51 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | LS479601 | LS479355 | LS479801 | |
| Indo53 | 1 | cubense | Indonesia | Musa sp. var. Pisang Siem | – | LS479357 | LS479803 | |
| Indo61 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | – | LS479366 | LS479812 | |
| Indo62 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | – | LS479367 | – | |
| Indo66 | 1 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479605 | LS479370 | LS479815 | |
| Indo77 | 1 | cubense | Indonesia | Musa sp. var. Pisang Kepok Pipik | LS479617 | LS479383 | LS479825 | |
| Indo89 | 1 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479627 | LS479397 | LS479837 | |
| Indo204 | 1 | cubense | Indonesia | Musa sp. var. Pisang Uli | LS479561 | LS479309 | LS479758 | |
| Indo222 | 1 | cubense | Indonesia | M. acuminata var. Cavendish | LS479576 | LS479324 | LS479773 | |
| 4JV11 | 1 | cubense | Jordan | M. acuminata var. Cavendish | LS479465 | LS479205 | LS479651 | |
| 4Leb1.2C | 1 | cubense | Lebanon | M. acuminata var. Cavendish | LS479466 | LS479206 | LS479652 | |
| NRRL 36102 | 1 | cubense | China | M. acuminata var. Cavendish | LS479468 | LS479209 | LS479655 | |
| 4Pak1.1A | 1 | cubense | Pakistan | M. acuminata var. Cavendish | LS479479 | LS479223 | LS479671 | |
| 4Phi2.6C | 1 | cubense | Philippines | M. acuminata var. GCTCV218 | LS479480 | LS479224 | LS479672 | |
| F. oxysporum | CAV794 | FOSC Clade 1 | cubense | Indonesia | Musa sp. var. Pisang Rubus | – | – | FJ664922 |
| CAV300 | FOSC Clade 1 | cubense | Indonesia | M. acuminata var. Valery | – | – | FJ664932 | |
| CAV1107 | FOSC Clade 1 | cubense | Vietnam | Musa sp. var. Cuoi Xiem | – | – | FJ664950 | |
| CAV299 | FOSC Clade 1 | cubense | Nigeria | M. acuminata var. Gros Michel | – | – | FJ664946 | |
| CAV602 | FOSC Clade 2 | cubense | Australia | M. acuminata var. Lady Finger | – | – | FJ664957 | |
| CAV189 | FOSC Clade 2 | cubense | Malawi | Musa sp. var. Harare | – | – | FJ664956 | |
| CAV194 | FOSC Clade 2 | cubense | Indonesia | Musa sp. var. Pisang Siem | – | – | FJ664955 | |
| 4,6,8FocCNPMF-R1 | FOSC Clade 4 | cubense | Brazil | Musa sp. var. Silk | LS479457 | LS479196 | LS479642 | |
| NRRL 34936 | FOSC Clade 3 | lycopersici | Solanum lycopersicum | LS479460 | LS479200 | LS479646 | ||
| NRRL 26406 | FOSC Clade 3 | melonis | Cucumis melo | LS479461 | LS479201 | LS479647 | ||
| NRRL 54002 | FOSC Clade 3 | Soil | LS479455 | LS479194 | LS479640 | |||
| NRRL 26381 | FOSC Clade 3 | lycopersici | S. lycopersicum | LS479456 | LS479195 | LS479641 | ||
| NRRL 25603 | FOSC Clade 1 | cubense | M. acuminata | – | – | AF008487 | ||
| NRRL 22550 | FOSC Clade 1 | pernicosum | Albizia julibrissin | – | – | AF008506 | ||
| NRRL 25357 | FOSC Clade 1 | Soil | – | – | AF008481 | |||
| NRRL 26035 | FOSC Clade 1 | canariensis | Phoenix canariensis | – | – | AF008485 | ||
| NRRL 20433 | FOSC Clade 2 | inflexum | Viciba faba | – | – | AF008479 | ||
| NRRL 25607 | FOSC Clade 2 | cubense | M. acuminata x M. balbisiana | – | – | AF008489 | ||
| NRRL 25609 | FOSC Clade 2 | cubense | M. acuminata x M. balbisiana | – | – | AF008490 | ||
| NRRL 26022 | FOSC Clade 2 | cubense | M. acuminata x M. balbisiana | – | – | AF008491 | ||
| NRRL 25598 | FOSC Clade 2 | glycines | Glycine sp. | – | – | AF008496 | ||
| NRRL 26178 | FOSC Clade 2 | melonis | Cucumis melo | – | – | AF008503 | ||
| NRRL 25420 | FOSC Clade 2 | vasinvectum | Gossypium hirsutum | – | – | AF008512 | ||
| NRRL 25369 | FOSC Clade 2 | Terminalia ivorensis | – | – | AF008482 | |||
| NRRL 26406 | FOSC Clade 3 | melonis | C. melo | – | – | AF008504 | ||
| NRRL 26379 | FOSC Clade 3 | radicis-lycopersici | S. esculentum | – | – | AF008508 | ||
| NRRL 22549 | FOSC Clade 3 | passiflorae | Passiflora edulis | – | – | AF008505 | ||
| NRRL 26033 | FOSC Clade 3 | radicis-lycopersici | S. esculentum | – | – | AF008507 | ||
| NRRL 26574 | FOSC Clade 3 | erythroxily | Erythroxylum coca | – | – | AF008495 | ||
| NRRL 26383 | FOSC Clade 3 | lycopersici | S. esculentum | – | – | AF008502 | ||
| NRRL 26380 | FOSC Clade 3 | lycopersici | S. esculentum | – | – | AF008509 | ||
| NRRL 26029 | FOSC Clade 3 | cubense | M. acuminata X M. balbisiana | – | – | AF008493 | ||
| NRRL 22555 | FOSC Clade 3 | tuberosi | S. tuberosum | – | – | AF008511 | ||
| NRRL 26203 | FOSC Clade 3 | lycopersici | S. esculentum | – | – | AF008501 | ||
| NRRL 26374 | FOSC Clade 3 | Homo sapiens | – | – | AF008483 | |||
| NRRL 25594 | FOSC Clade 4 | batatas | Ipomoea batatas | – | – | AY337717 | ||
| NRRL 26360 | FOSC Clade 4 | – | – | AY527522 | ||||
| F. phialophorum | 4,5FocIndo25 | 3 | cubense | Indonesia | M. acuminata var. Pisang Ambon | LS479464 | LS479204 | LS479650 |
| 4,5FocST4.98 | 3 | cubense | Spain | M. acuminata var. Dwarf Cavendish | LS479484 | LS479227 | LS479676 | |
| InaCC F826 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479505 | LS479251 | LS479700 | |
| InaCC F827 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479513 | LS479259 | LS479708 | |
| InaCC F830 | 3 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479536 | LS479282 | LS479731 | |
| InaCC F834 | 3 | cubense | Indonesia | M. acuminata var. Pisang Selendang | LS479557 | LS479305 | LS479754 | |
| InaCC F842 | 3 | cubense | Indonesia | Musa sp. var. Pisang Embuk | LS479582 | LS479331 | LS479780 | |
| InaCC F843 | 3 | cubense | Indonesia | Musa sp. var. Pisang Embuk | LS479583 | LS479332 | LS479781 | |
| 8InaCC F844 | 3 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479585 | LS479334 | LS479783 | |
| InaCC F845 | 3 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479586 | LS479335 | LS479784 | |
| InaCC F869 | 3 | cubense | Indonesia | M. acuminata var. Pisang Ambon Kuning | – | LS479362 | LS479808 | |
| InaCC F889 | 3 | cubense | Indonesia | M. acuminata var. Pisang Ambon Kuning | LS479622 | LS479391 | LS479832 | |
| InaCC F969 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479543 | LS479290 | LS479739 | |
| InaCC F970 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479544 | LS479291 | LS479740 | |
| 8InaCC F971 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479545 | LS479292 | LS479741 | |
| InaCC F972 | 3 | cubense | Indonesia | Musa sp. var. Pisang Wak | LS479546 | LS479293 | LS479742 | |
| InaCC F980 | 3 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479555 | LS479302 | LS479751 | |
| InaCC F981 | 3 | cubense | Indonesia | Musa sp. var. Pisang Kepok | – | LS479303 | LS479752 | |
| InaCC F982 | 3 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479558 | LS479306 | LS479755 | |
| InaCC F987 | 3 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479564 | LS479312 | LS479761 | |
| InaCC F995 | 3 | cubense | Indonesia | M. acuminata var. Pisang Kongkong | LS479570 | LS479318 | LS479767 | |
| 8InaCC F996 | 3 | cubense | Indonesia | M. acuminata var. Pisang Kongkong | LS479571 | LS479319 | LS479768 | |
| Indo64 | 3 | cubense | Indonesia | Musa sp. var. Pisang Susu | LS479603 | LS479368 | LS479813 | |
| NRRL 36101 | 3 | cubense | Australia | Musa sp. var. Mons Mari | LS479467 | LS479208 | LS479654 | |
| NRRL 36103 | 3 | cubense | Philippines | M. acuminata var. Cavendish | LS479469 | LS479210 | LS479656 | |
| NRRL 36109 | 3 | cubense | Australia | Musa sp. var. SH 3142 | LS479471 | LS479214 | LS479661 | |
| NRRL 36110 | 3 | cubense | Australia | Musa sp. var. Mons | – | – | LS479662 | |
| NRRL 36112 | 3 | cubense | South Africa | M. acuminata var. Cavendish | LS479473 | LS479216 | LS479664 | |
| 4,6Race1.0124 | 3 | cubense | Cuba | LS479483 | – | LS479675 | ||
| F. proliferatum | NRRL 62905 | FFSC | KU171687 | KU171707 |
KU171727 |
|||
| F. purpurascens | ATCC76244 | 2 | cubense | USA | M. acuminata var. Apple | – | LS479199 | LS479645 |
| InaCC F823 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479628 | LS479398 | LS479838 | |
| 8InaCC F886 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | – | LS479385 | LS479827 | |
| InaCC F913 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479492 | LS479236 | LS479685 | |
| InaCC F914 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479493 | LS479237 | LS479686 | |
| 8InaCC F966 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479539 | LS479286 | LS479735 | |
| InaCC F967 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479540 | LS479287 | LS479736 | |
| InaCC F968 | 2 | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479541 | LS479288 | LS479737 | |
| NRRL36107 | 2 | cubense | Honduras | Musa sp. var. Maqueno | – | LS479213 | LS479659 | |
| F. sacchari | NRRL 13999 | FFSC | – | – | AF160278 | |||
| F. sangayamense | 9InaCC F960 | FOSC Clade 5 Nov. | cubense | Indonesia | Musa sp. var. Pisang Kepok | LS479537 | LS479283 | LS479732 |
| InaCC F961 | FOSC Clade 5 Nov. | cubense | Indonesia | Musa sp. var. Pisang Kepok | – | LS479284 | LS479733 | |
| F. tardichlamydosporum | 4,6FocCNPMF-R2 | 6 | cubense | Brazil | Musa sp. var. Monthan | LS479458 | LS479197 | LS479643 |
| InaCC F956 | 6 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479532 | LS479278 | LS479727 | |
| InaCC F957 | 6 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479533 | LS479279 | LS479728 | |
| 8InaCC F958 | 6 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479534 | LS479280 | LS479729 | |
| InaCC F959 | 6 | cubense | Indonesia | M. acuminata var. Pisang Barangan | LS479535 | LS479281 | LS479730 | |
| NRRL 36105 | 6 | cubense | Honduras | Musa sp. var. Bluggoe | LS479470 | LS479211 | LS479657 | |
| NRRL 36106 | 6 | cubense | Australia | M. acuminata var. Lady finger | – | LS479212 | LS479658 | |
| NRRL 36108 | 6 | cubense | Tanzania | Musa sp. var. Ney Poovan | – | – | LS479660 | |
| NRRL 36111 | 6 | cubense | Australia | Musa sp. var. Bluggoe | LS479472 | LS479215 | LS479663 | |
| NRRL 36117 | 6 | cubense | Malaysia | Musa sp. var. Pisang awak legor | LS479476 | LS479220 | LS479668 | |
| F. tardicrescens | NRRL 36113 | 9 | cubense | Malawi | Musa sp. var. Harare | LS479474 | LS479217 | LS479665 |
| NRRL 37622 | 9 | pisi | Cicer sp. | LS479463 | LS479203 | LS479649 | ||
| NRRL 54005 | 9 | raphani | Raphanus sp. | LS479482 | LS479226 | LS479674 | ||
| NRRL 54008 | 9 | conglutinans | Raphanus sp. | LS479481 | LS479225 | LS479673 | ||
| F. verticilloides | NRRL 20956 | FFSC | Zea mays | – | – | FN552074 | ||
| Fusarium sp. | InaCC F872 | FFSC | Indonesia | Musa sp. var. Pisang Raja Nangka | – | LS479850 | LS479441 | |
| InaCC F940 | FIESC | Indonesia | M. acuminata var. Pisang Cere | – | LS479855 | LS479443 | ||
| InaCC F941 | FIESC | Indonesia | M. acuminata var. Pisang Cere | – | LS479856 | LS479444 | ||
| 9InaCC F950 | FFSC | Indonesia | Musa sp. var. Pisang Kepok | LS479870 | LS479852 | – | ||
| InaCC F951 | FFSC | Indonesia | Musa sp. var. Pisang Kepok | LS479871 | LS479853 | – | ||
| InaCC F952 | FFSC | Indonesia | Musa sp. var. Pisang Kepok | LS479872 | LS479854 | – | ||
| InaCC F962 | FFSC | Indonesia | M. acuminata var. Pisang Talas | – | LS479868 | LS479453 | ||
| InaCC F963 | FIESC | Indonesia | Musa sp. var. Pisang Awak | LS479875 | LS479859 | LS479445 | ||
| InaCC F964 | FIESC | Indonesia | Musa sp. var. Pisang Awak | LS479876 | LS479860 | LS479446 | ||
| InaCC F965 | FIESC | Indonesia | M. acuminata var. Pisang Talas | LS479877 | LS479863 | LS479448 | ||
| 9InaCC F974 | FSSC | Indonesia | Musa sp. var. Pisang Awak | LS479880 | LS479866 | LS479451 | ||
| InaCC F991 | FFSC | Indonesia | Musa sp. var. Pisang Kepok | LS479881 | LS479867 | LS479452 | ||
| 9InaCC F992 | FFSC | Indonesia | M. acuminata var. Pisang Mas Kirana | LS479882 | LS479869 | LS479454 | ||
| InaCC F993 | FFSC | Indonesia | M. acuminata var. Pisang Mas Kirana | – | LS479851 | LS479442 | ||
| Indo161 | FIESC | Indonesia | M. acuminata var. Pisang Talas | LS479873 | LS479857 | – | ||
| Indo167 | FIESC | Indonesia | Musa sp. var. Pisang Kepok | LS479874 | LS479858 | – | ||
| Indo 174 | FIESC | Indonesia | Musa sp. var. Pisang Awak | – | LS479861 | – | ||
| Indo175 | FIESC | Indonesia | M. acuminata var. Pisang Talas | – | LS479862 | LS479447 | ||
| Indo186 | FIESC | Indonesia | Musa sp. var. Pisang Kepok | LS479878 | LS479864 | LS479449 | ||
| Indo188 | FIESC | Indonesia | Musa sp. var. Pisang Awak | LS479879 | LS479865 | LS479450 | ||
InaCC: Indonesian Culture Collection, Research Center for Biology, Indonesian Institute of Sciences (LIPI) Cibinong, Indonesia; ATCC: American Type Culture Collection, U.S.A.; CAV: Forestry Agricultural Biotechnology Institutre (FABI), University of Pretoria South Africa; CBS: The Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; Indo: Collection of N. Maryani at Wageningen Plant Research, Wageningen University, The Netherlands; NRRL: Agricultural Research Service Culture Collection, USA; UMAF: Microbiology and Plant Pathology Laboratory Collection, University of Malaga, Spain.
Foc lineage/FOSC clade/Fusarium species complex.
rbp1: RNA polymerase II largest subunit; rpb2: RNA polymerase II second largest subunit; tef1: translation elongation factor-1alpha.
Collection of Wageningen Plant Research, Wageningen University, The Netherlands.
Ecosciences Precinct, Brisbane Australia.
Embrapa Cassava & Tropical Fruits, Brazil.
Pathogenic on Cavendish and Gros Michel (Tropical Race 4).
Pathogenic on Gros Michel (Race 1).
Non-pathogenic on Cavendish and Gros Michel.
Phylogenetic analyses
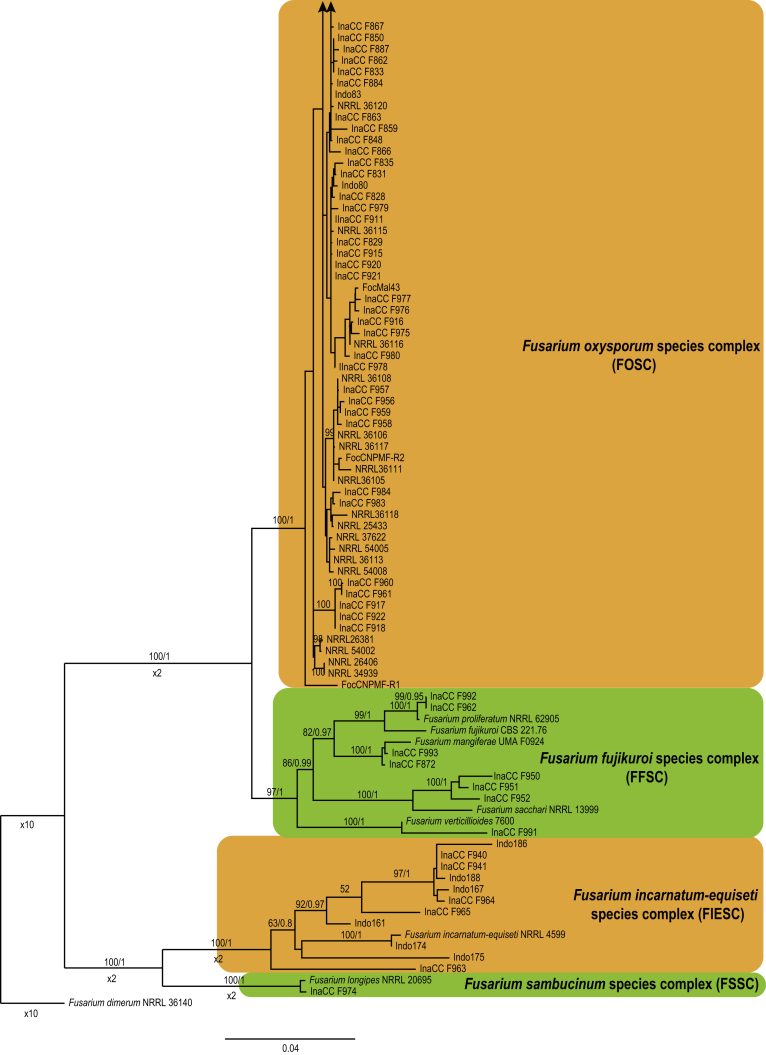
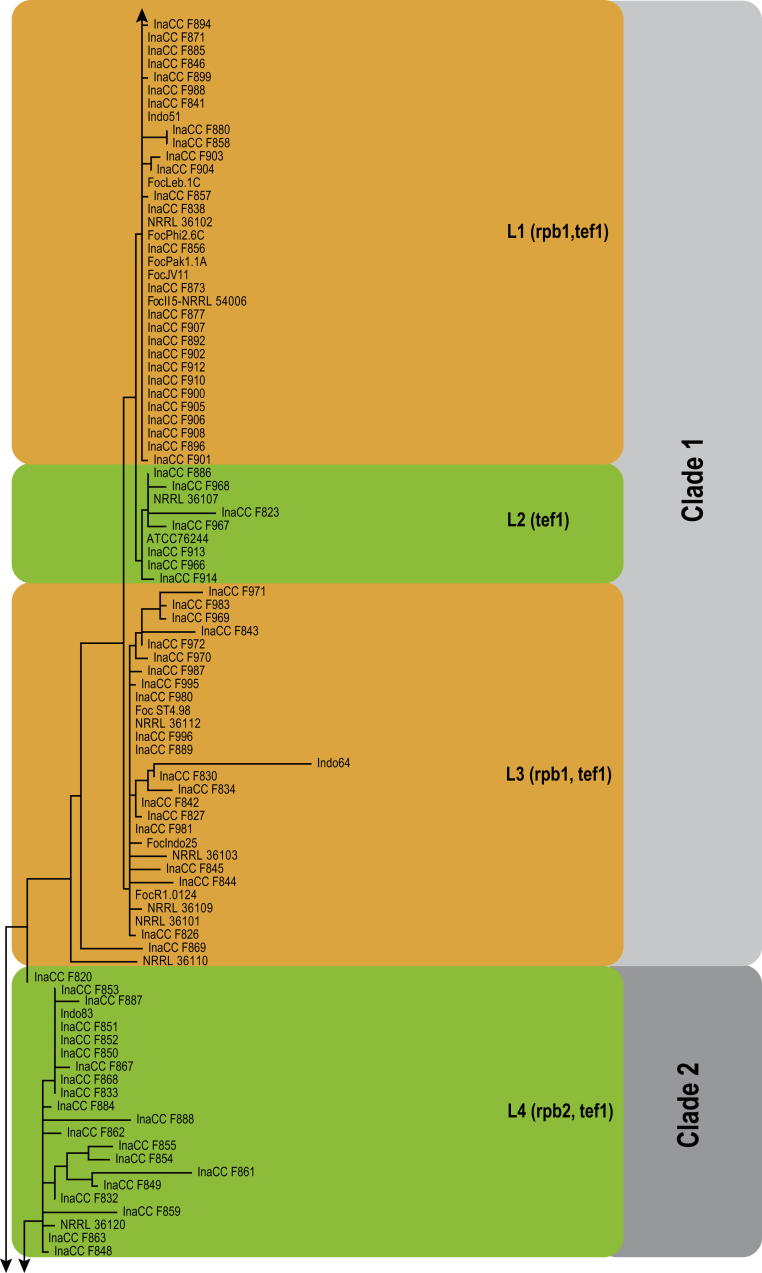
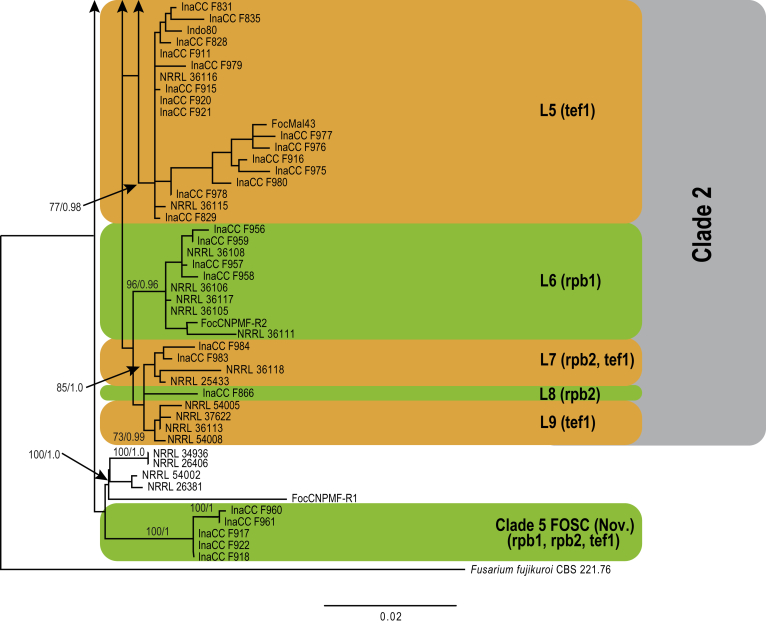
Approximately 632 bp were determined for tef1, 864 bp for rpb2 and 1 444 bp for the rpb1 gene regions. The congruency analyses revealed no conflicts in tree topologies, with only minor differences in branch support. Therefore, the sequences of the three loci were combined in a single dataset for subsequent analyses. For the BI and ML analyses, a GTR+I+G model was selected for all three gene regions and incorporated into the analyses. The ML tree topology confirmed the tree topologies obtained from the BI analyses, and therefore, only the ML tree is presented.
The combined tef1, rpb1 and rpb2 sequences dataset included 244 ingroup taxa and F. dimerum (NRRL 36140) as outgroup taxon. This dataset consisted of 2 909 characters, which yielded a single best ML tree with −InL = -9286.260647 (Fig. 4). The BI lasted for 11 M generations, and the consensus tree, with posterior probabilities, was calculated from 8 251 trees left after 2 750 trees were discarded as the “burn-in” phase.
Fig. 4.
Maximum likelihood tree inferred from the combined rpb1, rpb2 and tef1 genes sequence data set of 244 isolates. The bootstrap support values (BP) and Bayesian posterior probabilities (PP) are given at nodes. Coloured blocks indicate the various Fusarium species complexes included. The tree is rooted to Fusarium dimerum (NRRL 36140).
Phylogenetic inference of the three gene regions placed all isolates recovered from the symptomatic samples in the genus Fusarium (Fig. 4). Of these, 180 isolates clustered in the FOSC clade, one isolate clustered in the Fusarium sambucinum species complex (FSSC) closely related to F. longipes, 11 isolates clustered in the Fusarium incarnatum-equiseti species complex (FIESC), and eight isolates clustered in the Fusarium fujikuroi species complex (FFSC). The highest phylogenetic support was obtained using the tef1 and rpb1 gene regions. The rpb2 gene region displayed less resolution of the isolates, between the various Fusarium species complexes and within each complex. The clades representing FIESC and FSSC resolved in this study were highly supported (BS = 100 %; PP = 1). The FFSC resolved FOSC and other members of the FFSC into two highly supported clades (BP = 100 %; PP = 1 & BP = 97 %; PP = 1, respectively).
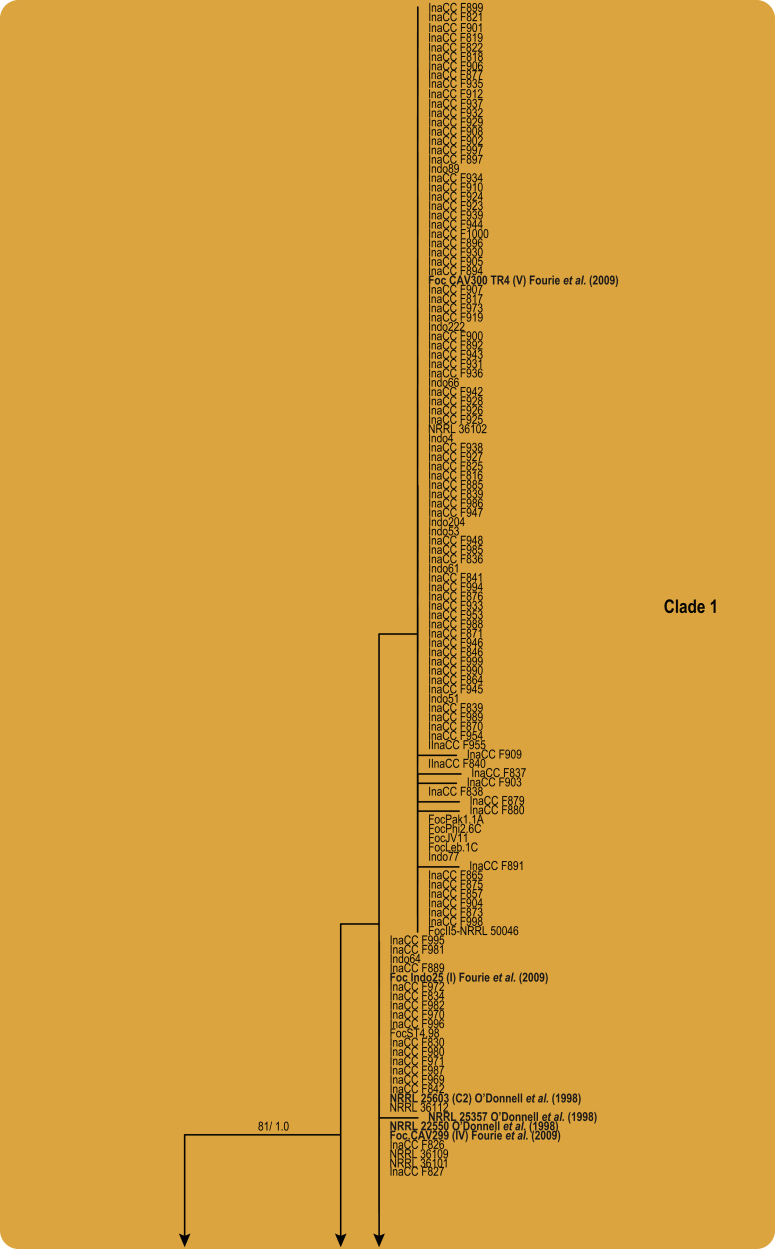
In the FOSC, using the single gene analyses of tef1, and after incorporation of the dataset of O'Donnell et al. (2004) and Fourie et al. (2009), two clades were resolved as in the previous study (O'Donnell et al. 2004; Fig. 5). None of the Indonesian isolates resided in Clade 3. A single isolate, representing FocCNPMF.R1 (Dita et al. 2010), clustered in the FOSC Clade 4. The phylogeny, however, revealed one new clade in the FOSC (BP = 100 %, PP = 1.0), assigned to FOSC Clade 5, comprising five isolates that were isolated from Pisang Kepok (ABB, 2n = 33) and Pisang Ambon (AAA, 2n = 33) in Central and South Kalimantan.
Fig. 5.
Maximum likelihood tree inferred from the tef1 gene sequence data set of 183 Indonesian isolates in the FOSC clade. Included are representatives of the studies by O'Donnell et al., 1998, O'Donnell et al., 2004 and Fourie et al. (2009), indicated in bold. The bootstrap support values >70 % (BS) and Bayesian posterior probabilities >0.95 (PP) are given at nodes. The tree is rooted to Fusarium fujikuroi (CBS 221.76).
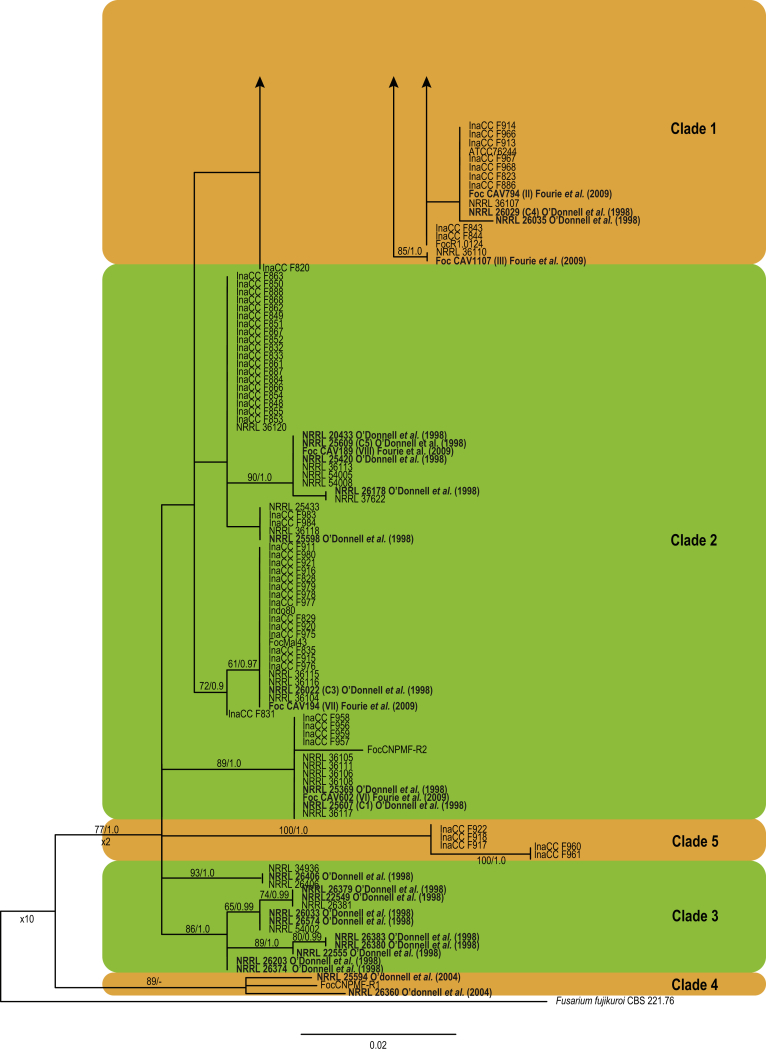
Further analyses of the Foc phylogeny using the combined tef1, rpb1 and rpb2 dataset included 216 ingroup taxa and F. fujikuroi (CBS 221.76.) as an outgroup taxon (Fig. 6). The majority of Indonesian isolates clustered in Clade 1, including eight previously established Foc lineages (Fig. 6; O'Donnell et al., 1998, Fourie et al., 2009), and the overall phylogeny revealed nine independent clonal lineages (Fig. 6). The Indonesian Foc isolates were equally distributed across the nine lineages except for L9 that did not include any Indonesian isolate. We did not identify significant correlation between the origin of the isolates and host genotypes.
Fig. 6.
Maximum likelihood tree inferred from the combined rpb1, rpb2 and tef1 genes sequence data sets. The bootstrap support values >70 % (BS) and Bayesian posterior probabilities >0.95 (PP) are given at nodes. Foc lineages are numbered based on the consensus from single and combine gene data sets represented by the coloured blocks. The tree is rooted to Fusarium fujikuroi (CBS 221.76).
Taxonomy
Based on phylogenetic inference and morphological observations, several novel Fusarium taxa could be identified in this study, and these are described below.
Foc Lineage L1
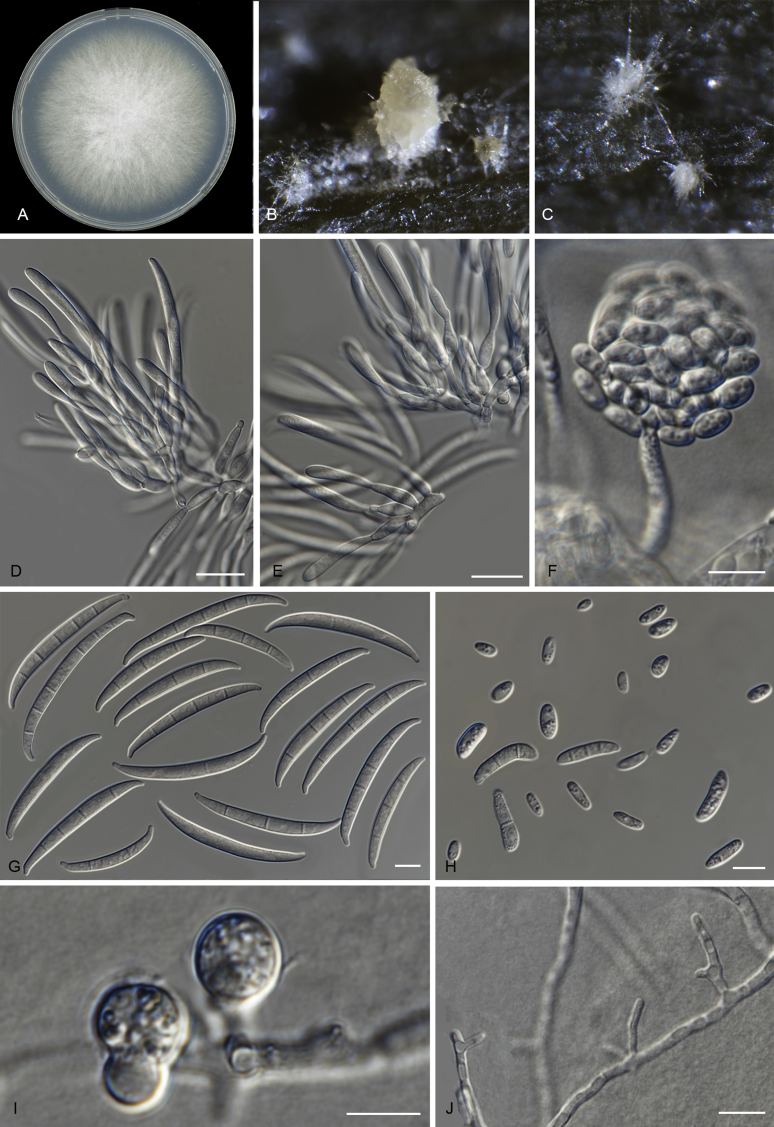
Fusarium odoratissimum N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826800. Fig. 7, Fig. 8.
Fig. 7.
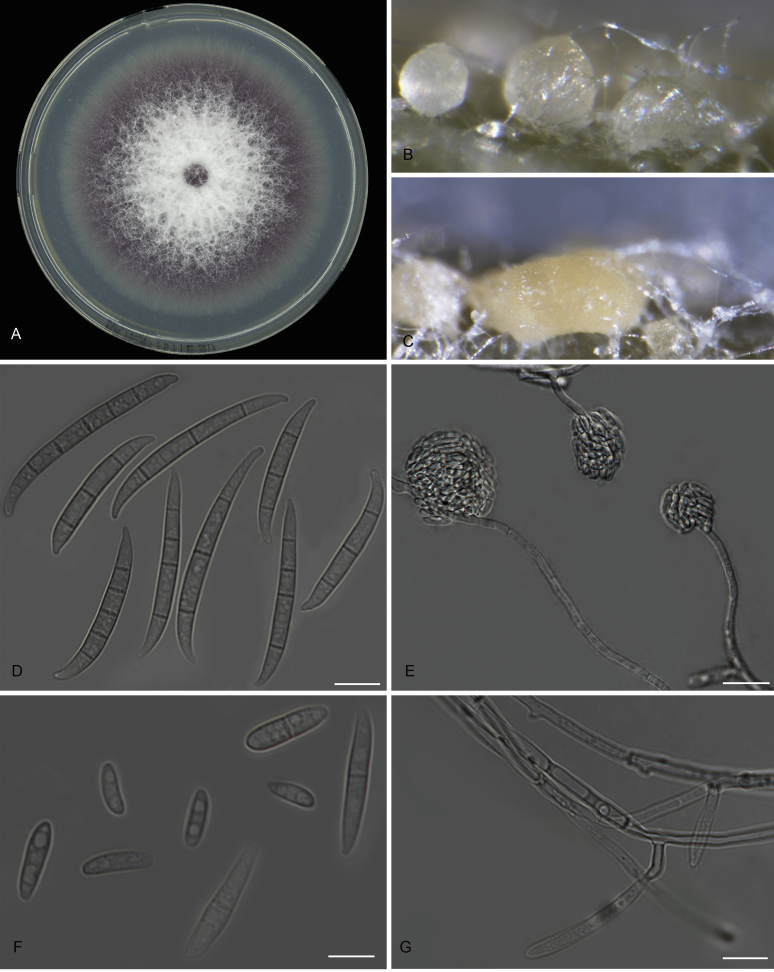
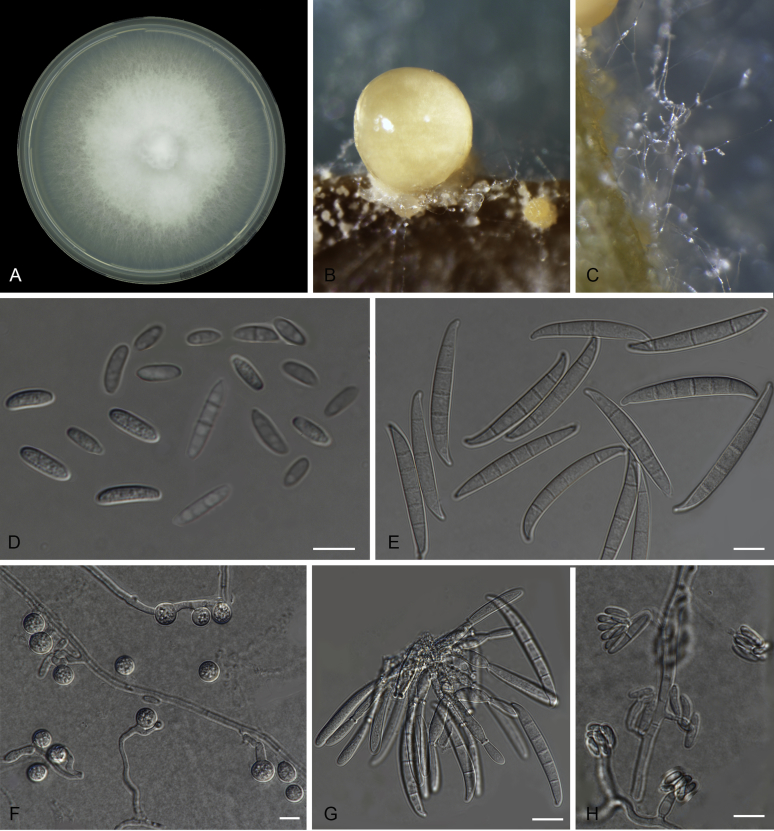
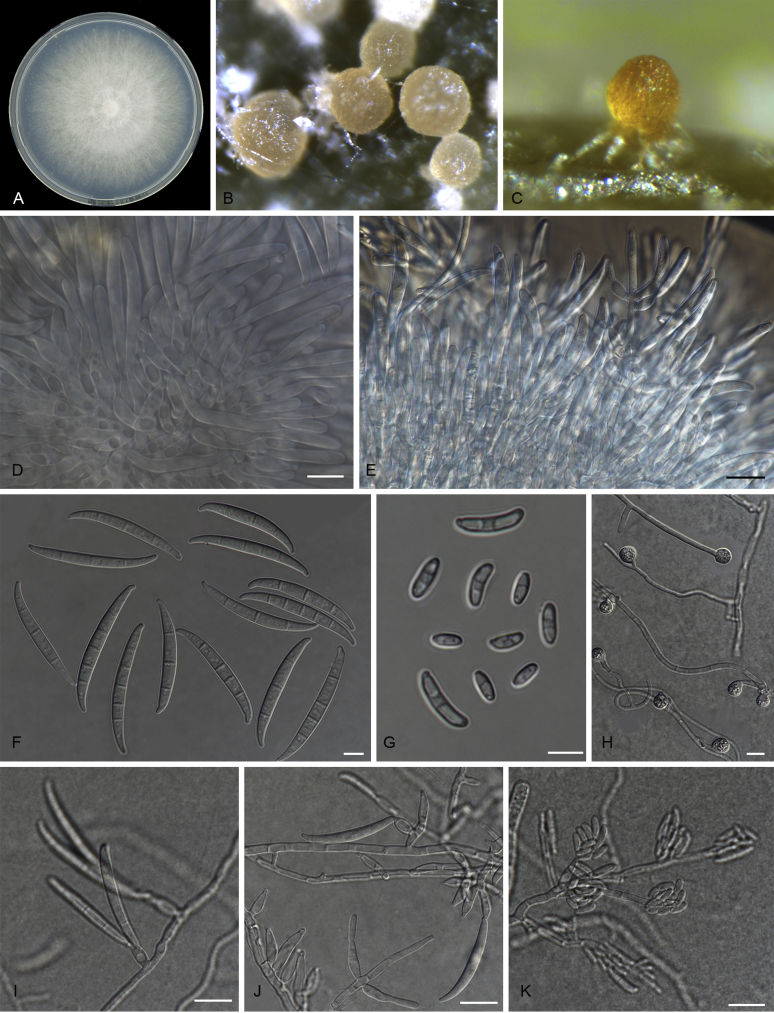
Fusarium odoratissimum (InaCC F817). A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D–E. Sporodochial branched conidiophores with monophialides. F. False head. G. Falcate-shaped macroconidia. H. Microconidia. I. Chlamydospores. J. Polyphialides. Scale bars D–J = 10 µm.
Fig. 8.
Fusarium odoratissimum (ex-type InaCC F822). A. Culture grown on PDA. B. Sporodochia on carnation leaves. C. Monophialides with initial conidia being formed. D. Falcate-shaped macroconida. E. Branched conidophores. F. Elliptical microconidia. G. Thick-walled chlamydospores. Scale bars C–G = 10 µm.
Etymology: Name refers to the strong odour associated with older cultures.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (44–)59–75(–79) × 6–8 μm (av. 67 × 7 μm), 0–6-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- or polyphialidic on sporodochia, or formed directly on hyphae (lateral phialides), 12–28 × 4–8 μm. Microconidia abundant on PDA and SNA, less frequent on CLA, oval to ellipsoid, (6–)8–16(–23) × (4–)6(–8) μm (av. 12 × 5 μm), 0–3-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA but formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores globose to subglobose, formed intercalarily or terminally, single or in pairs, (7–)9–13(–14) × (7–)8–11(–12) μm, rarely produced on SNA after 7 d, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.5–5.0 mm/d. Colony reverse, uniformly white and unpigmented. Colony surface dry, cottony, white, with filamentous margin. No exudates observed. Aerial mycelium abundant, cottony, with abundant sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange.
Geography and host: Kutai Timur, East Kalimantan, Musa sp. var. Pisang Kepok (ABB).
Pathogenicity: Pathogen on Gros Michel (AAA) and Cavendish (AAA).
Material examined: Indonesia, Kampung Salak Martadinata, Kutai Timur, East Kalimantan (117°26′850″E and 0°11′590″N), on infected pseudostem of Musa sp. var. Pisang Kepok (ABB), 16 Jun. 2014, N. Maryani (holotype preserved as metabolically inactive culture, InaCC F822).
Notes: Fusarium odoratissimum formed a small cryptic clade within the L1 cluster (Fig. 6), and can be distinguished by the septation of its macroconidia (0–6-septate) and microconidia (0–3-septate), characteristics not common for F. oxysporum (Leslie & Summerell 2006). This species also produces chlamydospores relatively more rapidly than was observed for other Fusarium isolates examined in this study. F. odoratissimum and all isolates in L1 produce a strong peculiarly stale odour in mature cultures, of which the causal volatiles remain to be characterised. Pathogenicity tests showed that F. odoratissimum and all isolates in L1 were able to infect Cavendish and Gros Michel bananas. Isolates in this lineage were thus classified as Foc-TR4.
Foc Lineage L2
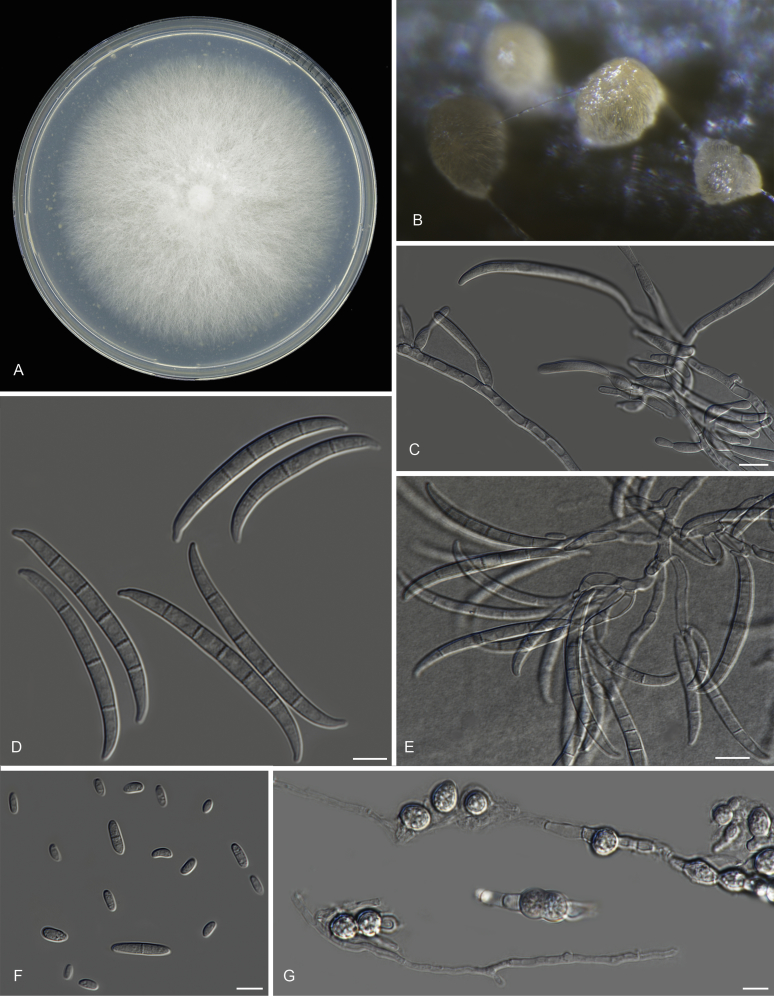
Fusarium purpurascens N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826801. Fig. 9.
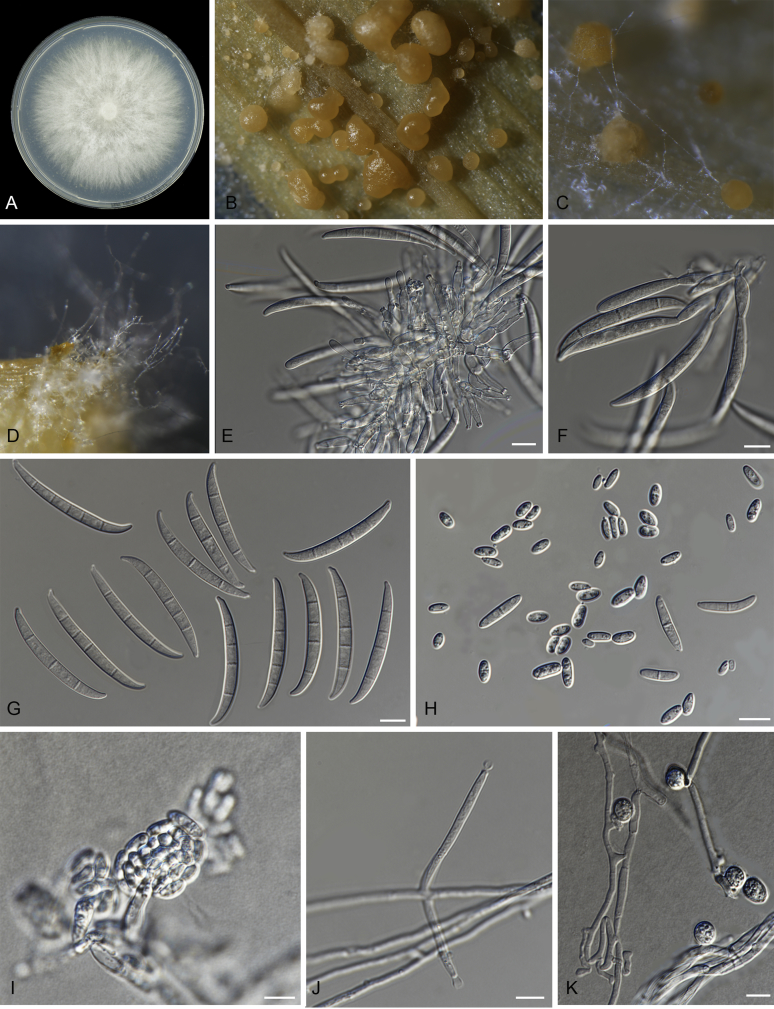
Fig. 9.
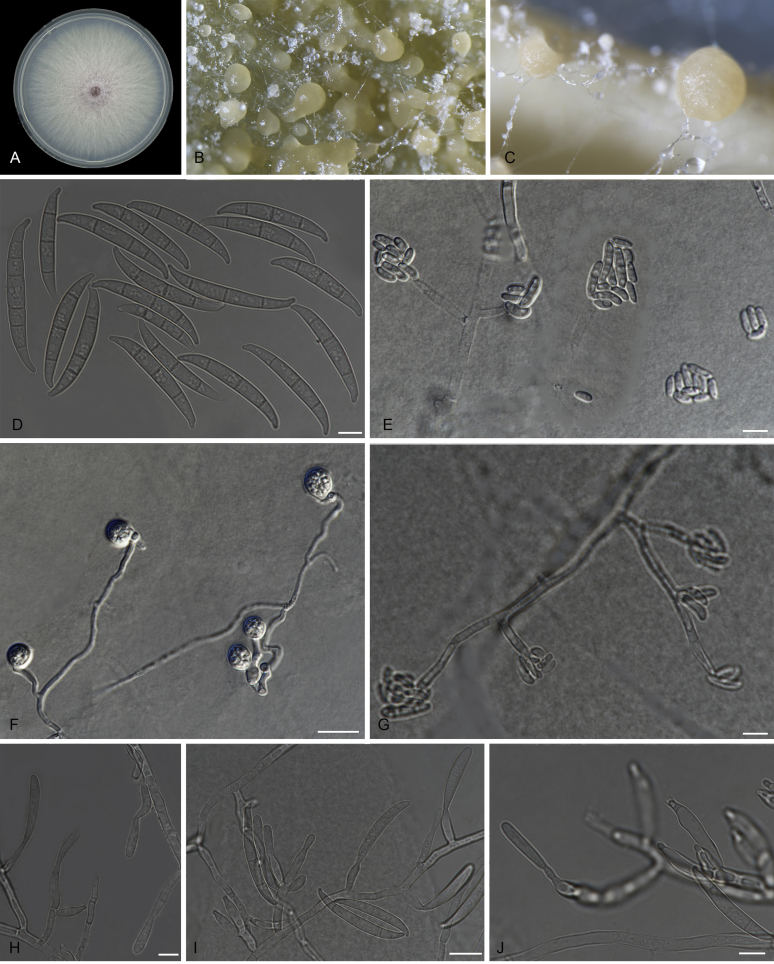
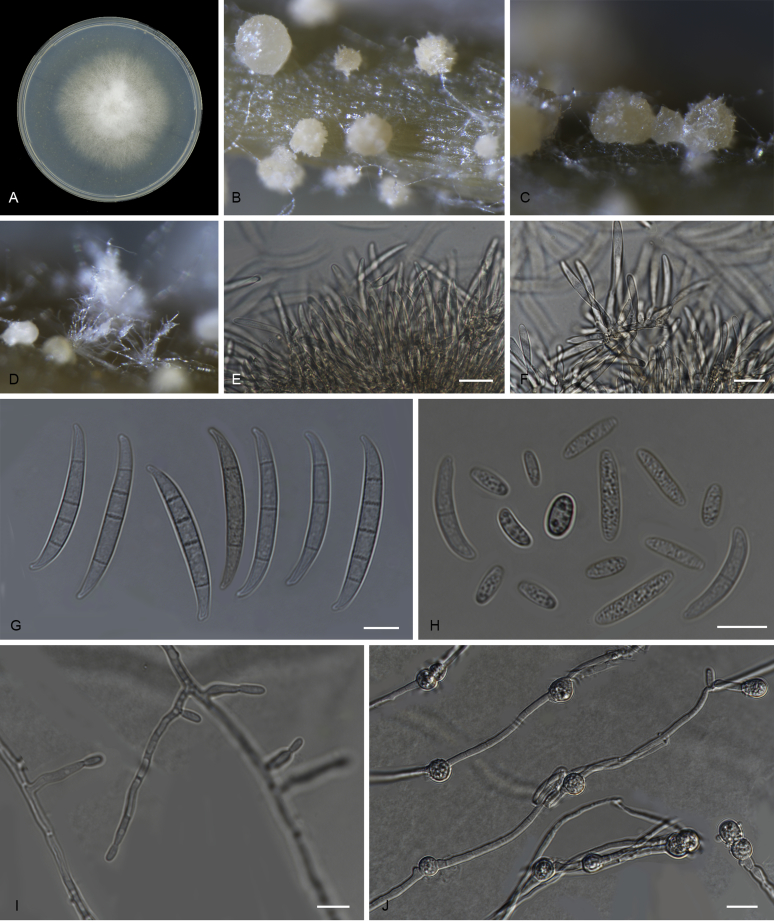
Fusarium purpurascens (ex-type InaCC F886). A. Culture grown on PDA. B–C. Sporodochia grown on carnation leaves. D. Falcate-shaped macroconidia. E. False heads. F. Microconidia. G. Monophialides. Scale bars D–G = 10 µm.
Etymology: Name reflects the purple pigmentation which was observed when cultivated on potato dextrose agar.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate (50–)55–63(–67) × (4–)6–7(–9) μm (av. 59 × 7 μm), 3–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- or polyphialidic on sporodochia, or formed directly from hyphae (lateral phialides), 5–45 × 3–8 μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (8–)18(–37) × (3–)5(–6) μm (av. 12 × 4 μm), 0–1-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA, and formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores not observed.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.4–4.8 mm/d. Colony reverse, livid purple. Colony surface dry, cottony, white, filamentous in the centre and livid purple towards the margin, forming exudate droplets. Aerial mycelium abundant, cottony, with moderate sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange.
Geography and host: Kutai Timur, East Kalimantan, Musa sp. var. Pisang Kepok (ABB).
Pathogenicity: Pathogen on Gros Michel (AAA).
Material examined: Indonesia, Kampung Salak Martadinata, Kutai Timur, East Kalimantan (117°26′684″E, 0°26′684″N), on infected pseudostem of Musa sp. var. Pisang Kepok (ABB), 17 Jun. 2014, N. Maryani (holotype preserved as metabolically inactive culture, InaCC F886).
Notes: Fusarium pupurascens exhibits the strongest purple colony colour on PDA of all the isolates with purple colonies. It is relatively slow-growing compared to other isolates clustered in lineage L1. No chlamydospores were observed for this species, in contrast to other L1 members, which readily produce chlamydospores in culture. Furthermore, F. purpurascens produces exudate droplets, something not observed among other L1 isolates. Older cultures become pigmented, a distinctive phenomenon rarely seen in L1. F. purpurascens and other isolates in this lineage were able to infect Gros Michel, and were therefore classified as Foc-Race1.
Foc Lineage L3
Fusarium phialophorum N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826802. Fig. 10.
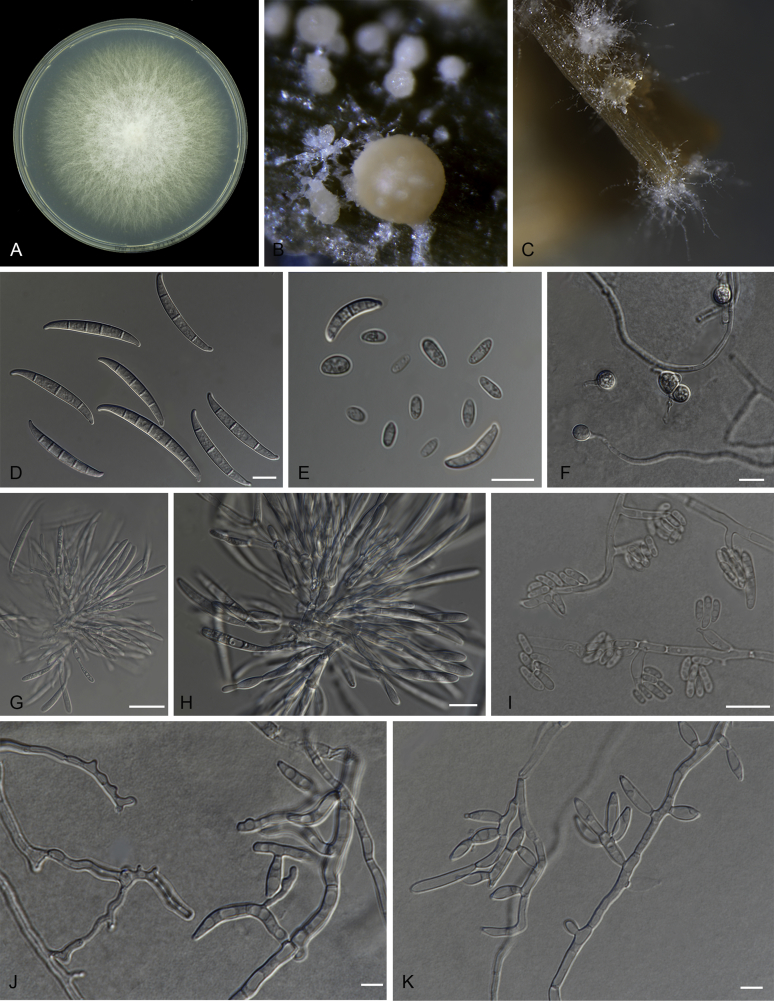
Fig. 10.
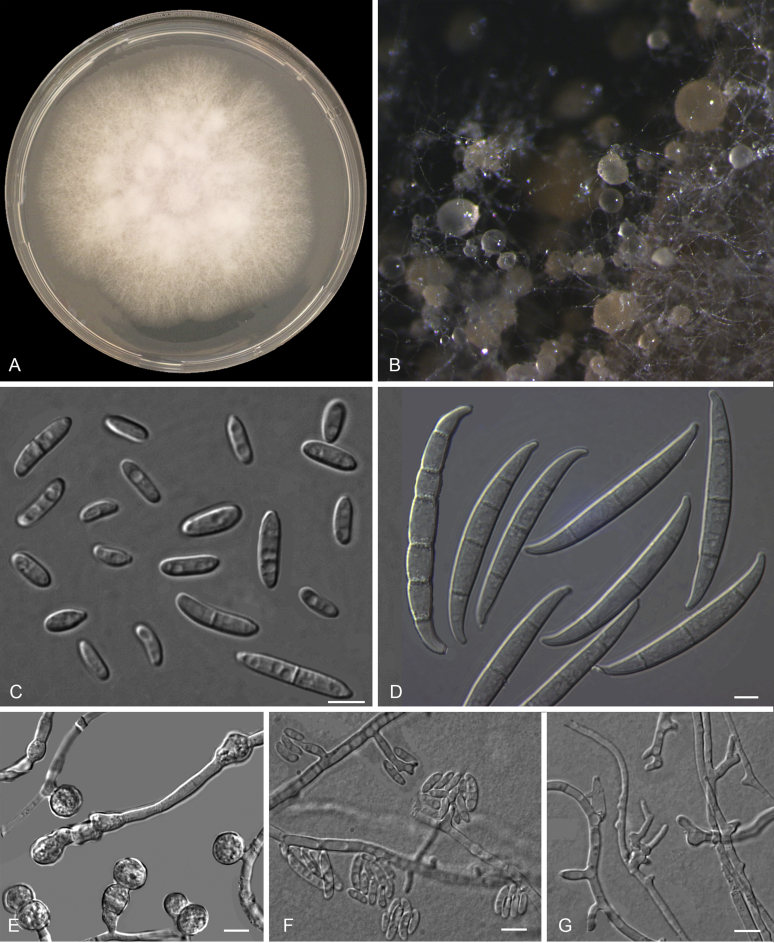
Fusarium phialophorum (ex-type InaCC F971) A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D. Aerial conidiophore on carnation leaves. E–F. Sporodochial phialides. G. Falcate-shaped macroconidia. H. Microconidia. I. False head. J. Lateral monophialides with long collaretes. K. Thick-walled chlamydospores. Scale bars E–K = 10 µm.
Etymology: Name refers to its elongated phialidic collarettes observed in culture.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (50–)54–60(–62) × (3–)4–5(–7) μm (av. 57 × 7 μm), 2–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells monophialidic on sporodochia, or formed directly from hyphae (lateral phialides) with elongated collarettes, 7–41 × 3–7 μm. Microconidia abundant on PDA, less frequent on CLA, ovoid to ellipsoid, (6–)7–16(–24) × (3–)4(–6) μm (av. 12 × 5 μm), 0–1-septate, arranged in false heads on branched or lateral conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA and formed abundantly on PDA, branched sparsely or forming short lateral conidiophores. Chlamydospores globose to subglobose, formed terminally, single or in pairs, (8–)9–12(–13) × (9–)10(–11) μm, rarely produced on SNA after 7 d, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.9–5.2 mm/d. Colony reverse, uniformly white and unpigmented. Colony surface dry, cottony, white, filamentous margin. No exudates observed. Aerial mycelium abundant, cottony, with high sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: Tanah Bumbu, South Kalimantan, Musa sp. var. Pisang Awak (ABB).
Pathogenicity: Pathogen on Gros Michel (AAA).
Materials examined: Indonesia, Kampung Betung, Tanah Bumbu, South Kalimantan (115°37′477″E, 3°37′45″S), on infected pseudostem of Musa sp. var. Pisang Awak (ABB), 20 Jun. 2014, N. Maryani (holotype preserved as metabolically inactive culture, InaCC F971).
Notes: Fusarium phialophorum has elongated phialidic collarettes, which are rarely found in other lineages. Polyphialidic conidiophores were not found, and chlamydospores were formed, but were rare. Isolates in this lineage were able to infect Gros Michel but not Cavendish, and were therefore classified as Foc-Race1.
Foc Lineage L4
Fusarium grosmichelii N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826803. Fig. 11.
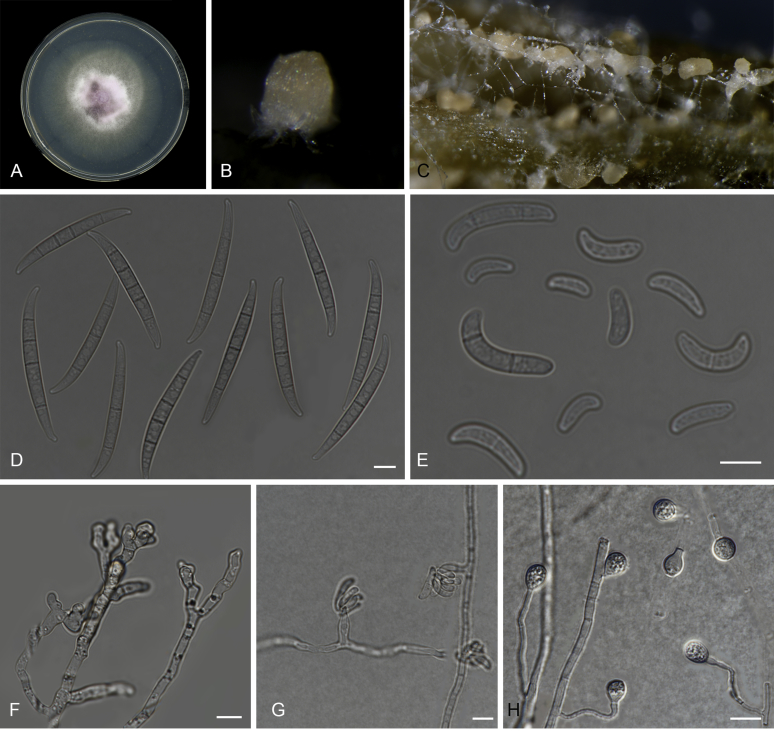
Fig. 11.
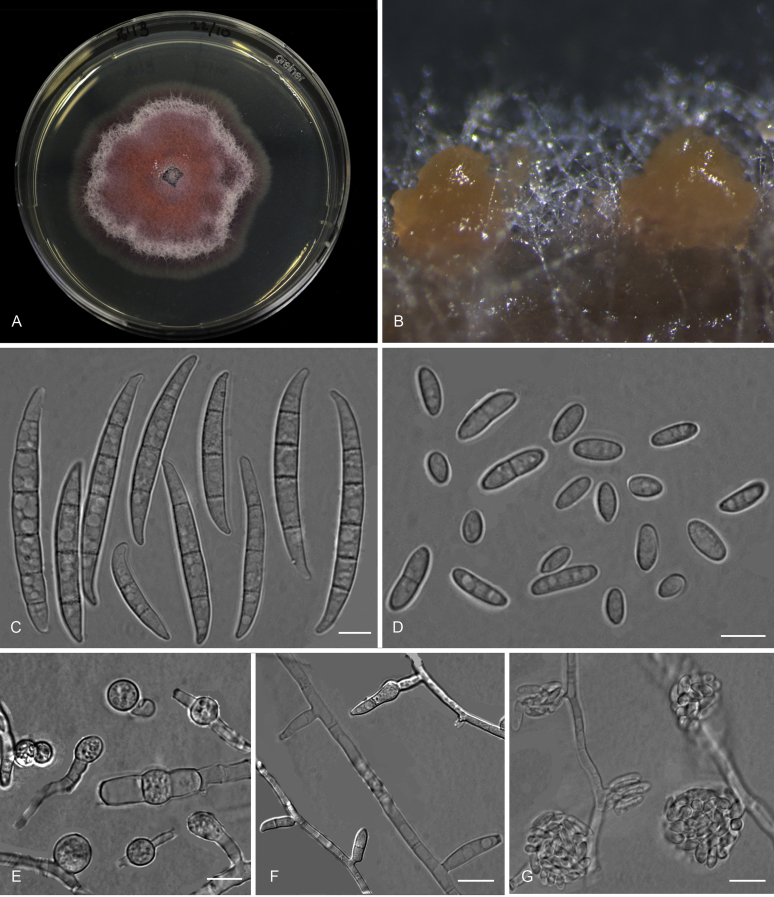
Fusarium grosmichelii (ex-type InaCC F833). A. Culture grown on PDA. B. Sporodochia on carnation leaves. C. Aerial conidiophores from stereo microscope. D. Falcate-shaped macroconidia. E. Microconidia. F. Chlamydospores. G–H. Sporodochial phialides. I. False heads. J. Polyphialides. K. Branched conidiophore. Scale bars D–F, H–K = 10 µm, G = 20 µm.
Etymology: Name reflects its association with the banana variety Gros Michel.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (47–)51–59(–64) × (5–)6–8(–9) μm (av. 55 × 7 μm), 3–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- or polyphialidic on sporodochia, on branched conidiophores, or formed directly from hyphae (lateral phialides), (8–)16–28(–36) × (3–)4–6(–7) μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (4–)9–17(–21) × (3–)4–6(–7) μm (av. 12 × 5 μm), 0–1-septate, arranged in false heads on branched or lateral conidiophores carried on hyphae. Chlamydospores globose to subglobose, formed terminally or intercalarily, single or in clumps, rarely produced on SNA after 7 d, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.7–5.0 mm/d. Colony reverse in the dark uniformly white and unpigmented. Colony surface dry, cottony white with filamentous margin. No exudates observed. Aerial mycelium abundant, cottony, with abundant sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: Bogor, West Java, Musa acuminata var. Pisang Ambon Lumut (AAA).
Pathogenicity: Pathogen on Gros Michel (AAA).
Materials examined: Indonesia, Suakarya Megamendung, Bogor, West Java (106°54′214″E, 6°41′185″N), on infected pseudostem Musa acuminata var. Pisang Ambon Lumut (AAA), 10 Jul. 2014, N. Maryani, (holotype preserved as metabolically inactive culture InaCC F833).
Notes: Fusarium grosmichelii is morphologically very similar to F. phialophorum, but differs in having a higher number of septa in its macroconidia (3–5-septate). F. grosmichelii and others in this lineage are morphologically similar to F. odoratissimum, but F. grosmichelii was not able to infect Cavendish. Most of the isolates in L4 were tested on Gros Michel, and were able to cause disease, and were thus classified as Foc-Race1.
Foc Lineage L5
Fusarium duoseptatum N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826804. Fig. 12.
Fig. 12.
Fusarium duoseptatum (ex-type InaCC F916). A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D. Falcate-shaped macroconidia. E. Microconidia. F. Polyphialidic conidiogenous cells. G. False heads. H. Chlamydospores. Scale bars D–H = 10 µm.
Etymology: Name reflects the fact that its microconidia are frequently 2-septate.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (50–)53–63(–68) × (5–)6–8(–9) μm (av. 58 × 7 μm), 3–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- or polyphialidic on sporodochia, or on aerial hyphae, or formed directly from hyphae as lateral phialides, (5–)9–25(–38) × (3–)4–7(–9) μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (9–)21(–33) × (2–)3(–6) μm (av. 15 × 5 μm), 0–2-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA, formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores globose to subglobose, formed laterally, intercalary or terminally, single or in pairs, (6–)8–10(–11) × (6–)7–9(–11) μm, abundantly produced on SNA after 7 d, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 3.8–4.1 mm/d. Colony reverse violet, mycelium becoming purple, and pigmented with age. Colony surface dry, cottony violet in the centre, and white towards the margin. No exudates observed. Aerial mycelium abundant, cottony, with moderate sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geographic and host: Kapuas, Central Kalimantan, Musa sp. var. Pisang Kepok (ABB).
Pathogenicity: Pathogen on Gros Michel (AAA).
Material examined: Indonesia, Serapat tengah, Kapuas Timur, Central Kalimantan (114°28′65″E, 3°6′9″S), on infected pseudostem of Musa sp. var. Pisang Kepok (ABB), 22 Jun. 2014, N. Maryani, (holotype preserved as metabolically inactive culture InaCC F916).
Notes: Fusarium duoseptatum has distinctive septation in its microconidia, being 0–2-septate, thus differing from F. grosmichelii, which is 0–1-septate. The former is relatively slow-growing compared to members of the most closely related lineage, L4, and forms pigmentation in the centre of colony that is not observed in isolates of L4. F. duoseptatum and most of the members of L5 were able to infect Gros Michel, and were therefore classified as Foc-Race1.
Foc Lineage L6
Fusarium tardichlamydosporum N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826805. Fig. 13.
Fig. 13.
Fusarium tardichlamydosporum (ex-type InaCC F958). A. Culture grown on PDA. B. Sporodochia on carnation leaves. C. Aerial conidiophore. D. Microconidia. E. Falcate-shaped macroconidia. F. Chlamydospores. G. Sporodochial phialides. H. False heads. Scale bars D–H = 10 µm.
Etymology: Name reflects the delayed chlamydospore production observed in this species.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (36–)37–43(–45) × (4–)5–6(–7) μm (av. 40 × 5 μm), 3–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells monophialidic on sporodochia, or on aerial hyphae, or formed directly on hyphae as lateral phialides, (3–)7–14(–19) × (2–)3–5(–8) μm. Microconidia abundant on PDA and SNA, ovoid to ellipsoid, (3–)5–9(–15) × (2–)5(–9) μm (av. 7 × 3 μm), 0–2-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA and formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores abundantly produced after 4 wk, globose to subglobose, (6–)7–10(–13) × (4–)6–9(–10) μm, formed terminally or intercalarily, single or in pairs, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.6–5.6 mm/d. Colony reverse sparsely dark purple in the centre, becoming white towards the margins, and purple slate, pigmented with age. Colony surface dry, cottony, with white filamentous margin, and lacking exudates. Aerial mycelium abundant, cottony, with abundant sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: Sikka Flores, Musa acuminata var. Pisang Barangan (AAA).
Pathogenicity: Pathogen on Gros Michel (AAA).
Materials examined: Indonesia, Desa Kota Uneng Kecamatan Alok, Sikka Flores, East Nusa Tenggara (112°12′16″E, 8°37′11″S), on infected pseudostem of Musa acuminata var. Pisang Barangan (AAA), 21 Aug. 2015, N. Maryani, (holotype preserved as metabolically inactive culture, InaCC F958).
Notes: Colonies of Fusarium tardichlamydosporum are relatively fast growing (av. 4.6–5.6 mm/d) compared to those of F. duoseptatum (av. 38–41 mm/d). Polyphialidic conidiophores were not observed in this species/lineage. Chlamydospores were produced, but only after 4 wk. F. tardichlamydosporum was able to infect Gros Michel, and is therefore classified as Foc-Race1.
Foc Lineage L7
Fusarium cugenangense N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826807. Fig. 14.
Fig. 14.
Fusarium cugenangense (ex-type InaCC F984). A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D. Falcate-shaped macroconidia. E. Microconidia. F. Chlamydospores. G. False heads. H. Monophialidic conidiogenous cells. I–J. Branched conidiophores. Scale bars D–J = 10 µm.
Etymology: Name reflects Cugenang, the location where this species was collected in Indonesia.
Macroconidia abundant on CLA, formed on sporodochia, on aerial conidiophores or on lateral phialides, falcate, (44–)47–54(–57) × (5–)6–7(–8) μm (av. 53 × 7 μm), 3–6-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells monophialidic on sporodochia, or on aerial hyphae, or formed directly from hyphae as lateral phialides, (5–)12–31(–45) × (3–)5–7(–8) μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (7–)8–11(–24) × (2–)7(–12) μm (av. 12 × 5 μm), 0–3-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA, and formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores rarely produced on SNA after 4 wk, globose to subglobose, (9–)10–14(–16) × (10–)11–14(–16) μm, formed terminally, single or in pairs, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 5.2–5.4 mm/d. Colony reverse purple at center to pale viscous grey, white towards the margins, becoming purple slate with age, and pigmented. Colony surface dry, cottony, dark purple to white with filamentous margin, lacking exudates. Aerial mycelium abundant, cottony, with profuse sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: Cianjur, West Java, Musa sp. var. Pisang Kepok (ABB).
Pathogenicity: Non-pathogenic on Gros Michel (AAA) and Cavendish (AAA).
Material examined: Indonesia, Cugenang, Cianjur, West Java (107°4′109″E, 6°47′867″S), on infected pseudostem Musa sp. var. Pisang Kepok (ABB), 10 Jul. 2014, N. Maryani, (holotype preserved as metabolically inactive culture, InaCC F984).
Notes: L7, including Fusarium cugenangense and other isolates, represents an Indonesian lineage with isolates that are closely related to other formae speciales (Fig. 6; e.g. NRRL 25433 F. oxysporum f. sp. vasinvectum). Polyphialidic conidiogenous cells were not observed in this species. This species has macroconidia with unique septation (3–6-septate) and microconidia (0–3-septate), which is rather uncommon for F. oxysporum species. This species causes a slight infection on Cavendish and Gros Michel, and testing on other cultivars such as Bluggoe (Pisang Kepok, ABB) are needed to fully classify strains as Foc-Race2.
Foc Lineage L8
Fusarium hexaseptatum N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826808. Fig. 15.
Fig. 15.
Fusarium hexaseptatum (ex-type InaCC F866). A. Culture grown on PDA. B. Sporodochia on carnation leaves. C. Microconidia. D. Falcate-shaped macroconidia. E. Thick-walled chlamydospores. F. False heads. G. Monophialides and polyphialides. Scale bars C–G = 10 µm.
Etymology: Name reflects the six conidial septa observed in its macroconidia.
Macroconidia abundant on CLA, less so on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (34–)45–71(–76) × (5–)6–8(–9) μm (av. 58 × 7 μm), 3–6-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- or polyphialidic on sporodochia, or formed directly from on hyphae (lateral phialides), 7–20 × 2–6 μm. Microconidia abundant on PDA and SNA, rare on CLA, ovoid to ellipsoid, (4–)8–23(–29) × (2–)7(–12) μm (av. 16 × 5 μm), 0–1-septate, arranged in false heads on branched conidiophores carried on hyphae. Aerial conidiophores rare on CLA and SNA and formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores abundantly formed in hyphae, globose to subglobose, (5–)14(–20) × (4–)6–12(–17) μm, formed terminally or intercalarily, single or in pairs.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.9–5.9 mm/d. Colony reverse, in the dark, white and becoming livid purple in the center of the colony. Colony surface with filamentous margin, dry, cottony, white becoming livid vinaceous in age. No exudates observed. Aerial mycelium abundant, cottony, with high sporulation. Sporodochia formed abundantly on CLA after 7 d, colourless to pale orange.
Geography and host: Sukabumi, West Java, Pisang Ambon Kuning (AAA).
Pathogenicity: Pathogen on Gros Michel (AAA).
Material examined: Indonesia, Parakan Lima, Sukabumi, West Java (107°5′869″E, 6°50′614″S), on infected pseudostem Musa acuminata var. Pisang Ambon Kuning (AAA), 7 Oct. 2014, N. Maryani, (holotype preserved as metabolically inactive culture, InaCC F866).
Notes: Fusarium hexaseptatum is the single species in L8. Macroconidia with 6 septa are abundantly observed in this lineage, whereas in L7 and L9, they are very rare. This lineage is distinguished from L7 and L9 by its ability to cause disease on Gros Michel, and therefore it was classified as Foc-Race1. F. hexaseptatum has chlamydospores that are relatively large compared to those in other lineages (av. 9 × 9 μm).
Foc Lineage L9
Fusarium tardicrescens N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826809. Fig. 16.
Fig. 16.
Fusarium tardicrescens (ex-tyoe CBS 102024). A. Culture grown on PDA. B. Sporodochia on carnation leaves. C. Falcate-shaped macroconidia. D. Microconidia. E. Thick-walled chlamydospores. F. Monophialides produce microconidia and macroconidia. G. False head. Scale bars C–G = 10 µm.
Etymology: Name reflects the slow growth rate in culture.
Macroconidia abundant on CLA and SNA, less abundant on PDA, formed on sporodochia on CLA and on aerial conidiophore on SNA and PDA, falcate, (52–)56–75(–89) × (5–)6–8(–9) μm (av. 66 × 7 μm), 2–6-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells mono- and polyphialidic on sporodochia formed directly from hyphae (lateral phialides), 7–32 × 2–6 μm. Microconidia abundant on PDA and SNA, less so on CLA, ovoid to ellipsoid, (7–)10–16(–20) × (2)–5(–7) μm (av. 13 × 4 μm), 0–1-septate, arranged in false heads on branched conidiophores carried on hyphae. Chlamydospores globose to subglobose, (5–)7–9(–10) × (5–)6–8(–10) μm, formed intercalarily or terminally, singly or in pairs, produced abundantly on SNA after 7 d, brown, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 2.9–3.9 mm/d. Colony reverse, in the dark, dark violet becoming dark livid and pigmented. Colony surface dry, cottony, dark purple becoming dark livid. No exudates observed. Aerial mycelium abundant, cottony, with abundant sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: NA.
Pathogenicity: NA.
Material examined: Malawi, Karonga, Misuku Hills, Musa sapientum cv. Harare, 1989, R.C. Ploetz (holotype preserved as metabolically inactive culture CBS 102024 = NRRL 36113).
Notes: Fusarium tardicrescens in L9 represents one of two lineages which clustered with other formae speciales. This lineage does not contain any Indonesian isolates. F. tardicrescens is the slowest growing species (av. 2.9–3.9 mm/d). F. tardicrescens causes moderate infection on both Cavendish and Gros Michel (Ordonez 2018).
Novel Clade/Taxa in FOSC
Fusarium kalimantanense N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826810. Fig. 17.
Fig. 17.
Fusarium kalimantanense (ex-type InaCC F917). A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D–E. Sporodochial phialides. F. Falcate-shaped macroconidia. G. Microconidia. H. Thick-walled chlamydospores. I. Monophialides producing macroconidia. J. Branched conidiophores. K. False heads. Scale bars D–K = 10 µm.
Etymology: Name reflects Kalimantan, the island in Indonesia from where this fungus was collected.
Macroconidia abundant on CLA, less abundant on PDA and SNA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (52–)56–63(–65) × (5–)6–7(–8) μm (av. 59 × 7 μm), 3–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells monophialidic on sporodochia, or on aerial hyphae, or formed directly from hyphae as lateral phialides, (9–)11–15(–16) × (2–)3(–5) μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (6–)8–15(–20) × (2–)3–4(–7) μm (av. 12 × 4 μm), aseptate, arranged in false heads on branched conidiophores borne on hyphae. Aerial conidiophore sparse on CLA and SNA and formed abundantly on PDA, branched sparsely or formed laterally. Chlamydospores rarely produced on SNA after 7 d, globose to subglobose, formed terminally or laterally, single or in pairs, (6–)7–10(–11) × (7–)8–9(–10) μm, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C with an average growth rate of 4.8–1.2 mm/d. Colony reverse rosy buff (pinkish) to white towards the margins, becoming fuscous black and pigmented with age. Colony surface dry, cottony, rosy buff (pinkish) to white, becoming purplish grey with age, filamentous margin, and lacking exudates. Aerial mycelium abundant, cottony, with moderate sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange to orange.
Geography and host: Katingan, Central Kalimantan, Musa acuminata var. Pisang Ambon (AAA).
Pathogenicity: Non-pathogenic on Gros Michel (AAA) and Cavendish (AAA).
Material examined: Indonesia, Pulau Malam, Katingan, Central Kalimantan (113°13′333″E, 1°36′374″S), on infected pseudostem Musa acuminata var. Pisang Ambon (AAA), 23 Jun. 2014, N. Maryani, (holotype preserved as metabolically inactive culture, InaCC F917).
Notes: Fusarium kalimantanense represents a new clade (Clade 5) in FOSC, which was previously considered to include only four clades (Fig. 5; sensu O'Donnell et al. 2004). This species has relatively fast-growing colonies compared to those of other members of FOSC in this study, and has a unique character in its aseptate microconidia. F. kalimantanense causes a slight infection on both Cavendish and Gros Michel. Further pathogenicity tests on other cultivars like Bluggoe (syn. Pisang Kepok, AAB) will be required to determine its race.
Fusarium sangayamense N. Maryani, L. Lombard, Kema & Crous, sp. nov. MycoBank MB826811. Fig. 18.
Fig. 18.
Fusarium sangayamense (ex-type InaCC F960). A. Culture grown on PDA. B–C. Sporodochia on carnation leaves. D. Aerial conidiophore. E–F. Sporodochial phialides. G. Falcate-shaped macroconidia. H. Microconidia. I. Short monophialides. J. Thick-walled chlamydospores. Scale bars D–J = 10 µm.
Etymology: Name reflects Sangayam, the location from where this species was collected in Indonesia.
Macroconidia abundant on CLA and SNA, rare on PDA, formed on sporodochia on CLA and on aerial conidiophores on SNA and PDA, falcate, (48–)52–60(–65) × (5–)6–7(–8) μm (av. 56 × 7 μm), 2–5-septate, with apical cells papillate, basal cells foot-shaped. Conidiogenous cells monophialidic, similar in sporodochia and on hyphae, polyphialidic, rare, (6–)11–31(–47) × (3–)4–6(–9) μm. Microconidia abundant on PDA and SNA, less frequent on CLA, ovoid to ellipsoid, (8–)9–17(–24) × (3–)4–6(–7) μm (av. 13 × 5 μm), 0–1-septate, arranged in false heads on branched conidiophores borne on hyphae. Aerial conidiophores rare on CLA, and formed abundantly on SNA and PDA, sparsely branched, and formed laterally. Chlamydospores rarely produced on SNA after 7 d, globose to subglobose, formed terminally or intercalarily, single or in pairs, (6–)7–10(–12) × (6–)7(–9) μm, rough-walled.
Culture characteristics: Colony on PDA showing optimal growth at 25 °C, with an average growth rate of 3.5–4.2 mm/d. Colony reverse uniformly white and unpigmented. Colony surface dry, cottony, white, with filamentous margin and lacking exudates. Aerial mycelium abundant, cottony, with moderate sporulation. Sporodochia formed abundantly on CLA after 7 d, pale orange.
Geography and host: Kota Baru, South Kalimantan, Musa sp. var. Pisang Kepok (ABB).
Pathogenicity: Non-pathogenic on Gros Michel (AAA) and Cavendish (AAA).
Material examined: Indonesia, Sangayam, Kota Baru, South Kalimantan (115°59′440″E, 2°20′420″S), on infected pseudostem of Musa sp. var. Pisang Kepok (ABB), 19 Jun. 2014, N. Maryani, (holotype preserved as metabolically inactive culture, InaCC F960).
Notes: Isolates of Fusarium sangayamense formed a subclade in the new FOSC Clade 5 (Fig. 6) with high support (BP = 100 % and PP = 1.0). F. sangayamense can be distinguished from F. kalimantanense based on the septation of its macroconidia (2–5-septate) and microconidia (0–1-septate). This species has polyphialidic conidiogenous cells, which are absent in F. kalimantanense. F. sangayamense was not able to infect Cavendish or Gros Michel.
Pathogenicity assays
The pathogenicity assay showed that all collected Foc isolates were able to cause typical Fusarium wilt symptoms on either Cavendish or Gros Michel, or in both varieties (Fig. 19). The positive control isolate FocII5-NRRL 54006 was lethal to both varieties, whereas all negative (water) controls remained free of disease. Isolates affecting Cavendish were classified as Foc-TR4 (Su et al. 1986), while those only infecting Gros Michel were classified as Foc-Race1 (Stover 1962a, Ploetz 1990). No fewer than 65 % of the isolates clustered in L1, which only comprised the strains that caused Fusarium wilt in Cavendish and hence, represented Foc-TR4. The rest of the isolates tested were able to infect Gros Michel and are therefore considered to be Foc-Race1 strains. Strains fitting this pathogenicity profile were equally distributed over all other lineages, except L7 and L9. L7 contains two Indonesian isolates that caused a slight discolouration of the corms of both varieties. Isolates in the new clade within FOSC were not able to cause disease in either tested banana variety. Isolates identified as other Fusarium species in the phylogenetic analyses were negative in all pathogenicity assays.
Fig. 19.
Pathogenicity assays. A. External wilting symptoms. B–C. Left panel Cavendish and right panel Gros Michel, corm symptom caused by Foc-Race1, Fusarium tardichlamydosporum. D–E. Left panel Cavendish and right panel Gros Michel, corm symptom caused by Foc-TR4, Fusarium odoratissimum.
Discussion
The Musa gene centre (Perrier et al. 2011), as with the wheat gene centre in the Middle-East (Banke et al. 2004, Stukenbrock et al. 2007) and that of potato in Central Mexico (Grünwald & Flier 2005), contains a myriad of endemic diseases that co-evolved with the host. It is therefore considered a typical hot-spot of pathogen diversity (Stukenbrock & McDonald 2008). The gene centre of Musa has been studied in detail since the previous century. The wild ancestor of edible banana, Musa acuminata, originated in South-East Asia and Melanesia, and Musa balbisiana originated in South Asia (Perrier et al. 2011), where Indonesia is the contact area between these two wild Musa species. Approximately 11–13 Musa acuminata sub-species are of Indonesian origin, found in Sumatra, Kalimantan, Java, and the Lesser Sundas (Daniells, 1995, Simmonds, 1962). Most of the Musa balbisiana sub-species found in Java, Sumatra, and Sulawesi originate from India (Ochse and Bakhuizen van den Brink, 1931, De Langhe, 2009). However, the genetic diversity of Musa pathogens in the centre of origin of Musa has remained virtually unsampled. Although a recent overview of Foc in Asia was published (Mostert et al. 2017), a need remained for a thorough taxonomic analysis of Foc in its centre of origin. Our results present the most comprehensive study of Foc in the Indonesian gene centre of banana to date. Isolates of Foc were recovered from all the samples that were collected in all areas surveyed. The results demonstrated that Fusarium wilt is widely distributed in Indonesia and could be found in every banana producing area surveyed. Past reports showing compatible results have spanned an area from Aceh province in the west to Papua province in the east (Nasir et al., 1999, Wibowo et al., 2011). In 2012, 1 700 of the 21 000 acres of cultivated banana suffered from Fusarium wilt in Indonesia, including large commercial Cavendish plantations (Jumjunidang et al. 2012). Factors making this disease difficult to control include traditional farming practices, limited quarantine restriction on movement of planting material, and limited knowledge on the dissemination of the pathogen(s). As a result, the disease is unwittingly distributed to new areas. Moreover, the abundant diversity of banana varieties in Indonesia allows farmers to easily change the varieties they grow, resulting in epidemiological contact that allows the pathogen to infect new cultivars in different areas.
Demographic factors could have played a significant role in the dissemination of this disease in Indonesia. Java is the most populated island and, therefore, banana production and the available cultivated varieties are the most numerous on this island, as is the pathogen. Mass migration of people from this over-populated island to less populated islands such as Kalimantan, Sumatra, and Papua from 1980 to 1990 could account for the dissemination of Fusarium wilt throughout Indonesia, since infected banana planting material was taken along (Nasir et al. 1999).
The high number of local banana varieties from which Foc was recovered indicate that co-evolution of this pathogen is occurring along with its host in this region. Nasir et al. (1999) reported that 15 local varieties in Sumatra were susceptible to Fusarium wilt, including the most popular varieties, Pisang Ambon Kuning (AAA, Gros Michel synonym), Barangan (AAA) and Pisang Raja Sereh (AAA). This finding was reconfirmed in this study. An increasing number of infected varieties was also reported by Hermanto et al. (2009) and Jumjunidang et al. (2012). Of the hundreds of banana cultivars identified in Indonesia, many appear to be resistant or partially resistant to Fusarium wilt, a prior finding that was also observed during the present survey. No wild banana or close relative surveyed in this study showed any symptoms of Fusarium wilt. In Africa, Ensete ventricosum, a member of the Musaceae, is susceptible to Foc-Race2 (Ploetz 2006). By contrast, Ensete glaucum growing on the outskirt forest of Flores, Indonesia, was found to be healthy. None of the wild M. acuminata varieties found during the surveys was susceptible to Fusarium wilt. This finding is in agreement with some reports and greenhouse experiments on the infection of Foc on wild M. acuminata. Musa acuminata var. malaccensis from the Malaysian Peninsula was reported to be experimentally resistant (Javed et al. 2004), as was its sister variety M. acuminata var. malaccensis from Sumatra. This study and our observations during surveys indicate that Indonesia is the primary gene centre of Foc, and the most likely place to find a diverse palette of disease resistance markers for Fusarium wilt in banana.
The high diversity of Foc isolates found in this study is unparalleled by the findings of any previous study (O'Donnell et al., 1998, Fourie et al., 2009) where a similar approach was used. The taking of larger numbers of samples in Indonesia inclusive of more banana cultivars, could result in an even higher diversity, as well as the discovery of yet more novel taxa belonging to FOSC. This accords with the view of Leslie & Summerell (2006), who stated that the most informative studies on the systematics and evolution of Fusarium species from natural ecosystems, as well as different agro-ecosystems, should incorporate native host populations, in order to allow discovery of the full existing species diversity (Leslie & Summerell 2006).
Employing rotations with alternative crops, such as corn, sugar cane, peanuts and coffee, was found to decrease disease incidence in some plantations in Sumatra, Java, and Kalimantan. However, this practice probably has allowed for other Fusarium species, pathogenic to the rotation crops, to become established in these plantations, explaining their recovery in this study. These species include F. mangiferae, F. proliferatum F. sacchari and F. verticillioides, which are members of the Fusarium fujikuroi species complex (FFSC) and are associated with several tropical crops (Marasas et al., 2006, Ploetz, 2006) such as mango, maize, rice and sugarcane (Hsuan et al. 2011). These crops were commonly found in the areas surveyed for Fusarium wilt on bananas during this study. Fusarium proliferatum and F. oxysporum have been reported from the roots of the wild banana, M. acuminata, from Malaysia (Zakaria & Rahman 2011), which is closely related to several other M. acuminata varieties present in Sumatra and Java (Nasution 1990). This study represents the first report of both F. longipes and F. incarnatum-equiseti from banana varieties displaying symptoms of Fusarium wilt, although disease symptoms could not be induced in the pathogenicity assays undertaken here. However, both species are well-known as soil inhabitants and saprobes with a wide global distribution in tropical regions (Leslie & Summerell 2006). They could, therefore, be secondary colonisers of the decaying vascular tissue collected during the survey. The majority of the isolates that clustered outside the FOSC clade are well-known endophytes of various plant hosts, saprobes, and soil inhabitants, and are known to be non-pathogenic to banana (Waalwijk et al., 1996, O'Donnell et al., 1998).
In the FOSC clade, the Indonesian isolates were equally distributed throughout the two previously known clades in FOSC (sensu O'Donnell et al. 2004). Several of these F. oxysporum isolates are known as endophytes of banana (O'Donnell et al. 1998), and are unable to induce disease on Cavendish or Gros Michel. Isolates obtained in this study that were found to be non-pathogenic to both banana cultivars tested were distantly related to the pathogenic isolates, and were more closely related to other formae speciales that are pathogenic to other crops. This finding supported the observations of Gordon & Okamoto (1992), who reported that Fusarium oxysporum f. sp. melonis, pathogenic to cucurbits, is only distantly related to non-pathogenic strains. This also supports the view that Foc and other formae speciales of F. oxysporum have a polyphyletic origin (Baayen et al., 2000, O'Donnell et al., 2009).
Nine Foc lineages were revealed in this study, albeit with varying levels of statistical support, and described as new species. This conclusion was based on combinations of the genealogical approaches described by Dettmann et al. (2003) and Laurence et al. (2014), with supporting evidence from the inclusion of eight previously established lineages of FOC (O'Donnell et al., 1998, Fourie et al., 2009). A lineage is recognised as independent in this system if it is found to be concordantly supported by the majority of the loci, or is well supported by at least one locus but not contradicted by any other locus. Two previously known clades of Foc were resolved in this study (Boehm et al., 1994, Bentley et al., 1995, O'Donnell et al., 1998, Fourie et al., 2009), with the majority of the isolates fell into in Clade1, Lineage1. This lineage, classified as Foc-TR4, was found on every island surveyed, including Papua and Flores and those that were previously thought to be free of Foc-TR4. This is in agreement with some reports on Fusarium wilt in Indonesia, which note that the majority of Foc strains isolated appeared to be Foc-TR4 (O'Neill et al., 2011, Jumjunidang et al., 2012). In terms of phylogenetic diversity, Foc-TR4 isolates were less diverse than Foc-Race1, which occurred in almost all lineages. The number of diverse banana varieties sampled could be the reason for the tremendous diversity of Foc-Race1 isolates found in this study. Many of the banana sampled belong to varieties Gros Michel (AAA) or Silk (AAB), both known to be highly susceptible to Foc-Race1 (Waite & Stover 1960).
The partial sequences of the three coding gene regions employed in this study, tef1, rpb1 and rpb2, are well-known to be robust for use in molecular-based identification of Fusarium species (O'Donnell et al. 2015), but are unable to distinguish all of the 24 VCGs (Puhalla, 1985, Ordonez et al., 2015) that are known to represent the widest genetic diversity of Foc. Direct VCG identification is a relatively objective but time-consuming test, and the results indicate genetic similarity rather than genetic differences (Kistler 1997). Therefore, VCGs represent good phenotypic characters for assessing diversity within populations, but genetic relationships among VCGs need to be assessed by other molecular tools.
The high diversity found, based on the number of isolates recovered from different banana varieties and the high number of lineages resolved in this study, support the hypothesis that the pathogen(s) co-evolved with the host in the host's centre of origin (Ploetz & Pegg 1997). The unique agro-ecosystems and variety of ecological niches found where banana cultivation is practiced in Indonesia provide a conducive environment for the pathogen to evolve. As mentioned above, subsistence farming in Indonesia has allowed for the dissemination of banana varieties with varying degrees of tolerance and resistance to Fusarium wilt. This practice may have created a suitable environment for the incumbent pathogen to evolve and to adapt to newly introduced banana varieties. The dynamics of host diversity in these agro-ecosystems will continue to select for new pathogens (Stukenbrock & McDonald 2008), a process that, in this study, yielded a diversity of species able to infect newly introduced banana cultivars.
Another scenario that could account for the high Foc diversity in Indonesia, irrespective of a lack of sexual reproduction, is horizontal gene transfer. Fusarium oxysporum has the ability to transfer specific chromosomes, sometimes containing unique pathogenicity genes, among non-pathogenic and pathogenic strains, resulting in new pathogenic lineages (Rep & Kistler 2010). This phenomenon is well recorded in Fusarium oxysporum f. sp. lycopersici, a pathogen of tomato (Ma et al. 2010). A recent study of the effector profile of different formae speciales of F. oxysporum, including Foc, indicated that these fungi have specific and unique effector profiles that reflect vertical and horizontal inheritance (van Dam et al. 2016). The endophytic character of some F. oxysporum strains, some of which are weak soil-borne pathogens (Stover 1962b), allows for relatively easy assimilation of pathogenicity genes from related pathogenic F. oxysporum strains via horizontal gene transfer (Vlaardingerbroek et al. 2016).
The race concept has been used extensively in F. oxysporum classification system by plant pathologists. Based on the results of the present study, it can be inferred that the Foc-TR4 isolates evolved recently from predecessors in Foc-Race1. Foc-Race1 displayed a higher phylogenetic diversity in this study than Foc-TR4. Once established, both races apparently co-evolved in the same region, meaning that possible horizontal gene transfer could be involved in the high diversity level seen in Foc-Race1, as well as in the emergence of Foc-TR4.
It was initially thought that the origin of pathogenic Foc is from non-pathogenic root inhabitants or endophytes of various wild M. acuminata plants in Java and Sulawesi that became pathogenic after their introduction to foreign banana germplasm (Buddenhagen 2007). Alternatively, native Foc-Race1 isolates may have been exposed to selection pressure through exposure to newly introduced banana varieties, as Foc-Race1 is known to infect diverse varieties like Silk (AAB), Pome (AAB), and Pisang Awak (ABB) (Waite & Stover 1960, Ploetz 2006). Isolates that clustered in the newly resolved subclade in the FOSC in this study were found to be non-pathogenic towards both Cavendish and Gros Michel. These isolates only caused initial discoloration in the corm, without any further disease development. They might be pathogenic on other germplasm, but until more banana varieties can be tested, this idea remains speculation.
Our study demonstrates that the Indonesian Foc population might be the most genetically diverse ever studied. Further genetic study of this population using deeper genomic coverage should now be conducted. Pathogenicity tests using more banana varieties could be used to assess the wide range of pathogenicity.
Our study gives an insight into the complexity of Fusarium wilt on banana in Indonesia. This is very important for disease management not only in Indonesia but also worldwide. As the pathogen continues to evolve, new lineages could arise and escape Indonesia. In striving to find banana resistance to Fusarium wilt, researchers should consider the high diversity of Indonesian Foc reported here as one of the main obstacles to overcome.
Acknowledgements
This research was supported by the KNAW-SPIN Project, “The Indonesian banana: Protecting a staple food from Panama disease collapse and exploiting its genetic diversity for discovery research”. Nani Maryani was also supported by a DIKTI (Directorate General of Higher Education, DIKTI) Scholarship, Ministry of Education, Indonesia. The authors would like to thank Dr Sarah M. Schmidt (Institute for Molecular Physiology, Heinrich-Heine-Universität Düsseldorf and Max Plank Institute for Plant Breeding Research, Köln, Germany), F. Ahmad (Research Centre for Biology, Indonesian Institute of Sciences, LIPI, Cibinong, Indonesia) and Muhammad Ilyas (Indonesian Culture Collection, INACC, LIPI, Cibinong, Indonesia) for participating in the sampling expeditions in Java, Sumatra, and Kalimantan. We also like to thank Dr Marcelo Sandoval-Denis (Westerdijk Fungal Biodiversity Institute) for helpful discussions and technical support and Dr Kerry O'Donnell (USDA) for the FOSC dataset.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Contributor Information
N. Maryani, Email: nani.maryanimartawi@wur.nl.
G.H.J. Kema, Email: gert.kema@wur.nl.
References
- Baayen R.P., O'Donnell K., Bonants P.J.M. Gene genealogies and AFLP analyses in the Fusarium oxysporum complex identify monophyletic and nonmonophyletic formae speciales causing wilt and rot disease. Phytopathology. 2000;90:891–900. doi: 10.1094/PHYTO.2000.90.8.891. [DOI] [PubMed] [Google Scholar]
- Banke S., Peschon A., McDonald B.A. Phylogenetic analysis of globally distributed Mycosphaerella graminicola populations based on three DNA sequence loci. Fungal Genetics and Biology. 2004;41:226–238. doi: 10.1016/j.fgb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Bentley S., Pegg K.G., Dale J.L. Genetic variation among a world-wide collection of isolates of Fusarium oxysporum f. sp. cubense analysed by RAPD-PCR fingerprinting. Mycological Research. 1995;99:1378–1384. [Google Scholar]
- Bentley S., Pegg K.G., Moore N.Y. Genetic variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense analyzed by DNA fingerprinting. Phytopathology. 1998;88:1283–1293. doi: 10.1094/PHYTO.1998.88.12.1283. [DOI] [PubMed] [Google Scholar]
- Boehm E., Ploetz R., Kistler C.H. Statistical analysis of electrophoretic karyotype variation among vegetative compatibility groups of Fusarium oxysporum f. sp. cubense. MPMI-Molecular Plant Microbe Interactions. 1994;7:196–207. [Google Scholar]
- Booth C. CAB, Commonwealth Mycological Institute; UK: 1971. The genus Fusarium. [Google Scholar]
- BPS . Badan Pusat Statistik (BPS); 2017. Statistics Indonesia.https://www.bps.go.id/ Produksi Tanaman Hortikultura. [Google Scholar]
- Buddenhagen I. Understanding strain diversity in Fusarium oxysporum f. sp. cubense and history of introduction of "Tropical Race 4" to better manage banana production. Proceedings of International Symposium on Recent Advances in Banana Crop Protection for Sustainable Production and Improved Livelihoods. 2007;828:193–204. [Google Scholar]
- Crous P.W., Gams W., Stalpers J.A. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology. 2004;50:19–22. [Google Scholar]
- Daniells J. Illustrated guide to the identification of banana varieties in the South Pacific. ACIAR Monograph. 1995;33:1–49. [Google Scholar]
- De Langhe E. Relevance of banana seeds in archaeology. Ethnobotany Research and Applications. 2009;7:271–281. [Google Scholar]
- Dettman J.R., Jacobson D.J., Taylor J.W. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003;57(12):2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. [DOI] [PubMed] [Google Scholar]
- Dita M.A., Waalwijk C., Buddenhagen I. A molecular diagnostic for tropical race 4 of the banana fusarium wilt pathogen. Plant Pathology. 2010;59:348–357. [Google Scholar]
- FAOSTAT . 2017. FAO statistics.http://www.fao.org/faostat/ [Google Scholar]
- Fisher N.L., Burgess L.W., Toussoun T.A. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology. 1982;72:151–153. [Google Scholar]
- Fourie G., Steenkamp E.T., Gordon T.R. Evolutionary relationships among the Fusarium oxysporum f. sp. cubense vegetative compatibility groups. Applied and Environmental Microbiology. 2009;75:4770–4781. doi: 10.1128/AEM.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia F., Alexander V., Nakasato T.G. 2018. A high throughput phenotyping method for the banana - Fusarium oxysporum f. sp. cubense pathosystem, with wider application for other Fusarium oxysporum – host relationships. In prep. [Google Scholar]
- Gerlach K.S., Bentley S., Moore N.Y. Characterisation of Australian isolates of Fusarium oxysporum f. sp. cubense by DNA fingerprinting analysis. Australian Journal of Agricultural Research. 2000;51:945–953. [Google Scholar]
- Gordon T., Okamoto D. Population structure and the relationship between pathogenic and nonpathogenic strains of Fusarium oxysporum. Phytopathology. 1992;82:73–77. [Google Scholar]
- Groenewald S., Van Den Berg N., Marasas W.F.O. The application of high-throughput AFLP's in assessing genetic diversity in Fusarium oxysporum f. sp. cubense. Mycological Research. 2006;110:297–305. doi: 10.1016/j.mycres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Grünwald N.J., Flier W.G. The biology of Phytophthora infestans at its center of origins. Annual Review Phytopathology. 2005;43:171–190. doi: 10.1146/annurev.phyto.43.040204.135906. [DOI] [PubMed] [Google Scholar]
- Hermanto C., Sutanto A., Jumjunidang Incidence and distribution of Fusarium wilt disease of banana in Indonesia. International ISHS-ProMusa Symposium on Global Perspectives on Asian Challenges. 2009;897:313–322. [Google Scholar]
- Hsuan H.M., Salleh B., Zakaria L. Molecular identification of Fusarium species in Gibberella fujikuroi species complex from rice, sugarcane and maize from Peninsular Malaysia. International Journal of Molecular Sciences. 2011;12:6722–6732. doi: 10.3390/ijms12106722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed M.A., Chai M., Othman R.Y. Study of Resistance of Musa acuminata to Fusarium oxysporum using RAPD markers. Biologia Plantarum. 2004;48:93–99. [Google Scholar]
- Jumjunidang, Edison, Riska Penyakit layu Fusarium pada tanaman pisang di provinsi NAD: Sebaran dan identifikasi isolat berdasarkan analisis vegetative compatibility group. Jurnal Hortikultur. 2012;22:164–171. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler H.C. Genetic diversity in the plant-pathogenic fungus Fusarium oxysporum. Phytopathology. 1997;87(4):474–479. doi: 10.1094/PHYTO.1997.87.4.474. [DOI] [PubMed] [Google Scholar]
- Koenig R., Ploetz R., Kistler H.C. Fusarium oxysporum f. sp. cubense consists of a small number of divergent and globally distributed clonal lineages. Phytopathology. 1997;87:915–923. doi: 10.1094/PHYTO.1997.87.9.915. [DOI] [PubMed] [Google Scholar]
- Komada H. Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Review of Plant Protection Research. 1975;8:114–124. [Google Scholar]
- Laurence M.H., Summerrell B.A., Burgess L.W. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology. 2014;118:374–384. doi: 10.1016/j.funbio.2014.02.002. [DOI] [PubMed] [Google Scholar]
- Leslie J.F., Summerell B.A. Blackwell Publishing Ltd; UK: 2006. The Fusarium Laboratory Manual. [Google Scholar]
- Ma L.J., Van Der Does H.C., Borkovich K.A. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasas W.F.O., Ploetz R.C., Wingfield M.J. Mango malformation disease and the associated Fusarium species. Phytopathology. 2006;96:667–672. doi: 10.1094/PHYTO-96-0667. [DOI] [PubMed] [Google Scholar]
- Mason-Gamer R., Kellogg E. Testing for phylogenetic conflict among molecular datasets in the tribe Triticeae (Graminae) Systematic Biology. 1996;45:525–545. [Google Scholar]
- Moore N.Y., Pegg K.G., Allen R.N. Vegetative compatibility and distribution of Fusarium oxysporum f. sp. cubense in Australia. Animal Production Science. 1993;33:797–802. [Google Scholar]
- Mostert D., Molina A., Daniells J. The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f. sp. cubense, in Asia. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0181630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir N., Pittaway P.A., Pegg K.G. A pilot study investigating the complexity of Fusarium wilt of bananas in West Sumatra, Indonesia. Australian Journal of Agricultural Research. 1999;50:1279–1283. [Google Scholar]
- Nasution R.E. Tokyo University of Agriculture; Japan: 1990. A Taxonomic Study of the Species Musa acuminata Colla with its Intraspecific Taxa in Indonesia. Ph.D dissertation. [Google Scholar]
- Nasution R.E. Rediscovery of two wild seeded bananas of Indonesia. Infomusa. 1993;2:16–17. [Google Scholar]
- Nirenberg H.I. A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany. 1981;59:1599–1609. [Google Scholar]
- O'Donnell K., Kistler C.H., Cilgenik E. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Sutton D.A., Rinaldi M.G. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Sutton D.A., Rinaldi M.G. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. Journal of Clinical Microbiology. 2004;42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K., Sutton D.A., Rinaldi M.G. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genetics and Biology. 2009;46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- O’Donnell K., Ward T.J., Robert V.A.R.G. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica. 2015;15:583–595. [Google Scholar]
- O'Neill W.T., Pattison A.B., Daniells J.W. Vegetaitive compatibility group analysis of Indonesian Fusarium oxysporum f. sp. cubense isolates. Acta Horticulturae. 2011;897:345–352. [Google Scholar]
- Ordonez N. Experimental Plant Sciences, Wageningen University; The Netherlands: 2018. A global genetic diversity analysis of Fusarium oxysporum f. sp. cubense, the Panama disease pathogen of banana. Ph.D dissertation. [Google Scholar]
- Ordonez N., Seidl M.F., Waalwijk C. Worse comes to worst: bananas and Panama disease—when plant and pathogen clones meet. PLoS Pathogens. 2015;11 doi: 10.1371/journal.ppat.1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochse J.J., Bakhuizen van den Brink R.C. Nijhoff; The Hague, NL: 1931. Vegetables of the Dutch East Indies: edible tubers, bulbs, rhizomes and spices included: survey of the indigenous and foreign plants serving as pot-herbs and side-dishes. [Google Scholar]
- Perrier X., De Langhe E., Donohue M. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of the National Academy of Sciences of the USA. 2011;108:11311–11318. doi: 10.1073/pnas.1102001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploetz R.C. Variability in Fusarium oxysporum f. sp. cubense. Canadian Journal of Botany. 1990;68(6):1357–1363. [Google Scholar]
- Ploetz R.C. Panama disease: return of the first banana menace. International Journal of Pest Management. 1994;40:326–336. [Google Scholar]
- Ploetz R.C. Panama disease, an old nemesis rears its ugly head: part 2, the cavendish era and beyond. Plant Health Progress. 2006 March: 1–17. [Google Scholar]
- Ploetz R., Pegg K. Fusarium wilt of banana and Wallace’s line: Was the disease originally restricted to his Indo-Malayan region? Australasian Plant Pathology. 1997;26(4):239–249. [Google Scholar]
- Puhalla J.E. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Canadian Journal of Botany. 1985;63:179–183. [Google Scholar]
- Rayner R.W. Commonwealth Mycological Institute; Kew, UK: 1970. A mycological colour chart. [Google Scholar]
- Rep M., Kistler H.C. The genomic organization of plant pathogenicity in Fusarium species. Current Opinion in Plant Biology. 2010;13:420–426. doi: 10.1016/j.pbi.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Telsenko M., Van Den Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds N. Longman ltd; UK: 1962. The Evolution of Bananas. [Google Scholar]
- Simmonds N., Shepherd K. The taxonomy and origins of the cultivated bananas. Journal of the Linnean Society of London, Botany. 1955;55:302–312. [Google Scholar]
- Smith F. A Cuban banana disease. Science. 1910;31:746–757. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover R.H. Oxford University Press; Oxford, UK: 1962. Fusarial wilt (Panama Disease) of bananas and other Musa species. [Google Scholar]
- Stover R.H. Studies on Fusarium wilt of bananas: IX competitive saprophytic ability of Fusarium oxysporum f. sp. cubense. Canadian Journal of Botany. 1962;40:1473–1481. [Google Scholar]
- Stukenbrock E.H., Banke S., Javan-Nikhah M. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Molecular Biology and Evolution. 2007;24(2):398–411. doi: 10.1093/molbev/msl169. [DOI] [PubMed] [Google Scholar]
- Stukenbrock E.H., McDonald B.A. The origins of plant pathogens in agro-ecosystems. Annual Review of Phytopathology. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- Su Hj, Hwang S.C., Ko W.H. Fusarial wilt of Cavendish bananas in Taiwan. Plant Disease. 1986;70:814–818. [Google Scholar]
- Tamura K., Peterson D., Peterson N. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmayor R.V., Jamaluddin S.H., Silayoi J.S.B. INIBAP; Montpellier, France: 1999. Banana Cultivar Names and Synonyms in Southeast Asia. [Google Scholar]
- Van Dam P., Fokkens L., Schmidt S.M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environmental Microbiology. 2016;18:4087–4102. doi: 10.1111/1462-2920.13445. [DOI] [PubMed] [Google Scholar]
- Vlaardingerbroek I., Beerens B., Rose L. Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environmental Microbiology. 2016;18:3702–3713. doi: 10.1111/1462-2920.13281. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Baayen R., De Koning J. Ribosomal DNA analyses challenge the status of Fusarium sections Liseola and Elegans. Sydowia. 1996;48:90–104. [Google Scholar]
- Waite B.H., Stover R.H. Studies on Fusarium wilt of bananas. VI. variability and the cultivars concept in Fusarium oxysporum f. sp cubense. Canadian Journal of Botany. 1960;38:985–994. [Google Scholar]
- Wibowo A., Subandiyah S., Sumardiyono C. Occurrence of Tropical Race 4 of Fusarium oxysporum f. sp. cubense in Indonesia. Journal Plant Pathology. 2011;27(3):280–284. [Google Scholar]
- Zakaria L., Rahman N.H.A. Endophytic Fusarium spp. from wild banana (Musa acuminata) roots. African Journal of Microbiology Research. 2011;5:3600–3602. [Google Scholar]