Abstract
Defective germline reprogramming in Miwi2- and Dnmt3l-deficient mice results in the failure to reestablish transposon silencing, meiotic arrest and progressive loss of spermatogonia. Here we sought to understand the molecular basis for this spermatogonial dysfunction. Through a combination of imaging, conditional genetics and transcriptome analysis, we demonstrate that germ cell elimination in the respective mutants arises due to defective de novo genome methylation during reprogramming rather than a function for the respective factors within spermatogonia. In both Miwi2-/- and Dnmt3l-/- spermatogonia the intracisternal-A particle (IAP) family of endogenous retroviruses is de-repressed, but in contrast to meiotic cells DNA damage is not observed. Instead we find that unmethylated IAP promoters rewire the spermatogonial transcriptome by driving expression of neighboring genes. Finally, spermatogonial numbers, proliferation and differentiation are altered in Miwi2-/- and Dnmt3l-/- mice. In summary, defective reprogramming deregulates the spermatogonial transcriptome and may underlie spermatogonial dysfunction.
Introduction
In mammals the germline is an acquired lineage derived from the proximal layer of the epiblast at the time of gastrulation1–6. This acquisition from the soma provides the germline with a major challenge – the necessity to undergo genome reprogramming to reset genomic DNA methylation patterns that underpin genomic imprinting and transposon silencing7–13. Genome demethylation in the mouse initiates at ~E9.0 as primordial germ cells (PGCs) migrate to genital ridges and is complete at ~E13.59,14. However, some IAP copies and young transposons, as well as a few genes escape full demethylation12,14–16. The key executor of de novo DNA methylation is DNMT3L that recruits and stimulates the DNMT3 de novo DNA methyltransferases17–24. The first wave of de novo methylation is rather indiscriminate leading to the bulk methylation of CpG islands and transposons13. Many IAP and LINE1 copies escape this round of methylation and become expressed threating the genomic integrity of the germline through transposition13. The PIWI proteins and their associated small non-coding PIWI-interacting RNAs (piRNAs) neutralize this threat by simultaneously destroying cytoplasmic transposon mRNAs and generating secondary antisense transposon-derived piRNAs that act to guide the nuclear PIWI protein MIWI2 (PIWIL4) to instruct methylation by an unknown mechanism25–27. The process of de novo DNA methylation is more or less complete at ~E18.516,28,29, even though the methylation of a few loci is finished only shortly after birth just prior gonocyte reentry into the cell cycle and developmental transition to spermatogonia30.
The failure of male germline reprogramming in Dnmt3l- or Miwi2-deficient mice results in sterility due to meiotic arrest in pachytene that originates from deregulated LINE1 and IAP elements and the ensuing DNA damage22,27,31,32. The loss of Dnmt3l or Miwi2 also results in the progressive loss of the germ cells in the adult testis27,32,33. For Miwi2-deficient mice the germ cell depletion has been shown to be a germ cell autonomous phenotype32. Thus, loss of either Dnmt3l or Miwi2 affects spermatogonia, the stem and mitotic cells of spermatogenesis. Whether the spermatogonial phenotype arises due to defective reprogramming or a direct function for either protein in spermatogonia remains unknown. There are conflicting data about Dnmt3l expression in the adult testes, however, most of the reports suggest that Dnmt3l transcript and protein is unlikely to be expressed in adult mouse testis22,34–38. The lack of Miwi2 expression was also reported for adult testis25, however, recently the Miwi2 promoter has been shown to be active in a subset of undifferentiated adult spermatogonia, although whether MIWI2 protein is expressed remains unknown39. These Miwi2-expressing spermatogonia are transit-amplifying cells during homeostatic spermatogenesis, but also merit a population of facultative stem cells that are critical for injury-induced regenerative spermatogenesis39. Here we sought to understand the mechanism by which MIWI2 and DNMT3L contribute to the maintenance of spermatogonia and spermatogenesis.
Results
MIWI2 but not DNMT3L is expressed in undifferentiated spermatogonia
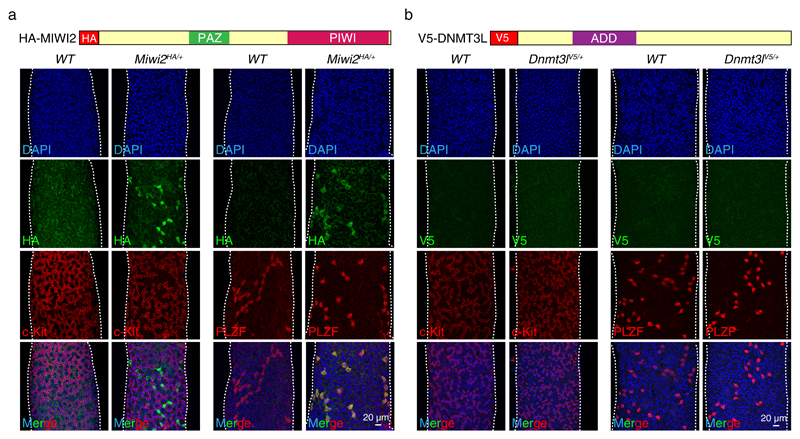
The loss of spermatogonia in the Miwi2-/- and Dnmt3L-/- mice could imply that defective germline reprogramming in the respective mutants disrupts spermatogonial function. Alternatively, MIWI2 and DNMT3L could be directly expressed in spermatogonia and underpin critical cellular functions within these cells. To understand the expression of MIWI2 protein in the adult testis we utilized the Miwi2HA allele that encodes a fully functional epitope-tagged HA-MIWI240. HA-MIWI2 was detected at the basal compartment of the seminiferous tubule in cells with spermatogonial morphology (Fig. 1a). We next co-stained Miwi2HA/+ seminiferous tubules with PLZF and c-Kit, markers of undifferentiated and differentiating spermatogonia41,42, respectively (Fig. 1a); and found that HA-MIWI2-expressing cells were PLZF positive and negative for c-Kit. To elucidate the expression of DNMT3L in the adult testis we generated the Dnmt3lV5 allele that encodes an N-terminal epitope-tagged V5-DNMT3L (Supplementary Fig. 1a, b). The introduction of the V5 epitope tag did not affect the function of DNMT3L protein as mice homozygous for the Dnmt3lV5 allele manifested normal spermatogenesis and were fertile (Supplementary Fig. 1c,d). While V5-DNMT3L could be readily detected in E16.5 fetal gonocytes (Supplementary Fig. 1e), it was not detected in spermatogonia or any other cell type of the adult testis (Fig. 1b). Additionally, the Dnmt3l transcript was very lowly expressed in juvenile undifferenitated spermatogonia (Supplementary Fig. 1f). In summary, MIWI2 but not DNMT3L protein is expressed in a subset of undifferentiated PLZF-positive spermatogonia.
Figure 1. MIWI2 but not DNMT3L is expressed in adult mouse undifferentiated spermatogonia.
(a, b) Representative images of seminiferous tubules from Miwi2HA/+ (a) and Dnmt3lV5/+ 8 weeks old mice (b) stained as indicated with anti-HA, anti-V5, anti-PLZF or anti-c-Kit antibodies.
MIWI2 function within adult spermatogonia is not required for spermatogenic homeostasis or regeneration
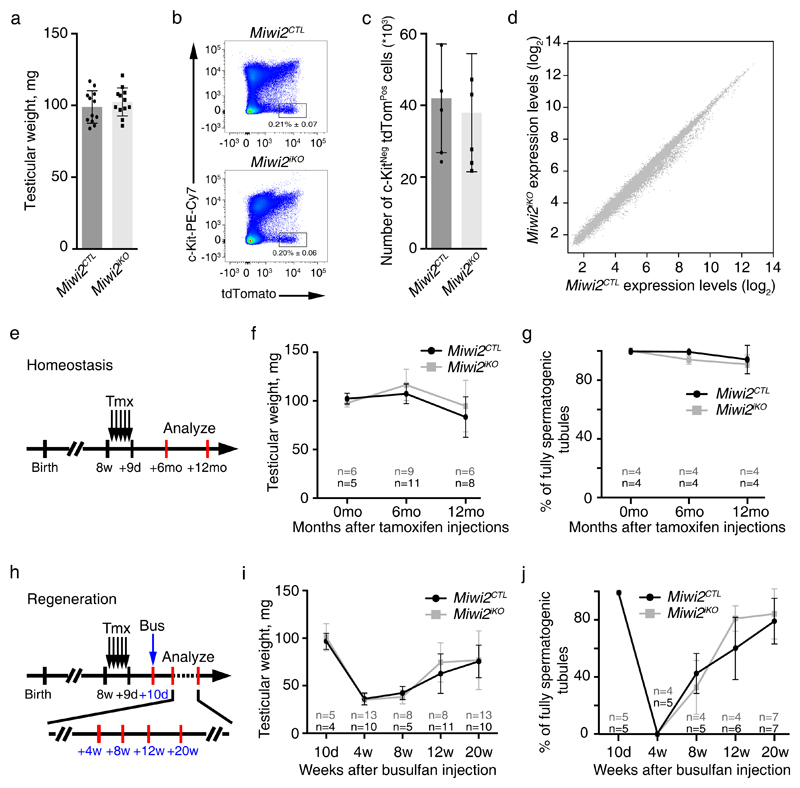
The fact that MIWI2 protein is expressed in a population of undifferentiated spermatogonia could indicate a possible direct molecular role for MIWI2 in these cells. To circumvent MIWI2’s critical role in germline reprogramming, conditional genetics must be employed. Miwi2 has been previously conditionally ablated using Stra8Cre that deletes in differentiating spermatogonia without any consequences to homeostatic mouse spermatogenesis43. However, Stra8Cre mediates gene deletion in differentiating spermatogonia just prior or upon entry into meiosis44–46 – that is after the attenuation of MIWI2 expression in the adult testis. We therefore utilized inducible conditional genetics to delete Miwi2 in adult mice. To this end we combined our Miwi2FL 27, Miwi2tdTom 39 and the tamoxifen (Tmx) inducible Rosa26ERT-Cre 47 alleles to generate experimental Miwi2FL/tdTom; R26ERT-Cre/+ and control Miwi2+/tdTom; R26ERT-Cre/+ mice. The Miwi2tdTom allele is a transcriptional reporter and a null allele of Miwi2, where the coding sequence of tdTomato followed by a poly(A) site has been inserted into the starting codon of Miwi239. The administration of Tmx to Miwi2FL/tdTom; R26ERT-Cre/+ and Miwi2+/tdTom; R26ERT-Cre/+ mice converts the Miwi2FL to Miwi2Null allele generating induced Miwi2 mutant (Miwi2iKO) and control (Miwi2CTL) mice, respectively. The presence of the Miwi2tdTom within the experimental design enables the isolation of Miwi2iKO and Miwi2CTL undifferentiated spermatogonia. Miwi2-tdTomato-expressing FACS-isolated spermatogonia from Miwi2iKO mice were shown to undergo a complete deletion of the Miwi2FL allele as determined by genotyping and RT-qPCR (Supplementary Fig. 2a,b). The acute deletion, analyzed 14 days after the last Tmx injection (Supplementary Fig. 2c), did not impact the testicular weight, spermatogenesis, the frequency or gene expression of adult undifferentiated Miwi2-tdTomato positive spermatogonia (Fig. 2a-d). We next sought to understand the long-term impact of Miwi2 deletion on spermatogenesis in aged cohort of Miwi2iKO and Miwi2CTL mice. The induced ablation did not impact spermatogenesis up to one year after deletion (Fig. 2e-g, Supplementary Fig. 2d), a time point where all germ cells are absent in the conventional Miwi2-/- mice27,32.
Figure 2. MIWI2 is not required for homeostatic or regenerative spermatogenesis.
(a) Testicular weight of Miwi2CTL and Miwi2iKO 11-week-old mice is shown. Error bars represent a standard deviation of the mean (n=6 animals). Individual data points are shown. (b) Representative FACS analysis plots of live CD45Neg CD51Neg gated testicular cells from Miwi2CTL and Miwi2iKO 11-week-old mice are shown. Numbers indicate the percentage of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos cells. (c) Enumeration of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos cells per testis from Miwi2CTL and Miwi2iKO 11-week-old mice is shown. Error bars represent a standard deviation of the mean (n=6 animals). Individual data points are shown. (d) Expression scatterplots showing relative average expression of transcripts between live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos CD9Pos cell populations from 11-week-old Miwi2CTL and Miwi2iKO mice. No significant changes in gene expression were observed. Analysis was done in biological triplicates (n=3 animals). (e) Scheme representing a time line for inducible Miwi2 deletion. Tamoxifen (Tmx) administration is depicted as black arrows and analysis time points are indicated with red bars at 6 months and 12 months after the last tamoxifen injection. (f) Testicular weight of Miwi2CTL and Miwi2iKO mice at the indicated time points is presented. Error bars represent a standard deviation of the mean. (g) Percentage of fully spermatogenic tubules in Miwi2CTL and Miwi2iKO at the indicated time points is presented. Error bars represent a standard deviation of the mean. (h) Scheme representing a time line for inducible Miwi2 deletion (tamoxifen (Tmx) administration is depicted as black arrows), testicular damage by busulfan (Bus) administration (blue arrow) and the analysis time points (red bars) of 4, 8, 12 and 20 weeks after the busulfan injection are indicated. (i) Testicular weight of Miwi2CTL and Miwi2iKO mice over the period of 20 weeks after busulfan injection is shown. Error bars represent a standard deviation of the mean. (j) Percentage of fully spermatogenic tubules in Miwi2CTL and Miwi2iKO mice over the period of 20 weeks after busulfan injection is presented. Error bars represent a standard deviation of the mean. Number of animals (n) per time point in f, g, i, j is indicated. Source data for a, c, f, g, i, j is provided.
The population of Miwi2-expressing spermatogonia is facultative stem cells and required for the efficient regenerative capacity of the testis upon injury39. Therefore, we sought to determine if MIWI2 may function in regenerative spermatogenesis. To do so, Miwi2iKO and Miwi2CTL mice were treated with busulfan, a DNA alkylating agent, at a dose that kills many spermatogonial cells, induces tissue damage and subsequent tissue repair48. The induced loss of MIWI2 did not impact the testicular regeneration upon busulfan-mediated damage (Fig. 2h-j, Supplementary Fig. 2e). In conclusion, MIWI2 function is not required in adult spermatogonia for homeostatic or regenerative spermatogenesis.
Defective reprogramming affects undifferentiated spermatogonia numbers, proliferation and differentiation in Miwi2-/- and Dnmt3l-/- mice
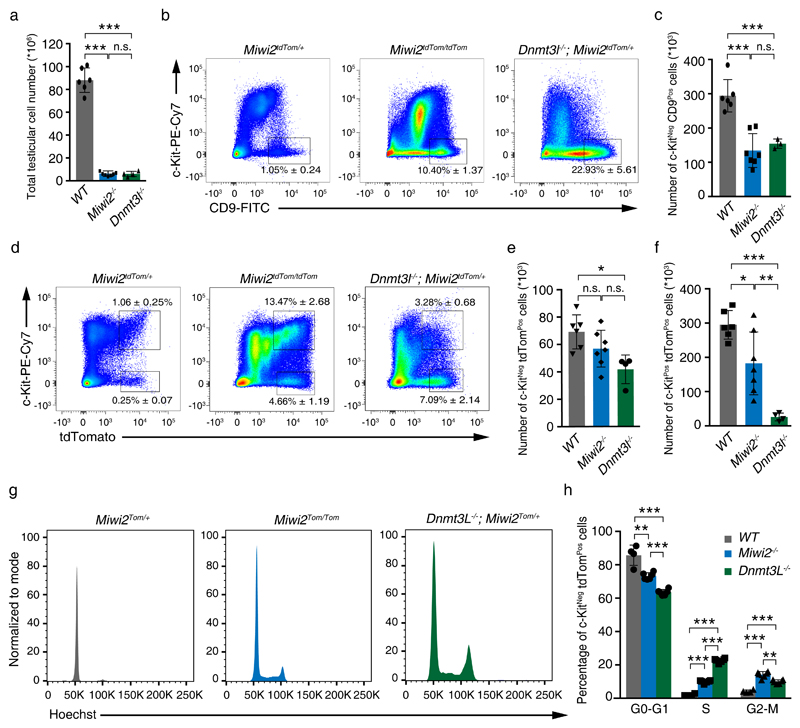
The fact that MIWI2 is not required for adult spermatogonial function and DNMT3L is not detected in spermatogonial populations indicates that the progressive loss-of-germ-cell phenotype in the respective mutants originates from defective reprogramming during fetal gonocyte development. We next sought to understand the impact of defective reprogramming on adult spermatogonial populations in Miwi2-/- and Dnmt3l-/- (Supplementary Fig. 3) mice. Firstly, reduced testicular cellularity is observed in 8 weeks old reprogramming mutants (Fig. 3a) that arises from the meiotic arrest phenotype in both mutants. Undifferentiated spermatogonial populations can be identified by CD45Neg CD51Neg c-KitNeg CD9Pos surface expression by FACS, this population contains Gfrα1 and Miwi2-expressing cells (Supplementary Fig. 4) that includes the stem, facultative stem and early transit-amplifying spermatogonial population39,49–51. Using this surface CD45Neg CD51Neg c-KitNeg CD9Pos stain (Supplementary Fig. 5a), a relative increase in the frequency of undifferentiated spermatogonia is observed, however, with the reduced testicular cellularity there is a reduction in the absolute numbers of undifferentiated spermatogonia in both Miwi2-/- and Dnmt3l-/- adult mice (Fig. 3b,c). Next, we analyzed the Miwi2-expressing (CD45Neg CD51Neg c-KitNeg Miwi2-tdTomHi; Supplementary Fig. 5a) undifferentiated spermatogonia population; Miwi2-/- mice tended to have slightly lower cell number of this population, whereas Dnmt3l-/- deficiency presented even lower number of these cells compared to the control mice (Fig. 3d,e). The expression of the Miwi2–tdTomato in CD45Neg CD51Neg c-KitPos (Supplementary Fig. 5a) cells identifies a nascent population of differentiating spermatogonia39. Interestingly, the absolute number of these differentiating spermatogonia are reduced in Miwi2-/- and severely depleted in Dnmt3l-/- in adult testis (Fig. 3d,f). In summary, Dnmt3l-deficiency has a greater impact on spermatogonia than the loss of Miwi2. Finally, we analyzed the cell cycle parameters of Miwi2-expressing undifferentiated spermatogonia (Supplementary Fig. 5b) and found that both Miwi2-/- and Dnmt3l-/- mice showed increased fractions of proliferating cells within this population (Fig. 3g,h). Together these results indicate that defective reprogramming affects undifferentiated spermatogonia cell numbers, proliferation and likely differentiation in Miwi2-/- and Dnmt3l-/- adult mice.
Figure 3. Defective reprogramming affects undifferentiated spermatogonia numbers, proliferation and differentiation.
(a) Total testicular cell numbers of Miwi2tdTom/+ (wild-type, WT), Miwi2tdTom/tdTom (Miwi2-/-) and Dnmt3l-/-; Miwi2tdTom/+ (Dnmt3l-/-) 8-week-old mice are shown. Error bars represent a standard deviation of the mean. Individual data points are shown. (b) Representative FACS plots of live CD45Neg CD51Neg gated testicular cells from 8-9-week-old mice of the indicated genotype is shown. Numbers indicate the percentage of live CD45Neg CD51Neg c-KitNeg CD9Pos cells. (c) Enumeration of live CD45Neg CD51Neg c-KitNeg CD9Pos cells per testis from 8-9-week-old mice of the indicated genotypes (as defined in panel a) is shown. Error bars represent a standard deviation of the mean. Individual data points are shown. (d) Representative FACS plots of live CD45Neg CD51Neg gated testicular cells from 8-9-week-old mice of the indicated genotype are shown. Numbers indicate the percentage of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos and live CD45Neg CD51Neg c-KitPos Miwi2-tdTomPos cells. (e-f) Enumeration of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos cells (e) and CD45Neg CD51Neg c-KitPos Miwi2-tdTomPos cells (f) per testis from 8-9-week-old mice of the indicated genotype (as defined in panel a) are shown. Error bars represent a standard deviation of the mean. Individual data points are shown. Number of animals (n) in a, c, e, f is 6 animals in WT, 7 animals in Miwi2-/-, 4 animals in Dnmt3l-/-, except panel c where 3 animals in Dnmt3l-/- is presented. (g) Representative cell cycle parameters as determined by DNA content in FACS analysis of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos gated testicular cells from 8-9-week-old mice of the indicated genotype are shown. (h) Enumeration of live CD45Neg CD51Neg c-KitNeg Miwi2-tdTomPos cells from 8-9-week-old mice of the indicated genotype (as defined in panel a) in G0-G1, S and G2-M cell cycle phases is shown. Error bars represent a standard deviation of the mean (n=4 animas in WT, 6 animals in Miwi2-/- and Dnmt3l-/-). Individual data points are shown. Significance in a, c, e, f, h was assessed using two-sided t-test. n.s. – not significant, * indicates a p value of < 0,01, ** indicates a p value of < 0,001, *** indicates a p value of < 0,0001. Source data for a, c, e, f, h is provided.
Selective IAP de-repression without DNA damage in Miwi2-/- and Dnmt3l-/- spermatogonia
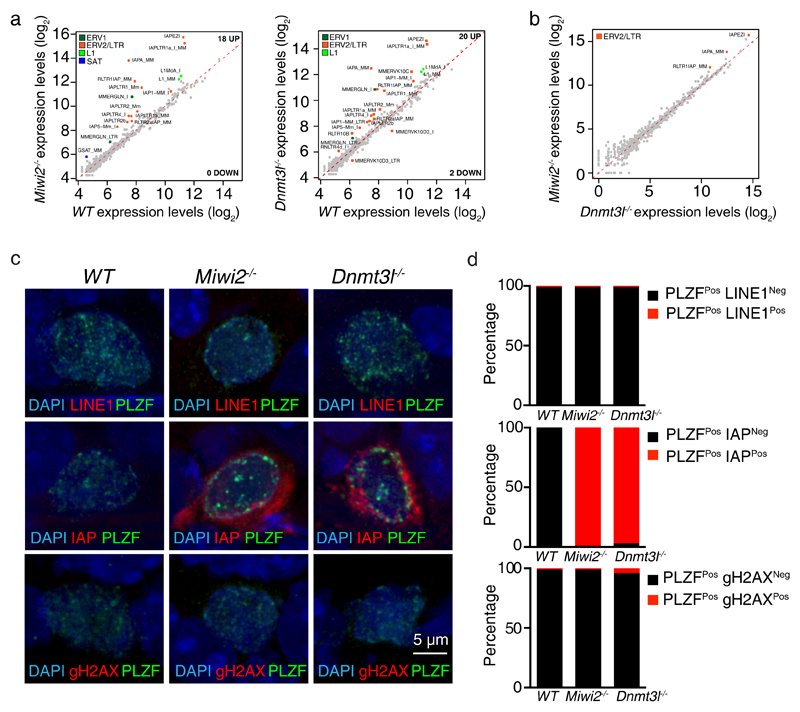
Given that aberrant genome methylation and transposon depression is common to both reprogramming mutants, we aimed to understand, which elements are de-repressed in Miwi2-/- and Dnmt3l-/- spermatogonia. To this end, we isolated (CD45Neg CD51Neg c-KitNeg Miwi2-tdTomHi undifferentiated spermatogonia by FACS from wild type, Miwi2-/- and Dnmt3l-/- juvenile postnatal day 14 (P14) (Supplementary Fig. 5a) mice and subjected the respective isolated RNA in biological triplicates to strand specific RNA-seq. We selected P14 for two reasons, firstly, the spermatogenic arrest is not present at P14 as the pachytene stage of meiosis has not been reached and consequent confounding effects are minimized. At this age the loss of Miwi2 and Dnmt3l does not alter overall undifferentiated spermatogonial cell numbers, but does modestly impact Miwi2-tdTomato expressing undifferentiated and differentiating spermatogonia (Supplementary Fig. 6). Secondly, purer populations of spermatogonia can be isolated from P14 testis in comparison to adult mice, especially when one of the genotypes presents a spermatogenic arrest. Strikingly, predominantly IAP family members were deregulated in both Miwi2-/- and Dnmt3l-/- spermatogonia transcriptomes (Fig. 4a). In fact, the deregulation of transposable elements was almost identical between the respective mutant spermatogonia (Fig. 4b). These results were confirmed by immunofluorescence staining of wild type, Miwi2-/- and Dnmt3l-/- testicular cross-sections with anti-PLZF to identify undifferentiated spermatogonia in combination with anti-ORF1p LINE1 or anti-GAG IAP antibodies. LINE1 ORF1p protein was not detected in any of the genotypes, whereas IAP GAG was expressed in both Miwi2-/- and Dnmt3l-/- spermatogonia (Fig. 4c and d). Thus, the deregulation of IAP and its potential transposition may underlie the progressive loss of spermatogonia in the reprogramming mutants. DNA damage is often used as a surrogate for transposition events32,52, however, we did not find evidence of DNA damage in spermatogonia in any of the genotypes (Fig. 4c and d). In summary, deficiency in Miwi2 and Dnmt3l results in nearly identical activation of the IAP family of ERVs without the induction of DNA damage.
Figure 4. Deregulation of IAP, but no DNA damage is found in Miwi2-/- and Dnmt3l-/- undifferentiated spermatogonia.
(a) Expression scatterplots showing relative average expression (arbitrary unit (AU)) of transposable elements between wild-type (WT) and Miwi2-/- as well as WT and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia. Significantly deregulated transposable elements (p < 0,01) with a fold change greater than 2 are highlighted. (b) Expression scatterplots showing relative average expression of transposable elements between Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia as presented in panel a. Analysis in a and b was done in biological triplicates (n=3 animals). (c) Representative images of undifferentiated spermatogonia in 8-week-old WT, Miwi2-/- and Dnmt3l-/- mouse testes stained with anti-LINE1 ORF1p and anti-PLZF (upper panel), anti-IAP Gag and anti-PLZF (middle panel) and anti-γH2AX and anti-PLZF (bottom panel) antibodies. (d) Enumeration of PLZFPos undifferentiated spermatogonia that are LINE1Neg or LINE1Pos (upper panel), IAPNeg or IAPPos (middle panel) and γH2AXNeg or γH2AXPos (bottom panel) (n>200 cells per genotype per staining). Source data for d is provided.
IAPs deregulate the Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes
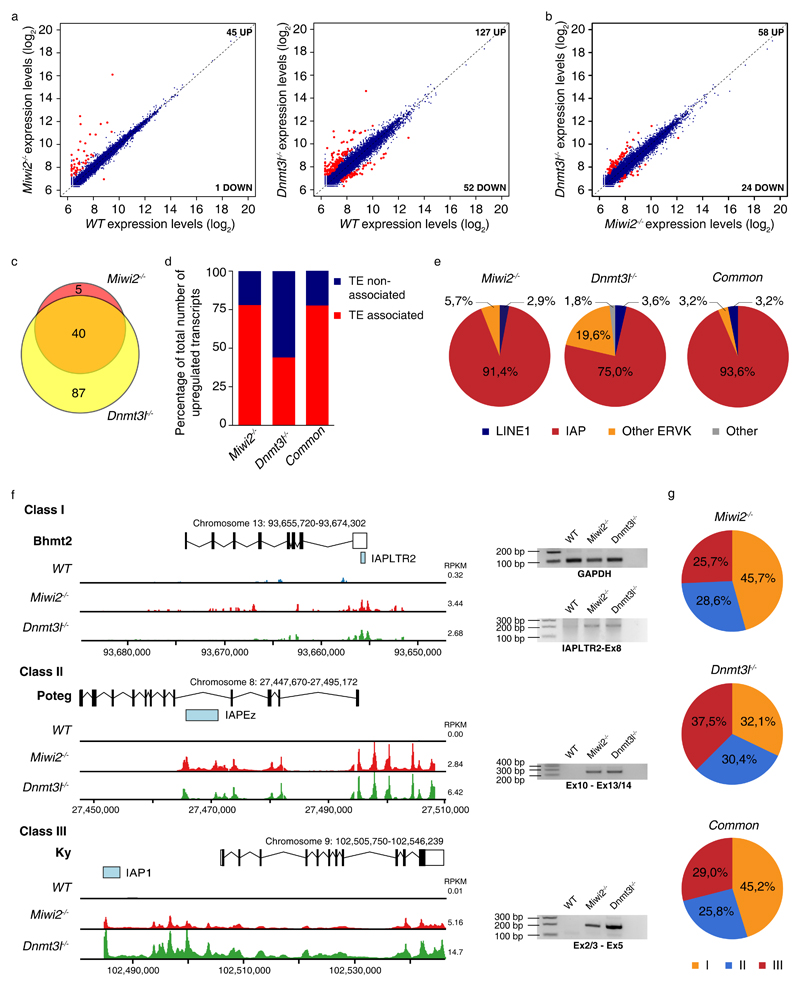
We next analyzed the expression of ENSEMBL-annotated genes in our spermatogonia RNA-seq datasets. Deregulated gene expression was observed in both of Miwi2-/- and Dnmt3l-/- transcriptomes (Fig. 5a, Supplementary Data Set 1). However, the magnitude of gene expression changes was distinct between Miwi2-/- and Dnmt3l-/- transcriptomes (Fig. 5a). The deregulation of gene expression was more severe in the Dnmt3l-/- spermatogonial transcriptome (Fig. 5b), however, the majority of upregulated genes in Miwi2-/- cells were also overexpressed in Dnmt3l-/- spermatogonia (Fig. 5c). Given that both factors regulate transposon silencing, we sought to understand if transposon de-repression could underlie this deregulated gene expression. With the manageable number of 132 upregulated genes we manually inspected if these upregulated transcripts in the respective mutants are associated with the presence of a nearby transposable element (TE). We found that a transposon acted as a promoter for the transcripts of 78% (35 genes) and 44% (56 genes) of the upregulated genes in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes, respectively (Fig. 5d). For the 40 genes that were commonly upregulated in the respective mutants, a TE association of 78% was observed (Fig. 5d). Within the TE-associated genes we examined the identity of the transposon, which was associated with the upregulated genes. IAPs dominated this association, where greater than 90% and 75% of upregulated genes in Miwi2-/- and Dnmt3l-/- spermatogonia, respectively, were driven by an IAP element (Fig. 5e, Supplementary Data Set 1). Other ERVs followed with LINE1 having a very minor to negligible association (Fig. 5e). Overall, we could define 3 classes of transcripts originating from IAPs (Fig. 5f). In class I the IAP was present in an intron and the TE partially overlapped with a single exon of the host gene. Class II was defined by an intronic IAP that drove the expression of the exons 3′of the insertion, such that a 5′ truncated chimeric transcript would be created. Class III was characterized by an upstream IAP initiating transcription prior to the first exon generating a chimeric IAP-full length genic transcript (Fig. 5f). Among the TE-associated upregulated genes in Dnmt3l-/- spermatogonia all 3 class were roughly equally represented; whereas a differential split was observed Miwi2-/- spermatogonia with ~46%, 29% and 26% encompassing class I to III, respectively (Fig. 5g). Representative loci for each of the classes and the validation of the transcripts by RT-PCR are presented in Fig. 5f (Supplementary Data Set 2). In summary, IAP elements greatly underlie the deregulated expression in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes.
Figure 5. IAPs deregulate gene expression in Miwi2-/- and Dnmt3l-/- undifferentiated spermatogonia.
(a) Expression scatterplots showing relative average expression in (arbitrary unit (AU)) of ENSEMBLE annotated genes between wild-type (WT) and Miwi2-/- as well as WT and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia. Significantly deregulated genes (p < 0,05) with a fold change greater than 2 are highlighted in red. (b) Expression scatterplots showing relative average expression of ENSEMBLE annotated genes between Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia as presented in panel a. Analysis in panels a and b was done in biological triplicates (n=3 animals). (c) Venn diagram indicating overlap of upregulated genes in Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia. (d) Percentage of upregulated genes in Miwi2-/-, in Dnmt3l-/- and upregulated genes that are common to both genotypes, whose expression was found associated with a transposable element (TE) activity, is indicated in red. Percentage of upregulated genes in corresponding groups, whose expression was not influenced by TE activity, is indicated in blue. (e) Distribution of TE groups, which were identified as drivers of upregulated genes in Miwi2-/-, in Dnmt3l-/- and upregulated genes that are common to both genotypes as identified in d. (f) Representative examples of class I, II and III IAP driven gene expression in Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia with RNA-seq tracks and RT-PCR validation of the respective transcripts presented (uncropped gel images are shown in Supplementary Data Set 2); for a given transcript the y-axis is equally scaled and the median RPKM of the transcript for the indicated genotype is shown (g) Distribution of a class I, II and III transposable element driven gene deregulation in a cohort of upregulated genes in Miwi2-/-, in Dnmt3l-/- and upregulated genes that are common to both genotypes.
Association of lowly-expressed de novo-identified transcripts with IAPs in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes
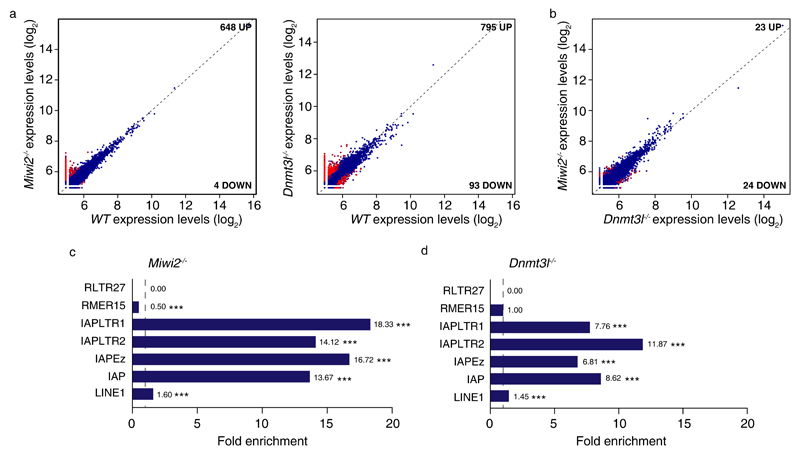
Having examined annotated genes, we next sought to understand if defective reprogramming resulted in the generation of novel transcripts within the spermatogonial transcriptomes. We reanalyzed the RNA-seq data now by de novo transcriptome assembly53 rather than mapping and quantifying reads to defined transcripts. The resulting datasets were filtered to include transcripts only greater than 200 bp in length. In this analysis, we found several hundred novel transcripts that were mostly upregulated in the respective mutants. The majority of the differentially expressed transcripts were expressed at very low levels (Fig. 6a, Supplementary Data Set 3). Interestingly, the loss of Miwi2 and Dnmt3l resulted in a very similar deregulation of these novel transcripts within the respective transcriptomes (Fig. 6b). Next we examined if the novel upregulated transcripts were associated with a transposable element. To this end we queried if a transposable element could be found within 1 kb from novel transcript’s transcription start site in comparison to a 10,000 randomly generated sized-matched transcript cohorts. We generated a fold enrichment by comparing the frequency of association for a given transposon group between random and observed (de novo-identified) transcript cohorts (Fig. 6c,d). IAPs by far gave the greatest and most significant fold enrichment for the novel low-expressed transcripts observed in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes. In summary, not only can IAPs deregulate annotated genes in reprogramming-defective spermatogonia, but they are also associated with the expression of low-expressed transcripts.
Figure 6. Increase in de novo-identified lowly expressed transcripts in Miwi2 or Dnmt3l-deficient undifferentiated spermatogonia.
(a) Expression scatterplots showing relative average expression (arbitrary unit (AU)) of transcripts identified by de novo transcriptome assembly between wild-type (WT) and Miwi2-/- as well as WT and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia. Significantly deregulated transcripts (p < 0,05) with a fold change greater than 2 are highlighted in red. (b) Expression scatterplots showing relative average expression of transcripts identified by de novo transcriptome assembly between Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia as presented in panel a. Analysis in a and b was done in biological triplicates (n=3 animals). (c, d) Fold enrichment of a transposable element occurrence within 1 kb of significantly upregulated transcripts identified by de novo transcriptome assembly in Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia as compared to a transposable element occurrence with 1 kb of a 10,000 randomly generated size-matched transcript cohorts. Significance in c and d was assessed using two-sided t-test. All values above 1 (indicated as a vertical dashed line) indicate a positive enrichment. *** indicates a p value of < 0,0001.
Defective de novo DNA methylation is associated with transcriptome rewiring
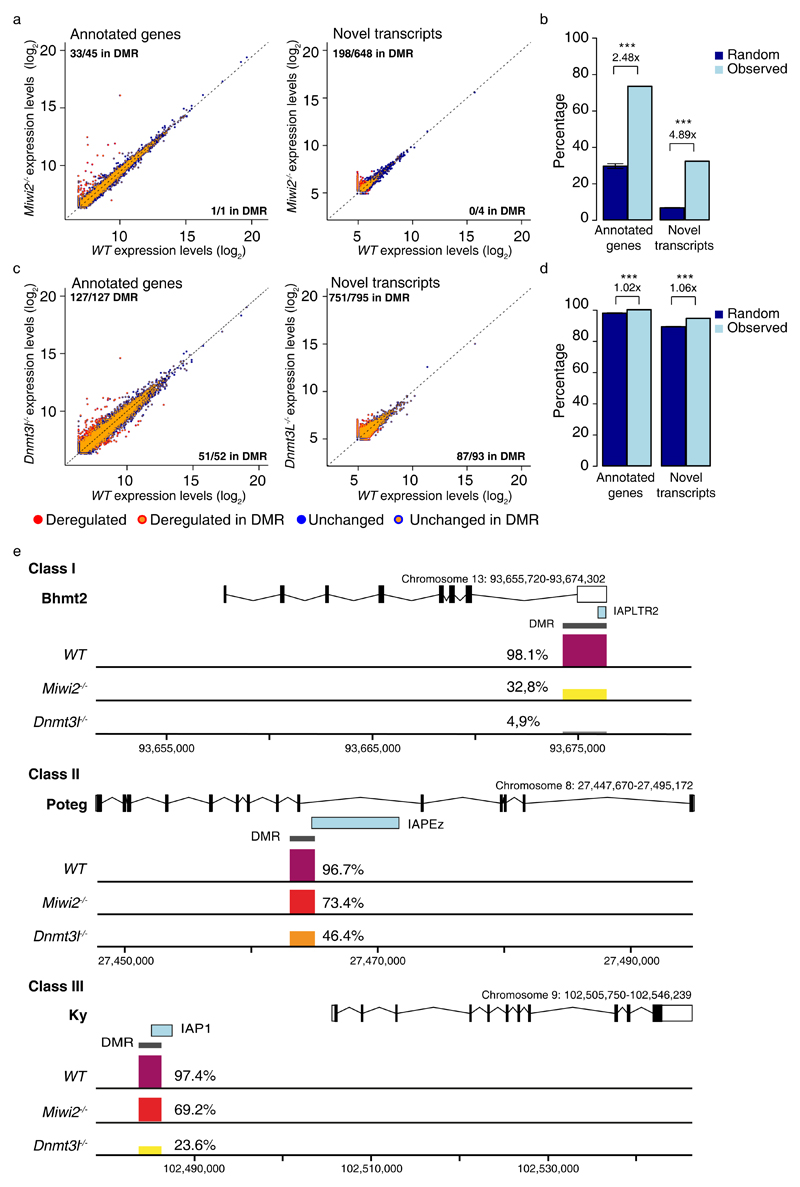
Next, we wanted to explore the underlying cause of the deregulated gene expression in both Miwi2-/- and Dnmt3l-/- transcriptomes. Again, a common defect in both genotypes is the defective methylation and expression of transposable elements. The activation of IAP as promoters in spermatogonia likely arises due to the defective de novo DNA methylation during germline reprogramming. To test this hypothesis we performed whole genome bisulfite sequencing (WGBS) on P14 Dnmt3l-/- spermatogonia and analyzed published data sets for wild-type and Miwi2-/- P14 spermatogonia that were purified and sequenced also in biological triplicates using the same methodology (Supplementary Fig. 7)40. As anticipated a genome-wide drastic reduction in CpG DNA methylation was observed in Dnmt3l-/- spermatogonia across all genomic classes, whereas Miwi2-deficency affected mostly transposable elements, particularly LINE1 and IAP elements (Supplementary Fig. 7). Given the repetitive nature of TEs, it can be difficult to get uniquely mapping reads that one can unambiguously assign to a transposon at a specific locus. We therefore decided to determine if there is an association between the deregulated loci and the differentially methylated regions (DMRs) identified in the WGBS of the respective genomes that are derived from uniquely mapping reads. The majority of deregulated genes (33/45) in Miwi2-/- spermatogonia were located within a DMR (Fig. 7a, Supplementary Data Set 4). Likewise, a large fraction of de novo-identified lowly-expressed transcripts were also associated with a DMR (Fig. 7a, Supplementary Data Set 4). To determine the significance of this enrichment, 10,000 randomly generated sized-matched transcript cohorts were inspected to overlap given DMRs as well. The test revealed a positive and significant correlation of the upregulated genes and de novo-identified upregulated transcripts to overlap with DMRs in the Miwi2-/- spermatogonia more often than given by chance (Fig. 7b). Nearly all deregulated genes and transcripts were associated with DMRs in Dnm3l-/- spermatogonia, given that the entire Dnm3l-/- spermatogonial genome is hypomethylated (Fig. 7c,d and Supplementary Fig. 7). Representative examples of DMRs associated with deregulated genes is shown in Fig. 7e in Miwi2-/- and Dnmt3l-/- genomes. In summary, defective de novo methylation is associated with the aberrant gene expression in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes.
Figure 7. Defective de novo DNA methylation is associated with deregulated gene expression in Miwi2-/- and Dnmt3l-/- undifferentiated spermatogonia.
(a) Expression scatterplots showing relative average expression (arbitrary unit (AU)) between wild-type (WT) and Miwi2-/- juvenile (P14) undifferentiated spermatogonia of ENSEMBLE annotated genes (right) and transcripts identified by de novo transcriptome assembly (left). Indicated in the figure is the methylation status in Miwi2-/- juvenile (P14) undifferentiated spermatogonia of the loci giving rise to the respective transcripts. (b) Percentage of significantly upregulated ENSEMBLE annotated genes and upregulated transcripts identified by de novo transcriptome assembly in Miwi2-/- juvenile (P14) undifferentiated spermatogonia that overlap DMRs is presented as “observed” genes in light blue. A percentage of a 10,000 randomly generated size-matched transcript cohorts that overlap DMRs is presented as “random” genes in dark blue. Error bars represent a standard deviation of the mean. *** indicates a p value of < 0,0001. (c) Expression scatterplots showing relative average expression between WT and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia of ENSEMBLE annotated genes (right) and transcripts identified by de novo transcriptome assembly (left) as presented in panel a. (d) Percentage of significantly upregulated ENSEMBLE annotated genes and upregulated transcripts identified by de novo transcriptome assembly in Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia that overlap DMRs as presented in panel b. (e) Representative examples of a DMR overlapping a transcription start site of IAP element in a case of class I, II and III gene expression upregulation. The percentage of DMR methylation in WT, Miwi2-/- and Dnmt3l-/- juvenile (P14) undifferentiated spermatogonia is indicated. Significance in b and d was assessed using two-sided t-test.
Discussion
The progressive loss of spermatogonia in Miwi2-deficient mice is a germ cell autonomous defect27,32. We demonstrate that, while MIWI2 is expressed in a subset of PLZF-positive undifferentiated spermatogonia in the adult testis, its function is not required for homeostatic or regenerative spermatogenesis. Thus, the spermatogonial dysfunction in Miwi2-/- mice must be a consequence of defective reprogramming. Likewise, we demonstrate that DNMT3L is not detected in germ cells of the adult testis and thus defective reprogramming must also underlie the loss of germ cells in Dnmt3l-/- mice. Our DNMT3L expression data are in contrast to Liao et al., who found that DNMT3L is expressed in postnatal testis38. Given that we do not find expression of the protein and could only detect very low mRNA levels in undifferentiated spermatogonia, we think the discrepancy likely arises from the antibody specificity in the other study. We found that amongst the ERVs IAPs were specifically deregulated with the LINE1 elements remaining silent in Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes. While both IAP and LINE1 elements are H3K9-Me2 by G9a (GLP) in spermatogonia45,52, this repression is only sufficient to silence LINE1s52. This likely reflects that the LTR of IAP contains a powerful promoter that can overcome G9a-mediated H3K9-Me2 and that CpG DNA methylation is the principal mechanism by which IAPs are silenced in spermatogonia52. Interestingly, DNA damage is not detected in Miwi2-/- or Dnmt3l-/- spermatogonia, which is in a stark contrast to abundant DNA damage observed in meiotic cells of the same genotype that manifest developmental arrest and apoptosis22,31,32. However, when LINE1 elements become de-repressed in the absence of CpG DNA methylation, G9a-mediated H3K9me2 and a functional piRNA pathway, the ensuing DNA damage rapidly annihilates the spermatogonial cells with the rapid loss of spermatogenesis52. Thus, the absence of DNA damage in Miwi2-/- and Dnmt3l-/- spermatogonia indicates that another mechanism other than transposon-induced DNA damage must underpin the spermatogonial dysfunction. Instead we find that the de-repressed IAP elements deregulate the Miwi2-/- and Dnmt3l-/- spermatogonial transcriptomes through inappropriately promoting gene expression and novel transcripts at low levels. Similarly, LINE1 and ERV elements were recently shown to deregulate gene expression at the zygotene stage in reprogramming mutants, although this deregulation does not impede developmental progression54. ERVs have been shown to act as promoters or enhancers of gene expression in other contexts as well55–61. For example, MuERV-L was identified as one of the first genes expressed during zygotic gene activation in late 1-cell-stage embryos56. Moreover, ERV LTRs have been co-opted to activate 2-cell-stage57,58 or 8-cell-stage embryo associated genes59. Likewise, ERVs have been shown to act as species-specific enhancers in the placenta60. In the examples presented above the genome is hypomethylated facilitating the co-opting of ERVs for the regulation of gene expression62–65. That said repressive mechanisms are in place to restrict ERV expression during pre-implantation development. The lysine-specific demethylase LSD1 (KDM1A) and TRIM28a (KAP1)-SETDB1 complex silence MuERV-L and MusD, IAP ERVs, respectively, during early embryonic development66–68. Lsd1-/- mice arrest at gastrulation with MuERV-L LTRs acting as promoters to drive expression of genes that are normally restricted to zygotic genome activation66. The loss of Trim28a or Setdb1 also results in early developmental arrest with deregulated of IAPs deregulating neighboring gene expression69–72. Indeed, Trim28- or Setdb1-deficient mESCs manifest proliferation and differentiation defects accompanied by reduced viability67,68,73. The aberrant gene expression in Miwi2-/- and Dnmt3l-/- cells encompasses not only annotated genes but also the appearance of numerous de novo IAP-associated transcripts. The affected loci in the reprogramming mutants are typically lowly or not expressed in wild type spermatogonia. We posit that the appearance of these not normally expressed genes and transcripts could be the basis for spermatogonial dysfunction. The deregulation of gene expression is more severe in Dnmt3l-/- compared to Miwi2-/- spermatogonia, which correlates with the more severe spermatogonial phenotype observed in Dnmt3l-/- mice. The observed transcriptomic deregulation in undifferentiated spermatogonia could affect basic spermatogonial stem cell properties such as a balanced self-renewal, cell cycle quiescence or survival that would ultimately lead to germ cell depletion. Indeed, a reduction in the overall numbers of undifferentiated spermatogonia that encompass the stem and primitive transit-amplifying spermatogenic populations is observed in the reprogramming mutants, coupled with the loss of spermatogonial cell cycle quiescence. Given that loss of quiescence in other stem cells such as the hematopoietic, neural or hair follicle stem cells has been associated with depletion of the stem cell pool perturbing tissue homeostasis74–77, this may be an important contributing factor to germ cell depletion in the respective reprogramming mutants. In summary, here we show that a defective germline reprogramming results in spermatogonial dysfunction and IAP-driven deregulation of the spermatogonial transcriptome.
Methods
Mouse strains
Miwi2FL, Miwi2tdTom, Miwi2HA, Gfrα1GFP mouse alleles used were previously reported27,39,40,78 and maintained on a mixed 129-C57Bl/6 genetic background. Rosa26ERT-Cre allele was obtained from Jackson laboratory as B6;129 gt(ROSA)26Sortm1(cre/Esr1)Nat/J.
The Dnmt3lV5 allele was created using CRISPR-Cas9 gene editing technology79. Briefly, the sequence encoding a BamHI restriction site and V5-tag was inserted after the starting ATG codon of Dnmt3l to generate the Dnmt3lV5 allele that results in an N-terminal V5-DNMT3L fusion protein. An sgRNA (5’ TTCTAGCCGATTACATCAA) targeting Dnmt3l starting ATG codon was used. The presence of the Dnmt3lV5 allele in mice was screened by digesting PCR products amplified by primers (Fw 5’ CATCATCCCAGGCCCTCATA, Rv 5’ TATGGAGCAAGTGGAGGCAA) flanking Dnmt3l ATG with BamHI.
Generation of the Dnmt3l– allele was achieved by recombineering with a targeting vector containing exons 1-7. In the targeting vector exon 2 was flanked with one loxP site at the 5’ of exon 2 and frt flanked neomycin resistance (frt-neoR) cassette with a second loxP site at the 3’ of exon 2 (Supplementary Fig. 3). Linearized targeting vector was electroporated in A9 mouse embryonic stem cells (mESCs). Genomic DNA, extracted from individually mESC-derived neomycin resistant clones, was digested with BamHI and used for screening by Southern blotting80 using 3’ external probe. Digestion of wild-type genomic DNA with BamHI resulted in 8,9 kb fragment. Integration of loxP flanked exon 2 with frt-neoR cassette introduced and additional BamHI site, reducing fragment size to 6,1 kb in targeted allele. Cre-mediated excision of exon 2 together with neoR cassette resulted in the Dnmt3l– allele represented by 4,2 kb fragment in Southern blotting. mESCs, containing recombined Dnmt3l locus were injected into C57BL/6 mouse eight-cell stage embryos. Dnmt3l-targeted mice were crossed with a mouse line harboring ubiquitously expressed Deleter-Cre recombinase. Cre-mediated recombination and excision of exon 2 together with neomycin resistance cassette resulted in the Dnmt3l– allele.
No blind experiments or randomization of samples or animals were employed in this study. All mice used in this study were male; and bred and maintained in EMBL Mouse Biology Unit, Monterotondo, and subsequently in the Centre for Regenerative Medicine, Edinburgh. All procedures were done in accordance to the current Italian legislation (Art. 9, 27. Jan 1992, nu116) under license from the Italian health ministry or the UK Home Office regulations, respectively.
Whole mount immunofluorescence on seminiferous tubules
Isolated 8-week-old mouse testes were dealbulginated and digested at 32°C for 8 minutes shaking in DMEM supplemented with Pen-Strep (Life technologies), NEAA (Life technologies), sodium pyruvate (Life technologies) and sodium lactate (Sigma) with 0,5 mg/ml collagenase (Sigma). After digestion, seminiferous tubules were washed with PBS and fixed in 4% paraformaldehyde (Sigma) at 4°C for 4 hours. After fixation, a few tubules were separated for subsequent steps of immunofluorescence. Tubules were permeabilized and blocked in 0.5% Triton-X100 (Sigma), 5% normal donkey serum (Sigma), 1% BSA (Sigma) and 0.1 M glycine (Sigma) for 3 hours at room temperature. Primary antibody incubation was done overnight at room temperature in PBS with 1% BSA (Sigma). The following antibodies were used at indicated dilutions: rabbit monoclonal anti-HA 1:200 (Cell Signaling 3724), mouse monoclonal anti-V5 1:100 (Invitrogen R960-25), rabbit polyclonal anti-PLZF 1:100 (Santa Cruz Biotechnology sc-22839), mouse monoclonal anti-PLZF 1:100 (Santa Cruz Biotechnology sc-28319), goat anti-c-Kit antibody 1:250 (R&D Systems AF1356). Appropriate donkey anti-mouse AlexaFluor488, donkey anti-rabbit AlexaFluor488, donkey anti-rabbit AlexaFluor546, donkey anti-mouse AlexaFluor546 and donkey anti-goat AlexaFluor546 secondary antibodies (Life technologies) were used in a dilution 1:1000 and incubated for 1 hour at room temperature in PBS with 1% BSA (Sigma) and 0,1% Triton X-100 (Sigma). DAPI (5 μg/μl) (Life technologies) was used to stain DNA. Samples were mounted with ProLong Gold antifade reagent (Life technologies). Leica TCS SP5 and SP8 confocal microscopes were used to acquire images. After acquisition images were processed with ImageJ and Adobe Photoshop CS5 computer programs. All images corresponding to the same experiment were acquired and processed afterwards applying the same settings. Testes from 2 adult mice were used per each genotype analyzed as biological duplicates. All the data was collected in 2 independent immunofluorescence experiments. Five images were acquired per each biological replicate per genotype.
Inducible Miwi2 deletion and histological analysis
To evaluate the impact of Miwi2 deletion on spermatogenesis, 8-week-old Miwi2Tom/+; Rosa26ERT-Cre (Miwi2CTL) and Miwi2Tom/FL; Rosa26ERT-Cre (Miwi2iKO) mice were administered with 10 mg/ml tamoxifen solution injected intraperitoneally with a dose of 75 mg/kg every second day until a total of 5 injections per animal were reached. To evaluate the impact of Miwi2 deletion on testicular regeneration, 8-week-old Miwi2CTL and Miwi2iKO mice were administered with tamoxifen as described previously and then injected with 4 mg/ml solution of busulfan intraperitoneally with a singular dose of 10 mg/kg ten days after the last tamoxifen injection. At the time points described in the text and figure legends mice were sacrificed and testicular weight recorded. Testes were fixed in Bouin’s fixative (Sigma) overnight at 4°C and paraffin embedded. A half of tamoxifen or tamoxifen and busulfan injected mouse testis was sectioned taking six 8-µm-thick representative sections throughout the length and stained with hematoxylin and eosin using routine methods. A picture of whole testis cross-section was reconstructed using the mosaic function of the Leica Microdissector 7000 microscope (LMD 7000). Total number and number of fully spermatogenic tubules in cross-sections were counted. An average of fully spermatogenic tubules in a testis of an animal of a respective genotype in a group was calculated.
Spermatogonial cell isolation
Isolated adult (Fig. 2,3, Supplementary Fig. 4) or postnatal day 14 (P14) (Fig. 4-7, Supplementary Fig. 5,6) testes from mice of the respective genotypes that also carried the Miwi2tdTom transcriptional allele in heterozygosity were dealbulginated and digested with collagenase (0,5 mg/ml, Sigma) at 32°C for 10 minutes shaking in DMEM media (Life technologies) supplemented with Pen-Strep (Life technologies), NEAA (Life technologies), sodium pyruvate (Life technologies) and sodium lactate (Sigma). After digestion seminiferous tubules were washed with DMEM and single cell suspension was prepared by further digesting seminiferous tubules with 0,05% trypsin (Life technologies) at 32°C for 8-10 minutes shaking. After digestion trypsin was neutralized by adding FCS (PAN Biotech), also DNase (Sigma) was added to a final concentration of 0,05 mg/ml. Cells were spun and resuspended in PBS with 3% FCS. For sorting and/or analysis of Miwi2CTL, Miwi2iKO, Miwi2tdTom/+, Miwi2tdTom/tdTom and Dnmt3l-/-; Miwi2tdTom/+ mice testicular single cell suspension was stained with the following antibodies: c-Kit-PE-Cy7 1:800 (eBioscience 25-1171), CD45-biotin 1:400 (eBioscience 13-0451), CD51-biotin 1:100 (Biolegend 104104), streptavidin-qDot605 1:50 (eBioscience 93-4317), CD9-FITC 1:50 (eBioscience 11-009) in PBS with 3% of FCS and 0,01% sodium azide (Sigma). SYTOX Blue (Life technologies) was used as a living dye. For the analysis of Miwi2td/Tom+; Gfra1GFP/+ mice single cell suspension was prepared and stained as described above, except for the use of CD9-FITC antibody, where CD9-APC antibody was used instead (eBioscience 17-0091-82). A target population was selected by excluding cell doublets, CD45Pos and CD51Pos cells by initially choosing to analyze cells with low side scatter and negative for live cell dye SYTOX Blue. Among live CD45Neg and CD51Neg cells, a target population was identified as c-KitNeg and having a live fluorescence of tdTomato (Miwi2-tdTomPos) and being CD9Pos (Supplementary Fig. 4,5a). Cells were sorted with an 85 μm nozzle using a FACSAria II SORP (BD Biosciences), reaching 98% of purity. The data was recorded using BD FACSDIVA software and later analyzed using FlowJo software. For cell cycle analysis of Miwi2tdTom/+, Miwi2tdTom/tdTom and Dnmt3l-/-; Miwi2tdTom/+ mice testicular single cell suspension was stained with the following antibodies: c-Kit-PE-Cy7 1:800 (eBioscience 25-1171), CD45-biotin 1:400 (eBioscience 13-0451), CD51-biotin 1:100 (Biolegend 104104), streptavidin-APC 1:400 (Life technologies S21374) in PBS with 3% of FCS and 0,01% sodium azide (Sigma) after incubating with 0,01 µg/µl Hoescht 33342 (Sigma) at 32°C for 20 minutes shaking. 7AAD (7-aminoactinomycin) (Sigma) was used as a living dye. Target population was selected as described above (Supplementary Fig. 5b). Cells were analyzed with FACSAria II SORP (BD Biosciences), the data was recorded using BD FACSDIVA software and later analyzed using FlowJo software.
Immunofluorescence
Testes were freshly embedded into OCT compound (Sakura), 8-μm-sections were cut, fixed in 4% paraformaldehyde (Sigma) 10 minutes at room temperature and then permeabilized for 10 minutes at room temperature in 0.1% Triton-X100 (Sigma). Subsequently sections were blocked for 30 minutes at room temperature in 10% normal donkey serum (Sigma), 1% BSA (Sigma) and 0.1 M glycine (Sigma). Primary antibody incubation was done overnight at 4°C in the blocking buffer. The following antibodies were used at indicated dilutions: mouse monoclonal anti-PLZF antibody 1:100 (Calbiochem OP128), rabbit L1 ORF1p 1:50052, rabbit anti-IAP Gag 1:500 (B. Cullen), rabbit polyclonal anti-γH2AX antibody 1:500 (Bethyl Laboratories IHC-00059), mouse monoclonal anti-V5 1:200 (Invitrogen R960-25). Appropriate donkey anti-mouse AlexaFluor488, donkey anti-rabbit AlexaFluor546 secondary antibodies (Life technologies) were used in a dilution 1:1000. DAPI (5 μg/μl) (Life technologies) was used to stain DNA. Samples were mounted with ProLong Gold antifade reagent (Life technologies). Leica TCS SP5 confocal microscope was used to acquire images. After acquisition images were processed with ImageJ and Adobe Photoshop CS5 computer programs. All images corresponding to the same experiment were acquired and processed applying the same settings. For evaluation of L1 ORF1p, IAP Gag and γH2AX expression in Miwi2- and Dnmt3l-deficient, as well as in wild-type undifferentiated spermatogonia, testes from three adult mice were used per each genotype as biological triplicates. All the data was collected in three independent immunofluorescence experiments. On average 20 images were acquired for each biological replicate per genotype and >230 PLZF expressing cells (approx. 80-100 per biological replicate) were manually inspected for expression of L1 ORF1p, IAP Gag and γH2AX expression in each genotype group. E16.5 fetal testes were used for immunofluorescence to validate V5-DNMT3L expression.
RT-(q)PCR
Sorted spermatogonia, as described above, from mice of respective genotypes, were lyzed in 1ml of QIAzol (QIAGEN). Extracted total RNA was used for reverse transcription reaction with SuperScript II or SuperScript IV first-strand cDNA synthesis system using random primers (Invitrogen) according to manufacturer‘s recommendations. cDNA from sorted adult spermatogonia was used for qPCR, which was performed using 2x SYBR green I master (Roche) in biological and technical triplicates and run on a LightCycler 480 (Roche). An expression level of a target transcript was normalized to the expression level of GAPDH gene using 2-ΔΔCt method.
cDNA from sorted postnatal day 14 spermatogonia was used to validate expression of selected genes in Miwi2- and Dnmt3l-deficient cells.
Primers used:
| GAPDH_Fw | 5‘TGTCGTGGAGTCTACTGGTG |
| GAPDH_Rv | 5‘TTCACACCCATCACAAACAT |
| Miwi2_Ex16_Fw | 5‘AGCGGCTGATATTGCAGATT |
| Miwi2_Ex17_Rv | 5‘GCTTCAAGGCATTGTCATCA |
| Bhmt2_Ex8_Fw | 5’TGTGTGTTGCTTGTCATGTTT |
| Bhmt2_IAP_Rv | 5’GAAGGCAGAGCACATGGAGT |
| Poteg_Ex10_Fw | 5’AACTGTGCTGGCAATTCACC |
| Poteg_Ex13-14_Rv | 5’CCGATTCTCTCAAAGGGTGG |
| Ky_Ex2-3_Fw | 5’CGACAACCAAGGAAACTTGC |
| Ky_Ex5_Rv | 5’GGGCATCTTTCCCTCCAG |
Gene expression analysis
Total RNA extracted from sorted spermatogonial cells from tamoxifen injected adult Miwi2CTL and Miwi2iKO mice at the time point noted in the text and figure legends was subjected to microarray gene expression analysis with GeneChip Mouse Gene 2.0 Array (Affymetrix). Three biological replicates were generated per genotype to ensure adequate power to detect statistically significant differences between the respective genotypes. Microarray data was normalized using the Robust Multi-array Average (RMA) expression measure. The linear model was then used to fit the data with the R package, limma. limma powers differential expression analyses for RNA-sequencing and microarray studies81.
RNA sequencing
Total RNA extracted from sorted juvenile P14 spermatogonial cells were subjected to DNase treatment and rRNA removal using RiboGone (Clonethech) protocol according to manufacturer’s instructions. DNase-treated and rRNA depleted RNA was subjected to library preparation using SMARTer Stranded RNA-Seq Kit (Clonetech) according to manufacturer’s instructions. 10 ng of total RNA was used as an input for library preparation. Libraries were sequenced using Illumina HiSeq2000 platform.
RNA sequencing data analysis
Paired FASTQ files for each replicate were mapped against the mouse genome (GRCm38) using the ENSEMBL version 79 GTF annotation using HiSat2 (2.0.4) for known splice sites. Mapped reads were quantified into count data for each sample using htseq-count (0.6.1). This count table was processed and normalized using DESeq2 for differential expression analysis across conditions in R/BioConductor. For novel transcript analysis, Trinity (2.2.0) was run on the raw FASTQ paired end data using default parameters for paired end data. Detected novel transcripts (FASTA) for each sample were compared against known ENSEMBL transcripts using BLASTN (E<= 1e-10). Sequences with BLASTN hits covering more than 60% of their total length were discarded, together with sequences less than 200 nt. Each recovered novel sequence was then mapped back to the mouse genome using exonerate (2.40) and converted to GTF format. These novel transcripts were added back to a unified GTF file containing both ENSEMBL and Novel transcripts and remapped using HiSat2 (as above) and converted to counts using htseq-count for subsequent analysis.
Transposable element expression analysis in RNA sequencing data
For TE analysis, the raw FASTQ files were mapped against known repeats from RepBase rodent repeats (rodrep) using Bowtie2 in paired-end mode. The repeat family and subfamily count for each sample was summed for each replicate into a master table of repeat hits per sample. This table of repeat counts was processed for differential repeat expression as above using DESeq2 in R/BioConductor. For repeat enrichment analysis of differentially expressed genes, each gene list was tested for the presence of repeats based on ENSEMBL RepeatMasker derived using bedtools intersect for genes against repeat BED files. The total number of hits to each repeat family was compared for enrichment against 10,000 random gene lists of the same size.
Statistics used in this study
The negative binomial wald test was used for differential expression of both known and novel transcripts using DESeq2. For repeat and DMR enrichment a two-sided t-test was performed (R/Bioconductor) for repeat or DMR presence in each gene list versus 10,000 random cohorts of genes of the same size. Elsewhere a two-sided t-test was used.
Whole genome bisulfite sequencing and alaysis was performed on genomic DNA isolated from spermatogonia isolated from P14 testes as described in 40, except that in 50 adjacent CpG running window probes percentage of methylation was calculated for probes that contained at least 3 reads per CpG.
Supplementary Material
Acknowledgements
The research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) – ERC grant agreement n° GA 310206. This study was technically supported by EMBL Genomic Core facility, as well as EMBL Monterotondo’s genome engineering, flow cytometry and microscopy core facilities.
Footnotes
Data availability
Mouse gene expression microarray data that support the findings have been deposited in www.ebi.ac.uk/arrayexpress with accession code E-MTAB-6612. Gene expression data that support the findings of this study have been deposited in www.ebi.ac.uk/ena with the accession code PRJEB23111. WGBS data that support the findings of this study have been deposited in www.ebi.ac.uk/arrayexpress with the accession code E-MTAB-6202 and E-MTAB-556140. Figures 2a,d,f,g,i,j, 3a,c,e,f,h, 4d, Supplementary Figures 1c, 3c, 6a,c,e,f have associated source data. Figures 5a,e,f, 6a, Supplementary Figure 7a have associated data sets. Data generated in immunofluorescence and flow cytometry experiments are available upon reasonable request.
Code availability
Computer code used to analyse RNA sequencing and whole genome bisulfite sequencing data is available upon request from A.J.E. and R.V.B. or W.R., respectively.
Author Contributions
L.V. contributed to the design, execution and analysis of the majority of experiments. R.V.B. generated the whole genome bisulfite libraries and performed the bioinformatic analysis of genomic methylation under the supervision of W.R. L.V. and I.I. performed the in vivo characterization of spermatogonia. L.V. and C.C. designed, generated and validated the Dnmt3lV5 allele. A.J.E. performed all gene expression and transposon bioinformatic analyses. D.O’C. conceived and supervised this study. L.V. and D.O’C. wrote the final version of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Chiquoine AD. The identification, origin, and migration of the primordial germ cells in the mouse embryo. The Anatomical record. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 3.Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Foundation symposium. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. discussion 84-91. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi T, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546:416–420. doi: 10.1038/nature22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki K, et al. The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Developmental cell. 2016;39:169–185. doi: 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Oestrup O, et al. From zygote to implantation: morphological and molecular dynamics during embryo development in the pig. Reproduction in domestic animals = Zuchthygiene. 2009;44(Suppl 3):39–49. doi: 10.1111/j.1439-0531.2009.01482.x. [DOI] [PubMed] [Google Scholar]
- 7.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 8.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 9.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi H, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome research. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang WW, et al. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molaro A, et al. Two waves of de novo methylation during mouse germ cell development. Genes & development. 2014;28:1544–1549. doi: 10.1101/gad.244350.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guibert S, Forne T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome research. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 16.Lees-Murdock DJ, De Felici M, Walsh CP. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics. 2003;82:230–237. doi: 10.1016/s0888-7543(03)00105-8. [DOI] [PubMed] [Google Scholar]
- 17.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 18.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 20.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 21.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 23.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 24.Barau J, et al. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909–912. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- 25.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Fazio S, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 28.Kafri T, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes & development. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 29.Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Kato Y, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 31.Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Hata K, Kusumi M, Yokomine T, Li E, Sasaki H. Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Mol Reprod Dev. 2006;73:116–122. doi: 10.1002/mrd.20387. [DOI] [PubMed] [Google Scholar]
- 34.La Salle S, et al. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol. 2004;268:403–415. doi: 10.1016/j.ydbio.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 35.Sakai Y, Suetake I, Shinozaki F, Yamashina S, Tajima S. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr Patterns. 2004;5:231–237. doi: 10.1016/j.modgep.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 36.La Salle S, et al. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol. 2007;7:104. doi: 10.1186/1471-213X-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shovlin TC, et al. Sex-specific promoters regulate Dnmt3L expression in mouse germ cells. Hum Reprod. 2007;22:457–467. doi: 10.1093/humrep/del379. [DOI] [PubMed] [Google Scholar]
- 38.Liao HF, et al. DNMT3L promotes quiescence in postnatal spermatogonial progenitor cells. Development. 2014;141:2402–2413. doi: 10.1242/dev.105130. [DOI] [PubMed] [Google Scholar]
- 39.Carrieri C, et al. A transit-amplifying population underpins the efficient regenerative capacity of the testis. The Journal of experimental medicine. 2017 doi: 10.1084/jem.20161371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasiliauskaite L, et al. A MILI-independent piRNA biogenesis pathway empowers partial germline reprogramming. Nature structural & molecular biology. 2017;24:604–606. doi: 10.1038/nsmb.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buaas FW, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 42.Yoshinaga K, et al. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development. 1991;113:689–699. doi: 10.1242/dev.113.2.689. [DOI] [PubMed] [Google Scholar]
- 43.Bao J, et al. Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ. 2014;21:783–796. doi: 10.1038/cdd.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Giacomo M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Molecular cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Hobbs RM, et al. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell stem cell. 2012;10:284–298. doi: 10.1016/j.stem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 48.Bucci LR, Meistrich ML. Effects of busulfan on murine spermatogenesis: cytotoxicity, sterility, sperm abnormalities, and dominant lethal mutations. Mutat Res. 1987;176:259–268. doi: 10.1016/0027-5107(87)90057-1. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Developmental cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Hara K, et al. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell stem cell. 2014;14:658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O'Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inoue K, Ichiyanagi K, Fukuda K, Glinka M, Sasaki H. Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice. PLoS genetics. 2017;13:e1006926. doi: 10.1371/journal.pgen.1006926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis MP, et al. Transposon-driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Rep. 2017;18:1231–1247. doi: 10.15252/embr.201744059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biology of reproduction. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 57.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Developmental cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goke J, et al. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell stem cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Chuong EB, Rumi MA, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franke V, et al. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome research. 2017;27:1384–1394. doi: 10.1101/gr.216150.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamoto Y, et al. DNA methylation dynamics in mouse preimplantation embryos revealed by mass spectrometry. Sci Rep. 2016;6 doi: 10.1038/srep19134. 19134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oda M, Oxley D, Dean W, Reik W. Regulation of lineage specific DNA hypomethylation in mouse trophectoderm. PLoS One. 2013;8:e68846. doi: 10.1371/journal.pone.0068846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schroeder DI, et al. Early Developmental and Evolutionary Origins of Gene Body DNA Methylation Patterns in Mammalian Placentas. PLoS genetics. 2015;11:e1005442. doi: 10.1371/journal.pgen.1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macfarlan TS, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes & development. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 68.Rowe HM, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 69.Cammas F, et al. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- 70.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowe HM, et al. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome research. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karimi MM, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell stem cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes & development. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Urban N, et al. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353:292–295. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim E, et al. Rb family proteins enforce the homeostasis of quiescent hematopoietic stem cells by repressing Socs3 expression. The Journal of experimental medicine. 2017;214:1901–1912. doi: 10.1084/jem.20160719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter D, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 77.Lay K, Kume T, Fuchs E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E1506–1515. doi: 10.1073/pnas.1601569113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uesaka T, et al. Conditional ablation of GFRalpha1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a Hirschsprung's disease phenotype. Development. 2007;134:2171–2181. doi: 10.1242/dev.001388. [DOI] [PubMed] [Google Scholar]
- 79.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morgan M, et al. mRNA 3' uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017;548:347–351. doi: 10.1038/nature23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.