Abstract
The failure to develop effective therapies for paediatric glioblastoma (pGBM) and diffuse intrinsic pontine glioma (DIPG) is in part due to their intrinsic heterogeneity. We aimed to quantitatively map assess the extent to which this was present in these tumours through subclonal genomic analyses, and to determine whether distinct tumour subpopulations may interact to promote tumorigenesis by generating novel subclonal patient-derived models in vitro and in vivo. Analysis of 142 sequenced cases revealed multiple tumour subclones, spatially and temporally co-existing in a stable manner as observed by multiple sampling strategies. We isolated genotypically and phenotypically distinct subpopulations which we propose co-operate to enhance tumorigenicity and resistance to therapy. Inactivating mutations in the H4K20 histone methyltransferase KMT5B (SUV420H1), present in <1% of cells, confer an abrogated DNA repair pathway and increased invasion/migration of neighbouring cells, in vitro and in vivo, through chemokine signalling and modulation of integrins. These data indicate that even rare tumour subpopulations may exert profound effects on tumorigenesis as a whole, and may represent a novel avenue for therapeutic development. Unravelling the mechanisms of subclonal diversity and communication in pGBM/DIPG will be an important step to overcoming barriers to effective treatments.
Keywords: glioma, heterogeneity, subclones, co-operativity, evolution
Introduction
Paediatric glioblastoma (pGBM) and diffuse intrinsic pontine glioma (DIPG) are a highly heterogeneous group of high grade glial tumours with no effective treatments 1. Integrated molecular profiling 2–7 has revealed unique, specific and highly recurrent mutations in genes encoding histone H3 variants to mark robust subgroups of pGBM and DIPG with distinct age of onset, anatomical distribution, clinical outcome, histopathological and radiological features 8,9. A paradigm shift away from extrapolating from inappropriate adult GBM data, and towards a more paediatric biology-specific approach to developing novel therapies, has been a major positive consequence of the discovery of these novel mechanisms of tumorigenesis 10–12.
Despite these advances in our understanding of the unique biological drivers of these diseases 13, an additional major challenge to improve outcomes for children with these tumours is likely to overlap with morphologically similar tumours in adults - their extensive intratumoral heterogeneity 14. This has been demonstrated spatially by the application of genomic analyses of topographically distinct areas of the tumour at resection 15, through longitudinal studies of tumour progression and recurrence 16, and through single-cell RNA sequencing of bulk primary tumour specimens 17. All of these analyses suggest the presence of multiple co-existing tumour subclones which may play important roles in the proliferative and invasive capacities of the tumour, as well as cell fate decisions in response to the tumour microenvironment and selective pressure associated with therapeutic intervention. To date, the relative contributions to the tumorigenic phenotype of these subclones is unclear, as is to what extent they interact during the tumour’s evolutionary history – key factors in understanding the implications for novel treatment strategies 18.
In adult GBM, multiple subclones may also be marked by differential, mutually exclusive gene amplification events present in an individual tumour 19–21, an observation also reported in isolated specimens of DIPG 22,23. In these examples, cells harbouring distinct receptor tyrosine kinase gene amplifications were found intermingled throughout tumour specimens in a manner which suggested an environment conducive to the co-existence of multiple cellular subpopulations 19–21. Two-dimensional mapping of these subclones across specimens showed some evidence of a predilection of certain subclones for perivascular niches, invasive tumour fronts, or on the periphery of necrotic areas 19,20. In evolutionary biology terms this stable co-existence in conjunction with a degree of specialisation appears to imply co-operativity 24. This posits a selective advantage for an interactive cellular network and promotes biological diversity within a tumour population as an important driver of the malignant phenotype in these cancers.
With pGBM and DIPG harbouring considerably fewer somatic mutations than adult GBM 13, we sought to investigate the possibility of tumour heterogeneity reflecting co-operation of subclones, in what we consider to be an ideal model system for cancers sharing these histologies. Through an integrated approach of single and multiple sequencing strategies of patient samples coupled with in vitro isolation of subclonal populations, we propose that biological diversity is selected for across time and space, with genotypically and phenotypically distinct tumour compartments working together to enhance key tumorigenic features such as invasion and migration.
Results
Paediatric GBM and DIPG are comprised of multiple subclones
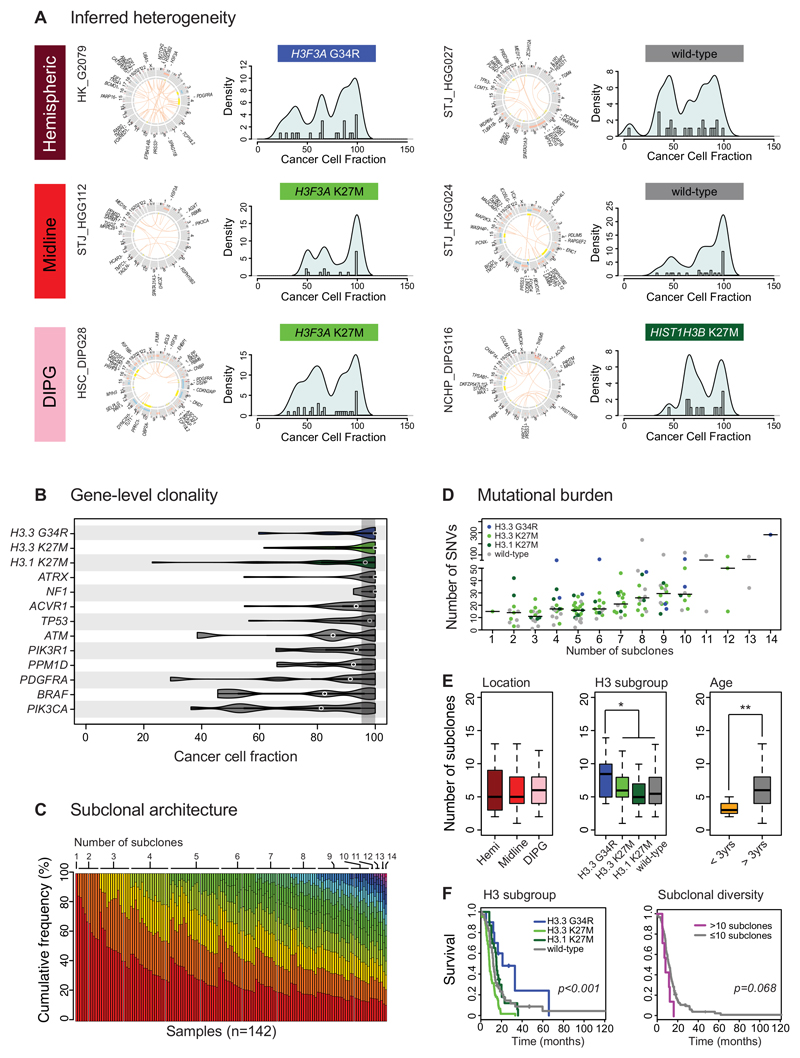
We re-analysed whole genome and exome sequencing from 142 published pGBM and DIPG specimens, for which matched germline data was available, from recently published studies 2–7. We calculated the cancer cell fractions (CCF) for all somatic single nucleotide variants (SNVs) and small insertions/deletions (InDels), taking into account the implied tumour cell percentage, overall ploidy, local copy number alterations and loss of heterozygosity 25,26 (Supplementary Table S1). In almost all cases, we observed a complex inferred subclonal architecture suggestive not of a single clonal expansion, but of multiple co-dominant subclonal populations, regardless of tumour location (n=93 DIPG, n=20 other midline, n=29 cerebral hemispheres) or histone mutation subgroup (n=10 H3.3 G34R (H3F3A), n=61 H3.3 K27M (H3F3A), n=23 H3.1 K27M (HIST1H3B, HIST1H3C), n=48 histone wild-type) (Figure 1A). Despite this variability in the fraction of any tumour harbouring a given mutation, at a gene level there were certain recurrent mutations that were found to be consistently clonal (H3F3A, HIST1H3B/C, ATRX, NF1), those predominantly clonal, but with some subclonal examples (ACVR1, TP53), and those frequently found in subclonal populations (ATM, PIK3R1, PPM1D, PDGFRA, BRAF, PIK3CA) (Figure 1B). These data provide important evidence for the likely timing of these mutations during tumour evolution. Using the EXPANDS package 27,28 we used the sequencing data to predict an absolute number of subclones present in each tumour sample, deriving a median number of 6 (range 1-14), with more than 85% of cases appearing to harbour 3-10 subclones (Figure 1C) (Supplementary Table S2). The percentage of clonal alterations ranged from 100% (n=1) to 5.2% (median=35.0%) (Supplementary Figure S1A). There was a direct relationship between the overall mutational burden (number of somatic coding SNVs) and number of subclones (Pearson r2=0.2188, p=4.36x10-9), though with several outliers. (Figure 1D). There were no differences in subclonal number between different anatomical sites (Figure 1E), despite the differing survival times of tumour location 29. pGBM with H3.3 G34R mutations had a significantly elevated number of subclones compared with other tumours (median=8.5, p=0.044, t-test), whilst there were significantly fewer in infant patients (<3 years at diagnosis; median=4, p=0.0108) (Figure 1E). Plotting the number of subclones against hazard ratios for overall survival in a similar manner to that described in a pan-cancer analysis 28, we identified cases harbouring more than 10 subclones to have the worst prognosis (relative risk = 3.3) (Supplementary Figure S1B). Although patients with H3.3 G34R mutations had a better prognosis (p=3.94x10-6, log-rank test), tumours with >10 subclones nonetheless showed a trend towards a shorter survival time, (p=0.068, log-rank test) (Figure 1F). In multivariate analysis including location, age and subgroup, only H3.3 K27M mutations (p=0.000082, Cox proportional hazards model), and a number of subclones greater than 10 (p=0.0082) were independent predictors of shorter survival (Supplementary Figure S1C).
Figure 1. Paediatric GBM and DIPG harbour a complex subclonal architecture.
(A) Inferred heterogeneity. Representative images (from n=142) of six cases of pGBM and DIPG from different anatomical locations and with different histone H3 mutation status. For each case, a CIRCOS plot is given highlighting somatic SNVs and InDels (outer ring), DNA copy number changes (red=gain, blue=loss) and loss of heterozygosity (yellow) on the inner rings, and intra-/inter-chromosomal translocations inside the circle (orange). The CCF for each somatic coding mutation is plotted as a histogram with a kernel density overplotted. In all cases, in addition to a peak of mutations present in 100% cells (clonal) there is a complex pattern of subclonal mutations (<95% CCF) forming multiple peaks at low frequencies within a given tumour. (B) Gene-level clonality. Violin plot of CCFs for a given series of gene mutations across all 142 independent cases of pGBM and DIPG (H3.3 G34R/V, n=10; H3.3 K27M, n=61; H3.1 K27M, n=23; ATRX, n=22; NF1, n=4; ACVR1, n=27; TP53, n=; ATM, n=5; PIK3R1, n=8; PPM1D, n=11; PDGFRA, n=7; BRAF, n=5; PIK3CA, n=15). The shaded area represents a CCF of 95-100% to indicate a clonal mutation. Purported drivers such as histone H3 mutations, ATRX and NF1 are almost wholly found to be clonal (though there are single outliers in some instances). Other genes such as PIK3CA, BRAF and PDGFRA are frequently found to be mutated in smaller subclonal compartments of the tumours. Kernel densities of CCFs are plotted for all samples harbouring a given mutation (number of independent cases listed on figure). (C) Subclonal architecture. The number of subclones present in 142 pGBM and DIPG is calculated from somatic mutation data using the EXPANDS package27, and ordered first by the number of subclones (coloured using a rainbow palette) and then by the proportion of the tumour defined by the major clone in each tumour. A single case was found to be clonal, with more than 85% cases harbouring between 3-10 subclones. (D) Mutational burden. Dotplot of the number of somatic coding SNVs (y axis) against the number of subclones (x axis), demonstrating a significant positive relationship (Pearson r2=0.2188, p=4.36x10-9, n=142 independent samples). The horizontal bar represents the median value. Individual tumours are coloured by their histone H3 mutation status, with outliers often seen to harbour H3.3 G34R (blue). (E) Clinical and molecular correlates of subclonal numbers. Boxplots highlighting no differences in the number of subclones on the basis of anatomical location, but an increased number in H3.3 G34R tumours (p=0.044, t-test), and a reduced number in infant cases (<3 years, p=0.0108, t-test) across all n=142 independent samples. The thick line within the box is the median, the lower and upper limits of the boxes represent the first and third quartiles, the whiskers 1.5x the interquartile range, and individual points outliers. (F) Prognostic implications. Kaplan-Meier curves demonstrating H3.3 G34R tumours have a longer overall survival than other pGBM and DIPG (p=3.94x10-6, log-rank test), however despite the association of this subgroup with an increased number of tumour subclones, an elevated subclonal diversity shows a clear trend towards shorter survival across all pGBM and DIPG (p=0.068, log-rank test). Comparisons we made including all n=142 independent samples. * p<0.05. **p<0.01.
The tumour cohort studied is heavily enriched in DIPG samples, and due to the unresectability of these lesions, were comprised of a mixture of pre-treated biopsy samples and post-treatment autopsy samples obtained post-mortem 2,3,5–7. We observed no systematic differences in subclonal architecture when comparing samples taken at these differing timepoints, regardless of diagnosis or histone mutation status (Supplementary Figure S1D). We were able to assess this directly for eight cases for which paired pre- and post-treatment sequencing data was available. By plotting the change in major subclonal tumour proportion over time, we observed changes in the proportion of individual subpopulations in response to therapy and tumour evolution; in all cases however, several significant populations remained unchanged, and both before and after treatment the tumour was inferred to harbour multiple subclones, suggesting either equivalent fitness of multiple subclones, or pressures restricting the ability of any given clone to sweep to fixation (Supplementary Figure S1E).
DIPG cells escape the pons early during tumour evolution
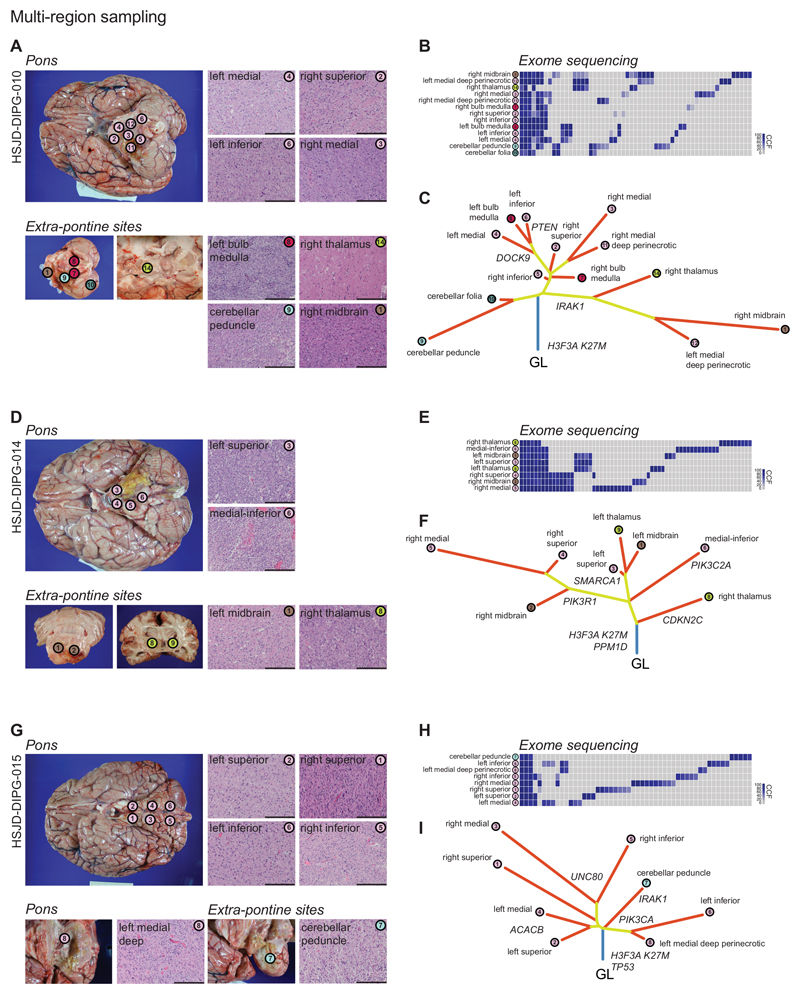
More direct evidence for the presence of multiple, genetically distinct subclones could be seen from sequencing 36 topographically distinct samples from 12 different patients (Supplementary Table S3). Comparing the CCFs from across a given tumour sample clearly demonstrated both the ubiquitous presence of presumed driver alterations (histone mutations, NF1) (Supplementary Figure S2A) but also a range of mutations private to only one portion of the tumour - of note, each distinct tumour region itself was inferred to harbour multiple subclones.
The collection of DIPG samples at autopsy represented a unique opportunity to evaluate the spatial heterogeneity of these tumours. For one case (QCTB-R091/R092), distinct low-grade and high-grade components were manually dissected and found to harbour key oncogenic mutations in one, and not the other region (e.g. PIK3CA H1047R in grade IV and not grade II) in addition to ubiquitous drivers such as ACVR1 (Supplementary Figure S2B). It has previously been shown that these diffusely infiltrating lesions may be found outside the pons and spread throughout the central nervous system at the time of death 30. Multi-sample sequencing strategies allowed us to again identify early driver events present throughout the tumour cells of an individual patient (H3F3A, HIST1H3B), as well as those occurring only at the point of escape of cells from the brainstem, such as mutations in WNK2, known to play a role in glioma cell migration and invasion 31 (Supplementary Figure S2C). Across multiple sites in multiple samples (Figure 2, Supplementary Figure S3, Supplementary Table S3), mapping SNVs and copy number aberrations (CNAs) revealed branching evolutionary trajectories. This was particularly evident in the most extensively sampled cases (Figure 2), where distinct branches highlighted the profound laterality of tumour evolution, whilst tumour cells found in midbrain, cerebellar and thalamic regions were seen to diverge early from the pontine mass. Whilst the difference in mutational profiles may be a result of invasive cells cycling more slowly, the presence of convergent or parallel evolution in key oncogenic drivers such as PIK3CA, NF1, MKI67, NOTCH1 and DMNT3A (Supplementary Figure S3) strongly suggests a predominantly early evolutionary divergence of cells which subsequently migrated outside the pons.
Figure 2. DIPGs infiltrate the brain through branching evolution and genotypic convergence.
(A) Multi-region sampling. Thirteen different tumour-harbouring regions of HSJD-DIPG-010 were sampled post-mortem, from within and outside the pons. Scale bar = 100μm. (B) Exome sequencing was carried out for all regions, with CCFs plotted as a heatmap for all variants found in at least one specimen, with anatomical location highlighted and colour-coded. (C) Phylogenetic trees were reconstructed using neighbour-joining algorithms based upon the nested subpopulation phylogenies calculated as part of EXPANDS, with clearly evident laterally-directed evolution and early escape from the pons of tumour cells found in distinct anatomical sites. (D-F) Eight different tumour-harbouring regions of HSJD-DIPG-014 subjected to the same analysis. (G-I) Eight different tumour-harbouring regions of HSJD-DIPG-015 subjected to the same analysis. Scale bar = 100μm.m
In vitro isolation of genotypically and phenotypically distinct subclones
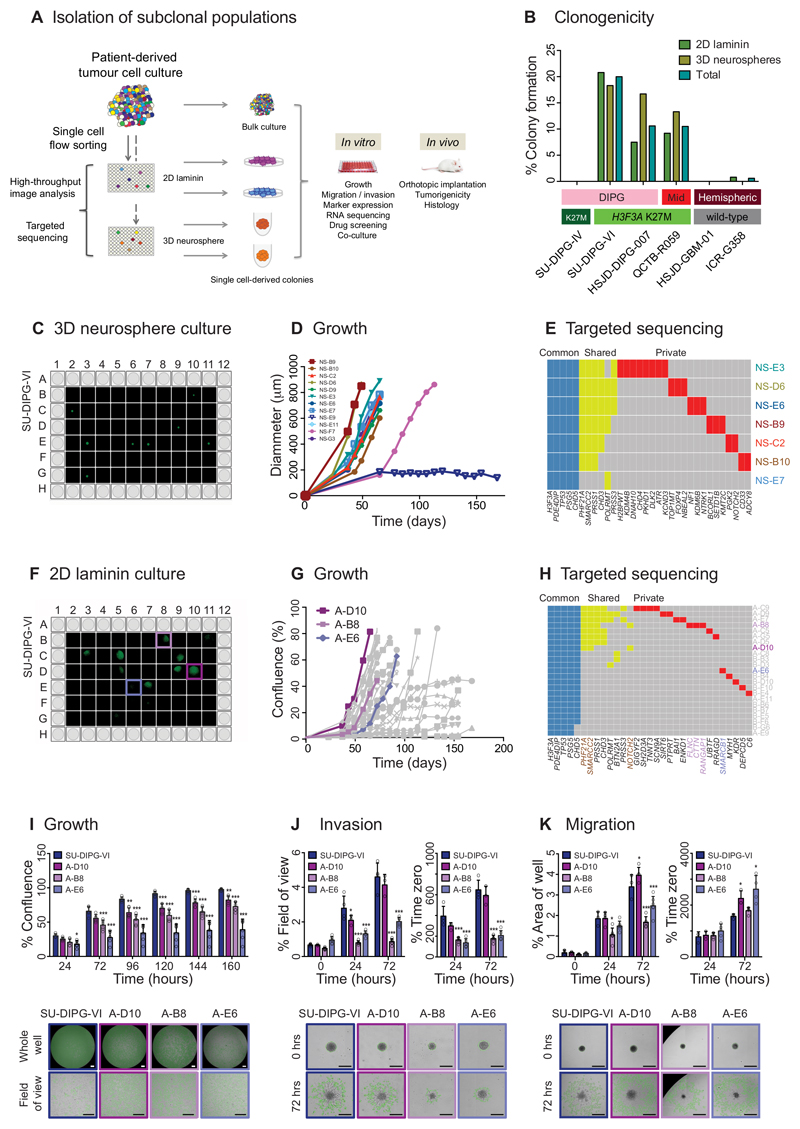
To determine whether the subclonal tumour cell populations present in pGBM and DIPG represented functionally distinct entities (rather than simply reflecting stochastic alterations occurring as a result of increasing genetic instability), we devised a methodology to isolate and expand single tumour cells under stem cell conditions, referred to as ‘stem-like’ cells, in both two- 32 (2D) and three-dimensional (3D) culture 33 for further analysis (Figure 3A). Using this approach, we identified three primary patient-derived H3.3 K27M mutant samples (two DIPG, one thalamic pGBM) (Supplementary Figure S4) from a well-characterised panel of six cultures (Supplementary Figure S5A,B) to readily form single cell-derived colonies in both 2D and 3D, at rates varying between 7.5-20.8% cells (Figure 3B). Colonies isolated from SU-DIPG-VI were identified using high-content image analysis (Figure 3C,F) and displayed highly variable growth characteristics in vitro when grown as 3D neurospheres (Figure 3D) and 2D on laminin (Figure 3G). When sequenced at high depth using a custom designed targeted panel (Supplementary Table S4), in addition to ubiquitously shared mutations (e.g. H3F3A, TP53), around half the colonies harboured a series of shared mutations not seen in the remainder (e.g. PRSS1, CHD3), whilst most were also found to contain a series of private events restricted to individual cell populations, including genes associated with cell shape and motility (FLNC, CTTN, RANGAP1) (Figure 3E,H). Individual laminin-grown colonies with fast (A-D10), intermediate (A-B8) and slow (A-E6) growth rates (Figure 3I) were seen to have significantly differing capacities for invasion into matrigel (Figure 3J) and migration on fibronectin (Figure 3K) in vitro. Thus, individual tumour samples contain a dynamic diversity of overlapping genotypic and phenotypic populations in the stem-like cell compartment.
Figure 3. Isolation of genotypically and phenotypically diverse single stem-like cell-derived subclones of paediatric GBM and DIPG.
(A) Isolation of subclonal populations. Experimental schema for disaggregation of heterogeneous mixtures of patient-derived tumour cells, flow sorting into single cells in 96-well plates, and allowing colonies to form as either 2D cultures adherent on laminin, or 3D neurospheres, all under stem cell conditions. Individual subclonal colonies are subjected to high-throughput phenotypic analysis and targeted resequencing, and further cultured for detailed in vitro and in vivo mechanistic comparison with heterogeneous bulk populations. (B) Clonogenicity. Percentage of single cells which formed colonies under 2D laminin and 3D neurosphere stem cell conditions are given for six pGBM and DIPG primary patient-derived cell cultures, labelled by anatomical location and histone H3 mutation subgroup. (C) 3D neurosphere culture from single cell-derived colonies from SU-DIPG-VI assessed by Celigo S imaging cytometer. (D) Growth of single cell-derived colonies over time, assessed as diammeter of neurosphere, labelled and colour-coded. (E) Targeted sequencing contingency plot of somatic mutations identified as common to all subclones (blue), shared amongst certain subclones (yellow) and private to individuals (red). (F) 2D laminin culture from single cell-derived colonies from SU-DIPG-VI assessed by Celigo S imaging cytometer. (G) Growth of single cell-derived colonies over time, assessed as diammeter of neurosphere, with subclones taken for later analysis highlighted: A-D10 (fast, purple), A-B8 (intermediate, pink) and A-E6 (slow, violet). (H) Targeted sequencing contingency plot of somatic mutations identified as common to all subclones (blue), shared amongst certain subclones (yellow) and private to individuals (red). Gene names are coloured to highlight private mutations in selected subclones, or common to A-D10 and A-B8 (brown). (I) Growth. Time-course for growth of selected subclones re-plated and grown over 160 hours, highlighting statistically significant differences among subclones and heterogeneous bulk cell populations of SU-DIPG-VI (blue). Representative images at 72 hours are provided from the Celigo S cytometer, with tumour cells marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (J) Invasion. Time-course of invasion of cells into a matrigel matrix over 72 hours, either as percentage of the total area in the field of view covered by invading cells, or as a percentage of time zero. Representative images given at 72 hours, with extent of tumour cell invasion marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (K) Migration. Time-course of tumour cell migration onto matrigel over 72 hours, either as percentage of the total area of the well covered by migrating cells, or as a percentage of time zero. Representative images given at 72 hours, with extent of tumour cell migration marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. * p<0.05. **p<0.01. ***p<0.001. All graphs represent mean +/- standard deviation.
Rare tumour subclones can harbour pathogenic variants driving differing phenotypes
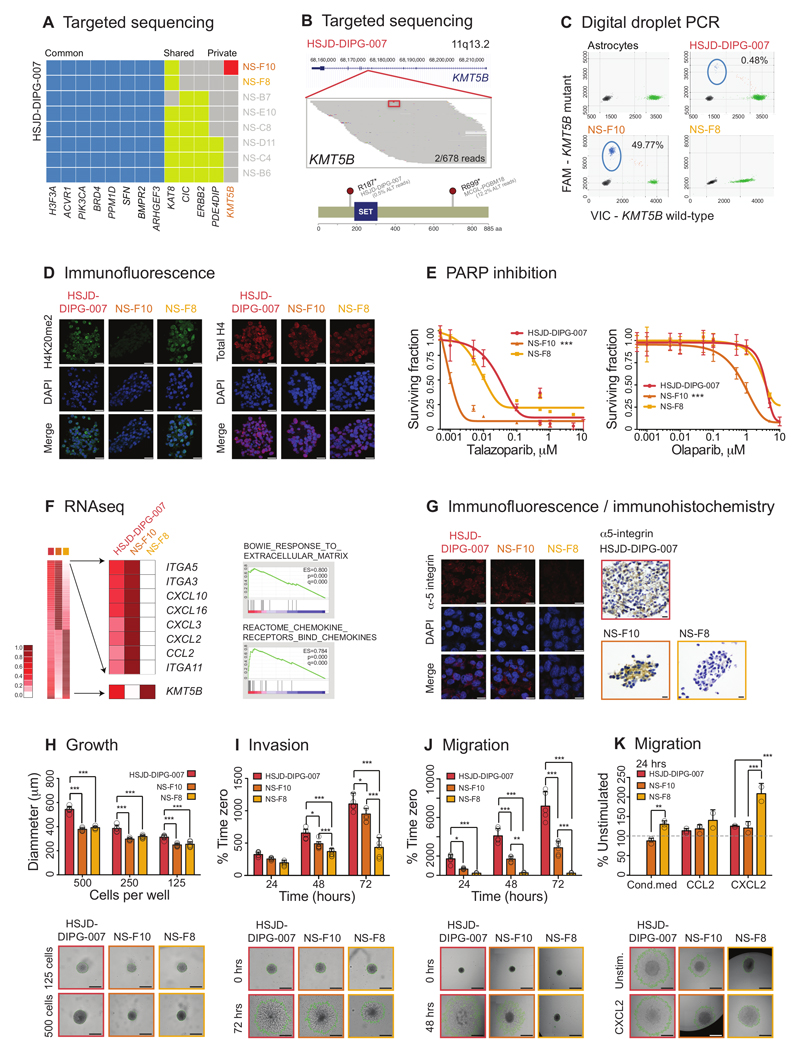
For HSJD-DIPG-007, we could utilise the ability to isolate these genetically and phenotypically distinct subclonal populations to investigate the role of individual genotypes without needing to artificially engineer the cells. We identified a single cell-derived neurosphere colony (NS-F10) to harbour a private mutation in the histone H4 methyltransferase KMT5B (SUV420H1) (Figure 4A), which was found to be present in the original bulk primary culture in only 2/678 reads (0.295%) (Figure 4B). This mutation results in the acquisition of a stop codon at amino acid position 187 (R187*), predicted to truncate the protein. Examining published sequencing datasets, we identified another case of pGBM from Schwartzentruber et al. 4, PGBM18, to harbour a subclonal R699* truncating mutation of KMT5B in 12.2% reads (Figure 4B), demonstrating this is not a unique observation. By digital droplet PCR, we confirmed that this mutation was present in (8060/16196) 49.77% of droplets from NS-F10 (assuming heterozygosity, this reflects presence in 99.64% cells), 108/22512 (0.48%) reactions of the original culture, and absent (not significantly different from normal human astrocyte control, 1/18484, 0.009%) from a ‘natural isogenic’ (confirmed by exome sequencing) counterpart subclone NS-F8 (Figure 4C). The KMT5B mutant (NS-F10) and wild-type (NS-F8) subclones did not show appreciable differences compared to each other, or the heterogeneous original bulk HSJD-DIPG-007 cells, in terms of morphology or immunophenotype (Supplementary Figure S5C). The methyltransferase encoded by the gene is predominantly involved in dimethylation, and to a lesser extent, trimethylation 34, of histone H4K20, and consequently by immunofluorescence we observed a reduction in H4K20me2 in NS-F10 compared to HSJD-DIPG-007 bulk cells and NS-F8 (Figure 4D). An unbiased drug screen of all three colonies against 80 chemotherapeutic and targeted agents (Supplementary Figure S6A-C) (Supplementary Table S5) revealed significantly enhanced sensitivity to multiple chemotypes of PARP inhibitors of the KMT5B mutant NS-F10 compared to wild-type NS-F8 and HSJD-DIPG-007 bulk cells (10-30 fold difference for olaparib, SF50 1nM vs 11nM and 31nM; 4.5 fold difference for talazoparib, SF50 1nM vs 3.833nM and 3.799nM, NS-F10 vs. NS-F8 and HSJD-DIPG-007 bulk population ANOVA p<0.001 in each case) (Figure 4E). Notably, when subclones were co-cultured, rather than a dilution effect dependent on the relative proportions, mixed cultures were similarly insensitive as the heterogeneous bulk population (Figure S6D). Thus the mutation appears to confer a loss of function to these cells, presumably due to an abrogated DNA repair process associated with loss of H4K20me2 (but not H4K20me3 or total H4, Supplementary Figure S6E) and recruitment of 53BP1 34.
Figure 4. Rare DIPG subclones with pathogenic somatic variants driving the cellular phenotype.
(A) Targeted sequencing. Contingency plot of common (blue), shared (yellow) and private (red) somatic mutations in single cell-derived neurospheres from primary patient-derived cell culture HSJD-DIPG-007. NS-F10 is the only subclone to harbour a mutation in KMT5B. (B) Pile-up representation of sequencing reads aligning to the KMT5B locus at 11q13.2. The R187* (c.559G>A) variant is highlighted in red (boxed for clarity) and is present in 2/678 reads of original heterogeneous sample. Cartoon representation of mutations identified in HSJD-DIPG-007 (c.559G>A, R187*, present in 0.47% total reactions by ddPCR) and MCGL-PGBM184 (c.2095G>A, R699*, present in 12.2% total reads by exome sequencing). Amino acid position labelled, and SET domain coloured blue. (C) Digital droplet PCR. Plot of assay for KMT5B wild-type (x axes) and R187* mutation (y axes) for normal human astrocytes, heterogeneous bulk cells, and subclones NS-F10 and NS-F8. Mutant reads are present in 49.77% droplets from NS-F10, equating to 99.64% cells harbouring a heterozygous mutation. They are absent from astrocytes and NS-F8, though are found in 0.48% droplets from the original bulk preparation. Taken from n=3 independent experiments. (D) Immunofluorescence. Heterogeneous bulk HSJD-DIPG-007 cells and subclones were stained using an antibody directed against H4K20me2 (green), or total H4 (red), with nuclei stained with DAPI (blue). Reduced expression of H4K20me2 is observed in KMT5B mutant NS-F10 cells. Representative images taken from n=3 independent experiments. Scale bar = 50μm. (E) PARP inhibition. Effect on cell viability (surviving fraction on y axes) of treatment of heterogeneous bulk cells and subclones with increasing concentrations of two different PARP inhibitors (x axes, log10 scale). ANOVA was used to test for significance of NS-F10 versus NS-F8 and HSJD-DIPG-007 bulk culture for talazoparib and olaparib. *** all p values <0.001. Data derived from n=3 independent experiments. (F) RNAseq. Heatmap of gene expression analysis from RNA sequencing data highlighting differential expression in KMT5B mutant NS-F10 subclones compared to wild-type NS-F8. The most highly elevated genes included a range of extracellular matrix remodellers (represented in gene set enrichment analysis by the gene set “BOWIE RESPONSE TO EXTRACELLULAR MATRIX”) and numerous secreted chemokines (gene set “REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES”). KMT5B itself is also differentially expressed. All cell preparations were sequenced n=1, and statistical comparisons made by Gene Set Enrichment Analysis using the Kolmogorov-Smirnov test (p) with multiple correction testing using the False Discovery Rate (q). (G) Immunofluorescence of bulk HSJD-DIPG-007 cells and subclones stained using an antibody directed against alpha-5 integrin (red). Nuclei are stained with DAPI (blue). Immunohistochemistry of embedded bulk HSJD-DIPG-007 cells and subclones stained using an antibody directed against alpha-5 integrin, and counterstained with haematoxylin. Representative images taken from n=3 independent experiments. Scale bar = 50μm. (H) Growth. Neurosphere growth of HSJD-DIPG-007 and derived subclones seeded with different cell densities showing significantly elevated growth in the heterogeneous bulk cells, but not among subclones. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (I) Invasion. Time-course of tumour cell invasion into matrigel over 72 hours, as a percentage of time zero using the Celigo S cytometer. Representative images given at 72 hours, with extent of tumour cell invasion marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (J) Migration. Time-course of tumour cell migration onto a fibronectin matrix over 72 hours, as a percentage of time zero using the Celigo S cytometer. Representative images given at 72 hours, with extent of tumour cell migration marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (K) Migration in response to stimulation with either conditioned media from HSJD-DIPG-007 heterogeneous bulk cells, or the chemokines CCL2 and CXCL2. Values are given as a percentage of unstimulated cells at 24 hours using the Celigo S cytometer. Representative images are given, with extent of tumour cell migration marked in green. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. All comparisons carried out by ANOVA, * p<0.05. **p<0.01. ***p<0.001. All graphs represent mean +/- standard deviation.
Distinct infiltrative phenotypes of genotypically divergent DIPG subclones in vivo
RNA sequencing analysis of the subclones revealed elevated gene expression in NS-F10 cells of a range of genes associated with remodelling the extracellular matrix (Figure 4F) (Supplementary Table S6). These included the fibronectin receptors alpha-3 and alpha-5 integrin, with differential protein expression validated by immunofluorescence and immunohistochemistry (Figure 4G). Although there was a slightly enhanced growth capability of the heterogeneous HSJD-DIPG-007 bulk cells (Figure 4H), NS-F10 and NS-F8 subclones were similar, though we did observe significant differences in invasion into matrigel (Figure 4I) and migration on fibronectin (Figure 4J), even after growth had been controlled for. This absence of alpha integrin expression likely underlies the inability of NS-F8 to migrate on fibronectin; of note, NS-F8 neurospheres also harboured a significantly reduced migratory capacity compared with NS-F10 on a range of other substrates including tenascin-C, laminin and matrigel (Supplementary Figure S6F). In all instances, the mixed population bulk HSJD-DIPG-007 cells were significantly more migratory than either subclone.
The KMT5B wild-type NS-F8 cells had significantly reduced invasive and migratory capacities, which could be reversed (unlike the KMT5B mutant NS-F10 cells) upon culture with conditioned media from the HSJD-DIPG-007 bulk cells (Figure 4K), suggesting the presence of expressed factors absent from the isolated NS-F8 cultures. These cells also differentially responded to the chemokine CXCL2 in terms of a significantly enhanced migration on fibronectin (Figure 4K). This C-X-C ligand was chosen as one of the most differentially expressed genes by RNA sequencing analysis in NS-F10 (and HSJD-DIPG-007 bulk) compared to NS-F8 (Figure 4F). Thus we have a model whereby paracrine signalling between subclones underlies the co-operative interactions observed in mixed populations.
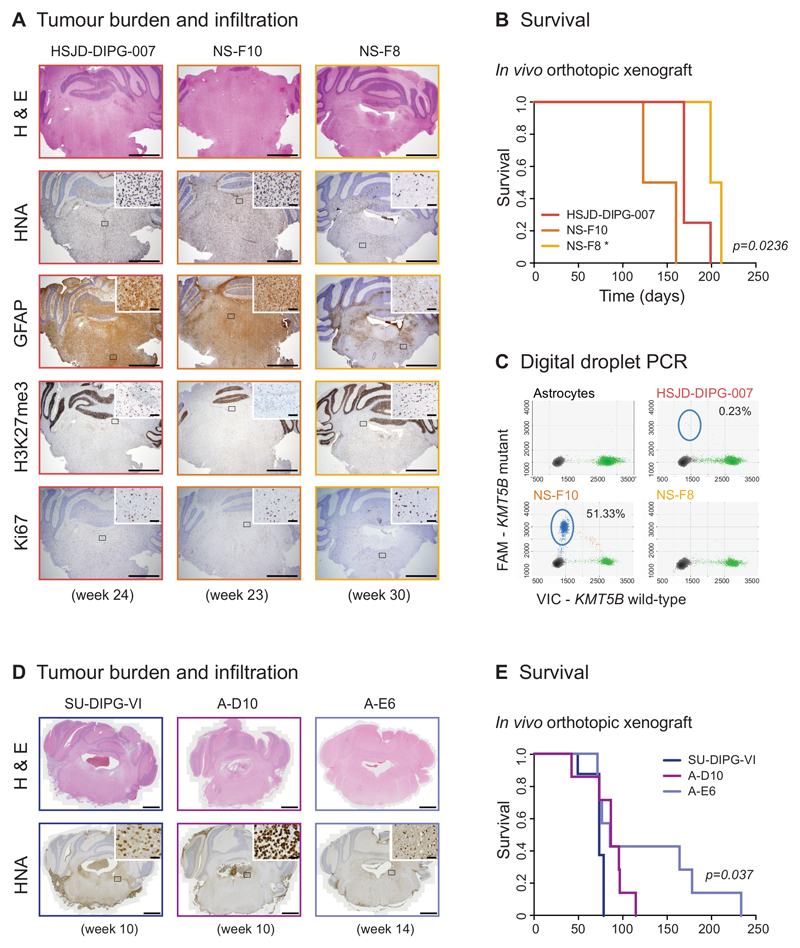
In line with in vitro data, phenotypic differences were also recapitulated in vivo, where both bulk cell populations and NS-F10 subclones formed diffusely infiltrating tumours within 23-24 weeks after orthotopic implantation in the pons of NOD-SCID mice, whereas NS-F8 lesions were significantly less infiltrative and conferred a lower tumour burden, even after 30-32 weeks, despite there being little difference in proliferative capacity and immunophenotype in the brains (Figure 5A). NS-F8 tumour-bearing mice also had a longer survival (median=205 days NS-F8 versus 141.5 days (NS-F10) and 169 days (bulk), p=0.0236, log-rank test) (Figure 5B). Tumours from heterogeneous bulk cells were confirmed by ddPCR to harbour a low subclonal frequency of KMT5B R187* mutation (0.23%) (Figure 5C), showing no significant selective pressure against the heterogeneous population. Thus even rare tumour cell subclonal populations may have different behaviours in vitro and in vivo of importance to key phenotypic features of DIPG which currently preclude effective treatments.
Figure 5. Distinct infiltrative phenotypes of genotypically divergent DIPG subclones in vivo.
(A) Tumour burden and infiltration. Heterogeneous HSJD-DIPG-007 bulk cells and NS-F10 and NS-F8 subclones were implanted directly into the pons of NOD-SCID mice and tumours allowed to form over 8 months. At weeks 23/24, bulk cells and NS-F10 formed diffusely infiltrating tumours throughout the brain, as seen by haematoxylin and eosin staining as well as immunohistochemistry with anti-human nuclei antigen (HNA), whereas NS-F8 had formed considerably less infiltrative lesions even at 30 weeks. Representative images from a total of n=4 mice per group. Scale bar = 1000μm (inset scale bar = 50μm). (B) Survival. Tumour-bearing animals implanted with NS-F8 subclones had significantly longer survival than heterogeneous HSJD-DIPG-007 bulk cells and NS-F10 (p=0.0236, log-rank test, n=4 mice per group). * p<0.05. (C) Digital droplet PCR. Plot of assay for KMT5B wild-type (x axes) and R187* mutation (y axes) for normal human astrocytes and tumours from mice implanted with heterogeneous bulk cells, and subclones NS-F10 and NS-F8. Mutant reads are present in 51.33% droplets from NS-F10 and 0.23% droplets from the original bulk preparation. Taken from n=3 independent experiments. (D) Tumour burden and infiltration. Heterogeneous SU-DIPG-VI bulk cells and A-D10 and A-E6 subclones were implanted directly into the pons of nude mice and tumours allowed to form over 8 months. At week 10, bulk cells and A-D10 formed highly cellular, infiltrating tumours, as seen by haematoxylin and eosin staining as well as immunohistochemistry with anti-HNA, whereas A-E6 had formed considerably less cellular lesions even at 14 weeks. Representative images from a total of n=8 mice per group. Scale bar = 1000μm (inset scale bar = 50μm). (E) Survival. Tumour-bearing animals implanted with A-E6 subclones had significantly longer survival than heterogeneous SU-DIPG-VI bulk cells and A-D10 (p=0.037, log-rank test, n=8 mice per group).
Notably, we observed similar results in a second model. A slower-growing subclone of SU-DIPG-VI in vitro, A-E6, formed a less cellular tumour (Figure 5D) and had an extended survival when grown orthotopically in vivo of more than 118 days longer than a rapidly proliferating, highly invasive subclone (A-D10) and 154 days longer than the unselected bulk culture (p=0.037, log-rank test) (Figure 5E).
DIPG subclones co-operate to enhance tumorigenic phenotypes
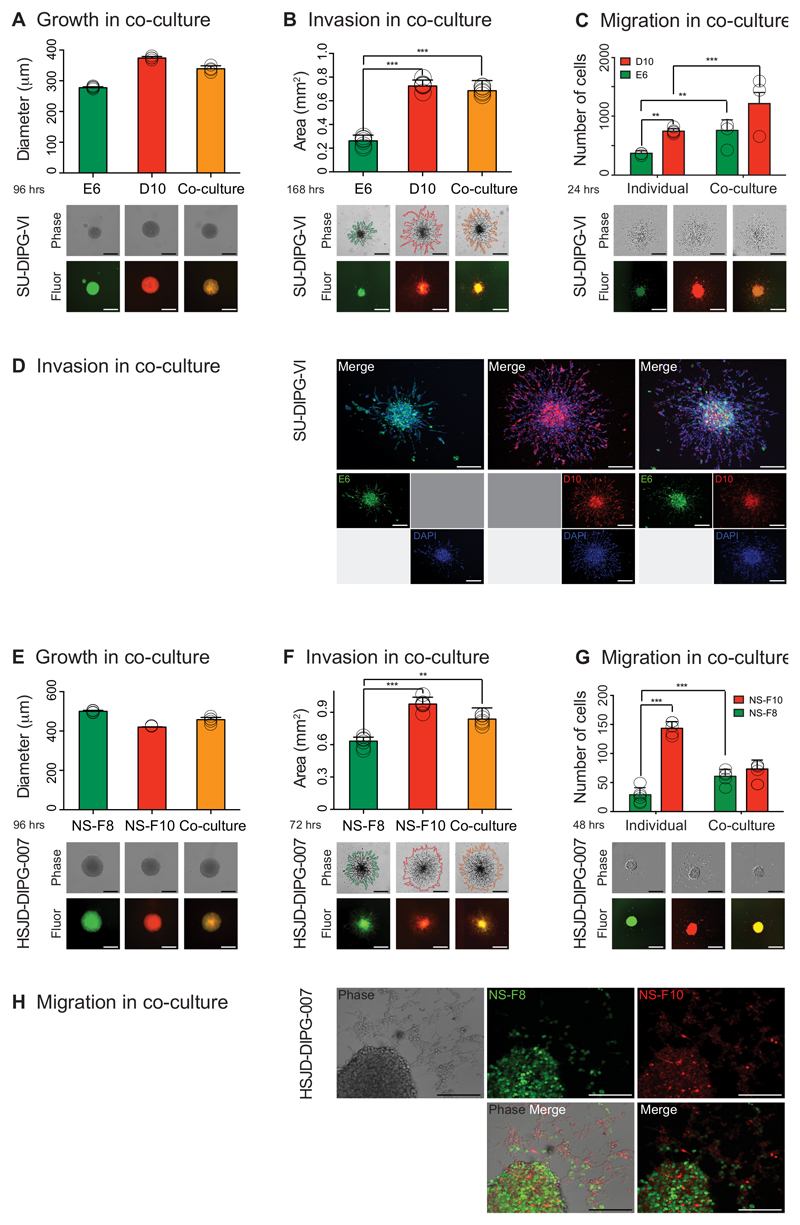
To explore the nature of these subclonal interactions, we differentially labelled and co-cultured genotypically and phenotypically distinct subclones from two DIPG samples – A-E6 and A-D10 from SU-DIPG-VI (Figure 6A-D), and NS-F8 and NS-F10 from HSJD-DIPG-007 (Figure 6E-H). When cultured in equal proportions having been re-plated as single neurospheres, although there was little difference in observed growth rates (Figure 6A,6 E), there was a marked enhancement of invasion and migration conferred on the poorly motile subclones by co-culture with their more invasive/migratory counterpart (Figure 6B, 6C, 6F, 6G). In both models, cell labelling allowed us to demonstrate that this was not a simple dilution effect of the mixture, but that the specific subclones otherwise lacking a pronounced ability to invade into matrigel (Figure 6D) or migrate on fibronectin (Figure 6H) were seen to have significantly enhanced phenotypes, clearly co-localised and moving in concert alongside their natural isogenic pairs (Supplementary Videos S1, S2). In vivo, co-cultured NS-F8/NS-F10 were found to retain their mixed proportions and infiltrate more extensively throughout the CNS than NS-F8 alone (Supplementary Figure S6G), conferring shorter survival on mice harbouring these orthotopic tumours (p=0.045, log-rank test) (Supplementary Figure S6H). Thus we conclude that there exists an actively maintained co-operative network of subclones within DIPGs that depend on strongly positive interactions to elicit the highly aggressive clinical phenotypes seen in children with this incurable disease.
Figure 6. DIPG subclones co-operate to enhance tumorigenic phenotypes.
Individual subclones of SU-DIPG-VI (A-D) and HSJD-DIPG-007 (E-H) were differentially labelled and cultured either as pure populations or mixed in equal ratios. (A) Growth of co-cultured (yellow) and mono-cultured E6 (green) and D10 (red) cells plated as single neurospheres after 96 hours, measured as diammeter of the sphere, with representative images provided from the Celigo S cytometer under phase contrast and fluorescence. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (B) Invasion of co-cultured (yellow) and mono-cultured E6 (green) and D10 (red) into matrigel over 72 hours, with area assessed by ImageJ software from representative images provided from the Celigo S cytometer under phase contrast and fluorescence. Co-cultures and D10 have significantly enhanced invasive capabilities compared to E6. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (C) Migration of mono- and co-cultured E6 (green) and D10 (red) on matrigel, assessed by the number of differentially labelled distant cells at 24 hours, with representative images provided from the IncuCyte Zoom live-cell analysis system under phase contrast and fluorescence. Cells from individual subclones have enhanced migratory properties when cultured together compared to alone. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (D) Confocal microscopy analysis of invasion of mono- and co-cultured E6 (green) and D10 (red) into matrigel after 4 days, with nuclei stained with DAPI. Poorly motile E6 cells are found to invade further and in greater numbers alongside D10 cells than when cultured alone. Representative images taken from n=3 independent experiments. Scale bar = 200μm. (E) Growth of co-cultured (yellow) and mono-cultured NS-F8 (green) and NS-F10 (red) cells plated as single neurospheres after 96 hours, measured as diammeter of the sphere, with representative images provided from the Celigo S cytometer under phase contrast and fluorescence. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (F) Invasion of co-cultured (yellow) and mono-cultured NS-F8 (green) and NS-F10 (red) into matrigel over 72 hours, with area assessed by ImageJ software from representative images provided from the Celigo S cytometer under phase contrast and fluorescence. Co-cultures and NS-F10 have significantly enhanced invasive capabilities compared to NS-F8. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (G) Migration of mono- and co-cultured NS-F8 (green) and NS-F10 (red) on fibronectin, assessed by the number of differentially labelled distant cells at 48 hours, with representative images provided from the IncuCyte Zoom live-cell analysis system under phase contrast and fluorescence. Cells from NS-F8 have enhanced migratory properties when cultured with NS-F10 compared to alone. Data derived and representative images taken from n=3 independent experiments. Scale bar = 500μm. (H) Confocal microscopy analysis of migration of mono- and co-cultured NS-F8 (green) and NS-F10 (red) on fibronectin after 3 days, with nuclei stained with DAPI. Poorly motile NS-F8 cells are found to migrate further and in greater numbers alongside NS-F10 cells than when cultured alone. Representative images taken from n=3 independent experiments. Scale bar = 200μm. All comparisons carried out by ANOVA, * p<0.05. **p<0.01. ***p<0.001. All graphs represent mean +/- standard deviation.
Discussion
Widespread intratumoral heterogeneity in human cancer has become a prevalent theme in high-throughput sequence analysis of tumour specimens, with critically important implications for the success of therapeutic targeting 35. Less attention has been given to the functional implications of this subclonal diversity and the interactions between distinct tumour subpopulations. Here we utilise pGBM and DIPG as cancer types with a relatively low mutational burden, yet a high degree of heterogeneity, to isolate these genotypically and phenotypically different compartments, and provide evidence that subclonal diversity is selected for due to co-operative interactions which promote tumorigenesis.
Single cell-derived colonies were established and expanded under stem cell culture conditions, though without marker pre-selection, in contrast to a recent approach 36. Phenotypic differences were observed in terms of morphology, growth, migration and invasion which could be linked directly to concurrent genotypic differences in the subclones. These properties, first identified through high-content screening during initial expansion, were maintained upon re-passaging in short-term culture, indicating inherently fixed characteristics in markedly different tumour cell subpopulations. Single nucleotide variants differing among single-cell derived colonies could be found at low frequencies in the original tumour mass, and thus were reflecting not an acquired artefact of the culture conditions but instead a propensity of genotypically distinct subclones to harbour stem-like properties, further evidenced through their tumorigenic capacity in vivo. It has previously been proposed that a branched Darwinian evolution model integrated with a hierarchy of multiple cancer stem cell populations may help explain the spatial and temporal characteristics of observed intratumoral heterogeneity 37, with evidence provided in leukaemia 38 and solid tumours 39–41.
In our models, the differing phenotypes of individual subclones were substantially less prominent than heterogeneous unsorted primary cultures, with enhanced growth, invasion and migration properties of mixed populations of cells supporting the interpretation of sequencing analyses suggesting that subclonal diversity is selected for spatially 42,43 and temporally 44 in these tumours. For such stable co-existence to be maintained during tumour evolution, a degree of co-operativity is implied. In our example, we isolate ‘natural isogenic’ subclonal populations differing by a key loss-of-function mutation in an H4K20 methyltransferase, in which the more migratory mutant cells are able to confer such properties on their wild-type counterparts, seemingly at least in part through expression of key chemokines such as CXCL2. A similar concept of ‘co-operative invasion’ was first identified in melanoma 45, whereby phenotypically distinct subpopulations of cells were found to co-migrate, a phenomenon also observed in DIPG in our genotypically distinct cells. Likewise, a recent elegant paper making use of lentiviral-transduced triple negative breast cancer cell line allowed for reconstruction of an aggressive phenotype in vivo using only two co-operating subclones – those overexpressing IL-11 and VEGFD 46. Such a mechanism obviates the need for clonal selection to drive tumorigenesis, and predicts for the maintenance of intratumoral heterogeneity we observe. Notably, the proportion of cells which harbour these more enhanced phenotypes may be low, and therefore remain unidentified in bulk tumour profiling studies, though still play a critical role in tumour development and maintenance.
pGBM harbouring H3.3 G34R/V mutations were found to harbour a higher mutational burden and a greater subclonal diversity than other tumour subgroups. Although the mechanisms are not known, this likely reflects an underlying DNA repair defect associated with the inability of the mutant H3K36 to be trimethylated, disrupting its important role in mismatch repair 47,48. Despite this, they do not have the mutational burden of hypermutator cases with biallelic mismatch repair deficiency, for whom immune checkpoint inhibitors appear to offer an exciting new therapeutic option 49. It is not clear therefore that G34R/V cases would benefit from a similar strategy. Unfortunately no H3.3 G34R/V cultures were available for our study, and the majority of our functional work was focused on H3.3 K27M mutant DIPG samples, which were more amenable to single cell-derived colony formation in our assay than other tumour genotypes (though it is not clear whether this reflects imperfect culture conditions for these subgroups). It has previously been shown that these diffusely infiltrating lesions may be found outside the pons and spread throughout the central nervous system at the time of death 30. Reconstructing phylogenies through sequencing of tumour cells spread throughout the brain at autopsy indicates an early escape of migratory cells from the pons prior to the rapid proliferative expansion occurring by the time of presentation and treatment. This has important implications for locally delivered therapies, and re-opens the debate for the use of whole brain irradiation upfront in children with DIPG. The later acquisition of convergent mutations in genes controlling key signalling pathways associated with proliferation at these distant sites also underlies the challenges in preventing tumour recurrence and/or metastasis at anatomically distinct sites in the central nervous system 50.
In summary these data demonstrate that pGBM and DIPG harbour a complex admixture of genotypically and phenotypically distinct stem-like cells driving a functionally-based intratumoral heterogeneity. Understanding how the derived subclones interact and adapt to the tumour microenvironment, and to therapy, will be a key requirement in order to maximise patient benefit from existing treatment options. Future strategies aimed at disrupting these interactions may represent a novel therapeutic approach in these diseases.
Online Methods
Published sequencing data
Raw data was obtained from the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) from five published sequencing studies, and provided under data access agreement from the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project (accession number EGAS00001000192 6,7), The Hospital for Sick Children (EGAS00001000575 2) and the McGill University – DKFZ Pediatric Brain Tumour Consortium (EGAS00001000226 4 and EGAS00001000720 3). We additionally included data from our own study (EGAS00001000572 5) and from four cases collected via the ICR (South West London MREC-approved study 10/H0803/126 with full consent) included in a recent International Cancer Genome Consortium (ICGC) study (EGAS00001001139 51), all of which were additionally part of a recent genomics meta-analysis by our group 29, processed data from which are housed at https://www.pedcbioportal.com. In total, we obtained whole genome (n=70) or exome (n=72) data from 142 pGBM and DIPG patients for whom matched germline data was available, six of whom additionally had data from paired longitudinal sampling. The median age was 6.8 years at diagnosis and the median survival 11.45 months (Supplementary Table S2).
Patients and samples
All patient material studied under South West London Research Ethics Committee approval. We obtained longitudinal paired samples from two patients from the Centre Hospitalier Régional et Universitaire Hautepierre, Strasbourg, France; five DIPG patients with multiple sampling taken at autopsy from the Hospital San Joan de Deu, Barcelona, Spain; five DIPG patients with multiple sampling taken at autopsy from Stanford Medical School, Stanford, CA, USA; three cases with multiple samples from the Queensland Children’s Tumour Bank, Brisbane, Australia; one case each with multiple samples from St Georges Hospital and Kings College Hospital, London, UK (Supplementary Table S3), all of which were collected locally after informed consent. The four previously sequenced patients were obtained from the Chinese University of Hong Kong, China (n=3) and University Hospital Sousse, Tunisia (n=1). DNA was extracted from frozen tissue by homogenisation prior to following the DNeasy Blood & Tissue kit protocol (Qiagen, Crawley, UK). DNA was extracted from FFPE material from either 20μm ribbons (n=2-4 per sample) or 5μm sections cut onto slides (n=10 per sample). Slides were hydrated through an ethanol series prior to manual microdissection into a tube using a sterile fine needle. All tissue was incubated overnight with proteinase K at 56°C with a further incubation for 3 hours the following morning, prior to following the QIAamp DNA FFPE tissue kit protocol (Qiagen, Crawley, UK) using 360μl of Buffer AL, 360μl of ethanol and eluted using 25μl of 10mM Tris buffer at pH8.5 for 7 minutes. Matched normal DNA was extracted from blood samples using the DNeasy Blood & Tissue kit (Qiagen, Crawley, UK). Concentrations were measured using a Qubit fluorometer (Life Technologies, Paisley, UK), with at least 400ng sent for exome sequencing at the Tumour Profiling Unit, ICR, London, UK using the 50Mb Agilent SureSelect platform (Agilent, Santa Clara, CA, USA), and paired-end-sequenced on an Illumina HiSeq2000 (Illumina, San Diego, CA, USA) with a 100bp read length. The average median coverage was 148x for the tumour exomes and 108x for tumour genomes.
Sequence analysis
For both published and newly generated raw sequencing data, reads were aligned to the hg19 build of the human genome using bwa v0.7.5a (bio-bwa.sourceforge.net), and PCR duplicates removed with PicardTools 1.5 (pcard.sourceforge.net). Somatic single nucleotide variants were called using the Genome Analysis Tool Kit v3.3-0 based upon current Best Practices using local re-alignment around InDels, downsampling and base recalibration with variants called by the Unified Genotyper (www.broadinstitute.org/gatk/). Structural variants were called from whole genome data using Breakdancer (http://breakdancer.sourceforge.net) filtered to remove commonly multi-mapped regions to identify somatic breakpoints separated by a minimum of 10kbp involving at least one Ensembl gene. Variants were annotated using the Ensembl Variant Effect Predictor v71 (www.ensembl.org/info/docs/variation/vep) incorporating SIFT (sift.jcvi.org) and PolyPhen (genetics.bwh.harvard.edu/pph2) predictions, COSMIC v64 (www.sanger.ac.uk/genetics/CGP/cosmic/) and dbSNP build 137 (www.ncbi.nlm.nih.gov/sites/SNP) annotations. Somatic variants used for further subclonal analysis (non-synonymous and synonymous) were covered by at least 10 reads in both tumour and normal sequences. Copy number was obtained by calculating log2 ratios of tumour/normal coverage binned into exons of known genes, smoothed using circular binary segmentation (www.bioconductor.org) and processed using in-house scripts. To infer the proportion of tumour cells in each sample to carry any given mutation, we calculated the cancer cell fraction (CCF) for each somatic variant25. Briefly, we determined the somatic allele-specific copy number profiles using read depth from whole genome / exome sequencing as above analysed by ASCAT26, which also provided for an estimate of the non-neoplastic cell contamination of the sample as well as the overall ploidy of the tumour. Loss of heterozygosity (LOH) was also calculated using ASCAT based upon a minor allele frequency <0.2. Allele-specific copy number, LOH and tumour cell purity were then used to calculate the cancer cell fraction (CCF) which estimates the percentage of tumour cells carrying each mutation25, and truncated to 100% where experimental variability in sequence reads produced a value greater than this figure. Intratumoral heterogeneity and the number and frequency of sub-populations within individual tumour samples were calculated with the EXPANDS algorithm using evolutionary biology principles including the Shannon and Simpson indices, and allowing for the concept that subclones may share a subset of variants that may be nested within each other 27. This used copy number-corrected variant allele frequencies of all somatic coding mutations clustered based on their cell-frequency probability distributions, and subject to pruning, to assign individual mutations to predicted sub-populations 27. For multi-region samples from the same patient, distance matrices derived from the cancer cell fractions of non-synonymous somatic coding mutations in each sample were used to construct phylogenies based upon neighbour-joining algorithms utilising the nested subpopulation calculated as part of EXPANDS, and visualized using the ape package (v3.1-4) in R. For paired longitudinal samples taken pre- and post-treatment, we fitted a kernel density estimate for the tumour variant allele frequencies at both timepoints and identified co-segregating clusters using a heatmap visualisation of the resulting biplot 52. A customised R function identified the x and y co-ordinates of each cluster centroid which served as an estimate of the number and relative composition of major subclones present in each sample. These were plotted pre- and post-treatment with coloured lines highlighting the inferred relationship between each cluster.
Cell culture
pGBM and DIPG patient-derived cultures were established either immediately after collection (biopsy, resection or autopsy) or from live cryopreserved tissue, with authenticity verified using short tandem repeat (STR) DNA fingerprinting 5 and certified mycoplasma-free. SU-DIPG-IV and SU-DIPG-VI has been published previously 11,33. Newly established cultures were first minced with the use of a sterile scalpel followed by gentle enzymatic dissociation with LiberaseTL (Roche Life Science) for 30 min at 37°C. Red blood cells were then lysed by using the AKC lysis buffer (Life Technologies), and tumour cell passed twice through a 70-μm filter. Cells were grown under stem cell conditions, either as two-dimensional (2D) adherent cultures on laminin 32, or as three-dimensional (3D) neurospheres 33. Cortical pGBM cultures ICR-G358 and HSJD-GBM-01 were cultured in a serum-free medium composed of the neural stem cell culture medium RHB-A (StemCells, Inc. Cambridge, UK) supplemented with human bFGF (20ng/mL), human-EGF (20ng/mL), human PDGF-AB (20ng/ml) (Miltenyi Biotec Ltd. Bisley, UK), and heparin (2ng/mL) (Stem Cell Technologies, Vancouver, BC, Canada). Thalamic H3.3 K27M pGBM QCTB-R059 and DIPGs HSJD-DIPG-007, SU-DIPG-IV and SU-DIPG-VI were cultured in a serum-free medium designated as “Tumor Stem Media (TSM)” as previously described 11, consisting of 1:1 Neurobasal(-A) (Invitrogen, Carlsbad, CA), and DMEM:F12 (Life Technologies), supplemented with HEPES, NEAA, Glutamaxx, Sodium Pyruvate (Life Technologies) and B27(-A) (Invitrogen, Carlsbad, CA), human bFGF (20ng/mL), human-EGF (20ng/mL), human PDGF-AA (10ng/mL) and PDGF-BB (10ng/mL) (Shenandoah, Biotech, Warwick, PA) and heparin (2ng/mL) (Stem Cell Technologies, Vancouver, BC, Canada).
Establishment of single cell colonies
Primary cultures were single cell flow sorted into the inner 60 wells of 96 well plates using a BD FACSAria I (SORP) instrument (BD) equipped with an automated cell deposition unit. Single cells were dropped in 100μl/well of the same media as described above, with the addition of penicillin and streptomycin (Life Technologies). Two 96-well flat bottom plates (Greiner Bio-one) were collected for 2D adherent culture and one 96 well round bottom ultra low attachment plate (Corning) was collected for 3D neurosphere culture. The outer 16 wells were filled in with 200μl/well of PBS to avoid evaporation of medium. 96 well plates were incubated at 37°C, 5% CO2, 95% humidity and cells re-fed twice weekly with 10-20μl of medium/well. Fully automated image analysis of single cell-derived colonies in 2D and 3D was carried out on a Celigo S cytometer (Nexcelom Inc.) 53. At indicated time points, 96 well plates were scanned and images acquired and growth assessed using the Confluence application for 2D adherent culture on laminin, and the Tumoursphere application for determining the diammeter of the neurospheres. Single cell-derived adherent colonies were collected when they reached approximately 80% confluency, while the neurospheres were collected at around 700-800μm diameter. On collection day, 10% of the cells were used to expand individual subclonal cultures, with the remaining 90% used for DNA extraction after overnight incubation with proteinase K and RNase A using the QIAamp DNA micro kit (Qiagen), and eluted using 25μl of 10mM Tris buffer pH8.5 for 5 minutes prior to quantitation. A minimum of 50ng DNA was used for targeted resequencing using a custom Agilent SureSelect panel of 435 genes recurrently mutated in pGBM / DIPG or including all members of the histone gene family (Supplementary Table S4).
High throughput / content image analysis
3D invasion assays were performed as previously described 53,54, with some modifications. Briefly, a total of 100μl medium was removed from ULA 96-well round-bottomed plates containing neurospheres of 250-300μm in diameter (given the different growth rate among the bulk cells and the single cell-derived colonies, cell densities were adjusted in order to obtain similar size neurospheres). 100μl of matrigel was gently added to each well (6 replicates) and plates were incubated at 37°C, 5% CO2, 95% humidity for 1hr. Once the matrigel solidified, 100μl/well of culture medium was added on top. Starting from time zero, and at intervals up to 72 hours, automated image analysis was carried out on a Celigo S imaging cytometer using the Confluence application. The degree of cell spreading in the matrigel was measured and the data plotted either as percentage of total area in the field of view covered by invading cells, or as percentage of initial size of each neurosphere at time zero (n=3). 3D migration assays were similarly performed as previously described 53,55, with some modifications. Briefly, flat-bottomed 96-well plates (Greiner Bio-one) were coated for 2hrs at room temperature with 50μl/well of fibronectin, laminin, tenascin, (Sigma-Aldrich) 10μg/ml in PBS with calcium and magnesium, or 125μg/ml matrigel (Corning) in culture medium in absence of growth factors. Once coating was completed, a total of 200μl/well of culture medium was added to each well. For stimulation assays, CCL2, CXCL2 (20ng/ml and 50ng/ml in TSM medium starved of all growth factors and B27 supplement) or medium harvested after five days culture of heterogeneous HSGD-DIPG-007 cells were used. A total of 100μl medium was removed from ULA 96-well round-bottomed plates containing neurospheres of 250-300μm in diameter, and the remaining medium including the neurosphere were transferred into the pre-coated plates. Starting from time zero, and at intervals up to 72 hours, automated image analysis was carried out on a Celigo S cytometer using the Confluence application. The degree of cell spreading on the different matrices was measured and data plotted either as percentage of total area in the well covered by migrating cells or as a percentage of the initial size of each neurosphere at time zero (n=3).
Digital droplet PCR
Digital droplet PCR was carried out on genomic DNA extracted from normal human astrocytes, heterogeneous HSJD-DIPG-007 bulk cells and subclones NS-F10 and NS-F8 using primers designed to detect KMT5B R187* (forward: GGCAATATTTCAAATCCACTGTCAGTT; reverse: GCAGGGTATACCATTTAAAGT CATTATCAATTTTTTTT) on a QX200 digital PCR platform (Bio-Rad). Reporter sequences were CAAACATTCGCAAATA (VIC, wild-type) and CAAACATTCACAAATAA (FAM, mutant). Briefly, the 20μL reactions consisted of 10 μL ddPCR™ Supermix for Probes (No dUTP, Bio-Rad), primers and probes at the same molar concentrations as used in qPCR, DNA up to 50ng, and molecular biology grade water. Each reaction was homogenised and partitioned into a theoretical maximum of around 23,000 droplets by creating an emulsion with Droplet Generation Oil for Probes (Bio-Rad). The 0.85nl droplets were then amplified using standard PCR cycling parameters and an annealing temperature of 60°C in accordance with the manufacturer’s recommendations. At endpoint, the fluorescence of each individual droplet was read on the Droplet Reader to identify presence or absence of mutant and wild-type target sequences. The QuantaSoft program (v1.4) fitted the droplet counts to the Poisson distribution in order to enumerate the DNA copies, from which the DNA concentration and mutant fraction could be calculated.
Drug screening
An in-house drug library encompassing 80 drugs either used in clinical practice or in late-stage development was used for screening. Each compound was dissolved in 100% dimethyl sulphoxide (DMSO) to give 5mM stocks and then diluted to 0.5, 0.05, 0.005 and 0.0005mM stocks in 96-well two-dimensional matrix plates. Daughter plates in 384-well format were prepared from these 96-well two-dimensional matrix racks using the Hamilton Microlab Star robotic platform. Compounds were stored under a nitrogen atmosphere using a StoragePod (Roylan Developments, Leatherhead, UK). Cells were seeded (1500 cells/well) into 384-well plates using a MultiDrop Combi Dispenser (Thermo Fisher Scientific, Leicestershire, UK) and allowed to form neurospheres as described above. Replicate cell plates were then loaded onto Microlab Star screening platform and drug plates were serially diluted in complete tumour stem cell medium before being added to the cell plates. The final drug concentrations used for each drug were 1000, 500, 100, 50, 10, 5, 1 and 0.5nM. The final DMSO concentration in all wells was 0.2% (v/v). Controls included 0.2% (v/v) DMSO (negative) and 10μM staurosporine (positive, Sigma-Aldrich). After incubation in drug-containing media for 5 days, cell viability was quantified with CellTiter-Glo (Promega) using a Victor X5 Multi-label plate reader luminescence protocol (Perkin Elmer, Waltham, MA, USA). Luminescence data from each well was normalised to the median signal from DMSO-containing wells to calculate the survival fraction. Plate-centred data from each screen were standardised by the use of a Z score statistic, where Z=0 represents no effect on viability and negative Z scores represent loss of viability. Z scores were calculated using the Median Absolute Deviation (MAD) of all effects in each cell line 56,57. Selective differential hits were validated individually using a wider range of concentrations using CellTiter-Glo as a read out of cell viability, in pure populations as well as mixed co-cultures, and surviving fractions calculated as before.
RNA sequencing
RNA was extracted by following the RNeasy Mini Kit protocol (Qiagen), quantified on a 2100 Bioanalyzer (Agilent Technologies), and sequenced on an Illumina GA-II genome analyser as 100bp paired end reads. RNA sequences were aligned to hg19 and organised into de-novo spliced alignments using bowtie2 and TopHat2 (https://ccb.jhu.edu/software/tophat). Raw read counts and fragments per kilobase per million reads mapped (FPKPM) were calculated for all known Ensembl genes in assembly v74 using bedtools (http://bedtools.readthedocs.org) and Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/).
Immunofluorescence
pGBM and DIPG cells were grown either adherent on laminin pre-coated 8 well-chamber slides (Cole Palmer) or in suspension as neurospheres in T75 cm2 tissue culture flasks. Cells on chamber slides were fixed with 4% paraformaldehyde at room temperature for 10 minutes and washed three times with phosphate buffered saline (PBS) solution. Neurospheres were collected into conical tubes, centrifuged for 10 min at 900rpm, washed once with PBS and after a further centrifugation, were fixed in 4% paraformaldehyde overnight 4°C and then embedded into agarose as previously described53. Paraffin-embedded neurospheres were sectioned using a microtome at 4μm thickness. Cells were permeabilised with 0.5% Triton X-100 solution for 10 minutes at room temperature and then blocked with appropriate serum according to the species of secondary antibody for 1 hour at room temperature. Primary antibodies directed against nestin (MAB5326 clone 10C2, Millipore, 1:400), SOX2 (3579, Cell Signalling, 1:400), GFAP (Z334, Dako, 1:50), CNPase (MAB326 clone 11-5B, Millipore, 1:200), TUJ-1 (MMS-435P, Covance, 1:2000), Olig-2 (Ab9610, Millipore, 1:200), and Musashi-1 (Ab5977, Millipore, 1:200) were added and incubated overnight at 4°C. Cells were then washed in PBS three times and incubated with Alexa Fluor488/555-conjugated secondary antibodies 1 hour at room temperature. For anti-H3K27me3, (9733, Cell Signalling, 1:100) and anti-alpha-5 integrin (ab15031, Abcam, 1:100), samples were incubated at 37°C for 20 minutes followed by a secondary antibody incubation at 37°C for 20 minutes. Nuclei were counterstained with DAPI, samples mounted with Vectashield (Vector Laboratories) and examined using a Leica DM2500 fluorescence microscope or a Zeiss LSM700 confocal microscope.
Co-culture experiments
Laminin-adherent cultures and neurospheres were dissociated and filtered through a 40μm cell strainer to remove residual clumps. Single cell suspensions were then incubated with CellTracker™ Red CMPTX (D10 and NS-F10) or CellTracker™ Green CMFDA (E6 and NS-F8 (Life Technologies) at final concentration of 5μM following the manufacturer’s instructions for suspension culture. Control unstained cells were incubated with an equivalent amount of DMSO. Once the staining protocol was completed, cells were washed once in complete medium, counted and seeded into 96 well round bottom ULA plates (Corning) at 1000 cells/well either in monoculture or in co-culture (50:50), and allowed to form a single neurosphere per well (n=6). One or two days post seeding, migration assays were performed on fibronectin- (NS-F10 and NS-F8) or matrigel- (D10 and E6) coated 96 well flat bottom plates (Essen Bioscience) and brightfield and fluorescent images images were acquired on an IncuCyte ZOOM® (Essen Bioscience). Images of a region of interest of identical size across all the replicates (n=6) were imported into ImageJ software and the number of cells migrated (at 12hrs for D10 and E6 and 48hrs for NS-F10 and NS-F8) were manually counted using the cell counter plugin and normalised to the cell ratio (100% for the monocultures and 50% for the co-cultures). Growth was assessed using the Celigo S as above, whilst invasion was measured as a measure of area covered using ImageJ upon image calibration using a 1mm graticule. Time lapse videos were also acquired using a Zeiss LSM700 confocal microscope with images acquired every 30 min.
In vivo orthotopic xenograft
All experiments were performed after review by the Animal Welfare and Ethical Review Board at Institute of Cancer Research, in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986, the United Kingdom National Cancer Research Institute guidelines for the welfare of animals in cancer research and the ARRIVE (animal research: reporting in vivo experiments) guidelines. A single cell suspension from heterogeneous bulk cells or subclones (HSJD-DIPG-007, NS-F10, NS-F8 or co-culture, in matrigel; SU-DPG-VI, A-D10 and A-E6, in media) was prepared immediately prior to implantation in four to eight NOD-SCID (HSJD-DIPG-007 and subclones) or nude (NCr-Foxn1nu) mice (SU-DIPG-VI and subclones) randomly allocated per group at P35. Animals were anesthetized with ketamine/xylazine (100 mg/kg / 5 mg/kg) and maintained under 1% isoflurane. The cranium was exposed via midline incision under aseptic conditions and 1 x 1 mm deep hole is drilled through the skull to the dura. Mice were placed in a stereotactic apparatus and 200,000 cells in 5μl were stereotactically implanted in the pontine area using a digital pump at infusion rate of 2 μL/min and 31-gauge Hamilton syringe. Coordinates used were 1.0 mm lateral to midline, 0.8 mm posterior to lambda, and –4 mm deep to cranial surface. At the completion of infusion, syringe needle was allowed to remain in place for a minimum of 2 min, then slowly manually withdrawn to minimize backflow of the injected cell suspension. Mice were followed for up to 8 months and were sacrificed upon deterioration of condition and tissue taken for further analysis. Mouse brains were collected and fixed in 10% buffered formalin solution for 48 hours prior to division into four parts and embedding in paraffin. 4μm-thick sections were cut and stained with haematoxylin and eosin (H&E). For immunohistochemistry, sodium citrate (pH 6.0) heat-mediated antigen retrieval was performed and staining was carried out using antibodies directed against human nuclear antigen (HNA) (MAB 4383, Millipore, 1:100), human GFAP (M0761 clone 6F2, Dako, 1:300), H3K27me3 (9733, Cell Signalling, 1:100),and Ki67 (M7240, DAKO, 1:100). All primary antibodies were diluted into 1% Tris buffer solution with 0.05% Tween-20, except Ki67 which was diluted into Dako antibody diluent and staining was performed using an autostainer. Anti-human GFAP was incubated for 30 min, anti-H3K27me3 and anti-HNA for 1 hour, all at room temperature. Envision™ detection system (DAKO K5007) was used for Ki-67 staining, whereas for all the others, Novocastra Novolink Polymer Detection Systems Kit (Leica Biosystem RE-7150) was used. Slides were then mounted using Leica CV Ultra mounting medium and assessed by an experienced pathologist (SP) blinded to cell identity.
Statistical analyses
Statistical analysis was carried out using R 3.3.0 (www.r-project.org) and GraphPad Prism 7. Comparisons between groups of continuous variables employed Student’s t-test or the analysis of variance (ANOVA) test. Univariate differences in survival were analysed by the Kaplan-Meier method and significance determined by the log-rank test. Multivariate analyses were carried out using the Cox proportional hazards model. All analyses were two-sided, and a p value of less than 0.05 after multiple testing correction was considered significant.
Supplementary Material
Acknowledgements
This work was supported by Cancer Research UK (grants C13468/A13982 and C13468/A23536), Abbie’s Army and the DIPG Collaborative, and the INSTINCT network funded by The Brain Tumour Charity, Great Ormond Street Children's Charity and Children with Cancer UK. We further acknowledge funding from the Brainchild Foundation (Australia), Children’s Hospital Foundation Queensland, the Xarxa de Bancs de Tumors de Catalunya (XBTC) sponsored by Pla Director d’Oncologia de Catalunya, ISCIII-FEDER (CP13/00189), McKenna Claire Foundation, and the US National Institutes of Health (grant K08NS070926). We acknowledge the support of the Queensland Children's Tumour Bank (QCTB) for provision of samples and clinical data. The QCTB is supported by the Children's Hospital Foundation (Queensland) and the Brainchild Foundation. The authors acknowledge NHS funding to the NIHR Biomedical Research Centre at The Royal Marsden and the ICR and Experimental Cancer Medicines Centre (ECMC) funding. This study makes use of data generated by the St. Jude Children’s Research Hospital – Washington University Pediatric Cancer Genome Project, Dr Cynthia Hawkins and the Hospital for Sick Children, and the McGill University-DKFZ Pediatric Brain Tumour Consortium. We would like to thank Louise Howell for confocal microcopy expertise, Maryou Lambros for technical advice, and Dr Hannes Vogel for neuropathology expertise and assistance with autopsy cases at Stanford University. We are indebted to the multi-disciplinary teams at the Royal Marsden Hospital, St George’s Hospital and Kings College Hospital for their continued assistance with prospective sample collection. Finally we thank the many children and families who contributed to this study through the donation of tumour tissue.
Footnotes
Life Sciences Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Code availability
All custom scripts for data processing are available upon request.
Data Availability
All new sequencing data are deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) under accession number EGAS00001001436.
Author contributions
M.V., A.B., A.M. and C.J. conceived the study and wrote the manuscript with contributions from all authors. M.V., A.B., V.M., K.K., S.P., M.C., K.R.T., H.P., C.J.L., A.G., T.F. and D.C. carried out experiments. M.V., A.B., A.M. and C.J. analysed data. L.V.M., E.Y.Q., W.J.I., A.S.M., H.-K.N., S.T, D.H.-B.B., N.E.-W., S.Z., S.V., H.C.M., L.R.B., A.J.M., S.A.-S., C.C., J.M., C.d.T., O.C., M.S., A.M.C. and A.M. prepared samples and provided clinical annotations. All authors approved the manuscript.
Competing Financial Interests Statement
The authors have no conflicts of interest pertaining to this manuscript.
References
- 1.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9:400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 2.Buczkowicz P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46:451–456. doi: 10.1038/ng.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fontebasso AM, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46:462–466. doi: 10.1038/ng.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 5.Taylor KR, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46:457–461. doi: 10.1038/ng.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu G, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gajjar A, et al. Pediatric Brain Tumors: Innovative Genomic Information Is Transforming the Diagnostic and Clinical Landscape. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:2986–2998. doi: 10.1200/JCO.2014.59.9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grasso CS, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21:555–559. doi: 10.1038/nm.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashizume R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14 doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prados MD, et al. Toward precision medicine in glioblastoma: the promise and the challenges. Neuro-oncology. 2015;17:1051–1063. doi: 10.1093/neuonc/nov031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Little SE, et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72:1614–1620. doi: 10.1158/0008-5472.CAN-11-4069. [DOI] [PubMed] [Google Scholar]
- 20.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20:810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Szerlip NJ, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paugh BS, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puget S, et al. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7:e30313. doi: 10.1371/journal.pone.0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314:1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nature communications. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andor N, Harness JV, Muller S, Mewes HW, Petritsch C. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. Bioinformatics. 2014;30:50–60. doi: 10.1093/bioinformatics/btt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andor N, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 2016;22:105–113. doi: 10.1038/nm.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackay A, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;32:520–537 e525. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caretti V, et al. Subventricular spread of diffuse intrinsic pontine glioma. Acta Neuropathol. 2014;128:605–607. doi: 10.1007/s00401-014-1307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moniz S, et al. Loss of WNK2 expression by promoter gene methylation occurs in adult gliomas and triggers Rac1-mediated tumour cell invasiveness. Human molecular genetics. 2013;22:84–95. doi: 10.1093/hmg/dds405. [DOI] [PubMed] [Google Scholar]
- 32.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell stem cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Monje M, et al. Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc Natl Acad Sci U S A. 2011;108:4453–4458. doi: 10.1073/pnas.1101657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen S, Schotta G, Sorensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic acids research. 2013;41:2797–2806. doi: 10.1093/nar/gkt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1258–1266. doi: 10.1158/1078-0432.CCR-14-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 39.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrissy AS, et al. Spatial heterogeneity in medulloblastoma. Nat Genet. 2017;49:780–788. doi: 10.1038/ng.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoffman LM, et al. Spatial genomic heterogeneity in diffuse intrinsic pontine and midline high-grade glioma: implications for diagnostic biopsy and targeted therapeutics. Acta Neuropathol Commun. 2016;4:1. doi: 10.1186/s40478-015-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikbakht H, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7:11185. doi: 10.1038/ncomms11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salloum R, et al. Characterizing temporal genomic heterogeneity in pediatric high-grade gliomas. Acta Neuropathol Commun. 2017;5:78. doi: 10.1186/s40478-017-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapman A, et al. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014;8:688–695. doi: 10.1016/j.celrep.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pai CC, et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nature communications. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfister SX, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell reports. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouffet E, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 50.Castel D, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130:815–827. doi: 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Cancer Genome Consortium PedBrain Tumor, P. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22:1314–1320. doi: 10.1038/nm.4204. [DOI] [PubMed] [Google Scholar]
- 52.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinci M, et al. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012;10:29. doi: 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vinci M, Box C, Eccles SA. Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp. 2015:e52686. doi: 10.3791/52686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinci M, Box C, Zimmermann M, Eccles SA. Tumor spheroid-based migration assays for evaluation of therapeutic agents. Methods Mol Biol. 2013;986:253–266. doi: 10.1007/978-1-62703-311-4_16. [DOI] [PubMed] [Google Scholar]
- 56.Brough R, et al. Functional viability profiles of breast cancer. Cancer discovery. 2011;1:260–273. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postel-Vinay S, et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene. 2013;32:5377–5387. doi: 10.1038/onc.2013.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.