Abstract
Targeted inhibitors of oncogenic Ras (rat sarcoma viral oncogene)-Raf signaling have shown great promise in the clinic, but resistance remains a major challenge: 30% of tumors with pathway mutations do not respond to targeted inhibitors, and of the 70% that do respond, all eventually develop resistance. Before cancer cells acquire resistance, they respond to initial drug treatment either by undergoing apoptosis (‘addiction’) or by surviving treatment albeit with reduced growth (‘tolerance’). As these drug-tolerant cells serve as a reservoir from which resistant cells eventually emerge, inhibiting the pathways that confer tolerance could potentially delay or even prevent recurrence. Here, we show that melanomas and other cancers acquire tolerance to Ras-Raf pathway inhibitors by activating autophagy, which is mediated by the cellular energy sensor AMP-activated protein kinase (AMPK). Blocking this AMPK-mediated autophagy sensitizes drug-tolerant melanomas to Ras-Raf pathway inhibitors. Conversely, activating AMPK signaling and autophagy enables melanomas that would otherwise be addicted to the Ras-Raf pathway to instead tolerate pathway inhibition. These findings identify a key mechanism of tolerance to Ras-Raf pathway inhibitors and suggest that blocking either AMPK or autophagy in combination with these targeted inhibitors could increase tumor regression and decrease the likelihood of eventual recurrence.
Introduction
The Ras (rat sarcoma viral oncogene)-Raf pathway is frequently activated in human cancers through mutations in Ras or its downstream effector, BRAF (v-Raf murine sarcoma viral oncogene homolog B). Given the central role that it plays in driving tumorigenesis, the Ras-Raf pathway has become a major focus for the development of targeted therapies. Although several strategies for inhibiting this pathway are being explored, the most successful strategy to date has been to develop targeted inhibitors of oncogenic forms of the BRAF protein. This has led to the development of vemurafenib and other targeted inhibitors that specifically inhibit the oncogenic forms of BRAF.1, 2, 3 Vemurafenib and other targeted drugs significantly prolong patient survival, but all tumors eventually develop resistance after a median time of 5–8 months.3, 4 Tumors that develop resistance are able to maintain MAPK phosphorylation—a downstream measure of Ras-Raf pathway signaling—even in the presence of vemurafenib.5, 6, 8, 9, 10, 11
Although all tumors eventually develop resistance and restore MAPK phosphorylation, they display significant differences in their initial responses to treatment. Vemurafenib causes some tumors to completely regress, most tumors only regress partially, if at all. Importantly, these differences in initial response cannot be explained by differences in the extent to which vemurafenib inhibits Ras-Raf pathway signaling as gauged by MAPK phosphorylation.5, 12, 13, 14
These observations in patients appear to be analogous to observations that have been made with populations of cancer cells in culture: inhibition of the Ras pathway triggers rapid apoptosis in some cancer cell lines (addiction), whereas other lines do not undergo apoptosis but rather survive pathway inhibition (tolerance).15 The phenomenon of drug tolerance can be distinguished from resistance because only the latter is associated with re-activation of Ras pathway signaling in the presence of the targeted inhibitor. Since resistant clones can only arise if some cancer cells survive the initial drug treatment, inhibiting the pathways that confer tolerance could improve patient outcomes by eliminating the reservoir of cells without which resistance would be unable to develop. However, while several mechanisms of resistance have been reported,5, 6, 7, 8, 9, 10, 11 how cells develop tolerance to Ras-Raf pathway inhibitors is not well understood.
Mutations in the Ras-Raf pathway provide a competitive advantage by enabling cancer cells to increase their glucose uptake in low nutrient conditions.16, 17 Consistent with this, vemurafenib and other inhibitors of Ras-Raf signaling reduce glucose uptake by cancer cells, which can be measured through positron emission tomography imaging with the glucose analog fluorodeoxyglucose.13, 18 It seems plausible that cancer cells might cope with this rapid reduction in glucose by activating some of the same pathways that normal cells use to adapt to the effects of nutrient starvation.
In normal cells, nutrient starvation triggers the activation of AMP-activated protein kinase (AMPK), which is a sensor of the cellular AMP:ATP ratio.19 Upon its activation, AMPK stimulates an internal scavenging program, termed autophagy, which provides essential nutrients by breaking down cellular components.20, 21 When autophagy is inhibited, cells are unable to survive even short bouts of nutrient starvation.22, 23 In this study, we examined whether autophagy might also promote tumor drug tolerance by enabling cancer cells to survive the starvation provoked by inhibition of the Ras-Raf pathway.
Results
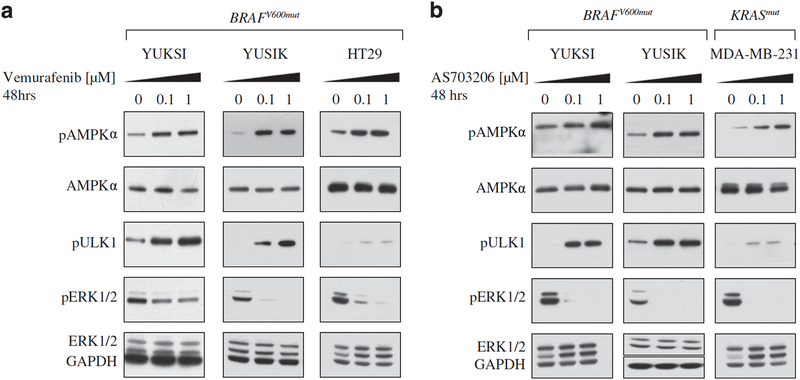
Ras-Raf pathway inhibitors activate AMPK signaling in cancer cells with Ras pathway mutations To examine whether inhibitors of the Ras pathway were provoking a starvation response, we assessed AMPK activation after 48 h of inhibitor treatment, in a panel of cancer cell lines with mutations in either Ras or Braf. Treatment with vemurafenib caused a dose-dependent increase in the phosphorylation of AMPK in two melanoma lines (YUKSI, YUSIK) and a colon cancer line (HT29) with BRAFV600 mutations (Figure 1a). Treatment with vemurafenib also increased the phosphorylation of ULK1, a downstream target of AMPK (Figure 1a).20, 24 As a control, vemurafenib treatment did not increase AMPK or ULK1 phosphorylation in cancer cells that lacked BRAF mutations (Supplementary Figure 1A). Consistent with the established role of the Ras pathway in promoting glucose uptake, we found that vemurafenib treatment also significantly reduced uptake of the fluorescent glucose analog 2-NBDG (Supplementary Figure 1B).
Figure 1. Vemurafenib and MEK inhibitors activate AMPK signaling in BRAF-mutant melanomas.
Western blots showing pAMPK, pULK1 and pERK1/2 levels in BRAF or KRAS- mutant cancer cells treated for 48 h with indicated doses of (a) vemurafenib or (b) AS703026. GAPDH is the loading control.
To determine whether these effects were unique to vemurafenib, we examined whether other inhibitors of the Ras-Raf pathway also induced AMPK signaling in cancer lines with BRAF or RAS mutations. Treatment with an MEK inhibitor (AS703026) induced the phosphorylation of both AMPK and ULK1 in YUKSI and YUSIK melanoma cells (Figure 1b). In addition, AS703026 treatment induced AMPK and ULK1 phosphorylation in the KRAS-mutant breast cancer line, MDA-MB-231 (Figure 1b). These findings indicated that Ras-Raf pathway inhibition in cancers with pathway mutations was sufficient to stimulate AMPK signaling.
Ras-Raf pathway inhibitors activate AMPK-dependent autophagy
AMPK promotes energy homeostasis by turning off anabolic processes that consume ATP and activating catabolic processes that generate ATP.25 One such catabolic process is autophagy,20, 21, 26 a ‘self-eating’ program in which cells engulf and break down their organelles and cytoplasmic proteins to generate energy. Autophagy is mediated by engulfment within double-membraned vesicles termed autophagosomes, which ultimately fuse with lysosomes resulting in degradation of their cargo.27 AMPK-mediated autophagy is an essential mechanism for restoring energy homeostasis in cells that are deprived of nutrients.
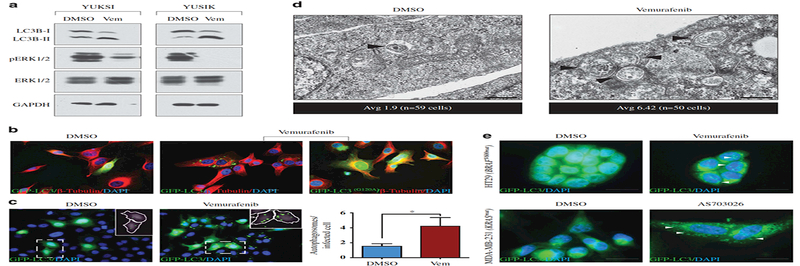
We next determined whether vemurafenib treatment induced autophagy. Using western blotting, we found that vemurafenib caused an increase in lipidated LC3B (microtubule- associated protein 1 light chain 3 beta) protein (LC3B-II), a marker of autophagy, in the BRAF- mutant YUKSI and YUSIK melanoma lines (Figure 2a). Consistent with this, direct visualization of autophagosomes through expression of a green fluorescent protein (GFP)-LC3B fusion protein also showed that vemurafenib treatment led to an increase in both autophagosome number and size; as a negative control, these changes were not observed when cells were infected with a mutant LC3BG120A-GFP that is unable to participate in autophagosome formation (Figures 2b and c; see Materials and methods for details). Electron microscopy further confirmed this finding: vemurafenib treatment led to an increase in double-membraned autophagic vesicles per cell with the average number of 6.42 (vemurafenib) vs 1.9 (DMSO) (Figure 2d). Induction of autophagy was not a unique effect of vemurafenib, but was also observed upon treatment with another BRAFV600 inhibitor (AZ628), as well as in response to the MEK inhibitor AS703026. All of these inhibitors stimulated autophagy, as gauged by an increase in GFP-LC3B labeled autophagosomes (Supplementary Figure 2A). As a negative control, vemurafenib did not inhibit MAPK phosphorylation or stimulate LC3B-II accumulation in cancer lines that did not harbor mutations in BRAF (Supplementary Figure 2B).
Figure 2. Vemurafenib and MEK inhibitors activate autophagy in BRAF-mutant melanomas.
(a) Western blot showing LC3B levels in BRAF-mutant melanoma cells treated with 1 μΜ vemurafenib or DMSO for 48 h. GAPDH is the loading control. (b) GFP-LC3B (wild type or G120A mutant) labeled YUKSI melanoma cells were treated with vemurafenib or DMSO for 48 h, and autophagosomes were visualized by fluorescence microscopy. Alexa Fluor 594-conjugated β- tubulin and DAPI were used to stain the cytoskeleton and the nuclei, respectively. Scale bar, 20 μm. (c) Fluorescence images showing GFP-LC3B labeled autophagosomes in YUKSI melanoma cells treated with vemurafenib or DMSO for 48 h. Inset shows identification of autophagosomes (green) inside cells (white outlines) by image segmentation analysis (see full Materials and methods). Data represent mean±s.e.m. (DMSO n=76 cells, Vem n=54 cells; *P<0.05 log-rank test. (d) Representative electron micrographs of DMSO- and vemurafenib- treated YUKSI cells at specified magnifications; black arrowheads mark double-membrane autophagosomes. Scale bar, 400 nm. Data below the images represent average autophagosomes per cell for respective treatments. (e) Fluorescence images showing GFP-LC3B labeled autophagosomes in HT29 colon cancer cells and MDA-MB-231 breast cancer cells treated with vemurafenib and AS703026, respectively. Scale bar, 20 μm.
Although the upregulation of autophagy can lead to an increase in autophagosome numbers, autophagosome numbers can also increase when their clearance is blocked.28 To distinguish between these possibilities, we used western blotting to assess the effects of vemurafenib treatment on p62 protein, an autophagosome marker that is often (though not always) decreased when autophagy is induced.28 Vemurafenib treatment reduced p62 levels (Supplementary Figure 2C); immunostaining for p62 in YUKSI cells showed co-localization of p62 with GFP-LC3B labeled autophagosomes upon vemurafenib treatment (Supplementary Figure 2D).
To confirm that autophagosomes were being cleared, we determined whether the GFP-LC3B autophagosomes stimulated by vemurafenib co-localized with the lysosome markers, LAMP1/LAMP2. We found that vemurafenib treatment led to a marked increase in the colocalization of GFP-LC3B and LAMP1/LAMP2, indicating that autophagosome clearance was not reduced by inhibitor treatment (Supplementary Figure 2D). Taken together, these observations indicate that the increase in autophagosome numbers observed upon treatment with vemurafenib is caused by an induction of autophagy, rather than by a block in autophagosome clearance.
To test whether induction of autophagy was specific to mutant BRAF inhibition or was a general consequence of Ras-Raf-MAPK inhibition, we inhibited MAPK signaling in other cancer lines with activating BRAF or RAS mutations. Autophagosomes accumulated in the BRAF-mutant HT29 colon cancer cell line in response to vemurafenib (Figure 2e, top panel). Autophagosomes also accumulated in the KRAS-mutant MDA-MB-231 breast cancer cell line in response to a MEK inhibitor (Figure 2e, bottom panel). However, MCF7 and T47D breast cancer cell lines, which lack Ras pathway mutations, did not activate autophagy upon pathway inhibition (Supplementary Figure 2E). Collectively, these findings indicated that cancer cells with activating BRAF or RAS mutations respond to MAPK pathway inhibitors by upregulating autophagy.
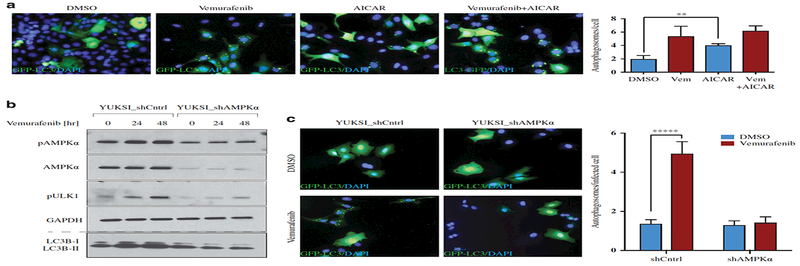
To further confirm that Ras pathway inhibition was activating autophagy via AMPK, we examined the kinetics of autophagy induction and AMPK activation in response to vemurafenib treatment. We observed LC3B-II accumulation and ULK1 phosphorylation (both of which are indicative of autophagy induction) after 24 h of vemurafenib treatment, which was concurrent with AMPK activation (Supplementary Figure 2F). Next, we treated YUKSI cells with an AMP mimetic (5-Aminoimidazole-4-carboxamide ribonucleotide, AICAR); AICAR induced AMPK and ULK1 phosphorylation (data not shown), and caused GFP-LC3B autophagosome accumulation (Figure 3a). Furthermore, we examined autophagy in YUKSI cells expressing a short hairpin RNA (shRNA) targeting AMPK and found that shRNA knockdown of AMPK abolished ULK1 activation and autophagosome formation in response to vemurafenib (Figures 3b and c). We also examined the ability of Ras pathway inhibitors to activate autophagy through AMPK in KRAS- mutant lung cancer. Using a cell line (T1) derived from the KRASmut/p53null lung adenocarcinoma mouse tumor model,29 we observed that treatment with MEK inhibitor activated AMPK and autophagy (Supplementary Figure 3A). However, in a KRASmut human NSCLC line NCI-H460 that has an inactivating mutation in LKB1, the upstream kinase of AMPK,30 Ras pathway inhibition did not activate AMPK or autophagy (Supplementary Figure 3A). Thus, Ras pathway inhibitors activate autophagy through AMPK.
Figure 3. AMPK signaling is required for the activation of vemurafenib-induced autophagy.
(a) Fluorescence images and quantification of GFP-LC3B labeled autophagosomes in YUKSI cells treated with DMSO, 1 μΜ vemurafenib, 1 mM AICAR, or both vemurafenib and AICAR. Data represent meants.e.m. across three fields of view ( · 75 cells in each condition; **P<0.01). (b) YUKSI melanoma cells expressing control or shAMPK were cultured with or without vemurafenib for 24 and 48 h. Levels of pAMPK, pULK1 and LC3B (examined on a 12% Bis-Tris gel) are shown. (c) Fluorescence images and quantification showing GFP-LC3B labeled autophagosomes in shCntrl or shAMPK expressing YUKSI cells treated with DMSO or vemurafenib. Data represent mean±s.e.m. (shCntrl/DMSO n=55 cells, shCntrl/Vem n=41 cells, shAMPK/DMSO n=36 cells, shAMPK/Vem n=61 cells; *****P<0.00001 log-rank test).
Autophagy is essential for cancer cells to tolerate Ras-Raf pathway inhibition
Although treatment with vemurafenib suppressed proliferation of the YUKSI and YUSIK melanoma lines, it did not appear to induce death in either of these lines, suggesting that they tolerated pathway inhibition. For instance, treatment with vemurafenib did not lead to an increase in either the sub-G0/G1 population or cleavage of caspase-3 (Supplementary Figures 3B and C). Since the findings above showed that vemurafenib treatment caused these drug- tolerant lines to induce AMPK signaling and autophagy, we next examined whether this induction was contributing to their ability to survive and tolerate BRAF inhibition.
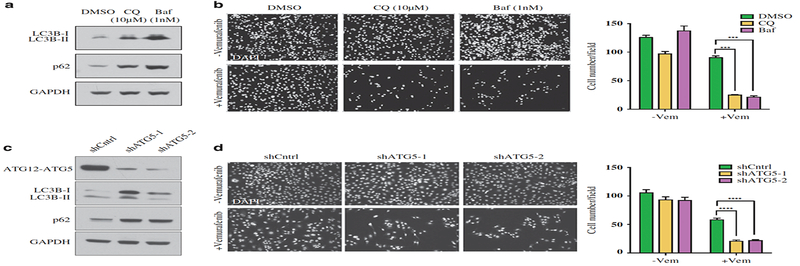
To address this, we treated YUKSI cells with chloroquine (CQ)28 or bafilomycin (Baf), which block autophagy by inhibiting autophagosome clearance (Figure 4a). Both CQ and Baf strongly sensitized YUKSI cells to vemurafenib (Figure 4b; Supplementary Figure 4A). We next used shRNA-mediated knockdown of a gene required for autophagy as an alternate strategy for autophagy inhibition. Two different shRNAs that targeted ATG5 blocked autophagic flux, and significantly abrogated vemurafenib-induced LC3B lipidation (Figure 4c; Supplementary Figure 4B). Compared with shCntrl cells, ATG5 knockdown cells were significantly more sensitive to vemurafenib (Figure 4d; Supplementary Figure 4C).
Figure 4. Autophagy inhibition sensitizes BRAF-mutant melanoma cells to vemurafenib in vitro.
(a) Western blots showing LC3B-II and p62 accumulation in YUKSI melanoma cells treated with autophagy blockers (CQ or Baf) for 72 h. (b) YUKSI melanoma cells were treated with DMSO or 1 μM vemurafenib, alone or in combination with autophagy blockers (CQ or Baf) for 72 h; Left—Cells remaining attached to the plate were fixed and DAPI stained to visualize the surviving population. Right—Data represent mean±s.e.m. across four different fields of view. (c) Western blots showing LC3B and p62 levels in YUKSI melanoma cells expressing a control (shCntrl) or two different shATG5 hairpins (shATG5–1 and shATG5–2). (d) shCntrl or shATG5 YUKSI melanoma cells were treated with DMSO or 1 μM vemurafenib, alone or in combination for 72 h; Left—Cells remaining attached to the plate were fixed and DAPI stained to visualize the surviving population. Right—Data represent mean±s.e.m. across eight different fields of view. ***P<0.001; ****P<0.0001.
To rule out the possibility that this effect was specific to ATG5, we performed similar studies with cells that express shRNAs targeting a different autophagy-essential gene, ATG7. Two different shRNAs that targeted ATG7 blocked clearance of autophagic flux, and also strongly sensitized YUKSI cells to vemurafenib treatment (Supplementary Figures 4D-F).
Blocking autophagy similarly sensitized the YUSIK BRAFV600 mutant melanoma line to vemurafenib, causing significantly reduced cell survival (Supplementary Figure 4G). Therefore, chemical or genetic ablation of autophagy sensitizes cells to vemurafenib, indicating that autophagy promotes tolerance to BRAF inhibition.
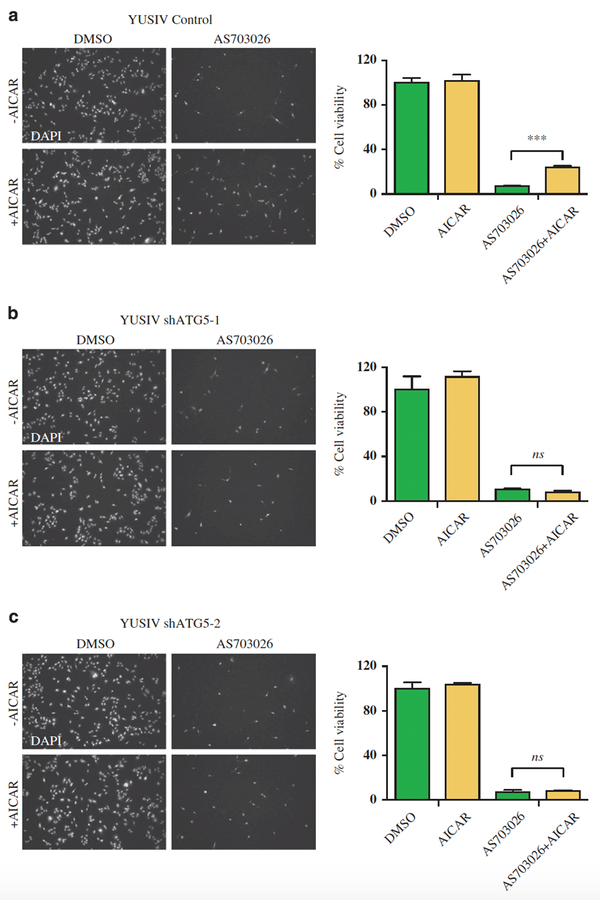
We next examined whether experimentally stimulating AMPK and autophagy would allow cells that are addicted to Ras-MAPK signaling to tolerate pathway inhibition. The YUSIV melanoma line is addicted to Ras pathway signaling and undergoes massive cell death at very low doses of pathway inhibitors (Supplementary Figure 5A). To induce AMPK signaling, we treated the YUSIV cells with the AMP mimetic AICAR, which, as expected, activated both AMPK and autophagy (Supplementary Figure 5B). Although AICAR treatment alone did not affect cell growth or viability, AICAR significantly increased the fraction of cells that survived upon treatment with the MEK inhibitor AS703026 (24% vs 7%; Figure 5a). Blocking autophagy with shRNAs against ATG5 prevented AICAR-induced tolerance to Ras pathway inhibition (Figures 5b and c); blocking autophagy with CQ had similar effects (Supplementary Figure 5C). AICAR also promoted tolerance to vemurafenib in a BRAFV600 mutant melanoma line (YUGEN8) previously reported to be addicted to the MAPK pathway (Supplementary Figure 5D).31 We observed that while YUGEN8 cells modestly activated AMPK upon vemurafenib treatment, co-treatment with vemurafenib and AICAR led to a much stronger AMPK activation, and moreover enabled cells to tolerate BRAF inhibition (Supplementary Figure 5E). These observations indicated that activation of AMPK promotes tolerance to Ras-Raf pathway inhibitors by inducing autophagy.
Figure 5. Autophagy promotes survival upon BRAFV600/MAPK inhibition.
YUSIV BRAFwt melanoma cells treated with 10 nM AS703026 with or without 1 mM AICAR for 72 h. Left—Cells remaining attached to the plate were fixed and DAPI stained to visualize the surviving population for (a) shCntrl, (b) shATG5–1 and (c) shATG5–2. Right—Data represent mean±s.e.m. across at least four different fields of view. ***P<0.001; ns, not significant.
Blocking autophagy synergizes with vemurafenib to inhibit melanoma tumor growth
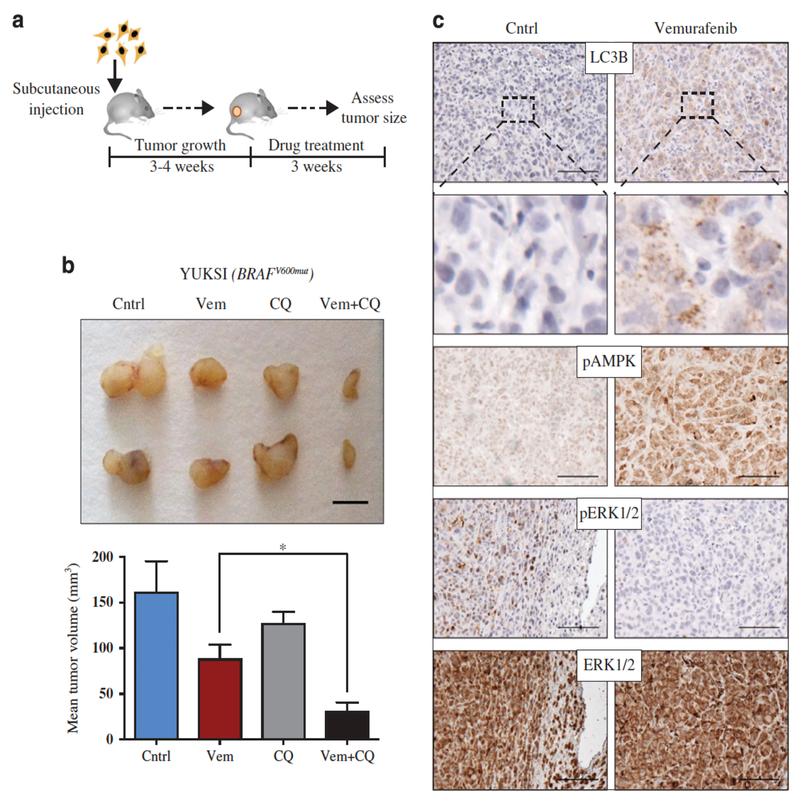
We next examined whether the effectiveness of vemurafenib could be improved by blocking autophagy in vivo. YUKSI melanoma cells were injected into mice and allowed to form palpable tumors (2–3 mm). Mice were then treated daily for 3 weeks with vemurafenib, CQ, a combination of both, or with a vehicle control (Figure 6a, schematic).
Figure 6. Autophagy inhibition synergizes with vemurafenib to prevent melanoma growth in vivo.
(a) YUKSI BRAFmut melanoma cells were injected into mice and allowed to form palpable tumors (4–5 mm) followed by indicated drug treatments (see full Materials and methods). (b) Top—Images of representative tumors from mice treated with control, vem, CQ or vem+CQ; scale bar 5 mm. Bottom—Data represent mean tumor volume±s.e.m. (n=5 mice per treatment group; *P<0.05). (c) Immunohistochemical staining showing levels of pAMPK, pERK1/2, ERK1/2 and LC3B puncta in tumors from vemurafenib- vs control-treated mice; scale bar 100 μm.
Consistent with the ability of YUKSI cells to tolerate MAPK pathway inhibition in vitro, vemurafenib reduced tumor growth but did not cause tumor regression, although ERK signaling was abrogated (Figures 6b and c). Also in agreement with the in vitro findings, vemurafenib activated AMPK and induced autophagy within tumors as evaluated by immunohistochemical staining for pAMPK and increased LC3B puncta (Figure 6c).
Although vemurafenib or CQ alone very modestly attenuated YUKSI tumor growth, combination treatment with CQ almost completely blocked tumor growth (Figure 6b). We stained tumor tissues with cleaved caspase-3 antibody and did not see an increase in apoptosis (data not shown). However, we noticed a significant increase in necrosis when autophagy was inhibited; vemurafenib-treated tumors in which autophagy had been inhibited had larger regions of necrosis, relative to vemurafenib-treated tumors in which autophagy had not been inhibited (Supplementary Figure 6A).
Blocking autophagy in YUKSI tumors by shRNA knockdown of ATG5 also significantly reduced tumor growth in response to vemurafenib, relative to tumors expressing a control shRNA (Supplementary Figure 6B). Treatment of heterogeneous tumors (created by co-injecting GFP- positive control cells with GFP-negative ATG5 knockdown cells) with vemurafenib caused selective depletion of shATG5 cells (Supplementary Figure 6C). Blocking autophagy also reduced tumor growth of another BRAF-mutant melanoma line (YUSIK) (Supplementary Figure 6D). Taken together, these observations indicate that autophagy increases the ability of BRAF- mutant melanomas to tolerate Ras pathway inhibition in vivo.
Discussion
Our findings show that cancer cells tolerate Ras pathway inhibition by activating AMPK and autophagy. Ras pathway inhibition activates autophagy in an AMPK-dependent manner, and blocking autophagy sensitizes cancers that would otherwise tolerate Ras pathway inhibitors. Conversely, cancer cells that would otherwise be sensitive to Ras pathway inhibitors acquire tolerance when AMPK activity is experimentally induced with an AMP mimetic. In this context, AMPK activation does not promote tolerance if autophagy is blocked, suggesting that autophagy is the key process activated downstream of AMPK to promote tolerance. Taken together, these findings provide a strong rationale for combining inhibitors of autophagy with inhibitors of the Ras pathway, a strategy being explored in several ongoing clinical trials.
Studies from the laboratories of E White and A Kimmelman have shown that KRAS/BRAF-driven lung and pancreatic cancers already have increased basal autophagy, which enables them to preserve mitochondrial function and resist metabolic stress in the tumor microenvironment. 32, 33, 34, 35 These investigators have shown that inhibiting this basal autophagy results in mitochondrial defects that impair the growth of KRAS/BRAF-driven tumors. BRAF-driven pediatric brain tumors also show high levels of autophagy, and increased sensitivity to autophagy inhibition.36 Recent studies from the laboratory of J Debnath have also suggested that the invasion of RAS-driven tumors may depend on pro-inflammatory cytokines whose secretion is dependent on a non-canonical form of autophagy.37 Since these studies demonstrate that autophagy is upregulated in KRAS/BRAF cancers, and is critical for cancer cell survival, one might have expected that inhibiting the MAPK pathway would lead to a reduction in autophagy. We find that this does not occur in the subset of cancers that are tolerant to Ras- Raf inhibition, which instead upregulate autophagy when the pathway is inhibited.
Prior research has also established a close connection between the Ras pathway and AMPK. Since AMPK reduces cellular proliferation upon nutrient deprivation, its activation often reduces cancer cell growth and is therefore being actively explored as a therapeutic strategy. Cantley and colleagues have shown previously that oncogenic BRAF blocks AMPK activation by inhibiting LKB1, and that vemurafenib can activate AMPK in BRAF-mutant melanomas.38 Our study extends these findings by showing that this AMPK activation induces autophagy, which is crucial for melanomas to tolerate BRAF inhibitors, and more generally for cancers with RAS or BRAF mutations to tolerate pathway inhibition. Additionally, our findings also suggest that AMPK activators, while potentially useful as single agents,39, 40 should be used with caution when applied in combination with Ras pathway inhibitors, whose effects they may well antagonize.
Our findings concerning tolerance to BRAF inhibitors complement those of Amaravadi and colleagues, who have shown that targeting autophagy also overcomes resistance to these inhibitors.41 Although we found that autophagy is activated by AMPK in tolerant cells, these authors have reported that AMPK does not have a role in activating autophagy in cells that are resistant to BRAF inhibitors. They have shown that, in drug-resistant melanomas, vemurafenib induces BRAF to bind the ER chaperone BIP, and that this results in ER stress and autophagy. We did not find any evidence that vemurafenib induced ER stress in our drug-tolerant melanoma lines. One possible explanation for this difference is that MAPK inhibition may be essential for vemurafenib to activate AMPK; since vemurafenib only inhibits the pathway in tolerant cells, AMPK may have a unique role in this context. Although the mechanisms involved are very different in these two contexts (Figure 3; Supplementary Figure 3D), blocking autophagy would appear to be a promising therapeutic strategy for both drug tolerance and resistance.
Materials and methods
Cell culture and reagents
Early passage human melanoma cell lines, YUKSI, YUSIK, YUSIT1, YUSIV, YUGEN8 and YUVON, were provided by Dr Ruth Halaban (Yale University School of Medicine), and were maintained in Opti-Mem with 5% FBS, GlutaMax, Penicillin and Streptomycin. HEK293T, HT29, MCF7, T47D, NCI-H460 and MDA-MB-231 cell lines were cultured in DMEM with 10% FBS. Mouse KRASmut/p53−/− NSCLC line (T1) was kindly provided by Dr Tyler Jacks (Koch Institute for Integrative Cancer Research). RAS and BRAF mutational status of the various lines is specified in the figures. GFP-LC3B baculovirus infection was performed following the manufacturer’s instructions (P36235; Life Technologies, Carlsbad, CA, USA). Lentivirus production, target cell infection and selection were performed as previously described.42 Vemurafenib (S1267; Selleck Chemicals, Houston, TX, USA) and AS703026 (S1475; Selleck Chemicals) were dissolved in DMSO, and CQ (C6628; Sigma, St Louis, MO, USA) was dissolved in PBS. Cells were treated with Raf/MAPK inhibitors for 48 h unless otherwise specified.
Electron microscopy
Transmission electron microscopy was performed using the standard procedure. Briefly, cells were fixed in a fixative solution containing 2.5 glutaraldehyde, 3 paraformaldehyde and 5% sucrose in 0.1 M sodium cacodylate buffer (pH 7.4). Cells were pelleted followed by treatment with 1% OsO4-veronal acetate, and cell pellet was dehydrated and embedded in Embed-812 resin. In all, 50 nm sections were cut using Reichert Ultracut E microtome (Leica Microsystems, Buffalo Grove, IL, USA), stained with uranyl acetate and lead citrate. Sections were examined using a FEI Tecnai spirit (Bio Twin, Hillsboro, OR, USA) at 80 KV and photographed with an AMT CCD camera. Autophagosomes/cell were quantified for DMSO (n=59 cells) and vemurafenib (n=50 cells) treatments.
Western blotting
Preparation of cell extracts and western blotting were performed using standard protocols. In all, 4–12 and 12% Bis-Tris gels were used for protein separation (NuPAGE; Life Technologies). The following antibodies from Cell Signaling Technology (Danvers, MA, USA) were used for immunoblotting: LC3B (#2775), pERK1/2 (#4377), ERK1/2 (#9102), p62 (#8025), ATG5 (#8540), pAMPKα (T172, #2535), AMPKα (#5832), pULKl (S555, #5869), ULK1 (#4773) and GAPDH-HRP (#3683).
Immunofluorescence
To visualize autophagosomes, GFP-LC3B labeled cells were fixed with 4% paraformaldehyde after indicated treatments, and washed with PBS before mounting. For additional staining, cells were permeabilized using 0.1% Triton X-100 and incubated with blocking solution (PBST with 10% goat serum and 3% BSA) for 1 h at room temperature. β-Tubulin Alexa Fluor 594 (CST #8058) and DAPI were used to stain cytoskeleton and nuclei, respectively. SQSTM1/p62 (CST #8025), LAMP1 (CST #9091) and LAMP2 (Abcam #25631) were detected using Alexa Fluor 555 anti-rabbit antibody following respective primary antibody incubations. Images were captured using Zeiss AxioPlan2 fluorescence microscope (Thornwood, NY, USA). Autophagosome quantification was performed using the CellProfiler software.43 In short, a CellProfiler pipeline was developed to identify cells based on the GFP signal. GFP-LC3B puncta were identified, and filtered based on intensity and size adjustments.
Flow cytometry
To measure glucose uptake, cells were cultured with DMSO or vemurafenib in glucose- and serum-free DMEM containing a fluorescent glucose analog, 2-NBDG (Cayman Chemicals #600470, Ann Arbor, MI, USA); uptake was assayed by flow cytometry following the manufacturer’s protocol, and analyzed by the FlowJo software (Ashland, OR, USA). For cell- cycle analysis, both floating and adherent cells were collected, fixed and stained with propidium iodide (EMD Biosciences # 537059, Billerica, MA, USA), and analyzed by the ModFit software (Verity Software House, Topsham, ME, USA). A total of 104 events were acquired in all cases.
Dose-response assays
Cells were seeded in 96-well plates, at a density of 3000 cells per well. Drugs were added at eight different doses with three replicates per dose. The same volume of DMSO was added in three replicates as a control. For each vemurafenib dose, CQ and Baf were used at 10 μM and 1 nM, respectively. Cell viability was measured after 72 h with CellTiterGlo reagent (Promega #G7572, Madison, WI, USA) following the manufacturer’s protocol.
Animal experiments and immunohistochemistry
NOD/SCID mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All animal studies were approved by the Animal Care and Use Committees of the Massachusetts Institute of Technology (Cambridge, MA, USA). One million cells (per mouse) were resuspended in 100 μl DMEM/Matrigel (BD Biosciences #354234, San Jose, CA, USA) mixture (1:1 volume) and subcutaneously injected into six- to eight-week-old female NOD/SCID mice (n · 5 mice per group). Once palpable tumors formed, mice were administered vemurafenib (20 mg/kg), and CQ (60 mg/kg), singly or in combination, and with vehicle control, everyday for 3 weeks, by intraperitoneal injections. Tumor sizes were recorded twice a week. Animals were randomized by weight. No blinding was applied in performing experiments. At the end of treatments, tumors were excised, fixed in 10% neutral-buffered formalin and embedded in paraffin. Serial sections (5 μm) were used for immunohistochemical analysis of pERK1/2, ERK1/2, pAMPK and LC3B (CST #3868). Briefly, tissue sections were deparaffinized using xylene and series of alcohols, followed by heating in 10 mM citrate buffer (pH 6.0) for antigen retrieval. Sections were blocked with blocking solution (Dako North America, Inc. #K4010, Carpinteria, CA, USA) for 1 h at room temperature, incubated with the primary antibody, washed, incubated with HRP-labeled secondary antibody, and stained using the DAB reagent. Nuclei were counterstained with Hematoxylin (Sigma #MHS16). Hematoxylin and eosin-stained sections were used to identify necrotic regions as previously described, marked by reduced tissue staining and patches of destroyed tumor architecture.44
Statistical analysis
All data unless otherwise specified represent mean±s.e.m. of at least three replicates. Statistical analyses were performed using GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Unpaired two-tailed Student’s t-test was performed for comparison between any two groups of data unless specified otherwise. P<0.05 was considered as significant. The assumption of normality was tested using a D’Agostino-Pearson omnibus test when n · 8. The assumption of equal variances was tested using a two-sample F Test. Welch’s correction was applied when unequal variances were found.
For animal experiments, we used power analysis to determine the minimum number of mice required to reach P<0.01. Assuming that difference in means/standard deviation · 2, · 4 mice are required per treatment group to reach this significance.
Supplementary Material
Acknowledgements
We thank Dr Tyler Jacks for providing us with mouse KRASmut/p53−/− NSCLC line, the Whitehead flow cytometry facility, and Nicki Watson and Wendy Salmon at the Whitehead Keck Imaging Facility for microscopy services. We thank Dr David Pincus and Dr Luke Whitesell for critical reading of the manuscript. This research was supported by grants from the Ellison Foundation (PBG) and Melanoma Research Alliance (#311800; PBG), a Yale SPORE in Skin Cancer funded by the National Cancer Institute (#1 P50 CA121974; RH), and funds from the NSFGRFP (#1122374; ESS).
Footnotes
Competing interests
The authors declare no conflict of interest.
Contributor Information
S Sanduja, Whitehead Institute for Biomedical Research, Cambridge, MA, USA.
Y Feng, Whitehead Institute for Biomedical Research, Cambridge, MA, USA.
R A Mathis, Whitehead Institute for Biomedical Research, Cambridge, MA, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
E S Sokol, Whitehead Institute for Biomedical Research, Cambridge, MA, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
F Reinhardt, Whitehead Institute for Biomedical Research, Cambridge, MA, USA.
R Halaban, Department of Dermatology, Yale University School of Medicine, New Haven, CT, USA.
P B Gupta, Whitehead Institute for Biomedical Research, Cambridge, MA, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA; Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA, USA; Harvard Stem Cell Institute, Cambridge, MA, USA.
References
- 1.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 2010; 467: 596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci USA 2008; 105: 3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011; 364: 2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT. Narrative review: BRAF opens the door for therapeutic advances in melanoma. Ann Intern Med 2010; 153: 587–591. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010; 363: 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010; 468: 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res 2008; 68: 4853–4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H et al. Melanomas acquire resistance to B- RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010; 468: 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 2011; 29: 3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011; 480: 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRbeta-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res 2011; 71: 5067–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012; 366: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalley KS. PLX-4032, a small-molecule B-Raf inhibitor for the potential treatment of malignant melanoma. Curr Opin Invest Drugs 2010; 11: 699–706. [PubMed] [Google Scholar]
- 14.Sondergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med 2010; 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin- mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flier JS, Mueckler MM, Usher P, Lodish HF. Elevated levels of glucose transport and transporter messenger RNA are induced by ras or src oncogenes. Science 1987; 235: 1492–1495. [DOI] [PubMed] [Google Scholar]
- 17.Kawada K, Nakamoto Y, Kawada M, Hida K, Matsumoto T, Murakami T et al. Relationship between 18 F-fluorodeoxyglucose accumulation and KRAS/BRAF mutations in colorectal cancer. Clin Cancer Res 2012; 18: 1696–1703. [DOI] [PubMed] [Google Scholar]
- 18.McArthur GA, Puzanov I, Amaravadi R, Ribas A, Chapman P, Kim KB et al. Marked, homogeneous, and early [18 F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J Clin Oncol 2012; 30: 1628–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy.Nat Rev Mol Cell Biol 2007; 8: 774–785. [DOI] [PubMed] [Google Scholar]
- 20.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 2011; 7: 643–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol 2005; 6: 439–448. [DOI] [PubMed] [Google Scholar]
- 23.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993; 333: 169–174. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Guan KL. Regulation of the autophagy initiating kinase ULK1 by nutrients: Roles of mTORC1 and AMPK. Cell Cycle 2011; 10: 1337–1338. [DOI] [PubMed] [Google Scholar]
- 25.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol 2010; 6: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohsumi Y Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell [DOI] [PubMed] [Google Scholar]
- 27.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: 463–477. [DOI] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001; 410: 1111–1116. [DOI] [PubMed] [Google Scholar]
- 30.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 2004; 101: 3329–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res 2010; 23: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov 2013; 3: 1272–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 2011; 25: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strohecker AM, White E. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy 2014; 10: 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011; 25: 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov 2014; 4: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov 2014; 4: 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell 2009; 33:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerezo M, Tichet M, Abbe P, Ohanna M, Lehraiki A, Rouaud F et al. Metformin blocks melanoma invasion and metastasis development in AMPK/p53-dependent manner. Mol Cancer Ther 2013; 12: 1605–1615. [DOI] [PubMed] [Google Scholar]
- 40.Petti C, Vegetti C, Molla A, Bersani I, Cleris L, Mustard KJ et al. AMPK activators inhibit the proliferation of human melanomas bearing the activated MAPK pathway. Melanoma Res 2012; 22: 341–350. [DOI] [PubMed] [Google Scholar]
- 41.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J et al. Targeting ER stress- induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 2014; 124: 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet 2005; 37: 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamrekelashvili J, Kruger C, von Wasielewski R, Hoffmann M, Huster KM, Busch DH et al. Necrotic tumor cell death in vivo impairs tumor-specific immune responses. J Immunol 2007; 178: 1573–1580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.