Statement of Significance:
Therapeutic antibody technologies have evolved over the last four decades. Although antibody engineering technologies may require further optimization, the methodology is generally established and, in many cases, validated clinically. One area of research that may potentially lead to a significant paradigm change is the development of single domain antibodies. This is extra matter in statement of significance more than 50 words for query purpose only. This is extra matter in statement of significance more than 50 words for query purpose only
I am pleased to introduce Antibody Therapeutics, the official journal of the Chinese Antibody Society (CAS) published by Oxford University Press. On an ordinary day in 2016, I received a text message from Dr. Shouye Wang (he later became the first CAS President) via WeChat, a popular social network among Chinese individuals worldwide. He invited me to a Boston-based group of Chinese scientists who were interested in antibody engineering and its use in the development of therapeutics. In 2011, I had started a group in Bethesda called the National Institutes of Health (NIH) Antibody Interest Group. Instantly, I saw the similarity in the goals of these two groups. Several months later, I was thrilled to learn that the Boston group had registered as a 501(c) nonprofit organization. They had decided to organize their first annual scientific meeting. This meeting would take place at the Cambridge Marriott near the Massachusetts Institute of Technology immediately before the Protein and Antibody Engineering Summit Conference in April 2017. I was invited to serve as the Chair of the Scientific Advisory Board. Before and during the first annual meeting, the level of momentum and excitement for academic collaboration grew to such a level that the society ambitiously decided to launch an international scientific journal. I was honored to learn that I had been invited to serve as the founding Editor-in-Chief. Enthusiastically, I accepted this invitation, suggesting that the new journal be named Antibody Therapeutics. Furthermore, I proposed that it be established as an international, peer-reviewed, open access journal to serve the global therapeutic antibody community. Over the past year, we spent many evenings and weekends discussing with colleagues and experts the formation of an editorial board and recruitment of assistant editors. We also dedicated ourselves to selecting a publisher, deciding the journal’s logo, testing the website, and preparing instructions for authors and reviewers. I must admit that it has taken considerably more effort than I had expected to launch a journal. Nevertheless, we are happy to share this new journal with the antibody community and warmly welcome you to join us by contributing your best research, method, or review articles to our journal!
Antibody therapeutics play an important role in the current cancer treatment landscape [1–4]. Antibodies to cell surface antigens are potentially effective in several ways: they can modulate cancer signaling pathways, recruit immune cells or proteins to the site of tumors, and deliver cytotoxins (Fig. 1). Therapeutic antibody technologies have dramatically evolved over the past four decades with advances in the fields of genetic engineering, protein engineering, cell engineering, and development of transgenic animals. Paul Ehrlich coined the term “magic bullets” a century ago. Monoclonal antibodies (mAbs) that specifically target disease cells have been proposed as the most promising candidates to be magic bullets against cancer, infectious diseases, and other diseases. In 1975, César Milstein and Georges Köhler at the Laboratory of Molecular Biology in Cambridge developed the hybridoma fusion technology to manufacture mAbs using the myeloma cells generated by Michael Potter at the National Cancer Institute in Bethesda [5,6]. Murine antibodies have immunogenicity and a short half-life. Thus, it was clear that development of mouse/human chimeric antibodies, humanized mouse antibodies, or human-derived antibodies was necessary for generation of effective antibody cancer therapeutics. Key advances in protein engineering technologies within the last two decades have transformed antibodies from research and diagnostic tools into a class of targeted therapeutics. Grafting of the complementarity-determining region (CDR) from a mouse antibody into a human antibody framework was the first clinically validated humanization technique [7–9]. The initial highlight success of this technique occurred when the CDR from the murine antibody, 4D5, was grafted into a human scaffold to create the therapeutic reagent trastuzumab [8]. Besides humanized antibodies, human mAbs have been discovered using transgenic mice or phage display technologies (Fig. 2) [10,11]. The technologies for generating human mAbs from transgenic mice were first described in the 1990s. Exogenous human immunoglobulin (Ig) loci were inserted into the mouse genome, and endogenous mouse Ig genes were disrupted [12,13]. More recently, human variable region gene segments, instead of the whole Ig loci, were introduced. This modification was made to promote more robust antibody response to tumor antigens [14]. Many human mAbs made from transgenic mice (e.g., panitumumab, golimumab, canakinumab, ustekinumab, ofatumumab, denosumab, ipilimumab, nivolumab, and alirocumab) or phage display-derived mAbs (e.g., adalimumab, belimumab, and raxibacumab) have been approved for marketing or are being tested in various stages of clinical trials. Using phage display technology, human single-chain Fv or Fab fragments can be isolated from a large library with a diversity of >1010 [15]. In addition to phage display, yeast and mammalian cell display technologies have also been developed. These technologies allow for recombinant antibody fragments to be expressed on the cell surface of eukaryote cells. This enables isolation of antigen-specific cells by flow cytometry and the discovery of high-affinity binders [16,17]. Although isolation of human mAbs directly from host B cells appeared very promising, until recently it has proven to be very difficult. Identification of broadly neutralizing human antibodies to HIV was successful after studying the B cell populations of nonprogressive infected individuals. Using state-of-the-art single B cell sorting and single-cell reverse transcription polymerase chain reaction (RT-PCR) cloning, the relevant Ig genes were isolated and identified [18,19]. However, it is still unclear whether the B cell sorting/cloning strategy could be effectively applied to the identification of therapeutic antibodies from cancer patients. A recent study offers hope for the use of cloned human antibodies as cancer therapeutics [20]. Alternatively, B cells have been immortalized and screened for specific binding properties [21]. Immortalization of human memory B cells using Epstein–Barr virus (EBV)-mediated transformation was reported as early as the late 1970s [22]. The drawback of this approach is that it involves screening large numbers of B cells to obtain very rare B cell clones with the desired specificity. Techniques developed more recently have helped to promote antibody-producing B cells in culture [23]. One of these techniques involves engineering B cells to express key proliferative factors, such as Bcl-6 and Bcl-xL, and encouraging B cell activation through the addition of CD40L and IL-21.
Figure 1.
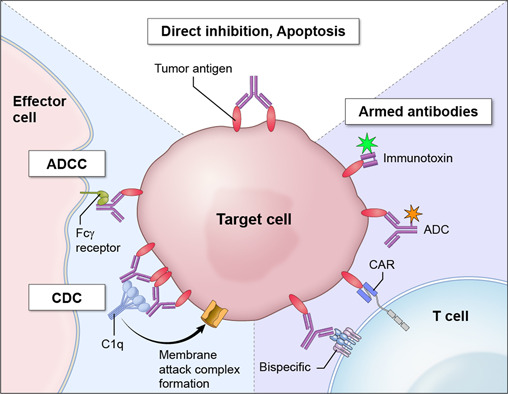
Mechanisms of antibody-based cancer therapies. Antibodies to cell surface antigens are potentially effective in several ways. First, antibodies directly inhibit tumor cell proliferation via inducing apoptosis or inhibiting key signaling pathways for cell growth. Second, antibodies can kill targeted tumor cells by initiating antibody-dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC). Third, antibodies can recruit immune cells such as T cells or chimeric antigen receptor (CAR) T cells. Finally, antibodies can be used to deliver cytotoxins including bacterial toxins (immunotoxins) or small toxic molecules (antibody drug conjugates) to kill tumor cells.
Figure 2.
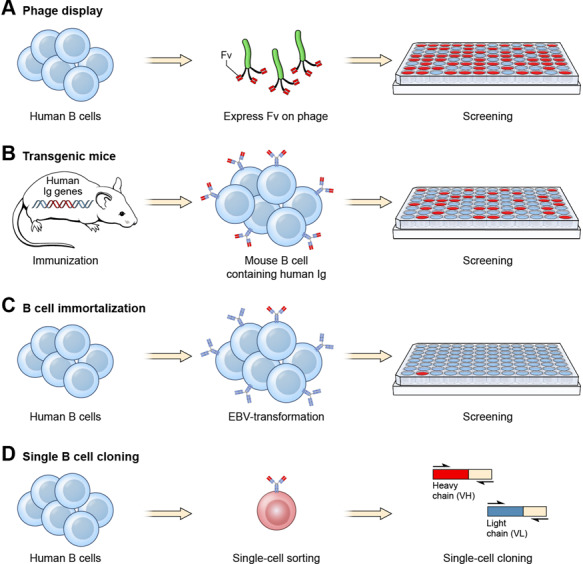
Examples of how human mAbs specific for tumor antigens can be isolated. Four major approaches have been developed to isolate human mAbs. (a) Phage display. A large phage display library is commonly used to isolate human scFv or Fab specific for an antigen [11]. After 3–5 rounds of panning, tumor-specific phage clones may be enriched and identified commonly by using enzyme-linked immunosorbent assay (ELISA). More recently, human single-domain antibodies are also isolated by phage display [28,29]. (b) Transgenic mice. Mice producing human Ig genes are immunized with a tumor antigen. Mouse hybridomas are created to produce stable mouse B cell lines. Mouse B cells expressing tumor-specific human mAbs are screened in ELISA using culture supernatant. (c) B cell immortalization. Human B cells are immortalized by EBV transformation. A large number of immortalized B cell clones are screened for antigen binding. (d) Single B cell sorting. Memory B cells are sorted by flow cytometry for the isolation of rare single B cells (<1%) for antigen binding followed by single-cell cloning by RT-PCR.
Although antibody engineering technologies may require further optimization, the methodology is well established and, in many cases, validated clinically. One area of research that may lead to a significant paradigm change in antibody engineering is the development of single-domain antibodies such as camelid VHH and shark VNAR (Fig. 3). These antibodies represent a new and very different class of products: small, easy to express and produce, stable, and capable of penetrating tissues to reach buried sites in tumor or viral antigens [24–26]. In recent years, my laboratory and collaborators have developed a group of human single-domain antibodies that target tumor-specific antigens such as mesothelin [27], GPC2 [28], and GPC3 with high affinity [29]. The single-domain antibodies have been used to inhibit Wnt, Yap, and other key cancer signaling pathways that are important for cell proliferation [28–31]. Interestingly, production of artificial single-domain proteins has also been pursued [32]. It would be interesting to see whether the new wave of single-domain proteins could eventually become a game changer in our toolbox of magic bullets for treating cancer and other diseases.
Figure 3.
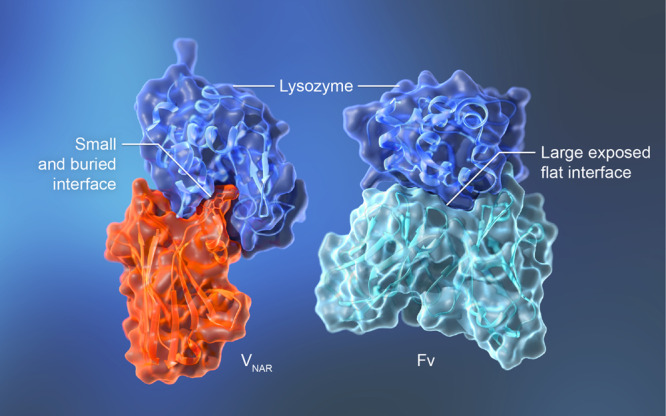
The complex of a single-domain antibody and a protein antigen reveals a buried binding site. (a) The nurse shark VNAR single domain in complex with lysozyme (PDB 1T6V) [24]. VNAR residues Arg100 and Tyr101 are shown in the ribbon diagram to illustrate their interaction with the substrate binding pocket of lysozyme. (b) The humanized HyHEL-10 Fv in complex with lysozyme (PDB 2EIZ) [38].
Despite more than four decades of research on targeted therapies, the solid tumors that kill over 90% of people still do not respond to most of our current therapies. The discovery of novel cancer antigens remains one of the greatest unmet medical challenges in humanity’s journey to find “magic bullets”. As the Cancer Moonshot Blue Ribbon Panel concluded, for most cancers, we know little about what tumor antigens can be safely and effectively targeted to discriminate cancers from normal tissues. My research has been focused on mesothelin and the cell surface glypican, GPC3. These are among the few “shared” tumor antigens that are being tested in clinical settings [33,34]. Tumor-specific mutations such as EGFRvIII [35] and KRAS [36] have provided great opportunities for targeted cancer therapies [37]. Genomics will continue to play an important role in target identification. Proteomics and glycomics will play a role in the identification of tumor-specific splice variants or isoforms and tumor-specific proteins that have unique conformations, glycosylation, or other post-translational modifications in the tumor microenvironment. In my opinion, this is the best time to launch a journal that aims to provide a multidisciplinary forum for all of our members in the therapeutic antibody community. The systems biology approach may change the paradigm in our journey to cure all cancers in the future. We hope that this journal will also serve as an engaging platform for networking, education, and career development among members of society and beyond.
I am honored to serve as the Editor-in-Chief of this new journal. I believe that antibody therapeutics is one of the fastest moving fields in modern medicine and an area of research that I have been focusing on. I am indebted to Dr. Shouye Wang for his faith in me; to Dr. Zhiqiang An for helping me as deputy editor; to Drs. Tiencen Hu, Songyu Wang, Hongyu Zhang, and Mi Deng for serving as assistant editors; to all of the members on the editorial board for their support; and to Nikul Patel and Margaret Graton (Oxford University Press) for maintaining high professional standards.
ACKNOWLEDGMENTS
I thank Zhiqiang An and Georgina Salazar at the University of Texas Health Science Center at Houston for their careful and critical reading of the manuscript. This work is supported by the Intramural Research Program of NIH, National Cancer Institute (NCI) (Z01 BC010891 and ZIA BC010891), the NCI Center for Cancer Research FLEX Program Technology Development Award, and the NIH Deputy Director for Intramural Research Innovation Award (to M.H.). I would like to thank Philippe Youkharibache (NCI) for creating the images that show the complexes of lysozyme with a conventional antibody Fv or a single-domain antibody (VNAR); Bryan Fleming (NCI), Gwen Buel (NCI), and Yen Phung (Georgetown University School of Medicine) for reading the manuscript; Alan Hoofring and Ethan Tyler (NIH Medical Arts Design Section) for making the figures; and Bradley Otterson (NIH Library) for editorial assistance. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The author declares no competing financial interest.

Mitchell Ho
Editor-in-Chief
REFERENCES
- 1. Carter, P. Improving the efficacy of antibody-based cancer therapies. Nat Rev Cancer 2001; 1: 118–29. [DOI] [PubMed] [Google Scholar]
- 2. Weiner, LM. Building better magic bullets—improving unconjugated monoclonal antibody therapy for cancer. Nat Rev Cancer 2007; 7: 701–6. [DOI] [PubMed] [Google Scholar]
- 3. Weiner, LM, Murray, JC, Shuptrine, CW. Antibody-based immunotherapy of cancer. Cell 2012; 148: 1081–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiner, LM, Dhodapkar, MV, Ferrone, S. Monoclonal antibodies for cancer immunotherapy. Lancet (London, England) 2009; 373: 1033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohler, G, Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975; 256: 495–7. [DOI] [PubMed] [Google Scholar]
- 6. Potter, M. The early history of plasma cell tumors in mice, 1954–1976. Adv Cancer Res 2007; 98: 17–51. [DOI] [PubMed] [Google Scholar]
- 7. Jones, PT, Dear, PH, Foote, Jet al. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature 1986: 321: 522–5. [DOI] [PubMed] [Google Scholar]
- 8. Carter, P, Presta, L, Gorman, CMet al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89: 4285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Queen, C, Schneider, WP, Selick, HEet al. A humanized antibody that binds to the interleukin 2 receptor. Proc Natl Acad Sci U S A 1989; 86: 10029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson, AL, Dhimolea, E, Reichert, JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov 2010; 9: 767–74. [DOI] [PubMed] [Google Scholar]
- 11. Clackson, T, Hoogenboom, HR, Griffiths, ADet al. Making antibody fragments using phage display libraries. Nature 1991; 352: 624–8. [DOI] [PubMed] [Google Scholar]
- 12. Green, LL, Hardy, MC, Maynard-Currie, CEet al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs. Nat Genet 1994; 7: 13–21. [DOI] [PubMed] [Google Scholar]
- 13. Lonberg, N, Taylor, LD, Harding, FAet al. Antigen-specific human antibodies from mice comprising four distinct genetic modifications. Nature 1994; 368: 856–9. [DOI] [PubMed] [Google Scholar]
- 14. Murphy, AJ, Macdonald, LE, Stevens, Set al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc Natl Acad Sci U S A 2014; 111: 5153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vaughan, TJ, Williams, AJ, Pritchard, Ket al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nature Biotechnol 1996; 14: 309–14. [DOI] [PubMed] [Google Scholar]
- 16. Boder, ET, Wittrup, KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature Biotechnol 1997; 15: 553–7. [DOI] [PubMed] [Google Scholar]
- 17. Ho, M, Nagata, S, Pastan, I. Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci U S A 2006; 103: 9637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scheid, JF, Mouquet, H, Feldhahn, Net al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009; 458: 636–40. [DOI] [PubMed] [Google Scholar]
- 19. Wu, X, Yang, ZY, Li, Y, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329: 856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bushey, RT, Moody, MA, Nicely, NL, et al. A therapeutic antibody for cancer, derived from single human B cells. Cell Rep 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson, PC, Andrews, SF. Tools to therapeutically harness the human antibody response. Nature Rev Immunol 2012; 12: 709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steinitz, M, Klein, G, Koskimies, Set al. EB virus-induced B lymphocyte cell lines producing specific antibody. Nature 1977; 269: 420–2. [DOI] [PubMed] [Google Scholar]
- 23. Kwakkenbos, MJ, Diehl, SA, Yasuda, Eet al. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nature Med 2010; 16: 123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stanfield, RL, Dooley, H, Flajnik, MFet al. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004; 305: 1770–3. [DOI] [PubMed] [Google Scholar]
- 25. Barelle, C, Gill, DS, Charlton, K. Shark novel antigen receptors—the next generation of biologic therapeutics? Adv Exp Med Biol 2009; 655: 49–62. [DOI] [PubMed] [Google Scholar]
- 26. De Genst, E, Silence, K, Decanniere, Ket al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 2006; 103: 4586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang, Z, Feng, M, Gao, Wet al. A human single-domain antibody elicits potent antitumor activity by targeting an epitope in mesothelin close to the cancer cell surface. Mol Cancer Ther 2013; 12: 416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li, N, Fu, H, Hewitt, SMet al. Therapeutically targeting glypican-2 via single-domain antibody-based chimeric antigen receptors and immunotoxins in neuroblastoma. Proc Natl Acad Sci U S A 2017; 114: E6623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feng, M, Gao, W, Wang, Ret al. Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2013; 110: E1083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao, W, Tang, Z, Zhang, Y-F, et al. Immunotoxin targeting glypican-3 regresses liver cancer via dual inhibition of Wnt signalling and protein synthesis. Nature Comm 2015; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao, W, Kim, H, Ho, M. Human monoclonal antibody targeting the heparan sulfate chains of glypican-3 inhibits HGF-mediated migration and motility of hepatocellular carcinoma cells. PloS One 2015; 10: e0137664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang, PS, Boyken, SE, Baker, D. The coming of age of de novo protein design. Nature 2016; 537: 320–7. [DOI] [PubMed] [Google Scholar]
- 33. Hassan, R, Ho, M. Mesothelin targeted cancer immunotherapy. Eur J Cancer 2008; 44: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ho, M, Kim, H. Glypican-3: a new target for cancer immunotherapy. Eur J Cancer 2011; 47: 333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wikstrand, CJ, Reist, CJ, Archer, GEet al. The class III variant of the epidermal growth factor receptor (EGFRvIII): characterization and utilization as an immunotherapeutic target. J Neurovirol 1998; 4: 148–58. [DOI] [PubMed] [Google Scholar]
- 36. Smit, VT, Boot, AJ, Smits, AM, et al. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res 1988; 16: 7773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson, LA, Scholler, J, Ohkuri, Tet al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med 2015; 7: 275ra222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakanishi, T, Tsumoto, K, Yokota, Aet al. Critical contribution of VH–VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10. Protein Sci 2008; 17: 261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]


