Summary
This study reports the role of RIPK1 in modulating HNSCC metastasis. We find RIPK1 is downregulated during the tumor progression and reduced RIPK1 expression correlates with promoter hypermethylation. In tumor-derived HNSCC cell lines, RIPK1 silencing enhances the tumor promoting properties while augmenting the apoptotic response to synthetic dsRNA.
Abstract
Head and neck squamous cell carcinoma (HNSCC) is a devastating disease for which new treatments, such as immunotherapy are needed. Synthetic double-stranded RNAs, which activate toll-like receptor 3 (TLR3), have been used as potent adjuvants in cancer immunotherapy by triggering a proapoptotic response in cancer cells. A better understanding of the mechanism of TLR3-mediated apoptosis and its potential involvement in controlling tumor metastasis could lead to improvements in current treatment. Using paired, autologous primary and metastatic HNSCC cells we previously showed that metastatic, but not primary tumor-derived cells, were unable to activate prosurvival NF-κB in response to p(I):p(C) resulting in an enhanced apoptotic response. Here, we show that transcriptional downregulation of receptor-interacting serine/threonine-protein kinase 1 (RIPK1) in metastatic HNSCC cells causes a loss of TLR3-mediated NF-κB signaling, resulting in enhanced apoptosis. Loss of RIPK1 strongly correlates with metastatic disease in a cohort of HNSCC patients. This downregulation of RIPK1 is possibly mediated by enhanced methylation of the RIPK1 promoter in tumor cells and enhances protumorigenic properties such as cell migration. The results described here establish a novel mechanism of TLR3-mediated apoptosis in metastatic cells and may create new opportunities for using double stranded RNA to target metastatic tumor cells.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the most frequent malignancy of the aerodigestive tract. However, due to limitations in treatment, over half of the 600 000 patients diagnosed with HNSCC die from metastatic complications (1). Although mortality rates from this disease have not improved dramatically in past decades, the molecular profiling of predictive biomarkers in select patient populations who are more responsive to certain chemotherapies, offer novel strategies for treating HNSCC with molecular-targeted agents (2). Beneficial clinical outcome has been associated with more ‘inflamed’ tumors. Among the novel targets, Toll-like receptor 3 (TLR3) expression has been shown to correlate with the efficacy of polyinosinic acid:polycytidylic acid (p(I):p(C)) to eliminate cancer cells, and stimulation of TLR3 yields antitumor effects in a number of studies (3,4). Furthermore, published data suggest that TLR3 agonists could be used to enhance cancer vaccine efficacy, by altering the tumor microenvironment to augment innate and adaptive immunity. TLR3 signaling may directly inhibit cell growth, migration, and induce apoptosis to also eliminate cancer cells (5–8).
In addition to TLR3 expression, numerous proteins play a role in defining the downstream signaling by mediating the pro-survival nuclear factor κB (NF-κB) and proapoptotic Interferon regulatory factor 3 (IRF3) signaling responses to p(I):p(C). A better understanding of the mechanism of TLR3-mediated apoptosis and its potential involvement in controlling tumor metastasis could lead to improvements in current treatment. Using paired cell lines derived from autologous primary and metastatic HNSCC, we previously showed that the cells derived from metastatic tumors, but not primary tumors, were unable to activate NF-κB, while the proapoptotic IRF3 signaling remained intact. Consequently, stimulation of the cells from metastatic tumors with p(I):p(C) resulted in an enhanced apoptotic response due to the imbalance in downstream signaling (9). Here, we describe the molecular factors causing increased cell death in response to TLR3 stimulation.
While investigating the upstream mediators of the TLR3-NF-κB signaling pathway we found that receptor interacting protein kinase 1 (RIPK1) was downregulated in metastatic HNSCC. Although the main role of RIPK1 has been attributed to TNFα mediated NF-κB activation and necroptosis, it has been shown that RIPK1 is also required for the TLR3-NF-κB pro-survival signaling (10–12). Additionally, it has been shown that RIPK1 expression prevents TLR3-RIPK3-MLKL mediated necroptosis thus, downregulation of RIPK1 could lead to an enhanced apoptotic and necroptotic response to TLR3 signaling (13,14). Here, we show that downregulation of RIPK1 is produced by epigenetic modifications (DNA methylation) in HNSCC cells and this loss results in a clinically exploitable enhanced apoptosis by p(I):p(C) treatment. The results described here establish a novel mechanism of TLR3 mediated apoptosis in metastatic cells and may create new opportunities for using double stranded RNA to target metastatic tumor cells.
Materials and methods
Cell lines and tissues
The primary and metastatic HNSCC cell lines were derived from the primary and metastatic lymph nodes and characterized at the University of Pittsburgh as described before (9,15). The cell lines were authenticated within the last 6 months by HLA typing and STR DNA profiling as described before (16), and monitored regularly to be free of mycoplasma contamination. All cell lines were cultured in DMEM, (Lonza, Rockland, ME) containing 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin/streptomycin (Lonza) at 37ºC in a humidified 5% CO2 atmosphere. Pairs of primary and metastatic HNSCC tissues from same patients were obtained during surgery as per University of Pittsburgh IRB-protocol 99-069.
Antibodies and immunohistochemistry
IHC was performed on foundation fixed, paraffin-embedded prospective tissue microarrays (0.6mm cores) containing paired primary and metastatic tumors and stained with RIPK1 antibody (1:1000, BD Transduction Laboratories). Antibodies against the cleaved PARP were purchased from Cell Signaling Technology (Beverly, MA) and actin from Santa Cruz Biotechnology (cat.# sc-47778 Dallas, TX). Polyinosinic:polycytidylic acid [p(I):p(C)] was from GE Healthcare (Piscataway, NJ) and dissolved in phosphate-buffered saline before use. 5-Aza-2′-deoxycytidine was ordered from Sigma (cat# A3656).
Western blotting analysis
To prepare whole cell lysates, cells were washed in ice-cold phosphate-buffered saline, scraped and collected in lysis buffer (20mM HEPES pH 7.4, 1 % Triton-X 100, 150mM NaCl, 1.5mM MgCl2, 12.5mM b-glycerophosphate, 2mM EGTA, 10mM NaF, 2mM DTT, 1mM Na3VO4, 1mM PMSF plus 1× protease inhibitors). Equal amounts of protein extracts were subjected to 8% SDS–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. Membranes were blocked with 5% nonfat dry milk and incubated with primary antibody and subsequently with horseradish peroxidase-conjugated secondary antibody for 1h at room temperature. After washing, membranes were exposed onto film and then developed.
Lentiviral vectors
Doxycycline inducible lentiviral vectors were generated by performing LR recombination between pENTR/D-TOPO FLAG-RIPK1-HA and pInducer 20 destination vector (17). Lentiviruses were packaged in 293T and pseudotyped with VSV G protein as before (9). Transduction of PCI-15B and OSC-19 cells were carried out overnight at 37ºC in the presence of 1 μg/ml polybrene. Cells were selected with 500 μg/ml G418 or 1 μg/ml Puromycin to establish stable cell lines.
Quantitative PCR analysis of gene expression
Total RNA was purified using Trizol reagent (Invitrogen) and treated with DNase I (DNA Free kit, Ambion, Foster City, CA). Total RNA (1 µg) was used for reverse transcription using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) and subjected to real-time PCR using a CFX96 real time system (Bio-Rad) according to manufacturer’s instructions. Primers for RIPK1 (forward 5′-CTGGGCTTCACACAGTCTCA-3′ reverse 5′-GTCGATCCTGGAACACTGGT-3′) and RPL32 were as previously reported (18). PCR amplification of each gene was normalized to that of RPL32.
DNA methyltransferase inhibition assay
Cells were seeded 2×105 per well in 12 well plates. After 24h, the cells were treated with 0, 1, 5, 10 or 15 uM 5-Aza-2′-deoxycytidine. The media was changed every 24h for 72h and replaced with fresh 5-Aza-2′-deoxycytidine. After 72h incubation, the cells were harvested for RT-qPCR and western blot analysis.
Anoikis-recovery assay
Twenty-four well plates were coated with 200 µl poly-HEMA (10mg/ml) diluted in 75% ethanol three times allowing the reagent to dry before reapplying. After the wells had dried 5×105 cells were plated and incubated in the poly-HEMA coated wells for 48h, then transferred to six-well plates for recovery. Cells were recovered for 24 and 96h before they were fixed with a 50/50 mixture of methanol and water containing 4% crystal violet.
Migration assay
Two-dimensional wound-healing (scratch) assay
PCI-15A-shCTRL and PCI-15A-shRIPK1 were seeded 2×105 cells per well in 12 well plates and were incubated until reaching ~100% confluence (24h). The cell monolayer was then ‘scratched’ once with a p-200 pipet tip causing an ~1mm wound in the monolayer. Microscopic phase contrast images were taken immediately after and 12h post-wound in addition to an image of the grid of a hemacytometer to measure healing of the scratch.
Three-dimensional transwell migration assay
To assay the three-dimension migration of our PCI-15A-shCTRL and PCI-15A-shRIPK1 cell lines, we seeded cells in BDbiocoat 8.0 micron fibronectin inserts (pre-coated with human fibronectin, which promotes cell attachment) and quantified the number of cells migrated and normalized these values to BDbiocoat 8.0 micron control inserts according to the manufacturers protocol.
Datasets and statistical analysis
Datasets
The Cancer Genome Atlas was used for both datasets in this study (promoter methylation and expression) (19). For the TCGA expression data, we downloaded level 3 RNAseqV2 data for the N = 30 patient samples with both normal solid tissue and primary solid tumor. After consolidating the RSEM values for RIPK1 and actin from each of these samples into an excel database, we normalized the RSEM value of RIPK1 to that of actin. Similarly, the TCGA methylation data was downloaded and beta values were consolidated into a spreadsheet according to patient-matched normal solid tissue and primary solid tumor.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001 represent statistical significance by two-tailed paired Student’s t test analysis. Wherever applicable, plots show mean with standard error bars. When correlating the expression and methylation data we used the Pearson’s correlation coefficient using an n = 20 (df = 18) one-tailed test for negative correlation with a critical value of −0.468.
Results
Downregulation of RIPK1 in metastatic head and neck cancer cells
We have previously observed that there is an enhanced apoptotic response to TLR3 ligands in cell lines obtained from metastatic tumors compared to those obtained from the matched primary tumors. This enhanced apoptosis resulted from a loss in the TLR3-NF-κB prosurvival signaling pathway, while the death promoting TLR3-IRF3 pathway remained intact (9). Next, we evaluated the expression of another protein in TLR3-NF-κB signaling, RIPK1, which is an essential adaptor molecule for this signaling pathway. Immunoblotting of whole-cell lysates from paired primary (PCI-15A and PCI-6A) and matched metastatic (PCI-15B and PCI-6B) tumor derived cell lines showed a marked decrease of RIPK1 protein expression in metastatic cells (Figure 1A). This finding validated previous reports suggesting reduction in RIPK1 levels during tumorigenesis (20–22). Additionally, we analyzed the mRNA levels in the autologous pair of PCI-15 cells and found that mRNA levels of RIPK1 are also downregulated between metastatic (PCI-15B) and primary (PCI-15A) cell lines, suggesting that the downregulation of RIPK1 could occur at the transcriptional level (Figure 1B).
Figure 1.

Downregulation of RIPK1 in metastatic head and neck cancer cell lines. Whole-cell lysates from autologous paired primary tumor derived cell lines (PCI-15A and PCI-6A) and metastatic tumor derived cell lines (PCI-15B and PCI-6B) were immunoblotted with anti-RIPK1 and anti-actin antibodies (A). (B) qRT-PCR analysis of RIPK1 mRNA expression levels in PCI-15A and PCI-15B total RNA. Following the normalization of each sample with the internal control RPL32, RIPK1 mRNA levels are shown as expression percent with respect to PCI-15A. Each bar represents mean and standard error from triplicate samples.
Downregulation of RIPK1 expression in head and neck cancer correlates with disease progression in vivo
To validate our in vitro evidence suggesting that RIPK1 is downregulated in HNSCC, we probed the tumor microarrays generated from resected primary and matched metastatic tumors with RIPK1 antibody. As shown in Figure 2A (upper panel), there was a decrease in the expression of RIPK1 between the primary (left image) and the matched metastatic tumor (right image). Semiquantitative analysis of RIPK1 expression levels in these tumor microarrays showed a significant decrease in the RIPK1 protein levels in metastatic tumors compared to primary tumors (Figure 2A, lower graph). To further validate this observation, we utilized the publically available Cancer Genome Atlas (TCGA) HNSCC mRNA expression data (19). Comparison of RIPK1 mRNA expression between tumor and normal marginal tissues from the same patients showed reduced RIPK1 expression in tumor tissues compared to that of normal tissues (Figure 2B). This suggests that RIPK1 expression is not only downregulated during the primary to metastatic transition, but also during the transition from normal to primary tumor lesion formation.
Figure 2.
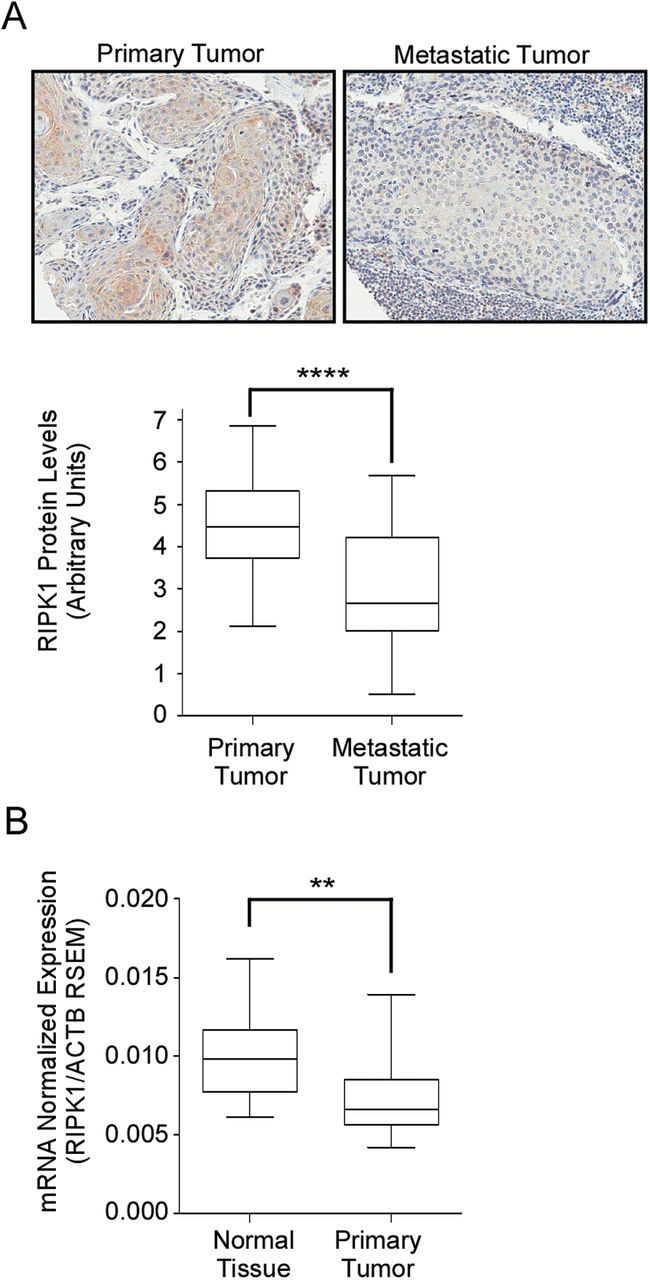
Downregulation of RIPK1 expression in head and neck cancer correlates with disease progression. Immunohistochemical analysis of resected primary (n = 90) and lymph node metastatic (n = 32) tumor specimens from patients with HNSCC was performed for RIPK1 expression. A representative H&E stained sample is shown in (A), upper panel. The lower graph shows the quantitation of RIPK1 expression (B) RIPK1 mRNA expression from RNAseq2 data in HNSCC patient paired normal solid tissue and primary solid tumors (n = 30) obtained from The Cancer Genome Atlas. This data was provided as RNA-Seq gene expression estimation with read mapping uncertainty (RSEM) and the RSEM of RIPK1 was normalized to the RSEM of Actin.
Ectopic expression of RIPK1 modulates double stranded RNA-mediated apoptosis
After observing that RIPK1 expression is downregulated in tumor cells, we sought to determine if the downregulation of RIPK1 was responsible for the enhanced apoptosis that we observed in metastatic HNSCC. We used shRNA to reduce RIPK1 expression in cells derived from primary HNSCC tumors (PCI-15A) to the level comparable to that from cells derived from metastatic tumors (PCI-15B) (Figure 3A, lanes 5–8 versus lanes 1–4, middle panel). Reciprocally, we stably restored the expression of RIPK1 in PCI-15B (Figure 3B, lanes 5–8 versus lanes 1–4, middle panel). As shown in Figure 3A treatments with increasing concentration of p(I):p(C) showed significant increase in the apoptotic response in PCI-15A-shRIPK1 cells compared to PCI-15A-shCTRL cells (Figure 3A, lanes 5–8 versus lanes 1–4, top panel) as measured by cleaved-PARP (c-PARP) expression. Similarly, in Figure 3B we found that when we restored the expression of RIPK1 in the PCI-15B cells there was reduced apoptotic response to p(I):p(C) (Figure 3B, lanes 5–8 versus lanes 1–4, top panel). Similar observations were made in another metastatic HNSCC derived cell line OSC-19, where we restored the expression of RIPK1 and observed reduced apoptotic response to p(I):p(C) (Figure 3C). To further validate these findings, we stably expressed RIPK1 under a tetracycline-inducible promoter in PCI-15B cells. Treatment of these (PCI-15B-iRIPK1) cells with doxycycline led to a dose-dependent increase in RIPK1 expression (Figure 3D, lanes 1–6, middle panel), which correlated with the dose-dependent reduction in apoptotic response to p(I):p(C) (Figure 3D, lanes 1–6, top panel, quantitation at the bottom). Together these results suggest that RIPK1 expression levels control the apoptotic response to TLR3 ligands in HNSCC and ectopic changes in RIPK1 expression contribute to the changes in the apoptotic response to p(I):p(C) treatment.
Figure 3.
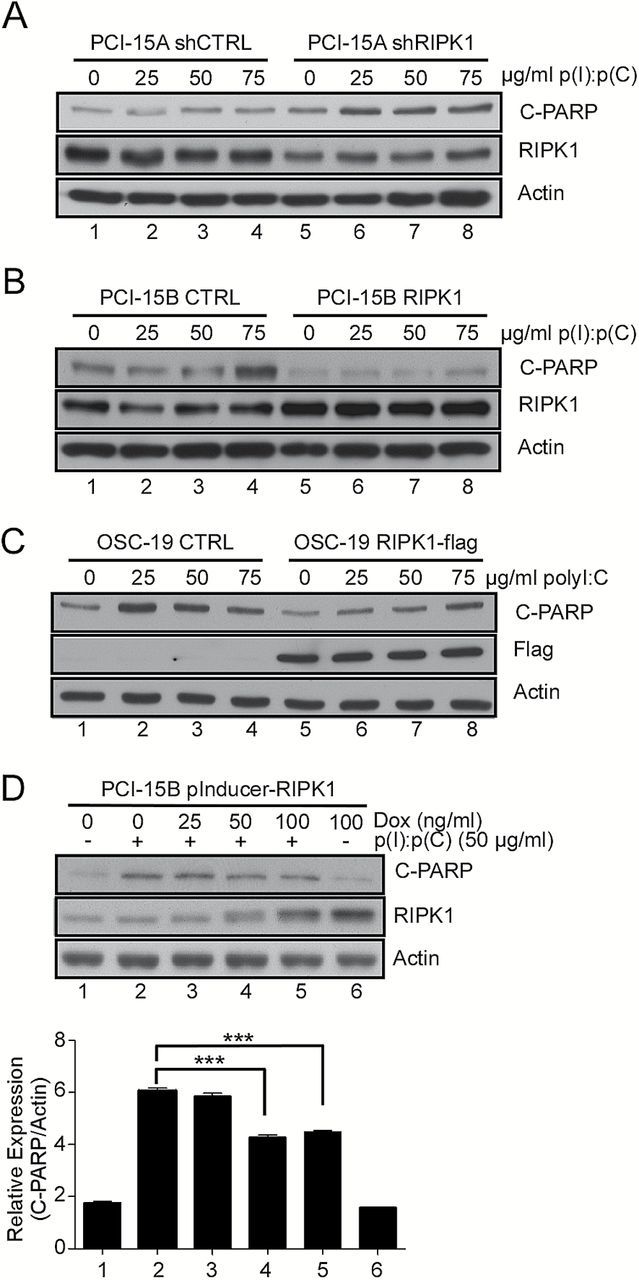
Ectopic changes in RIPK1 expression modulates dsRNA-mediated apoptosis. (A) PCI-15A primary cell lines were stably transduced with RIPK1 shRNA. Whole-cell lysates from these cells treated with increasing amounts of p(I):p(C) were analyzed by immunoblot using antibodies for cleaved PARP and actin as a loading control. (B) RIPK1 expression was restored in PCI-15B metastatic HNSCC cell line by transducing with a RIPK1 expression construct followed by the similar analysis as in (A). (C) Similar to (B) RIPK1 was ectopically expressed in another metastatic HNSCC cell line OSC-19. Whole-cell lysates from indicated cells treated with increasing amounts of p(I):p(C) were similarly analyzed by immunoblot as in (B). (D) The PCI-15B metastatic cell line was stably transduced with a dox-inducible RIPK1 expression vector. Whole-cell lysates were prepared from these cells treated with an increasing concentration of dox in the presence and absence of 50 μg/ml p(I):p(C). Cleaved PARP levels were analyzed (C, bottom graph) using the ImageJ gel quantification software package normalizing to actin levels. Bars represent the mean and standard error from triplicate readings of the representative blot provided in (C, upper panel).
RIPK1 promoter methylation controls its expression in tumors
As we observed a decrease in both the RIPK1 protein and mRNA in our autologous primary and metastatic HNSCC cell lines, we reasoned that the downregulation may be at the transcriptional level. Since it is well known that abnormal DNA methylation is widespread in cancer and plays an important role in the development of tumors, we hypothesized that promoter methylation may contribute to the RIPK1 downregulation (23). In addition to the mRNA data shown in Figure 2B, the TCGA database also provides DNA methylation status of HNSCC tumors analyzed with the Illumina Infinium HumanMethylation450 platform. We analyzed this data from N = 30 samples, and found that there was an increase in RIPK1 promoter methylation (as quantified by the probe-specific β value) in tumor samples compared to the normal tissue (Figure 4A). In this analysis, we included promoter methylation β values from all available CpG islands for RIPK1 as well as related gene (RIPK2) and a housekeeping gene (actin). This showed that although alterations in promoter methylation may be widespread in tumor samples, the changes in promoter methylation were specific to the RIPK1 promoter, and that this method has the sensitivity to detect relevant changes. Average β value calculated from the above 30 samples showed a statistically significant difference in promoter methylation for the probe position −868 from the RIPK1 transcription start site (Figure 4B), which showed a significant negative correlation between increased promoter methylation verses decreased RIPK1 mRNA expression (Figure 4C). To experimentally validate these finding in our system, we treated the HNSCC cell line PCI-15B with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. As expected, treatment with 5-aza-2′-deoxycytidine increased RIPK1 mRNA and protein expression in a dose-dependent manner (Figure 4D and E) without changing the expression of other housekeeping genes. These results indicated that the reduced RIPK1 expression in tumor samples can partly be attributed to the increased methylation of RIPK1 promoter.
Figure 4.

Promoter methylation correlates with RIPK1 expression in tumor samples. (A) RIPK1 CpG island methylation data from HNSCC patient paired normal solid tissue and primary solid tumors (n = 30) analyzed with Illumina Infinium HumanMethylation450 was obtained from The Cancer Genome Atlas. Graphic representations of the average beta values in normal versus primary samples for each of the CpG islands are provided (A, right). The beta value is the ratio of the methylated probe intensity and the overall intensity and therefore 1.0 (red) relates to higher methylation and 0 (green) to lower methylation. A significant difference was observed in one of the RIPK1 CpG islands −868 from the transcription start site (B). (C) Correlation between the change in methylation status at −868 from paired normal and primary samples to the matched change in expression between paired normal and primary samples (same patient TCGA methylation vs RNAseq2 data). Statistical values represent Pearson’s correlation coefficient using an n = 20 (df = 18) one-tailed test for negative correlation with a critical value of −0.468. PCI-15B metastatic cells were treated with an increasing concentration of the DNA methyltransferase inhibitor (5-aza-2′-deoxycytidine) and the mRNA and protein levels were quantitated by qRT-PCR (D) and immunoblotting (E), respectively. Bars represent the mean and standard error from triplicate readings of the representative blot provided in (E, upper panel)
Modulation of RIPK1 expression changes tumor-promoting properties
Our observation suggested that loss of RIPK1 expression during tumor progression promoted p(I):p(C)-mediated apoptosis, which is counterintuitive as one of the hallmarks of cancer is the resistance to apoptotic stimuli. Therefore, after establishing the mechanistic basis for the reduced RIPK1 expression in HNSCC tumors, we focused on its functional consequences related to tumorigenesis. First, we sought to determine the contribution of RIPK1 downregulation on cell migration. We used two approaches, a two-dimensional wound-healing assay (Figure 5A), and a three-dimensional transwell migration assay (Figure 5B) using the PCI-15A shRIPK1 cells. We observed that wound-healing was significantly more efficient in the PCI-15A shRIPK1 cells compared to the shCTRL cells indicating that decreased RIPK1 expression increases the migration of PCI-15A cells (Figure 5A). Similarly, when the same cell line (PCI-15A-shRIPK1) was used in a transwell migration assay, there was an enhanced migration of the PCI-15A-shRIPK1-cells through the fibronectin-coated membrane compared to the control cells (ECM). Another feature of the enhanced metastatic ability of cancer cells is resistance to anoikis, a form of programmed cell death that is induced in anchorage-dependent cells following detachment from the surrounding extracellular matrix. Using an anoikis-resistance assay, we found that compared to control cells, PCI-15A cells expressing shRNA to RIPK1 were less capable of reattaching and recovering after being cultured in an anoikis inducing environment (Figure 5C). These data again suggest that despite being more prone to p(I):p(C)-mediated apoptosis, reduced RIPK1 expression in HNSCC cells promotes several other protumorigenic properties of the cell.
Figure 5.

Modulation of RIPK1 expression changes tumor-promoting properties. (A) Cell migration of PCI-15A primary cell lines stably expressing RIPK1 shRNA in a 2D wound-healing assay. 2D migration was quantified as the measured (mm) difference between the scratched gap at 0 and at 24h (A, lower). Cell migration was also measured in these cells using a 3D transwell migration filter assay (B), where 3D migration was quantified based on the number of cells that migrated through a porous membrane coated with fibronectin. (C) PCI-15A shRIPK1 cells were more anoikis-resistant than control cells. Cells were plated in poly-HEMA coated wells for 48h to prevent attachment. After 48h, the cells were collected and transferred to uncoated plates to allow for reattachment and recovery. After 24 and 96h of recovery, the wells were fixed with methanol, stained with crystal violet and imaged.
Discussion
RIPK1 has been recently established as a multifunctional protein controlling both cell death pathways as well as inflammatory response (13,14,17). Here, we describe another important contribution of this protein in tumorigenesis. While examining the differences in toll-like receptor signaling between primary and metastatic HNSCC, we have shown previously that there is an enhanced apoptotic response to p(I):p(C) in HNSCC cells derived from metastatic tumors compared to the primary tumor. This differential p(I):p(C) sensitivity was linked to the inability of TLR3-mediated NF-κB activation in the respective cells. The metastatic tumor derived cells had reduced induction of NF-κB-target genes compared to primary tumor derived cells when stimulated with p(I):p(C). However, this was not due to the complete loss of NF-κB activity in these cells. Indeed upon pretreatment with IL-1β prior to p(I):p(C) stimulation activated NF-κB in these cells leading to the rescue of the p(I):p(C)-mediated apoptosis phenotype (9). RIPK1 is an important adaptor for TLR3-pathway, where it is involved only in the NF-κB activation in a kinase activity independent manner. We therefore compared the expression changes of RIPK1 in cell lines and tumor samples to gain further mechanistic insight in this phenomenon. Our results show that a loss of RIPK1 expression during tumorigenesis, specifically in metastatic tumor derived cells, is responsible for the enhanced apoptosis by p(I):p(C) treatment. We found that reduction in RIPK1 expression during tumorigenesis is due to the promoter methylation at specific sites, which can be reversed by the treatment with DNA methyltransferase inhibitor. Based on the ENCODE ChIP-seq results the specific site, −868 at the RIPK1 promoter is occupied by the transcription factor ARID3A in various cell lines. It is therefore possible that the enhanced methylation at −868 found in tumor cells reduces ARID3A binding to RIPK1 promoter resulting in reduced RIPK1 transcription. Taken together our results reveal several important aspects of RIPK1 function in the context of tumorigenesis. First, our in vivo and in vitro data suggests despite promoting p(I):p(C)-mediated cell death, downregulation of RIPK1 is correlated with tumor progression. Second, ectopic downregulation of RIPK1 enhances pro-tumorigenic properties of cells, suggesting its crucial function in maintaining cellular integrity. Third, the differential levels of RIPK1 in various stages of tumor progression may significantly influence the outcome of prosurvival NF-κB signaling, which is prominently activated in various tumors.
Double-stranded RNAs have been used as potent adjuvants to chemotherapeutics by triggering growth arrest and apoptosis in cancer cells (5–7,24). However, there have been limitations in its efficacy to eliminate cancer in certain patients thus studies have been initiated to understand the molecular basis. Interestingly, in these studies, TLR3 expression itself has been suggested as a potential biomarker for treatment as its expression was shown to positively correlate with the efficacy of p(I):p(C) to eliminate breast cancer and human neuroblastoma (5). More relevant to HNSCC, recently it was been shown that TLR3 agonists, in the presence of IAP (inhibitor of apoptosis proteins) inhibitors, can have cooperative cytotoxic effects and limit cell growth of nasopharyngeal carcinomas (6). The positive effects of p(I):p(C) treatment have also been shown to inhibit cell proliferation and induce apoptosis in oral squamous cell carcinoma (7). In this context, our finding that the expression levels of RIPK1 controls the apoptotic response to p(I):p(C), might provide an important indicator for its in vivo efficacy. The expression levels of RIPK1 in tumor cells can potentially be used as a novel biomarker to determine the p(I):p(C) treatment efficacy.
In summary, our results suggest that epigenetic changes during tumor progression promotes downregulation of RIPK1 expression allowing the tumor cell to evade anoikis, and may contribute to tumorigenesis by enhancing the ability of the tumor cell to migrate. However, because of the dual role RIPK1 plays in augmenting both cell death and survival signaling, these findings could suggest a therapeutically exploitable means to provide better efficacy for the p(I):p(C) adjuvant therapy.
Funding
This work was supported in part by AI118896 from NIAID/NIH (S.N.S.). K.D.M. was supported by the National Institutes of Health training grant (T32AI049820). This project used the UPCI core facilities and was supported in part by award P30CA047904.
Acknowledgements
We thank Sandra Gibson for her assistance in our acquisition of the HNSCC cell lines. Additionally, we would like to thank Drs. Kathy Shair and Laura Wasil for their assistance with the anoikis-recovery assay, Dr. Uma Chandran for her suggestions on data mining and Drs. Adriana Forero, Rolando Cuevas and Jianzhong Zhu for their expert advice on protocols.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- HNSCC
head and neck squamous cell carcinomas
- NF-κB
nuclear Factor κB
- P(I):p(C)
polyinosinic acid:polycytidylic acid
- TLR3
toll-like receptor 3
References
- 1. Leemans C.R., et al. (2011) The molecular biology of head and neck cancer. Nat. Rev. Cancer, 11, 9–22. [DOI] [PubMed] [Google Scholar]
- 2. Patel A.N., et al. (2012) Treatment of recurrent metastatic head and neck cancer: focus on cetuximab. Clin. Med. Insights Ear Nose Throat, 5, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salaun B., et al. (2011) TLR3 as a biomarker for the therapeutic efficacy of double-stranded RNA in breast cancer. Cancer Res., 71, 1607–1614. [DOI] [PubMed] [Google Scholar]
- 4. Sharma S., et al. (2013) TLR3 agonists and proinflammatory antitumor activities. Expert Opin. Ther. Targets, 17, 481–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu W.M., et al. (2013) Toll-like receptor 3 expression inhibits cell invasion and migration and predicts a favorable prognosis in neuroblastoma. Cancer Lett., 336, 338–346. [DOI] [PubMed] [Google Scholar]
- 6. Vérillaud B., et al. (2012) Toll-like receptor 3 in Epstein-Barr virus-associated nasopharyngeal carcinomas: consistent expression and cytotoxic effects of its synthetic ligand poly(A:U) combined to a Smac-mimetic. Infect. Agent. Cancer, 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park J.H., et al. (2012) Poly I:C inhibits cell proliferation and enhances the growth inhibitory effect of paclitaxel in oral sqaumous cell carcinoma. Acta Odontologica Scandinavica, 70, 241–245. [DOI] [PubMed] [Google Scholar]
- 8. Chen L., et al. (2012) TLR3 dsRNA agonist inhibits growth and invasion of HepG2.2.15 HCC cells. Oncol. Reports, 28, 200–6. [DOI] [PubMed] [Google Scholar]
- 9. Umemura N., et al. (2012) Defective NF-kappaB signaling in metastatic head and neck cancer cells leads to enhanced apoptosis by double-stranded RNA. Cancer Res., 72, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meylan E., et al. (2004) RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol., 5, 503–507. [DOI] [PubMed] [Google Scholar]
- 11. Hsu H., et al. (1996) TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity, 4, 387–396. [DOI] [PubMed] [Google Scholar]
- 12. Stanger B.Z., et al. (1995) RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell, 81, 513–523. [DOI] [PubMed] [Google Scholar]
- 13. Dillon C.P., et al. (2014) RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell, 157, 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rickard J.A., et al. (2014) RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell, 157, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 15. Lin C.J., et al. (2007) Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck, 29, 163–188. [DOI] [PubMed] [Google Scholar]
- 16. Jie H.B., et al. (2015) CTLA-4⁺ regulatory T cells increased in cetuximab-treated head and neck cancer patients suppress NK cell cytotoxicity and correlate with poor prognosis. Cancer Res., 75, 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meerbrey K.L., et al. (2011) The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA, 108, 3665–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu J., et al. (2010) High-throughput screening for TLR3-IFN regulatory factor 3 signaling pathway modulators identifies several antipsychotic drugs as TLR inhibitors. J. Immunol., 184, 5768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Network C.G.A. (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 517, 576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamarajan P., et al. (2012) Receptor-interacting protein (RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and tumorigenesis. Cancer, 118, 5800–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hur G.M., et al. (2006) The death domain kinase RIP has an important role in DNA damage-induced, p53-independent cell death. J. Biol. Chem., 281, 25011–25017. [DOI] [PubMed] [Google Scholar]
- 22. Ramnarain D.B., et al. (2008) RIP1 links inflammatory and growth factor signaling pathways by regulating expression of the EGFR. Cell Death Differ., 15, 344–353. [DOI] [PubMed] [Google Scholar]
- 23. Sharma S., et al. (2010) Epigenetics in cancer. Carcinogenesis, 31, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calzolari A., et al. (1997) Immunohistochemical vs molecular biology methods. Complementary techniques for effective screening of p53 alterations in head and neck cancer. Am. J. Clin. Pathol., 107, 7–11. [DOI] [PubMed] [Google Scholar]


