Supplemental Digital Content is available in the text
Keywords: body mass index, cytotoxic T-lymphocyte-associated antigen 4, homeostatic model assessment for insulin resistance, polycystic ovary syndrome, single nucleotide polymorphism
Abstract
The autoimmune and gene etiology are implicated in the pathogenesis of polycystic ovary syndrome (PCOS). The cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) is important for negative regulation of T-cell activation, and CTLA4 gene has been identified as a risk factor for some autoimmune diseases. However, none studies have been performed about the association between PCOS and the CTLA4 gene before. Here, we aimed to investigate the association of CTLA4 with PCOS in the Chinese Han population though a case–control association analysis of 606 individuals. The tagging variants rs733618 and rs231775 in the CTLA4 gene were detected using polymerase chain reaction-denaturing gradient gel electrophoresis method. Further analysis found the rs733618 was significantly different between case and control groups in either genotypic or allelic distribution (P = .01 and .009, respectively) while rs231775 not. Moreover, rs733618 was significantly associated with higher body mass index in the dominant model (P = .003) and with higher waist/hip ratio in the recessive model (P = .02). Interestingly, rs733618 was only found to have significant association with homeostatic model assessment for insulin resistance (HOMA-IR) in both dominant and recessive model (P = .009 and .0065, respectively). This is the first study to investigate the association of CTLA4 gene with PCOS. The CTLA4 gene is suggested to correlated with PCOS, and influence PCOS through regulating obesity and the HOMA-IR in a novel way.
1. Introduction
Polycystic ovary syndrome (PCOS) is the most prevalent metabolic disease with various symptoms affecting about 10% women of reproductive age worldwide.[1] The clinical features of PCOS are polycystic ovarian morphology, hyperinsulinemia, and hyperandrogenism.[2] Patients with PCOS may succumb to overweight and potential health risks of type II diabetes mellitus and female subfertility, etc.[3,4] Till now the etiology of PCOS has yet to be determined, although it is believed to be hereditary in nature and caused by multiple factors.[5] Since the onset of PCOS is characterized by low-grade inflammation and detected auto-antibodies,[6,7] genetic factors interacting with immunologic factors are intriguing to be implicated in the pathogenesis of this syndrome.
Recently, both mouse models and human study results demonstrated that reduced capability of Treg generation may possibly lead to PCOS.[8] And cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) is a specific marker of Treg and can compete with CD28 to bind to CD80 and CD86 on antigen-presenting cells, such as macrophages. Through blocking the engagement of CD28 on T cells, CTLA4 can prevent T-cell activation, and thereby reducing the levels of interferon-γ, interleukin (IL)-6, and IL-1β.[9] Therefore, loss of CTLA4 in natural Treg cells may possibly affect the immune response and occurrence of PCOS. The complete CTLA4 gene is located on chromosome 2q33.2 spans chr2: 203,867,787-203,873,959 (GRCh38) and contains 4 exons. CTLA4 is considered as a possible risk gene for many T-cell-mediated autoimmune diseases, such as rheumatoid arthritis.[10] However, the roles of CTLA4 gene in the PCOS are yet to be known.
Thus, to understand the relationship of the CTLA4 gene with PCOS, a case–control association study is performed. Here, polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) method was used to genotype the single nucleotide polymorphisms (SNPs) in the CTLA4 gene within the Chinese Han ethnic patients with PCOS and healthy population. Furthermore, the association of CTLA4 gene with PCOS or the clinical features of PCOS was analyzed.
2. Materials and methods
2.1. Patients
Totally 294 patients with PCOS were recruited from gynecology clinic of the First Affiliated Hospital of Xinjiang Medical University from January 2015 to December 2016. The diagnosis was based on the 2003 Rotterdam consensus criteria.[11] All recruited patients had 2 items of the following: clinical or biochemical hyperandrogenism, that is, hirsutism, acne, or the value of testosterone >68 ng/dL; more than 10 ovarian basal follicles; and a history of menstrual disorders, that is, anovulatory infertility, amenorrhea/oligomenorrhea, or frequent menstruation. Patients with other hyperandrogenism-related diseases, such as hypothyroidism, androgen-secreting tumors, and congenital adrenal hyperplasia, etc, were excluded. A total of 312 healthy women who had body examination at the same gynecology clinic during the same period were recruited as controls. Those healthy women had no hypertension or family history of disorders, such as diabetes. All participants were Chinese Han ethnic without thyroid abnormalities, cardiovascular system disorders, hepatic or renal dysfunction, and other endocrine metabolic diseases. The medical ethical committee of the First Affiliated Hospital of Xinjiang Medical University approved the study and all the participants in the project provided written informed consent.
2.2. SNP selection and genotyping analysis
The criteria of a minor allele frequency ≧0.05 and r2≧0.80 in the Chinese Han population were used to select tagging SNPs in the CTLA4 gene based on the 1000 genomes SNP databases (http://www.1000genomes.org). Those selected potentially functional SNPs were located within 2 kb upstream of the 5′ untranslated region and 2 kb downstream of the 3′ untranslated region of CTLA4 gene.
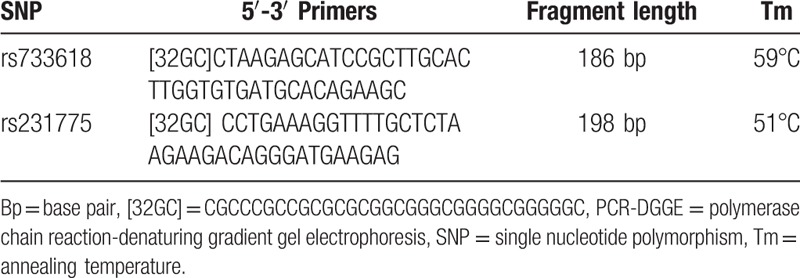
A total of 2 mL EDTA anti-coagulated blood sample was drawn from each participant to isolate and store genomic DNA using the previous methods.[12,13] The 50 μL PCR mixture containing 1.5 mmol/L magnesium chloride, 200 μmol/L for each deoxyribonucleotide triphosphate, 5 μg template DNA, 100 pmol/L specific DGGE primer pairs (Table 1), and 2.5 U Taq DNA polymerase (Promega, Fitchburg, WI) was processed under 94°C for 5 minutes, 30 cycles including 95°C for 1 minute, annealing temperature for each SNP for 1 minute and 72°C for 1 minute; and then 72°C for 10 minute.
Table 1.
PCR-DGGE primers for each detected SNP.
The Bio-Rad Decode system was used to perform DGGE to genotype polymorphisms of these 2 variants in CTLA4 gene according to the general methods.[10] Briefly, 30 μL PCR products containing loading buffer were used for the gel electrophoresis under the conditions of 60°C, 1×Tris-borate-EDTA buffer and 130 voltage. The denaturing gradience is 20% to 40% and 10% to 40%, and electrophoresis time is 7 and 6 hours for rs733618 and rs231775, respectively. At the end of electrophoresis, the gels were stained with ethidium bromide and photographed under an ultraviolet transilluminator (Fig. 1). The PCR products with special gel staining patterns were purified for the Sanger sequencing. The genotype for either variant was identified by DGGE gel staining pattern.
Figure 1.
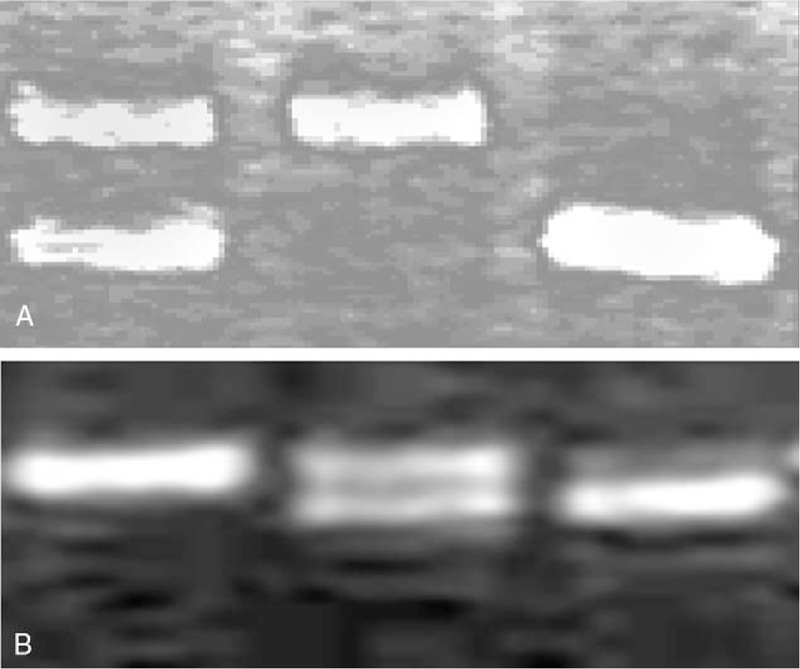
Denaturing gradient gel electrophoresis gel images of rs733618 (A) and rs231775 (B) genotyping results. For each image, there are 3 lanes, that is, lane 1, 2, and 3 from left to right. Lane 1 indicated TC and AA for rs733618 and rs231775, respectively; lane 2 indicated TT and GA; and lane 3 indicated CC and GG.
2.3. Clinical tests
Anthropometric measurements for every participant were taken after participants refrained from exercise and food for at least 3 hours. Body weight in minimal clothes and without shoes and height was measured with a height and weight measurement instrument. Waist and hip circumferences were measured with an anthropometric tape. The waist/hip ratio (WHR) and body mass index (BMI) were calculated based on the following formula: WHR = Waist, cm/Hip, cm and BMI, kg/m2 = Weight, kg/(Height, m × Height, m). Along with the anthropometric measurements, the menstrual and fertility history of participants were recorded. Blood pressure was measured with a traditional sphygmomanometer.
About 5 mL blood were extracted from the veins of each participant between 8:00 am to 9:00 am after an overnight fast during the 3rd to 5th days of one menstrual cycle or during any day of the enstruation cycle for amenorrhea women. The total cholesterol (TC), triglycerides (TG), plasmaestradiol (E2), total testosterone (T), luteinizing hormone (LH), and follicle-stimulating hormone (FSH) were measured by Axsym Automated Immunoassay Analyzer (Abbott, IL) according to our previous study.[14] Fasting plasma insulin was measured by an insulin human direct enzyme-linked immunosorbent assay kit (Invitrogen, London, UK), and glucose was detected by the hexokinase method. The homeostatic model assessment for insulin resistance (HOMA-IR) was calculated based on the following formula: HOMA - IR, mmol⋅mIU/L2 = (Fasting insulin, mmol/L × Fasting glucose, mIU/L)/22.5.
2.4. Statistical analysis
The difference of the clinical characteristics between patients with PCOS and healthy women was detected using a Mann–Whitney test or Student t test. The genotypes of rs231775 and rs733618 in CTLA4 were calculated by manual counting. Hardy–Weinberg equilibrium (HWE) for each SNP was confirmed using Fisher exact test. The Power and Sample Size Program was used to calculate the power. A Cochran–Armitage trend test or Chi-squared test was used to detect the difference of the genotypic or allelic distribution between patients with PCOS and healthy controls.[15,16] The software THESIAS was used to perform the haplotype analysis.[12,17] The global test for the association of the haplotypes with PCOS was the (n − 1)df test. Here, n refers to the number of haplotypes. Odds ratios (ORs), 95% confidence interval (CI) and other statistical analyses were implemented using the R software version 3.1.2.
3. Results
Based on the 1000 genomes project database, 16 CTLA4 SNPs from 103 Chinese Han population met the general criteria and constructed 2 linkage blocks (Fig. S1). From each block, 1 tag SNP was selected, that is, rs231775 and rs733618. The rs231775 is also known as +49A/G which causes A-G transition polymorphism in exon 1 of the CTLA4 gene and results in a Thr to Ala (T17A) substitution at the 17th site of the protein. The -1722 (T/C) variant is the alternative name of rs733618, which is located in the promoter region of CTLA4 genes and allele T may increase CTLA4 gene transcription level.[18]
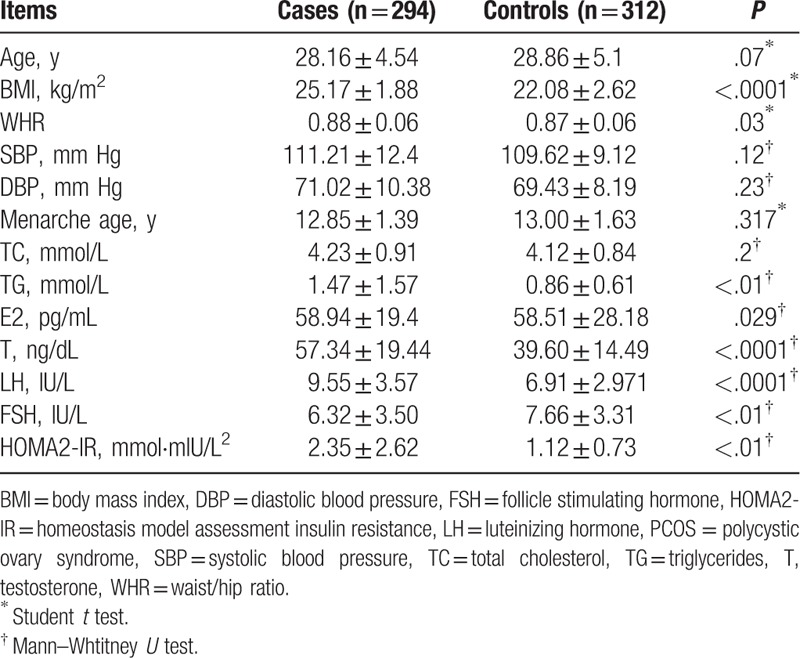
The clinical characteristics of all 606 participants are presented in Table 2. Among them, 294 patients with PCOS and 312 healthy women had no significant difference in age, menarche age, blood pressure, and total cholesterol. However, the mean levels of BMI, WHR, LH/FSH, E2, LH, T, and HOMA-IR were significantly decreased in healthy women than those in patients with PCOS.
Table 2.
The basic clinical data of patients with PCOS and healthy controls.
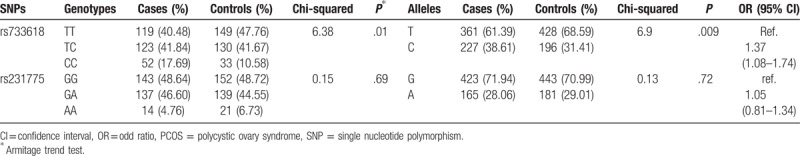
The distributions of genotype/allele frequency in both patients with PCOS and controls are shown in Table 3. The genotype frequencies for both rs231775 and rs733618 in CTLA4 gene in the control group have no significant deviation from HWE (P = .17 and .60, respectively). The rs733618 has significant difference between the patients with PCOS and the controls in both the genotypic and allelic frequency distributions (P = .011 and .0086, respectively), while the rs231775 has not (P = .69 and .72, respectively). Furthermore, the rs733618 may play roles on PCOS in recessive model (Table S1). Power analysis results suggested that the present study sample size of 294 patients and 312 controls had about 81% power to detect associations in both the genotypic and allelic models under the minor allele with about 3% frequency, an OR of 1.5, and a threshold of significance at P = .05.
Table 3.
Genotypes and alleles frequencies of rs231775 and rs733618 polymorphisms among PCOS and control groups.
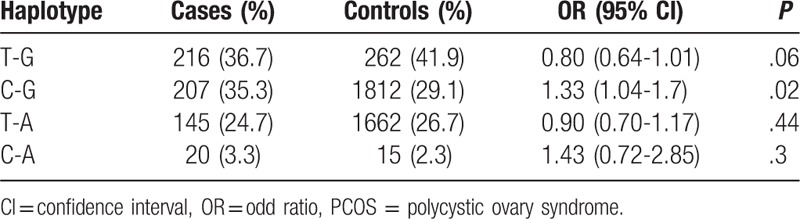
Haplotypes built up with these 2 SNPs are listed in Table 4. The value r2 for 2 variants of rs231775 and rs733618 is 0.103. Haplotype association analysis showed that CTLA4 were not significantly associated with PCOS (P-global = .064). However, the rs733618 to rs231775 haplotype of C-G had significant effects on PCOS (OR = 1.33, 95% CI 1.04–1.7, P = .02).
Table 4.
Analysis of haplotype constructed by rs733618-rs231775 in patients with PCOS and control individuals.
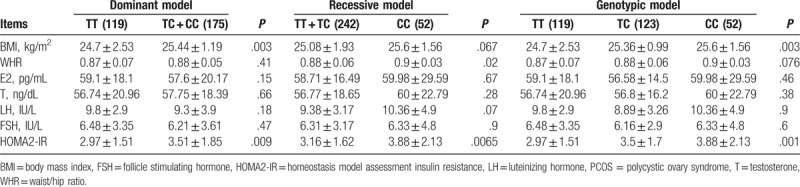
Furthermore, we analyzed the association of the rs733618 in CTLA4 gene with clinical parameters in the patients with PCOS using dominant and recessive genetic models (Table 5). Both WHR and BMI are obesity indices suggested by World Health Organization (WHO). A criterion for overweight is BMI ≥ 25 kg/m2 and WHR ≥ 0.8. Our study suggests that the patients with PCOS containing the allele C of rs733618 are more obese than those who do not contain the allele C since the rs733618 C allele is significantly associated with higher BMI in the dominant model (P = .003). Moreover, rs733618 C allele is significantly associated with higher WHR in the recessive model (P = .02). However, the allele C of rs733618 was found to have no significantly different distribution of the levels of testosterone, FSH, LH, and E2 (P > .05) in both dominant and recessive models, although it is significantly associated with higher HOMA2-IR in both models.
Table 5.
Relationship between rs733618 and key phenotypic traits in PCOS women.
4. Discussion
The PCOS is a complex endocrine syndrome resulting from the interaction of genes and environmental factors. And an autoimmune etiology is suggested to be the pathogenesis of PCOS,[1] the proinflammatory state is involved in the progress of PCOS, chronic low-grade inflammation was reported to promote the development of ovarian and metabolic dysfunction in PCOS. Moreover, reduced capability of Treg generation has been found to lead to PCOS in humans and estrogen exposed PCOS mouse model.[8] CTLA4, a specific Treg markers, is an important costimulatory molecule indulged in playing suppressive function of Treg cells. Therefore, CTLA4 is supposed to affect the immune response and play roles in the pathogenesis of PCOS. However, the association of CTLA4 and PCOS has not been studied before.
To our knowledge, the present study is the first one to determine the relationship of CTLA4 gene with PCOS in the Chinese Han population. Here, we investigated the CTLA4 gene structure constructed by genotyped SNPs in 1000 genome project database and selected 2 tags SNPs of rs231775 and rs733618, which are potential functional SNPs. Then, we used PCR-DGGE to genotype these 2 tag SNPs in 294 patients and 312 controls. PCR-DGGE is a more efficient and less cost method to detect various variants in a DNA fragment compared with other methods, such as TaqMan.[10] This method has been widely used to analyze the known and unknown gene mutation among different biology samples.[19]
Furthermore, through a case–control association analysis of the 2 SNPs with PCOS, it was found that the variant rs733618 instead of rs231775 was significantly associated with PCOS. And the rs733618 may play roles on PCOS in recessive model. Although the global haplotypes constructed by these two SNPs in CTLA4 were not significantly associated with PCOS (P-global = .064), the rs733618-rs231775 haplotype of C-G had significant effects on PCOS (OR = 1.33, 95% CI 1.04–1.7, P = .02).
Interestingly, the variant rs733618 was found to be significantly associated with higher BMI in the dominant model and with WHR in the recessive model. These results support that CTLA4 is linked to obesity. A recent mouse model study demonstrates that the treatment of CTLA4Ig, a fusion protein of CTLA4, can decrease both adipose tissue weight and adipocyte size in high fat diet mice.[9] Since the allele C of rs733618 is significantly associated with overweight, it is inferred that the allele C of rs733618 may downregulate the expression of CTLA4. Under the lower expression of CTLA4, the body may be easier to increase adipose tissue weight and keep low grade of inflammatory state, which are the main clinical characters of PCOS. However, the variant rs733618 is not significantly associated with the levels of testosterone, FSH, LH, and E2 in the body, which suggests that there is no interaction between CTLA4 and hormonal factors. Moreover, other studies about the correlation between Treg percentage and hormonal variables (FSH, LH, estradiol, and T) obtained similar results that no significant correlation was found except LH and the number of Tregs is independent of any effects from steroid hormones.[8,20] Furthermore, the present study indicated the allele C of rs733618 in CTLA4 was significantly associated with the levels of HOMA-IR in both dominant and recessive models. The HOMA score is a good indicator of insulin resistance. Both obesity and insulin resistance are frequent events in hyperandrogenic women. It has been found that obesity leads to a decrease of anti-inflammatory Treg cells in adipose tissue with insulin resistance increased.[21,22] Furthermore, some potential therapeutic strategies of targeting Tregs are being made to treat type 2 diabetes associated with obesity.[23] These findings suggest that CTLA4 may be involved in the metabolic systems inducing obesity and insulin resistance through regulating T-cell activation. And moreover, insulin resistance is a crucial factor in the occurrence and development of PCOS. Current results suggest that CTLA4 may have an important role in the pathogenesis of PCOS.
The present study is the first preliminary analysis about the association of 2 functional tag SNPs in CTLA4 gene with PCOS and its main clinical characters. However, there are several limitations. Firstly, the PCOS diagnosis in the present study is based on the 2003 Rotterdam consensus instead of a new criterion, that is, the Androgen Excess Society guideline. The main difference between these 2diagnosis criteria is that hyperandrogenism is necessary for the later and unnecessary for the former since for the 2003 Rotterdam consensus 2 items are required from 3 conditions: clinical and/or biochemical hyperandrogenism, oligoovulation or anovulation, and polycystic ovaries while for AES guidance, both clinical and/or biochemical hyperandrogenism and ovarian dysfunction and/or polycystic ovaries are required. Thus, some patients without hyperandrogenism were recruited in the present study. However, these patients did really have polycystic ovaries and should not be excluded. Secondly, Treg level or some immunologic markers, such as CTLA4 protein level, are not measured. Finally, here only 2 tagging SNPs of CTLA4 are evaluated in the Chinese Han population, and a comprehensive analysis of other SNPs across different ethnic population is not performed. Therefore, further extensive studies on the effects of CTLA4 on PCOS involving gene sequencing data in different ethnic populations may help to understand the role of CTLA4 in the pathogenesis of PCOS, and to identify some potential variants as biomarkers of PCOS.
5. Conclusion
In the present study, the CTLA4 gene is suggested to be involved in the genetic predisposition to the PCOS. The variant rs733618 requires more evidence to validate to be a potential biomarker for PCOS, since it is not only significantly associated with PCOS but also with BMI in the dominant model, WHR in the recessive model and HOMA-IR in both models. It is supported that CTLA4 may be involved in the metabolic systems inducing obesity and insulin resistance through regulating T-cell activation. Further biochemistry and cellular biology and animal model studies on the CTLA4 are required to gain more insights on the effects of CTLA4 on PCOS.
Acknowledgments
The authors thank the anonymous scientists for their helpful suggestions and all participants in this study.
Author contributions
Conceptualization: Lei Zhao, Rong Ma.
Methodology: Jing Su, Yan Li.
Resources: Ting Qiu.
Software: Guanglong Su.
Writing – original draft: Jing Su, Yan Li.
Writing – review & editing: Lei Zhao, Jing Wang.
Supplementary Material
Footnotes
Abbreviations: AES = the Androgen Excess Society, BMI = body mass index, CI = confidence interval, CTLA4 = cytotoxic T-lymphocyte-associated antigen 4, DBP = diastolic blood pressure, FSH = follicle-stimulating hormone, HOMA-IR = homeostatic model assessment for insulin resistance, HWE = Hardy–Weinberg equilibrium, IL = interleukin, LH = luteinizing hormone, OR = odds ratios, PCOS = polycystic ovary syndrome, PCR-DGGE = polymerase chain reaction-denaturing gradient gel electrophoresis, SBP = systolic blood pressure, SNPs = single nucleotide polymorphisms, T = testosterone, TC = total cholesterol, TG = triglycerides, WHR = waist/hip ratio.
This article does not contain any studies with animals performed by any of the authors. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent was obtained from all individual participants.
Jing Su and Yan Li contributed equally to this work (JS performed the genotyping analysis, YL performed the statistics analysis).
This study was funded by the natural science foundation of Xinjiang Uygur autonomous region (no: 2015211C066).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Franks S. Polycystic ovary syndrome. New Eng J Med 1995;333:853–61. [DOI] [PubMed] [Google Scholar]
- [2].Group TREA-SPCW. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19–25. [DOI] [PubMed] [Google Scholar]
- [3].Huang G, Coviello A. Clinical update on screening, diagnosis and management of metabolic disorders and cardiovascular risk factors associated with polycystic ovary syndrome. Curr Opin Endocrinol Diabetes Obes 2012;19:512–9. [DOI] [PubMed] [Google Scholar]
- [4].Shang K, Jia X, Qiao J, et al. Endometrial abnormality in women with polycystic ovary syndrome. Reprod Sci 2012;19:674–83. [DOI] [PubMed] [Google Scholar]
- [5].Ehrmann DA. Polycystic ovary syndrome. New Eng J Med 2005;352:1223–36. [DOI] [PubMed] [Google Scholar]
- [6].Repaci A, Gambineri A, Pasquali R. The role of low-grade inflammation in the polycystic ovary syndrome. Mol Cell Endocrinol 2011;335:30–41. [DOI] [PubMed] [Google Scholar]
- [7].Palacio JR, Iborra A, Ulcova-Gallova Z, et al. The presence of antibodies to oxidative modified proteins in serum from polycystic ovary syndrome patients. Clin Exp Immunol 2006;144:217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Krishna MB, Joseph A, Subramaniam AG, et al. Reduced Tregs in peripheral blood of PCOS patients - a consequence of aberrant Il2 signaling. J Clin Endocrinol Metab 2015;100:282–92. [DOI] [PubMed] [Google Scholar]
- [9].Fujii M, Inoguchi T, Batchuluun B, et al. CTLA-4Ig immunotherapy of obesity-induced insulin resistance by manipulation of macrophage polarization in adipose tissues. Biochem Biophys Res Commun 2013;438:103–9. [DOI] [PubMed] [Google Scholar]
- [10].Lei C, Dongqing Z, Yeqing S, et al. Association of the CTLA-4 gene with rheumatoid arthritis in Chinese Han population. Eur J Hum Genet 2005;13:823–8. [DOI] [PubMed] [Google Scholar]
- [11].Rotterdam EA-SPCWG. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19–25. [DOI] [PubMed] [Google Scholar]
- [12].Cai L, Deng SL, Liang L, et al. Identification of genetic associations of SP110/MYBBP1A/RELA with pulmonary tuberculosis in the Chinese Han population. Hum Genet 2013;132:265–73. [DOI] [PubMed] [Google Scholar]
- [13].Jiang SY, Li LL, Yue J, et al. The effects of SP110's associated genes on fresh cavitary pulmonary tuberculosis in Han Chinese population. Clin Exp Med 2016;16:219–25. [DOI] [PubMed] [Google Scholar]
- [14].Li S, Zhao L, Wan XH. A missense variant rs4645843 in TNF-alpha gene is a risk factor of polycystic ovary syndrome in the uygur population. Tohoku J Exp Med 2017;243:95–100. [DOI] [PubMed] [Google Scholar]
- [15].Cai L, Yuan W, Zhang Z, et al. In-depth comparison of somatic point mutation callers based on different tumor next-generation sequencing depth data. Sci Rep 2016;6:36540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang T, Liu CL, Li LL, et al. A new method for identifying causal genes of schizophrenia and anti-tuberculosis drug-induced hepatotoxicity. Sci Rep 2016;6:32571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tregouet DA, Escolano S, Tiret L, et al. A new algorithm for haplotype-based association analysis: the stochastic-EM algorithm. Ann Hum Genet 2004;68(Pt 2):165–77. [DOI] [PubMed] [Google Scholar]
- [18].Perez-Garcia A, Osca G, Bosch-Vizcaya A, et al. Kinetics of the CTLA-4 isoforms expression after T-lymphocyte activation and role of the promoter polymorphisms on CTLA-4 gene transcription. Hum Immunol 2013;74:1219–24. [DOI] [PubMed] [Google Scholar]
- [19].Turaki AA, Bomer M, Silva G, et al. PCR-DGGE analysis: unravelling complex mixtures of badnavirus sequences present in yam germplasm. Viruses 2017;9:E181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chapman JC, Min SH, Freeh SM, et al. The estrogen-injected female mouse: new insight into the etiology of PCOS. Reprod Biol Endocrinol 2009;7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Priceman SJ, Kujawski M, Shen S, et al. Regulation of adipose tissue T cell subsets by Stat3 is crucial for diet-induced obesity and insulin resistance. Proc Natl Acad Sci U S A 2013;110:13079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhong J, Rao X, Braunstein Z, et al. T-cell costimulation protects obesity-induced adipose inflammation and insulin resistance. Diabetes 2014;63:1289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Becker M, Levings MK, Daniel C. Adipose-tissue regulatory T cells: critical players in adipose-immune crosstalk. Eur J Immunol 2017;47:1867–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


