Abstract
p53 has been implicated in the pathophysiology of Huntington’s disease (HD). Nonetheless, the molecular mechanism of how p53 may play a unique role in the pathology remains elusive. To address this question at the molecular and cellular biology levels, we initially screened differentially expressed molecules specifically dependent on p53 in a HD animal model. Among the candidate molecules, wild-type p53-induced gene 1 (Wig1) is markedly upregulated in the cerebral cortex of HD patients. Wig1 preferentially upregulates the level of mutant Huntingtin (Htt) compared with wild-type Htt. This allele-specific characteristic of Wig1 is likely to be explained by higher affinity binding to mutant Htt transcripts than normal counterpart for the stabilization. Knockdown of Wig1 level significantly ameliorates mutant Htt-elicited cytotoxicity and aggregate formation. Together, we propose that Wig1, a key p53 downstream molecule in HD condition, play an important role in stabilizing mutant Htt mRNA and thereby accelerating HD pathology in the mHtt-p53-Wig1 positive feedback manner.
Introduction
Since the causal gene Huntingtin (Htt) was identified (1), many efforts have been made to identify drug target for Huntington’s disease (HD). At present, mutant allele-specific targetting of Htt is a hot topic in the translational studies (2–12).
In the nucleus, mutant Htt with expanded polyQ aberrantly interacts with several nuclear proteins and disturbs their functions (13,14). Among these nuclear proteins that interact with Htt, roles for p53 in HD pathology have been explored in several independent laboratories (15–18). p53 is overrepresented in HD animal models and patient brains, and depletion of p53 ameliorates cellular pathology and behavioral deficits in an HD animal model (16). Therefore, p53 and related pathways may be drug targets for HD. However, exploring good targets in the pathways except for p53 itself will be realistic, as p53 is involved in so many vital cell functions (19,20).
In this study, we focus on cell biological and biochemical approaches to identify p53 downstream molecule(s) that can be a target for drug discovery in HD. Among such downstream molecules, we focus the RNA binding protein Wig1 (21). Wig1 is abundantly expressed in the brain and is believed to be neuronal (21–23), especially located in the nucleus (24). We report that Wig1 is overrepresented in HD cell and animal models as well as patient brains. Furthermore, Wig1 stabilizes mutant Htt mRNA to a greater extent than wild-type Htt, and mediates mutant Htt-elicited cytotoxicity and aggregate formation.
Results
Genes involving the p53 pathway are aberrantly expressed in HD models and patient brains
We previously showed that genetic deletion of p53 ameliorates pathological and behavioral deficits of a transgenic (Tg) animal model for HD [Htt-N171-82Q-Tg mice in which an N-terminal fragment of mutant Htt (amino acids 1–171) with expanded 82 polyQ stretch is overexpressed (25), designated mHD in this study] by crossbreeding with p53 knockout mice (KO). Thus, to address a crucial mediator for HD pathology in the p53 pathway, we compared gene expression profile of the forebrain from wild-type (wt), mHD, KO and the crossbred animals (mHK) with a mini-array including 113 key genes involving the p53 pathway (Fig. 1A). First, we selected the genes whose expression is upregulated at least 1.5-fold in mHD compared with wt, and downregulated at least 1.3-fold in mHK compared with mHD (Fig. 1B). Among 26 out of 113 genes under the criteria, we paid further attention to eight genes whose promoters contain p53-binding site(s) implicating direct influence of p53 on the targets (Fig. 1B) (26–33). These targets are: Bax (Bcl2-associated X protein); Bbc3/Puma (Bcl2 binding component 3/p53-upregulated modulator of apoptosis); Wig1 [wild-type p53-induced gene1; in mouse called as Zmat3 (zinc finger matrin type 3)]; Sesn2 [Sestrin 2; also called Hi95 (Hypoxia-induced gene 95)]; Mdm2 (mouse double minute 2); Hif1-α (Hypoxia inducible factor 1, alpha subunit); Cx3cl1 (Chemokine C-X3-C motif ligand 1) and Pten (Phosphatase and tensin homolog).
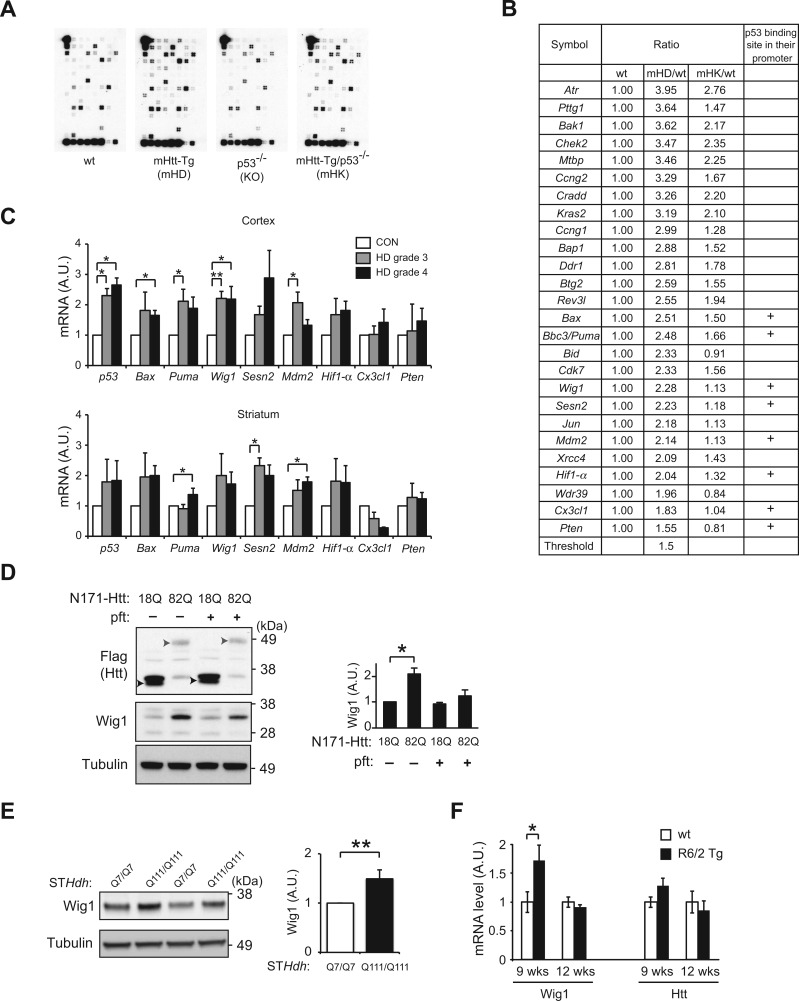
Figure 1.
Screening of p53 downstream targets in HD using p53 microarray. (A) Differential expression of genes involved in the p53 signaling pathways among wild-type (wt), mHD, KO and mHK mice by using the Oligo GEArray for mouse p53 signaling pathway. Array images are shown. (B) Comparison of gene expression profiles between wt, mHD and mHK mice at 5 months old reveal a group of genes which show significantly different expression levels compared with wt mice including eight p53 target genes carrying p53 binding site. (C) Quantitative RT-PCR results using cortex and striatum of HD patients (grades 3 and 4) and healthy controls (CTL) reveal significant upregulation of p53, Bax, Puma, Wig1 and Mdm2 in the cortex of HD patients, and Puma, Sesn2 and Mdm2 in the striatum of HD patients compared with control group (Grade 3, n = 3; Grade 4, n = 5; age-matched controls, n = 5). (D) Expression of Wig1 in HD cell model. Protein level of Wig1 is significantly higher in N171-Htt-82Q-expressing differentiated PC12 cells compared with N171-Htt-18Q-expressing PC12 cells, which was blocked by pifithrin-α (pft, 10 μM), a p53 inhibitor. Upper arrow head and lower arrow head indicate N171-Htt-82Q and N171-Htt-18Q, respectively. (E) Upregulation of Wig1 in striatal cells that express endogenous full-length mutant Htt. Protein level of Wig1 is significantly higher in the mutant Htt-expressing striatal cells (STHdhQ111/Q111) than wt Htt-expressing striatal cells (STHdhQ7/Q7). (F) Quantitative RT-PCR results using the striatum of R6/2 transgenic (Tg) and wt mice. Wig1 mRNA levels are upregulated in R6/2 Tg striatum at 9 weeks but not at 12 weeks of age, when compared with those in wt mice. No significant difference, but a trend of increase, is observed in the levels of mouse-derived endogenous Htt mRNA between Tg and wt mice. Data are represented as mean ± s.e.m. (*P < 0.05, **P < 0.01, Student’s t-test). See also Supplementary Material, Tables S1 and S2.
We next questioned whether the expression of p53 and these eight downstream molecules were altered in the brains of HD patients (Supplementary Material, Table S1). We observed a general trend supporting the upregulation of p53 and the target molecules in both cerebral cortex and striatum of HD patients (Fig. 1C and Supplementary Material, Table S2). Among them, p53, Bax and Wig1 were significantly higher in the cortex of HD, whereas Puma and Mdm2 were significantly elevated in the striatum of HD (Fig. 1C). We then selected Wig1 for further study, given that it is more preferentially expressed in the brain (21–23). Unfortunately, no commercial antibodies against Wig1 could be reliably used in autopsied brains due to significant background signals. Thus, we validated a possible upregulation of Wig1 at the protein level in a HD cell model (16). Wig1 protein expression was significantly augmented in neuronal PC12 cells expressing N171-Htt-82Q, which was blocked by pifithrin-α, a representative p53 inhibitor (Fig. 1D). We next examined the level of Wig1 by using the striatal cell lines expressing endogenous either full-length wt or mutant Htt (STHdhQ7/Q7 or STHdhQ111/Q111, respectively) (34). The level of Wig1 protein expression was significantly upregulated in STHdhQ111/Q111 cells compared STHdhQ7/Q7 cells (Fig. 1E). We next measured the expression levels of Wig1 and Htt in the striatum of the R6/2 transgenic (Tg) mice, a representative polyQ pathology model of HD. We observed significant upregulation of Wig1 in R6/2 brains at 9 weeks, but not 12 weeks of age, compared with controls (Fig. 1F). We observed an increase in the levels of mouse-derived endogenous Htt at 9 weeks in accordance with a significant upregulation of Wig1 at the same time point (Fig. 1F) but the increase was within a suggestive range.
Wig1 preferentially affects the levels of mutant Htt
To address a role for Wig1 in HD pathology, we knocked down Wig1 by using three independent shRNAs in PC12 cells (Supplementary Material, Fig. S1A). We then exogenously expressed either wt or mutant N-terminal Htt fragment (N171-Htt-18Q or -82Q) and tested how knockdown of Wig1 may affect the levels of these exogenous Htt. Interestingly, we observed a significant decrease in the level of Htt fragments, in particular mutant Htt (Fig. 2A and Supplementary Material, Fig. S1B). The absolute expression levels of exogenous N171-Htt-18Q and N171-Htt-82Q were comparable (Supplementary Material, Fig. S1C).
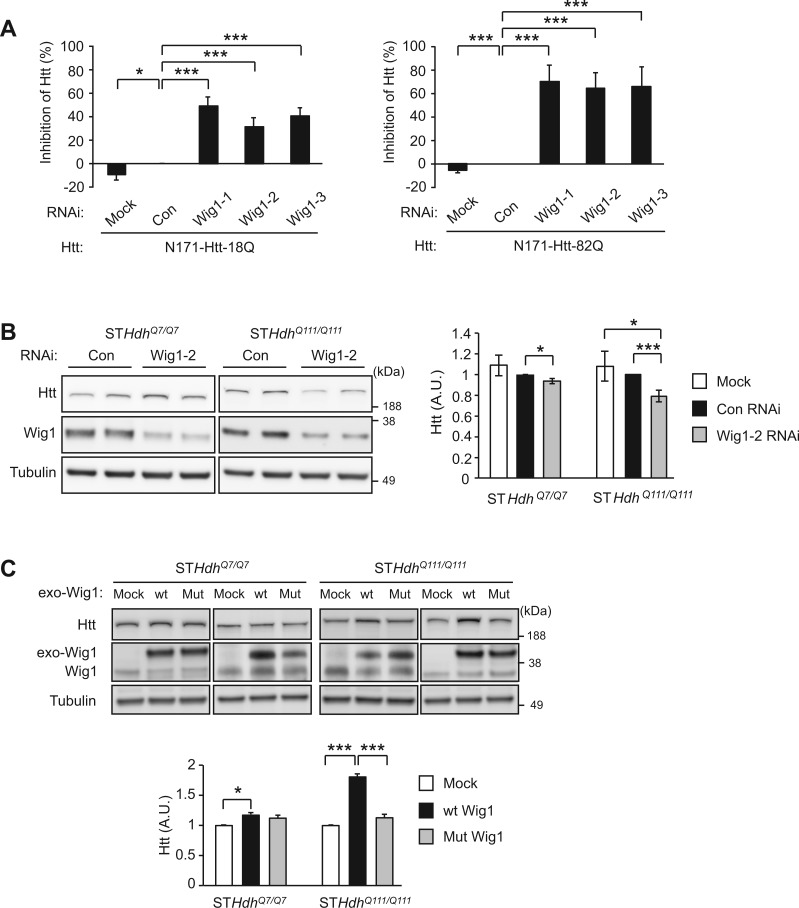
Figure 2.
Upregulation of Htt protein expression level by Wig1. (A) Knockdown of Wig1 in differentiated, either wt or mutant Htt fragment (N171-Htt-18Q or N171-Htt-82Q, respectively)-expressing PC12 cells results in significant decrease in Htt fragments at the protein level, in particular mutant Htt fragemnts. (B) Knockdown of Wig1 in striatal cell lines expressing either wt or mutant full-length Htt (STHdhQ7/Q7 or STHdhQ111/Q111, respectively) results in significant decrease in Htt protein level, in particular mutant Htt protein. (C) Overexpression of Wig1 in STHdhQ7/Q7 or STHdhQ111/Q111 striatal cell lines significantly augments the mutant Htt protein level, which is suppressed by expression of RNA binding-defective mutant Wig1 (Wig1-H88A). The effect of overexpressed wt Wig1 in STHdhQ7/Q7 cells on wt Htt level is lesser extent than that of mutant Htt. Data are represented as mean ± s.e.m. (*P < 0.05, ***P < 0.001, Student’s t-test). See also Supplementary Material, Fig. S1.
We hypothesized that modulation of the levels of Wig1 expression either by knockdown or overexpression would influence Htt levels in an allele-specific manner compared with control conditions. We first examined the effects of Wig1 knockdown on endogenous full-length Htt in STHdhQ7/Q7 and STHdhQ111/Q111 cells. When 20–25% Wig1 knockdown is achieved with lentiviral-shRNA, a significant reduction in the levels of Htt was observed (Fig. 2B and Supplementary Material, Fig. S1D). The extent of the reduction is markedly greater in mutant Htt compared with wt, which is consistent with the observation in PC12 cells expressing N-terminal Htt fragment (Fig. 2A and B).
We then questioned the effects of overexpression of Wig1 in the striatal cells. We observed a significant augmentation in the levels of Htt, in particular that for mutant Htt (Fig. 2C), which consistently mirrored the influences of Wig1 knockdown. In contrast to wt Wig1, a mutant with impairment in RNA binding capability (Wig1-H88A) (35,36), did not affect the levels of Htt (Fig. 2C). Taken together, these results suggest that RNA-binding of Wig1 protein may underlie the stabilization of Htt mRNA.
Wig1 preferentially binds to mutant htt mRNA
Binding of Wig1 protein with RNA is well known (24,35,36). Thus, we hypothesized that this protein binds directly to Htt mRNA, which can eventually affect the level of Htt protein. To address this, we utilized an RNA-binding protein immunoprecipitation (RIP) combined with real-time reverse transcription polymerase chain reaction (RT-PCR) with the striatal cells. We precipitated a protein-RNA complex with an anti-Wig1 antibody, and the levels of RNA associated with this immunoprecipitates were examined. We did not observe any signal of mRNAs for unrelated molecules, such as Actin and GAPDH, whereas the signal for Htt mRNA was observed (Fig. 3A). The Htt signal is specifically associated with Wig1 protein, as we did not observe Htt signal from the precipitates with IgG (Fig. 3A). In contrast, we did not observe any evidence that Wig1 and Htt interact at the protein levels (Supplementary Material, Fig. S2A).
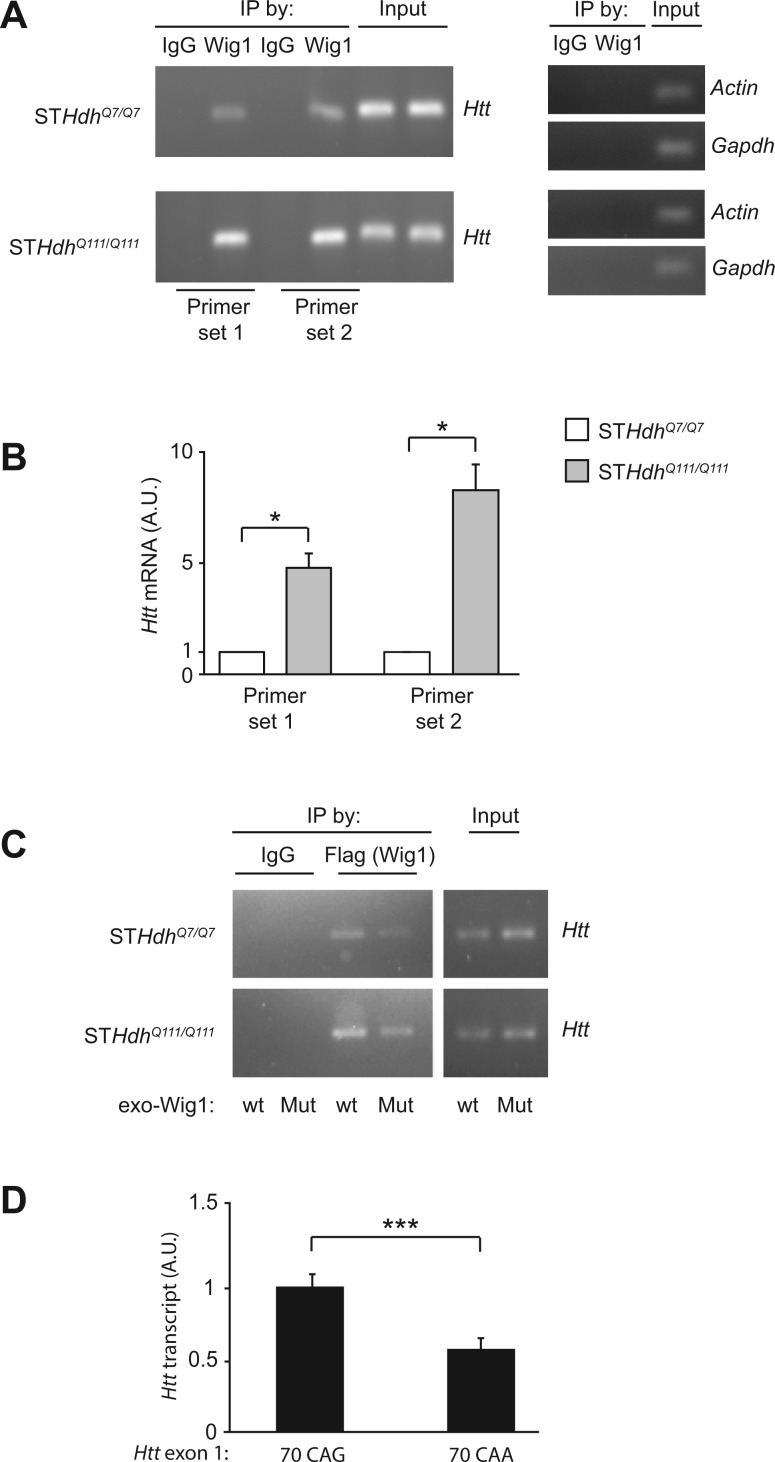
Figure 3.
Preferential binding of Wig1 toward mHtt mRNA. (A) Examination of binding of Wig1 with Htt mRNA by RIP assay. Wig1 proteins preferentially bind mutant Htt mRNA compared with its binding to wt Htt mRNA. (B) Quantitative RT-PCR analysis of Wig1 protein-Htt mRNA binding from RIP assay. Binding of Wig1 protein with mutant Htt mRNA is more enriched than Wig1 protein-wt Htt mRNA binding. (C) Attenuated Wig1 protein-Htt mRNA binding with RNA binding-defective mutant Wig1 (Wig1-H88A). (D) Quantitative RT-PCR analysis of binding between the GST-Wig1 protein and Htt exon 1 transcript in the RNA-protein binding assay. Wig1 protein binds with the Htt exon1 transcript containing 70 CAG more efficiently than the 70 CAA-carrying transcript. Data are represented as mean ± s.e.m. (*P < 0.05, ***P < 0.001, Student’s t-test). See also Supplementary Material, Table S2 and Fig. S2.
Provided that Wig1 protein binds with Htt mRNA, does this RNA–protein interaction underlie preferential increases in the level of mutant rather than wt Htt protein? To address this question, we compared the Wig1 protein-Htt mRNA interaction between the striatal cells expressing wt Htt (STHdhQ7/Q7) and those expressing mutant Htt (STHdhQ111/Q111). By conducting quantitative real-time PCR for Htt in the immunoprecipitates with the anti-Wig1 antibody (used in experiments of Fig. 3A), we found that mutant Htt mRNA from STHdhQ111/Q111 was significantly enriched by 4–8-folds compared with wt Htt mRNA from STHdhQ7/Q7 (Fig. 3B). Thus, we conclude that Wig1 binds to Htt mRNA, in particular mutant over wt Htt mRNA. The basal level of Htt mRNA was not different between STHdhQ7/Q7 and STHdhQ111/Q111 (Supplementary Material, Fig. S2B). Note that the influence of Wig1 in these experiments is more robust than that in the experiments shown in Fig. 2B. We believe that one of the main reasons to account for the quantitative differences underlies relative low knockdown efficiency of lentivirus-mediated Wig1 siRNA as shown in Supplementary Material, Fig. S1D. Furthermore, compared with wt Wig1, RNA binding-defective mutant Wig1 (Wig1-H88A) showed significantly less interaction with Htt mRNA (Fig. 3C). We then hypothesized that the preferential binding of Wig1 to mutant over wt Htt mRNA might be due to a hairpin structure formed by the expanded CAG repeats. To test this idea, we designed RNA-protein binding assays in vitro: we tested the levels of interaction between the purified GST-Wig1 protein and the Htt exon 1 transcript with 70 CAG repeats in comparison with the interaction between the protein and the Htt exon 1 transcript with 70 CAA that did not form a hairpin structure (37). GST-Wig1 bound with the 70 CAG repeat-containing transcript to a significantly greater extent compared with the transcript with 70 CAA repeats (Fig. 3D and Supplementary Material, Fig. S2C). In summary, these results suggest that Wig1 protein preferentially influences the mutant Htt allele through interaction with a hairpin structure formed by the expanded CAG repeats possibly in a repeat length-dependent manner.
Wig1 mediates and facilitates mutant Htt-elicited cytotoxicity and aggregate formation in a HD cell model
We questioned how Wig1 influences mutant Htt-induced cytotoxicity and aggregate formation in neurons, probably via its action on stabilizing the mutant Htt. To address this question, we used rat primary cortical neurons that transiently expressed N171-Htt-18Q or -82Q (16), in which we further knocked down endogenous Wig1 with the established shRNAs used earlier (Fig. 2A and Supplementary Material, Fig. S1A and B). First, we observed a significant reduction of N171-Htt-82Q-elicited cytotoxicity by knocking down Wig1 (Fig. 4A). Furthermore, the knockdown of Wig1 was also effective to decrease the level of mutant Htt-associated aggregate formation (Fig. 4B).
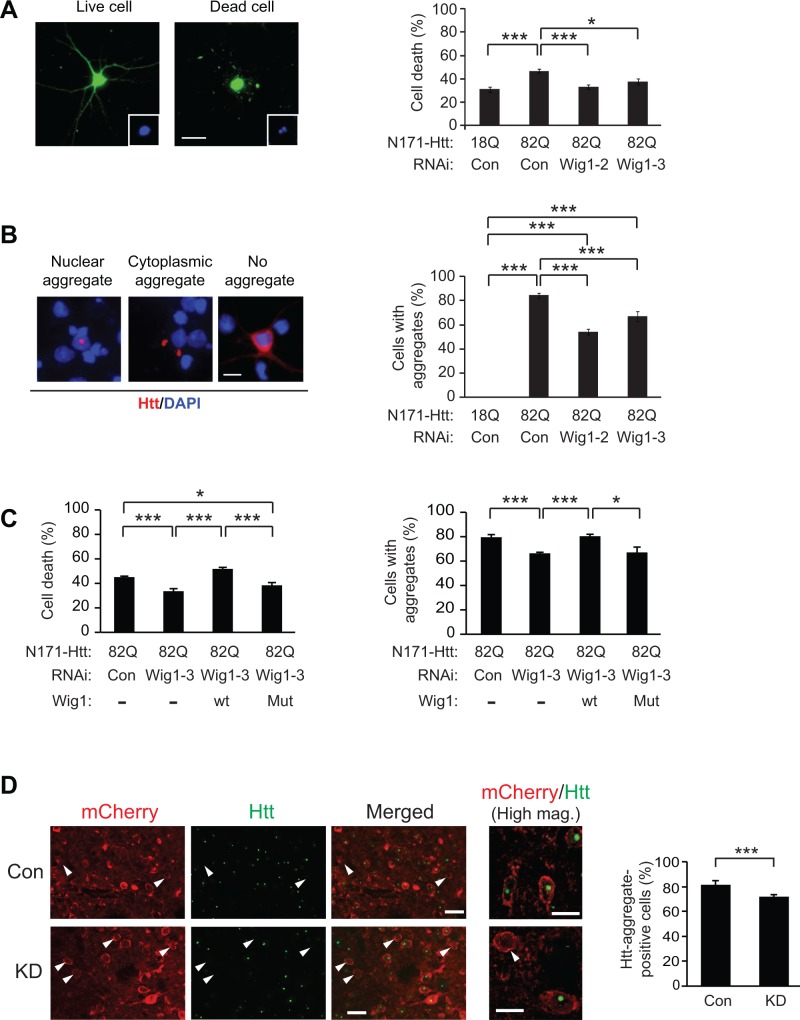
Figure 4.
Role for Wig1 in mHtt-elicited pathology in HD models. (A) Suppression of mutant Htt-elicited cell toxicity by knockdown of Wig1 in rat cortical primary neurons. Green, GFP. Inset shows DAPI-stained nucleus. The scale bar represents 20 µm. (B) Amelioration of mutant Htt-elicited aggregate formation by knockdown of Wig1 in rat cortical primary neurons. Red, Htt aggregates; blue, DAPI nuclear staining. The scale bar represents 10 µm. (C) RNA-binding region in Wig1 is critical for its facilitation of mutant Htt-elicited cytotoxicity and aggregate formation. Expression of wt Wig1 in rat cortical primary neurons expressing mutant Htt fragment (N171-Htt-Q82) and RNAi against Wig1 completely suppresses Wig1 RNAi-mediated amelioration of cell death (the left panel) and aggregate formation (the right panel), whereas RNA binding-defective mutant Wig1 (Wig1-H88A) does not. All the graphs represent at least three independent experiments. (D) Reduction of the Htt-positive aggregate formation by knockdown of Wig1 in the striatum of R6/2 mice. Htt-positive aggregates were labeled with an antibody against Htt MAB5374 (EM48; green) at 9 weeks of age. White arrowhead indicates the AAV-infected striatal cells that do not contain Htt-positive aggregates. Con, control AAV; KD, Wig1 KD AAV. Scale bars represent 20 μm. High magnification (High mag.): Scale bars represent 10 μm. Data are represented as mean ± s.e.m. (*P < 0.05, ***P < 0.001, Student’s t-test). See also Supplementary Material, Figs S1 and S3.
We then addressed whether RNA-binding ability of Wig1 has a pivotal role in mediating these HD-associated cellular phenotypes. The beneficial effects of Wig1 knockdown on N171-Htt-82Q-elicited cytotoxicity and aggregate formation were suppressed by introduction of wt but not by RNA binding-defective mutant Wig1 (Wig1-H88A) (Fig. 4C). These results support that RNA-binding ability of Wig1 is important for its influences on HD phenotypes. To address a role for Wig1 in HD pathology in vivo, we knocked down Wig1 in the striatum of the R6/2 model by stereotaxic injection of a viral vector-mediated Wig1 RNAi (Supplementary Material, Fig. S3). The knockdown of Wig1 significantly decreased the number of cells that carried Htt-positive aggregates (Fig. 4D). These results are, in essence, consistent with the previous publication in which p53 KO mice were crossbred with HD animal models [Bae et al. (16)].
Discussion
Here, we show that Wig1 protein interacts with Htt mRNA, preferentially with mutant Htt and mediates HD-associated pathological cellular phenotypes, such as cytotoxicity and aggregate formation. Accordingly, Wig1 upregulates mutant Htt protein and contributes to HD cellular pathology. Although p53 has been implicated in the pathophysiology of HD, the mechanism of how p53 play a unique role in the pathology remains elusive. The main goal and scope of this study is to elucidate the role of Wig1, a downstream target of p53, in HD pathology at the molecular and cellular levels.
What is the molecular mechanism whereby Wig1 binds to mutant over wild-type Htt? Expanded CAG repeats in the Htt mRNA can form a hairpin structure, which is a double-stranded-like RNA conformation (38). Given that Wig1 is known to preferentially interact with double-stranded RNA (24,35), we propose a working hypothesis in which Wig1 protein preferentially binds to mutant over wt Htt mRNA through interactions with the hairpin-like, double-stranded conformation of the expanded CAG repeats, based on the RNA-protein binding assay in vitro shown in Fig. 3D.
Several lines of evidence have shown that p53 reportedly upregulates Htt, whereas mutant Htt further stabilizes p53 (15–18,39). The present data on Wig1 provide a key mechanism of how p53 augments the levels of Htt, in particular mutant Htt. Thus, p53-Wig1-mutant Htt noxious cycle may underlie a key pathology of HD. The validation of our proposed mechanism in multiple HD models, in particular full-length mutant Htt knock-in mouse models would be crucial for a future perspective.
Because wild-type Htt has numerous physiological functions (14,40), it is important to consider approaches that can target mutant Htt, but not wild-type Htt, selectively (or at least preferentially). Allele-specific silencing of Htt may be possible, either by targeting the expanded CAG tract directly (5,7,9) or by targeting genetic polymorphisms linked to CAG expansion (6,8,10,41). We propose that Wig1 is an important mediator in HD pathology that regulates the levels of Htt, especially mutant Htt (by preferential binding to mutant Htt mRNA). Many mechanism-oriented efforts of developing novel therapeutic strategies have been taken place for HD (2,3,42–45). Drugs that block the interaction of Wig1 protein and mutant Htt mRNA may also provide a basis for a novel therapeutic strategy to alleviate this devastating disease in the near future.
Materials and Methods
Mice
The mice used in this study were: wild-type, p53 KO mice, an animal model for HD [Htt-N171-82Q-Tg mice in which an N-terminal fragment of mutant Htt (amino acids 1–171) with expanded 82 polyQ stretch is overexpressed] (mHD) (25), and the crossbred animals between KO and mHD (mHK) as previously described (16). The mice were analyzed at 5–5.5 months of age. Mice were sacrificed and brains were stored at −80°C until use. For RNA extractions, forebrain region was dissected, snap-frozen and were stored at −80°C until processed. R6/2 Tg and control wild-type male mice (Jackson Laboratory) were analyzed at 9 or 12 weeks of age.
Human brain samples
Human postmortem brain specimens from the cortex and striatum (HD Grade 3, n = 3; HD Grade 4, n = 5; age-matched controls, n = 5) were generously provided by the Harvard Brain Tissue Resource Center (Supplementary Material, Table S1).
RNA extraction and Oligo GEArray p53 microarray analysis
RNA was extracted from forebrain region of mice using RNeasy Kit (Qiagen) according to manufacturer’s instructions. We used Oligo GEArray of Mouse (OMM-027) p53 Signaling Pathway microarray (SuperArray Bioscience) that is designed to profile gene expression of a panel of 113 key genes involved in the p53 pathways. For analysis, 3 μg of total RNA was reverse transcribed. Then, biotin-labeled cRNA was synthesized from cDNA with the use of the TrueLabeling-AMP Linear RNA Amplification kit (SuperArray Biosciecnce). cRNA was purified using the Array-Grade cRNA cleanup kit (SuperArray Biosciecnce), quantified and hybridized overnight to gene-specific probes involved in the p53 signaling pathway that were spotted on the GEArray membranes (2 μg of cRNA/membrane). After incubation with streptavidin-AP conjugate, the array image was developed with CPD-Star chemiluminescent substrate (chemiluminescent detection kit; Superarray) and recorded with x-ray film. The images were scanned, and data analyzed by GEArray Expression Analysis Suite Software (SuperArray Biosciecnce). The numerical data were then further evaluated with Microsoft Excel. Data evaluation included background correction (subtraction of minimum value) and median normalization. Data filtering criteria is mentioned in the results.
RNA-binding protein immunoprecipitation
RIP was performed according to the manufacturer’s instructions (EZ-Magna RIP Kit, Millipore). In brief, experiments involve immunoprecipitation (IP) of endogenously formed complexes of RNA-binding proteins and coisolation of any RNA species associated with that RNA-binding protein. The following antibodies were used in IP: anti-Wig1 (Santa Cruz) and anti-Flag (M2, Sigma-Aldrich). After purification of these RNA species, semiquantitative RT-PCR or quantitative RT-PCR (for primer information see Supplementary Material, Table S2) was performed to identify bound mRNAs.
Quantitative real-time PCR
Total RNA was isolated from frozen human brain tissue samples (Supplementary Material, Table S1) or dissected mouse striatal tissues using RNeasy Mini Kit (Qiagen) and followed by cDNA synthesis using SuperScript First-Strand System for RT-PCR (Invitrogen) using Oligo(dT)20 primers according to the manufacturer’s instructions. Quantitative real-time PCR was performed by using 7900HT Sequence Detector System (Applied Biosystems). Specific primers for the target genes were designed by Primer3 program (http://frodo.wi.mit.edu). Primer sequences are listed in Supplementary Material, Table S3. Quantitative RT-PCR was carried out using SYBR GreenER qPCR SuperMix for ABI PRISM (Invitrogen) for the 7900HT in a final volume of 10 μl. The PCR conditions for 7900HT were set as: 50°C for 2 min, 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min and a dissociation step of 95°C for 15 s, 60°C for 15 s and 95°C for 15 s. The expression level of β-Actin and GAPDH genes were used for normalization. We analyzed the data with SDS 2.4 (Applied Biosystems).
Htt exon 1 transcript production
A Plasmid containing Htt exon 1 sequences with 70 CAG repeats, a T7 promoter at the 5′ end, and a Bts I restriction site at the 3′ end was obtained from Dr Kevin M. Weeks (University of North Carolina). The identical plasmid except for the presence of 70 CAA repeat sequences was obtained by de novo synthesis (Blue Heron Biotechnology). Plasmids were linearized with Bts I restriction enzyme (New England Biolabs), and linearization was confirmed by agarose gel electrophoresis. Linearized template sequences were in vitro transcribed using T7 RNA polymerase (Roche) and purified by phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5, Ambion) extraction, and subsequent ethanol precipitation at −80°C. Transcripts were resuspended in 0.5X TE buffer, and RNA purity was confirmed by gel electrophoresis.
RNA-protein binding assay
Transcripts were denatured at 95°C for 2 min, snap-cooled on ice for 2 min and refolded at 37°C for 30 min in 50 mm Tris–HCl (pH 8.0), 75 mm KCl and 3 mm MgCl2. Purified GST-Wig1 or GST (40 μg) was incubated in 20 μl of glutathione-sepharose 4B resin (GE Life Sciences) equilibrated in binding buffer [50 mm Tris–HCl (pH 7.4), 150 mm NaCl and 5 mm MgCl2] for 30 min at 4°C. After washing three times to remove any unbound proteins, immobilized GST-Wig1 or GST was incubated with refolded transcripts (1 ng) at room temperature for 20 min. The resin was washed with the binding buffer five times to remove unbound Htt RNA transcripts. Then, the resin was incubated in binding buffer containing proteinase K (Ambion) at 55°C for 30 min, and Htt transcripts were recovered by phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.5, Ambion) extraction, and subsequent ethanol precipitation at −80°C overnight. The recovered transcripts were used for cDNA synthesis using a SuperScript III First-Strand Synthesis System (Invitrogen) and Htt gene-specific primer (5′-TCA GCT TTT CCA GGG TCG CCA TG-3′). These cDNAs were used for qRT-PCR analysis using Htt primer three pairs (Supplementary Material, Table S2) and followed earlier described qRT-PCR method.
Production and purification of fusion protein
Wig1 full-length cDNA was subcloned into the Sal I and Not I sites of the pGEX-6P-2 vector (GE Life Sciences). GST-Wig1 fusion protein expressed in BL21(DE3) cells was affinity-purified using glutathione-sepharose 4B columns (GE Life Sciences).
Immunoblotting
At the indicated time points after transfection, cells were pelleted and lysed in RIPA buffer [50 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 5 mm dithiothreitol (DTT), 1 mm EDTA, 1% Triton X-100, protease inhibitor mixture (Roche)] with brief sonication. Protein concentration was measured with BCA protein assay reagent (Pierce). Equal amounts of protein were loaded and separated by SDS-PAGE. The following antibodies were used: anti-Wig1 (M-20, Santa Cruz; GTX107246, GenTex), anti-Tubulin (DM1A, Sigma-Aldrich), anti-Flag (M2, Sigma-Aldrich), anti-Htt (MAB2166, Millipore) and anti-HAP1 (N-18, Santa Cruz) antibody. Quantitative densitometric measurement of immunoblots was performed using Image J program (NIH) using β-tubulin as a loading control. Note that all antibodies against Wig1 did not reliably work in biochemical and histological analyses with brain tissues.
Coimmunoprecipitation
Coimmunoprecipitation followed by western blotting were performed as previously described (46,47). Cells were lysed in IP buffer (50 mm Tris pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.1 mg/ml bovine serum albumin (BSA), protease inhibibitor cocktail) or at the indicated time points after transfection, cells were pelleted and lysed in RIPA buffer [50 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 5 mm DTT, 1 mm EDTA, 1% Triton X-100, protease inhibitor mixture (Roche)]. Co-immunoprecipitates or the RIPA lysates were resolved by SDS-PAGE and analyzed by western blotting. The following antibodies were used: anti-Wig1 (M-20, Santa Cruz), anti-Tubulin (DM1A, Sigma-Aldrich), anti-Flag (M2, Sigma-Aldrich), anti-Htt (MAB2166, Millipore) and anti-HAP1 antibody (N-18, Santa Cruz). Quantitative densitometric measurement of immunoblots was performed using Image J program (NIH) using tubulin as a loading control.
Cell cultures, transfection and lentiviral infection
PC12 cells were maintained in DMEM with 10% fetal bovine serum (FBS) and 5% horse serum (HS) (Invitrogen). Differentiation was initiated by adding 50 ng/ml of NGF with culture medium changed to DMEM with 1% FBS and 1% HS. NGF was supplemented daily after differentiation. Transfection of Htt expression constructs was carried out with Lipofectamine 2000 (Invitrogen) for PC12 cells before differentiation. Pfithrin-α (p53-specific inhibitor, 10 µM) was added in the medium along with NGF. PC12 cells were harvested after 96 h after transfection. STHdhQ7/Q7 or STHdhQ111/Q111cells (gifts from Marcy Macdonald, Harvard) were grown in DMEM with 10% FBS and G418 (400 µg/ml) at 33°C. After 24 h, the cells were transfected with cDNA constructs using lipofectamine 2000 (Invitrogen, for STHdh cells) as per the manufacture’s instructions, or infected with 106 particles of lentivirus expressing control or Wig1-2 shRNA for each 6-well plate.
Culture and transfection of primary cortical neurons
Embryonic day 18 (E18) cortical cultures were prepared from Sprague-Dawley rat embryos. The cerebral cortices were obtained by removing brainstem, thalamus, striatum and hippocampus. The dissociated neurons were grown on poly-D-lysine (200 µg/ml)-coated 18 mm diameter glass coverslip in Neurobasal medium with B27 supplement and 0.5 mm L-glutamine (Invitrogen). The cell density per each 12-well was 3 × 105 cells. On 4 days in vitro, neurons were cotransfected with DNA using Lipofectamine 2000 (Invitrogen) following the manufacturers’ protocols. The ratio of plasmid to Lipofectamine 2000 was 1:1. Transfection efficiency was consistently 3–5%.
Plasmids and shRNAs
Knockdown of target genes is carried out by using a pSuper vector. For Wig1, three shRNA constructs are made, and their target sequences are as follows:
Wig1 shRNA1, 5′-GCATCCTTGCCTCTGGCAG-3′ (for mouse and rat)
Wig1 shRNA2, 5′-CTCTGCAATGTCACCTTGA-3′ (for human, mouse and rat)
Wig1 shRNA3, 5′-GAGACTTCGTCTGGCCGAA-3′ (for mouse)
For expression of N-terminal, 171 amino acids of Htt under the control of CMV promoter, pAAV-Htt N171-18Q-IRES-hrGFP or pAAV-Htt N171-82Q-IRES-hrGFP were used.
Lentivirus system
The vector contains an H1 promoter for shRNA (control or Wig1 shRNA2) expression and an ubiquitin promoter for expression of EGFP marker. Human embryonic kidney (HEK)293FT cells are cotransfected with a pFUGW-shRNA-EGFP plasmid along with Δ8.9 and VSVg constructs using Lipofectamine 2000 (Invitrogen) (47). Seventy-two hours after transfection, medium is collected and filtered. Viral particles in the filtered medium are concentrated by ultracentrifugation. Lentiviral pellet is dissolved in Neurobasal media (Invitrogen). Viral titers are determined by counting EGFP-positive cells after infection with serial dilutions of the viral solution.
AAV virus preparation
HEK293-AAV cells (Stratagene) were cotransfected with a transgene plasmid along with two packaging plasmids for AAV1 using standard calcium phosphate precipitation method. Cells were harvested 72 h posttransfection and disrupted by three times of freeze and thaw cycles. AAV viral particles were isolated and concentrated by series of ammonium sulfate precipitations, applied to a discontinuous gradient of iodixanol (Optiprep density gradient medium; Sigma-Aldrich), and centrifuged at 350 000g for 60 min. An AAV-containing 40% iodixanol fraction was carefully collected from an ultracentrifuge tube (Beckman), and concentrated using a centrifugal filter device (Ultracel-100K; Amicon). Stock viral titers: 4.0 × 1013 particles/ml for rAAV2/1-U6-shControl-CMV::mCherry; 2.1 × 1013 particles/ml for rAAV2/1-U6-shWig1-2-CMV::mCherry.
Cytotoxicity assay and aggregate formation assay
Neuronal death was quantified at 108 h after transfection in a blinded manner as previously described (48). Briefly, cells with both degenerated neurites and shrunken nuclei were considered dead. Morphology of neurites and nuclei was monitored by GFP signal and DAPI staining, respectively. For mHtt aggregate formation, N171-82Q was transfected into mouse embryonic neurons and anti-Huntingtin (EM48, Millipore) mouse monoclonal antibody was used. Nuclear and cytoplasmic aggregates were scored 4.5 days after transfection. Aggregates were visualized by immnunohistochemistry with anti-Huntingtin (EM48, Millipore) mouse monoclonal antibody. Cellular compartmentalization was determined by overlaying GFP and DAPI images.
Stereotaxic virus injection
Mice (5.5 week of age R6/2 Tg male mice; Jackson Laboratory) were deeply anesthetized with avertin (16 mg/Kg, intraperitoneal injection; Sigma-Aldrich) and placed into a stereotaxic apparatus (David Kopf Instruments). The head was leveled using Bregma and Lambda reference points and a craniotomy was performed. Diluted AAV virus (1.5 × 1012 particles/ml) was injected at two sites in the dorsal striatum of each hemisphere using a pulled glass micropipette controlled by an auto-nanoliter injector (Nanoject II, Drummond) for 1.0 μl per hemisphere: coordinates from Bregma; 0.5 μl at +0.9 AP, ± 1.9 ML, −3.0 DV; 0.5 μl at +0.4 AP, ± 2.1 ML, −3.4 DV. After each injection, the micropipette was left in place for 10 min and then slowly withdrawn. The skin was sutured and closed. Mice were recovered from anesthesia under a heat lamp, and then were returned to their home cage. Mice were sacrificed at the age of 9 weeks for further analyses.
Evaluation of Htt aggregates in vivo
The AAV-injected R6/2 mice at 9 weeks were processed for immunohistochemical examination. Brain cryosections (20 μm thick) were incubated for 24 h at 4°C with primary antibody: MAB5374 (EM48, mouse IgG, 1:200 dilution) in 2% normal goat serum (NGS)/3% BSA in 1XPBST. Next, sections were washed and incubated for 1.5 h at room temperature with secondary antibody (Alexa Flour 488-conjugated goat anti-mouse IgG antibody, Molecular Probes) in 1% NGS/1XPBST. Stained sections were washed and coverslips were mounted on tissue sections with Prolong Gold antifade reagent with DAPI (Invitrogen). The control or Wig1 knockdown AAV-infected mCherry-positive striatal cells were examined for the presence of EM48-positive Htt-aggregates (Control AAV, n = 4; Wig1 KD AAV, n = 8) using an epifluorescence microscope (Zeiss, Axio Observer, D1). At least 400 cells per hemisphere were obtained and percentage of cells containing Htt-aggregates was calculated.
Statistical analysis
All data are presented as the mean ± s.e.m. Statistical analyses between two groups were performed by using a two-tailed Student’s t-test (paired or unpaired as appropriate) and a value of P < 0.05 was considered statistically significant.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors thank Dr Pamela Talalay, Dr Hanna Jaaro-Peled, Dr Lindsay Hayes, Dr Nao J. Gamo, Mr Travis Faust and Ms Yukiko Lema for critical reading and organizing the article. They also thank Dr Solomon Snyder for initial guidance to B.-I.B. They thank Dr Kevin M. Weeks for providing the pT7-Htt exon1-70 CAG plasmid.
Conflict of Interest statement. None declared.
Funding
This work was supported by the National Institute of Health (MH-084018, MH-094268, MH-069853, MH-085226, MH-088753, MH-092443, MH-105660, and DA-040127) to A.S.; Stanley Foundation, RUSK, S&R Foundation, Brain & Behavior Research Foundation, and Maryland Stem Cell Research Fund to A.S.; Brain & Behavior Research Foundation to S.-H. K. and N.S.
References
- 1. The Huntington’s Disease Collaborative Research Group. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 2. Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L, Davidson B.L. (2005) RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. U. S. A., 102, 5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G. et al. (2007) Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U. S. A., 104, 17204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P. et al. (2009) Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol., 65, 276–285. [DOI] [PubMed] [Google Scholar]
- 5. Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I, Corey D.R. (2009) Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol., 27, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.P., Zamore P.D., Aronin N. (2009) Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr. Biol., 19, 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gagnon K.T., Pendergraff H.M., Deleavey G.F., Swayze E.E., Potier P., Randolph J., Roesch E.B., Chattopadhyaya J., Damha M.J., Bennett C.F. et al. (2010) Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry, 49, 10166–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M, Hayden M.R. (2011) Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther., 19, 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu J., Liu J., Yu D., Chu Y, Corey D.R. (2012) Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res., 40, 11270–11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Frank Bennett C. et al. (2013) Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res., 41, 9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sah D.W., Aronin N. (2011) Oligonucleotide therapeutic approaches for Huntington disease. J. Clin. Invest., 121, 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson C.D., Davidson B.L. (2010) Huntington’s disease: progress toward effective disease-modifying treatments and a cure. Hum. Mol. Genet., 19, R98–R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cha J.H. (2007) Transcriptional signatures in Huntington’s disease. Prog. Neurobiol., 83, 228–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zuccato C., Valenza M., Cattaneo E. (2010) Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev., 90, 905–981. [DOI] [PubMed] [Google Scholar]
- 15. Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E, Thompson L.M. (2000) The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U. S. A., 97, 6763–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bae B.I., Xu H., Igarashi S., Fujimuro M., Agrawal N., Taya Y., Hayward S.D., Moran T.H., Montell C., Ross C.A. et al. (2005) p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron, 47, 29–41. [DOI] [PubMed] [Google Scholar]
- 17. Feng Z., Jin S., Zupnick A., Hoh J., de Stanchina E., Lowe S., Prives C, Levine A.J. (2006) p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene, 25, 1–7. [DOI] [PubMed] [Google Scholar]
- 18. Ryan A.B., Zeitlin S.O., Scrable H. (2006) Genetic interaction between expanded murine Hdh alleles and p53 reveal deleterious effects of p53 on Huntington’s disease pathogenesis. Neurobiol. Dis., 24, 419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobs W.B., Kaplan D.R, Miller F.D. (2006) The p53 family in nervous system development and disease. J. Neurochem., 97, 1571–1584. [DOI] [PubMed] [Google Scholar]
- 20. Gottlieb E., Vousden K.H. (2011) p53 regulation of metabolic pathways. Cold Spring Harb. Perspect. Biol., 2, a001040.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varmeh-Ziaie S., Okan I., Wang Y., Magnusson K.P., Warthoe P., Strauss M, Wiman K.G. (1997) Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene, 15, 2699–2704. [DOI] [PubMed] [Google Scholar]
- 22. Gillardon F., Spranger M., Tiesler C, Hossmann K.A. (1999) Expression of cell death-associated phospho-c-Jun and p53-activated gene 608 in hippocampal CA1 neurons following global ischemia. Brain Res. Mol. Brain Res., 73, 138–143. [DOI] [PubMed] [Google Scholar]
- 23. Shimizu M., Miyazaki I., Higashi Y., Eslava-Alva M.J., Diaz-Corrales F.J., Asanuma M., Ogawa N. (2008) Specific induction of PAG608 in cranial and spinal motor neurons of L-DOPA-treated parkinsonian rats. Neurosci. Res., 60, 355–363. [DOI] [PubMed] [Google Scholar]
- 24. Mendez-Vidal C., Wilhelm M.T., Hellborg F., Qian W, Wiman K.G. (2002) The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res., 30, 1991–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A. et al. (1999) Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet., 8, 397–407. [DOI] [PubMed] [Google Scholar]
- 26. Wu X., Bayle J.H., Olson D, Levine A.J. (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev., 7, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 27. Miyashita T., Reed J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80, 293–299. [DOI] [PubMed] [Google Scholar]
- 28. Shiraishi K., Fukuda S., Mori T., Matsuda K., Yamaguchi T., Tanikawa C., Ogawa M., Nakamura Y., Arakawa H. (2000) Identification of fractalkine, a CX3C-type chemokine, as a direct target of p53. Cancer Res., 60, 3722–3726. [PubMed] [Google Scholar]
- 29. Stambolic V., MacPherson D., Sas D., Lin Y., Snow B., Jang Y., Benchimol S, Mak T.W. (2001) Regulation of PTEN transcription by p53. Mol. Cell, 8, 317–325. [DOI] [PubMed] [Google Scholar]
- 30. Yu J., Zhang L., Hwang P.M., Kinzler K.W., Vogelstein B. (2001) PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell, 7, 673–682. [DOI] [PubMed] [Google Scholar]
- 31. Budanov A.V., Shoshani T., Faerman A., Zelin E., Kamer I., Kalinski H., Gorodin S., Fishman A., Chajut A., Einat P. et al. (2002) Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene, 21, 6017–6031. [DOI] [PubMed] [Google Scholar]
- 32. Wilhelm M.T., Mendez-Vidal C, Wiman K.G. (2002) Identification of functional p53-binding motifs in the mouse wig-1 promoter. FEBS Lett., 524, 69–72. [DOI] [PubMed] [Google Scholar]
- 33. Sermeus A., Michiels C. (2011) Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis., 2, e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E, MacDonald M.E. (2000) Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet., 9, 2799–2809. [DOI] [PubMed] [Google Scholar]
- 35. Mendez Vidal C., Prahl M, Wiman K.G. (2006) The p53-induced Wig-1 protein binds double-stranded RNAs with structural characteristics of siRNAs and miRNAs. FEBS Lett., 580, 4401–4408. [DOI] [PubMed] [Google Scholar]
- 36. Vilborg A., Glahder J.A., Wilhelm M.T., Bersani C., Corcoran M., Mahmoudi S., Rosenstierne M., Grander D., Farnebo M., Norrild B. et al. (2009) The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc. Natl. Acad. Sci. U. S. A., 106, 15756–15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Busan S., Weeks K.M. (2013) Role of context in RNA structure: flanking sequences reconfigure CAG motif folding in huntingtin exon 1 transcripts. Biochemistry, 52, 8219–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Mezer M., Wojciechowska M., Napierala M., Sobczak K, Krzyzosiak W.J. (2011) Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res., 39, 3852–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ehrnhoefer D.E., Sutton L, Hayden M.R. (2011) Small changes, big impact: posttranslational modifications and function of huntingtin in Huntington disease. Neuroscientist, 17, 475–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cattaneo E., Zuccato C., Tartari M. (2005) Normal huntingtin function: an alternative approach to Huntington’s disease. Nat. Rev. Neurosci., 6, 919–930. [DOI] [PubMed] [Google Scholar]
- 41. Warby S.C., Montpetit A., Hayden A.R., Carroll J.B., Butland S.L., Visscher H., Collins J.A., Semaka A., Hudson T.J, Hayden M.R. (2009) CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet., 84, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tanaka M., Machida Y., Niu S., Ikeda T., Jana N.R., Doi H., Kurosawa M., Nekooki M., Nukina N. (2004) Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat. Med., 10, 148–154. [DOI] [PubMed] [Google Scholar]
- 43. Zala D., Colin E., Rangone H., Liot G., Humbert S., Saudou F. (2008) Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum. Mol. Genet., 17, 3837–3846. [DOI] [PubMed] [Google Scholar]
- 44. Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R, Yang X.W. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron, 64, 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. An M.C., Zhang N., Scott G., Montoro D., Wittkop T., Mooney S., Melov S, Ellerby L.M. (2012) Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell, 11, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hara M.R., Agrawal N., Kim S.F., Cascio M.B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J.H., Tankou S.K., Hester L.D. et al. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol., 7, 665–674. [DOI] [PubMed] [Google Scholar]
- 47. Sen N., Hara M.R., Ahmad A.S., Cascio M.B., Kamiya A., Ehmsen J.T., Agrawal N., Hester L., Dore S., Snyder S.H. et al. (2009) GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron, 63, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nucifora F.C., Jr, Sasaki M., Peters M.F., Huang H., Cooper J.K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V.L. et al. (2001) Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science, 291, 2423–2428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.