Abstract
The role of post-transcriptional gene regulation in human brain development and neurodevelopmental disorders remains mostly uncharacterized. ELAV-like RNA-binding proteins (RNAbps) are a family of proteins that regulate several aspects of neuronal function including neuronal excitability and synaptic transmission, both critical to the normal function of the brain in cognition and behavior. Here, we identify the downstream neuronal transcriptional and splicing networks of ELAVL2, an RNAbp with previously unknown function in the brain. Expression of ELAVL2 was reduced in human neurons and RNA-sequencing was utilized to identify networks of differentially expressed and alternatively spliced genes resulting from haploinsufficient levels of ELAVL2. These networks contain a number of autism-relevant genes as well as previously identified targets of other important RNAbps implicated in autism spectrum disorder (ASD) including RBFOX1 and FMRP. ELAVL2-regulated co-expression networks are also enriched for neurodevelopmental and synaptic genes, and include genes with human-specific patterns of expression in the frontal pole. Together, these data suggest that ELAVL2 regulation of transcript expression is critical for neuronal function and clinically relevant to ASD.
Introduction
One of the unfortunate consequences of the intricacies of the highly evolved human brain has been the emergence of neurodevelopmental disorders such as autism spectrum disorder (ASD). There is a strong genetic component to the etiology of ASD and similar disorders. Exome studies have uncovered hundreds of genes linked to ASD with no single gene accounting for greater than a few percent of cases (1–4). These results indicate that most individuals with ASD have heterogeneous genetic insults that converge upon common biological pathways during brain development. Understanding the molecular mechanisms of this convergence will be essential for the development of better methods of diagnosis and broadly applicable therapeutics.
RNA splicing has recently emerged as playing a key role in nervous system development and numerous diseases (5–13). In particular, altered splicing occurs in ASD (14–16) suggesting that investigating the genes involved in RNA splicing within the human brain as well as the specific transcripts altered in ASD will provide a foundation for exploring the relationship between alternative splicing and the origin of neurodevelopmental disorders. Several splicing factors have been implicated in ASD; in particular, the gene encoding RBFOX1 (previously known as A2BP1) has been repeatedly linked to ASD at both the genetic as well as functional level (1,14,17–20). As there are few differentially expressed genes (DEGs) in ASD brains compared with controls (14,21), these findings suggest that post-transcriptional alterations might have a greater impact on ASD than overall gene expression levels.
We previously determined the alternatively spliced transcripts occurring downstream of RBFOX1 in human neurons (18). Within the introns flanking the RBFOX1 regulated alternatively spliced exons, we identified a significant enrichment of motifs for another RNA-binding protein (RNAbp), ELAVL2 (ELAV-like neuron-specific RNA-bp2; also known as HuB), a gene that is differentially expressed when RBFOX1 levels are reduced (Supplementary Material, Figure S1) (18). These results suggested that RBFOX1 and ELAVL2 might coordinately regulate RNA processing in human neurons.
There is little known about the function of ELAVL2 in the brain. Previous work has identified bound mRNAs to ELAVL1 in the brain (22) and the bound mRNAs and alternative splicing downstream of ELAVL3 or ELAVL4 in the brain (23), but the molecular pathways downstream of ELAVL2 are presently unknown. Therefore, among the four neuronal ELAV-like (nELAVL) family members expressed in the brain, ELAVL2 remains the only one for which the downstream pathways have yet to be determined. Recent work in mouse hippocampus has suggested that ELAVL2 has an activity-dependent function hypothesized to be post-transcriptional either in the form of RNA stabilization or translational control (24). We previously identified a human-specific frontal pole co-expression network enriched for ELAVL2-binding motifs as well as increased expression of ELAVL2 in the frontal pole on the human lineage (25). ELAVL2 has also been associated with schizophrenia in a Japanese cohort (26). Taking these data together with the strong association of RBFOX1 regulated networks with ASD risk (1) and the common dysregulated pathways in ASD and schizophrenia (27), we hypothesized that ELAVL2 might regulate critical gene expression pathways involved in human neurodevelopment.
To investigate this hypothesis, we reduced ELAVL2 levels in primary human neurons (phNs) via RNAi-mediated knockdown (kd) and carried out RNA-sequencing (RNA-seq) similar to our approach for studying RBFOX1 (18). We identify a number of alternatively spliced transcripts downstream of ELAVL2 that overlap with RBFOX1 targets as well as targets of FMR1, a gene encoding the RNAbp FMRP that causes the neurodevelopmental condition Fragile X Syndrome, which often includes ASD as a comorbidity (28–31). Alternatively spliced as well as DEGs downstream of ELAVL2 correspond to a number of known ASD risk genes and genes that encode synaptic proteins, consistent with the predicted model of synaptic deficits being associated with ASD (32). More strikingly, we identify co-expression modules containing genes with conserved regulation downstream of both ELAVL2 and RBFOX1 representing genes involved in ASD, FMRP-regulation, and/or synaptic function. Finally, ELAVL2 targets that are also specifically changed in the human frontal pole compared with non-human primates have higher connectivity in co-expression networks compared with other genes. These data are the first to identify the molecular pathways downstream of ELAVL2 and the results suggest that the ELAVL2-regulated pathways are involved in normal human brain function and their disruption may play a role in neurodevelopmental disorders including ASD.
Results
ELAVL2 is co-expressed with other RNAbps during human cortical development
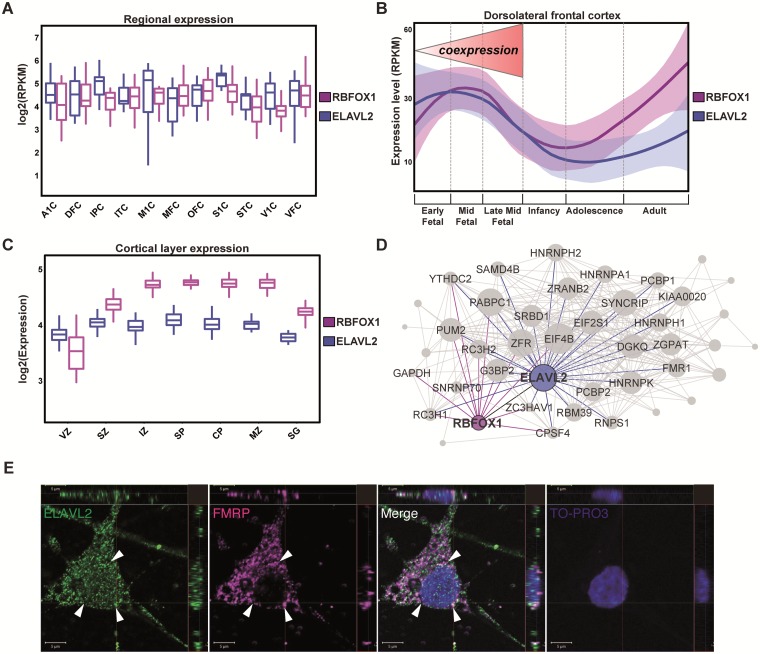
Positive correlations between gene expression can suggest involvement in similar cellular or molecular pathways and/or protein interactions (33). To explore the relationship between ELAVL2 and other RNAbps such as RBFOX1 we examined the dynamic expression of both genes in silico during the development of human cortical regions using a database of human brain gene expression throughout the lifespan (www.brainspan.org). Notably, we found positive co-expression between ELAVL2 and RBFOX1 during fetal cortical development (Fig. 1A), suggesting a potential shared biological relationship. To explore this relationship further, as the frontal cortex is involved in cognitive and neurodevelopmental disorders, we asked whether ELAVL2 and RBFOX1 are co-expressed in this particular region. We found that ELAVL2 and RBFOX1 are significantly co-expressed in frontal cortex (Spearman rank correlation test, ρ = 0.61, P = 0.00014) with an even higher co-expression during fetal development (Spearman rank correlation test, ρ = 0.72, P = 0.0017), suggesting an important role for these genes in influencing the development of this brain region during this critical time period (Fig. 1B). We identified similar co-expression in the ventricular zone, a neurogenic niche important for neuronal proliferation and differentiation (Fig. 1C). This is interesting as alternative splicing has been recently implicated in neurogenesis (15,34,35) and a previous study found expression of ELAVL2 within the mouse ventricular and intermediate zones (36). However, we should note that RNA-seq of the developing mouse cortex observed significantly less expression of RBFOX1 in the ventricular zone and subventricular zone compared with ELAVL2 (37), perhaps suggesting species-specific differences in co-expression.
Figure 1.
ELAVL2 expression in human brain and phNs. (A) Expression distribution of RBFOX1 and ELAVL2 in fetal cortical regions (A1C, primary auditory cortex; DFC, dorsolateral frontal cortex; IPC, inferior parietal cortex; ITC, inferior temporal cortex; M1C, primary motor cortex; MFC, medial frontal cortex; OFC, orbital frontal cortex; S1C, primary sensory cortex; STC, superior temporal cortex; V1C, primary visual cortex; VFC, ventral frontal cortex). (B)ELAVL2 and RBFOX1 expression comparison in dorsolateral prefrontal cortex throughout the lifespan. The fetal developmental period in which RBFOX1 and ELAVL2 are strongly co-expressed is indicated. (C) Expression distribution of ELAVL2 and RBFOX1 in cortical layers (VZ, ventricular zone; SZ, subventricular zone; IZ, intermediate zone; SP, subplate; CP, cortical plate; MZ, marginal zone; SG, subpial granular layer). (D) Co-expression sub-module of RNAbps in prefrontal fetal cortex. Interactions represent potential protein interactions between RNAbps predicted by co-expressed genes. Each link corresponds to a weight calculated according to the correlation of the gene sets between nodes (see Methods section). RNAbps are preselected as nodes in the network. Shown are links between ELAVL2 (blue), RBFOX1 (purple), FMR1 (green) and other RNAbps (grey). (E) Co-localization confocal images of ELAVL2 and FMRP in phNs. Arrows indicate the perinuclear localization of both RNAbps. TO-PRO-3 is used to identify nuclei. Scale bar, 5 μm.
In order to assess additional potential RNAbp interactions in the fetal frontal pole, we used the weighted topological overlap (wTO) method (38) to infer ELAVL2, RBFOX1 and FMR1 partners in fetal frontal cortical development. As co-expression correlations can sometimes infer biological interactions, such as protein-protein interactions between the protein products of two genes (33), wTO aims to highlight potential pathways between genes of interest, e.g. RNAbps, based on the interaction of co-expressed genes (see Methods section). The wTO method therefore requires a predetermination of which genes will be the nodes in a network module. In this case, we assigned all RNAbps as potential nodes and the resultant association of genes with these RNAbps should identify novel and as well as previously-supported interactions with RNAbps in the developing frontal cortex. Nodes with the greatest number of connections to other genes are considered “hub” genes and can be thought as driving much of the structure of the network. Using this approach, we found that ELAVL2 is one of the major hubs of the module and, in addition, network prediction showed strong interaction between ELAVL2, RBFOX1 and FMR1 (Fig. 1D). PUM2, an FMRP target involved in embryonic development that interacts with RBFOX2 and ELAVL1 (31,39,40), is strongly associated with ELAVL2, RBFOX1 and FMR1, reflecting novel pathways not yet characterized. We also found that HNRNPA1, a gene implicated in neuronal death and Alzheimer’s disease (41), HNRNPK, a gene implicated in neuronal differentiation and previously linked with the nELAVL family (13,42,43), and HNRNPH1, an RBFOX1 target gene whose binding site is enriched near RBFOX1 target exons (18), are all strongly connected with ELAVL2. RC3H2 and G3BP2 are RNAbps that are targets of nELAVL proteins (23,43) and hubs of the module strongly linked with ELAVL2 and RBFOX1. Co-expression between RNAbps has been also found at the proteomic level with higher co-expression in fetal brain compared with adult brain (Supplementary Material, Figure S2) (44,45), suggesting a role for those RNAbps in the development of human brain. Taken together, our genomic approach identified ELAVL2, RBFOX1 and FMR1 as important hubs in an RNAbp network expressed during frontal cortical development.
ELAVL2 targets are involved in synaptic function and neurodevelopmental disorders
To assess the role of ELAVL2 in regulating target genes in human brain development, we used phNs that can recapitulate human in vivo brain gene expression patterns (25,46–48). We first assessed the expression of ELAVL2 in the phN system using an ELAVL2-specific antibody (Supplementary Material, Figure S3) (24). ELAVL2 is expressed in MAP2 and NeuN positive phNs (Fig. 1E and Supplementary Material, Figure S4), supporting the use of these cells as a model to study ELAVL2 function. ELAVL2 is expressed throughout the cell including the nucleus; however, we noted colocalization of ELAVL2 and FMRP in the perinuclear region of phNs (Fig. 1E). These data are in line with previous work demonstrating an enrichment of FMRP in perinuclear structures in neurons (49).
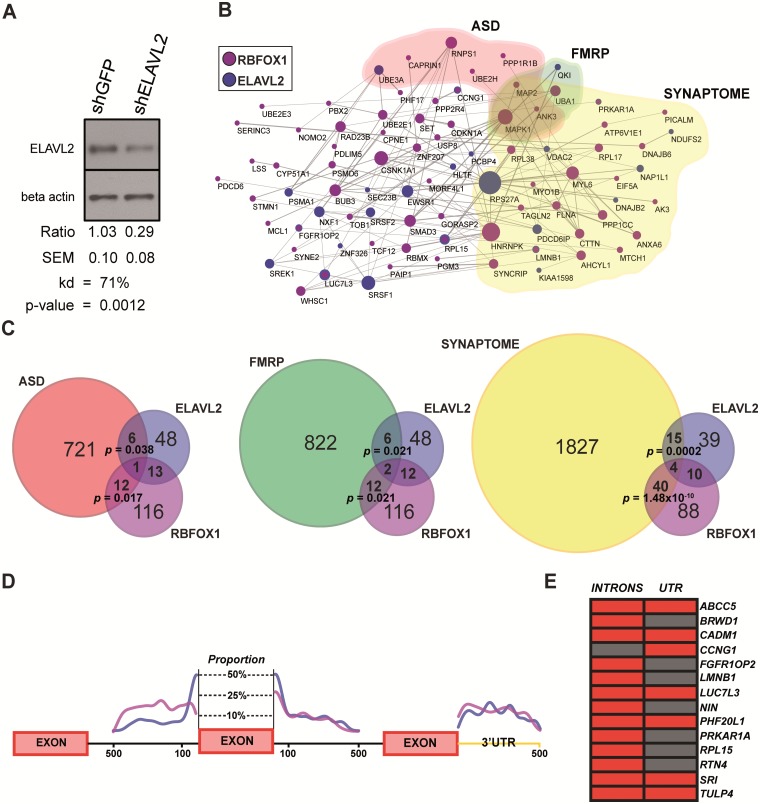
To examine the genetic network downstream of ELAVL2 in human neurons, we used a lentiviral short-hairpin in the microRNA context to kd ELAVL2 expression and differentiated phNs for 4-weeks. ELAVL2 protein levels were significantly reduced using this method (Fig. 2A), reflecting haploinsufficiency intended to alter, but not eliminate, ELAVL2 function. Although ELALV2 transcript levels were unchanged with kd (Supplementary Material, Table S2), changes in translation in the absence of changes in transcription are frequently observed for shRNAs constructed in the microRNA context (50). We carried out RNA-seq of five biological replicates in phNs with either ELAVL2 kd or a control hairpin.
Figure 2.
ELAVL2 kd alters alternative splicing events in phNs. (A) Characterization of ELAVL2 kd by immunoblotting. Protein expression of ELAVL2 was significantly reduced by 71% using shELAVL2. Samples were normalized to beta-actin and compared with shGFP control (t-test, P = 0.0012, n = 4 with each experiment performed at least in duplicate). (B) Protein–protein interaction network of ELAVL2 alternative spliced genes (blue) and RBFOX1 alternative spliced genes (purple). Also shown are genes associated with ASD (red); genes associated with FMRP-regulated pathways (green); and genes associated with synaptic function (yellow). (C) Overlaps of ELAVL2 and RBFOX1 alternatively spliced genes with ASD, FMRP-regulated pathways and synaptic genes. (D) Position enrichment of the ELAVL2 (blue) and RBFOX1- (purple) binding sites in exon-flanking intronic regions and the 3′UTR of alternative spliced genes for all of the alternatively spliced genes in either dataset. (E) Co-occurrence of binding sites in introns or UTRs of the 14 common alternatively spliced targets of ELAVL2 and RBFOX1. Red corresponds to a co-occurrence; Grey corresponds to a non-occurrence.
To identify potential target genes, we conducted an in silico prediction of alternatively spliced events regulated by ELAVL2. Due to differences between the current method and prior computational analyses, in order to make direct comparisons, we also carried out the same approach for the data from the RBFOX1 study as previously described (18). In total, we identified 68 alternatively spliced genes in ELAVL2 kd and a total of 142 alternatively spliced genes in the reanalysis of RBFOX1 kd. As expected, a significant 18% (26 genes; hypergeometric test, P = 4.49 × 10−11) overlap with the previous analysis was observed, consistent with typical published overlaps between different splicing algorithms (51). Almost all of the alternative splicing events detected in the presence of ELAVL2 and RBFOX1 kd belong to the cassette exon category with many of the events also categorized as mutually exclusive exon skipping or the use of alternative 3’ or 5’ splice sites (Supplementary Material, Table S1).
To improve our understanding of the functional relationships between splicing events driven by ELAVL2 and RBFOX1, we highlighted alternatively spliced events representing known protein–protein interactions. We found an overrepresentation of known protein-protein interactions among the alternatively spliced genes emphasizing common pathways in which ELAVL2 and RBFOX1 might be involved (Fig. 2B). Among the most interesting pathways, we found that RNPS1, an RNAbp recently associated with ASD (52), and HNRNPK are affected by RBFOX1 splicing and are strongly associated with ELAVL2 during brain development (Fig. 1D), suggesting co-regulation of similar pathways between those four important neuronal RNAbps. We also identified significant overlaps of alternatively spliced genes driven by ELAVL2 and RBFOX1 involved in ASD, FMRP-related pathways and synaptic function (Fig. 2C), implicating a role for ELAVL2 in neurodevelopmental disorders such as ASD and intellectual disability (ID) (Supplementary Material, Table S1). Taken together, these data highlight novel and well-known targets of ELAVL2 and RBFOX1, strengthening links to ASD and brain development.
To further validate the alternative splicing prediction, we examined the enrichment of ELAVL2 and RBFOX1-binding motifs in the flanking region of the alternatively spliced exons. Interestingly, we found a similar pattern of enrichment for ELAVL2- and RBFOX1-binding sequences in the local intronic space (500 nt), with a higher specificity in the downstream intronic sequence, and in the 3′-untranslated region (3′-UTR) when analyzing all of the alternatively spliced genes in either dataset (Fig. 2D). These data are consistent with previous findings in which RBFOX1 and nELAVL family members show preferential binding to the downstream intronic region (17,18,23). Moreover, we found a similar distribution of binding site enrichment in the 3′-UTR of the alternatively spliced genes suggesting a role for ELAVL2 along with RBFOX1 in mRNA stabilization and trafficking as previously hypothesized (17,23,53,54). When we examined the co-occurrence of ELAVL2- and RBFOX1-binding sites in the 14 genes alternatively spliced by both factors, we found that 13/14 exhibit co-occurrence in intronic regions and 7/14 exhibit co-occurrence in the 3’UTR (Fig. 2E). As recent work has indicated that RBFOX1 regulates transcript stability (53), we therefore investigated whether ELAVL2 also regulates transcript stability. Using our reanalyzed RBFOX1 phN dataset, we first confirmed regulation of transcript stability by RBFOX1 (Wilcoxon ranked test, P = 1.2 × 10−04). However, we did not find that ELAVL2 expression in phNs affects transcript stability (Wilcoxon ranked test, P = 0.9). Taken together, these results support a role for ELAVL2 in regulating alternative splicing and both converging and parallel pathway regulation with RBFOX1.
Identification of genes differentially expressed downstream of ELAVL2
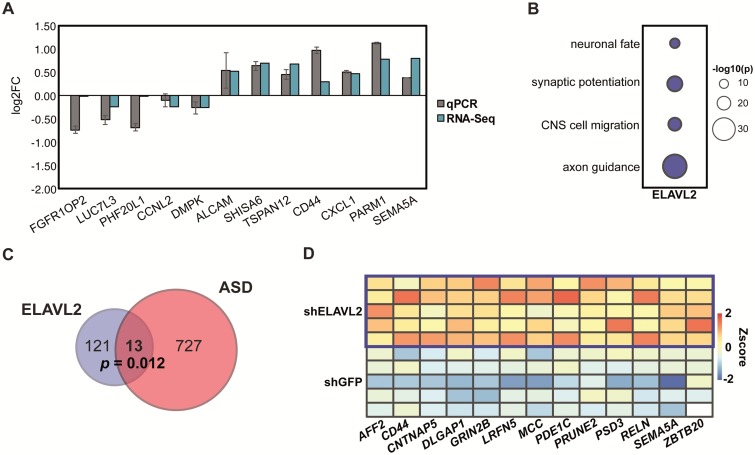
Because alternative splicing can affect other mechanisms of gene expression, such as transcription, to gain insight into the potential function of ELAVL2 on the gene regulatory networks important for brain development, we determined the set of DEGs in the kd phNs and their association with neurodevelopmental disorders. We found in total 134 DEGs with ELAVL2 kd and 99 DEGs with RBFOX1 kd (absolute log2FC > 0.3, false discovery rate (FDR) < 0.05; Methods and Supplementary Material, Table S2). To validate the ELAVL2 DEGs, we performed qRT-PCR on 12 selected genes including 3 differentially spliced genes from Figure 2E (Fig. 3A). Even though the differentially spliced genes did not reach our cutoff for differential expression, we could observe a significant change in expression, suggesting our statistical cutoff is sufficiently stringent for removing false positives but may allow for false negatives. We next ascertained whether these DEGs are enriched for any biologically relevant functional categories. Notably, we found that ELAVL2 DEGs are enriched for pathways implicated in neurodevelopment such as axon guidance, neuronal migration and synaptic function (Fig. 3B).
Figure 3.
ELAVL2 kd alters gene expression in phNs. (A) Validation of 12 DEGs detected by RNA-seq using qRT-PCR. RNA-seq values are shown in blue and qPCR values are shown in grey. Standard error of the mean is indicated by the black bars. (B) Gene ontology enrichment of DEGs downstream of ELAVL2. (C)ELAVL2 DEGs are enriched for ASD genes. (D) Expression of the 13 ASD genes that are upregulated with ELAVL2 kd.
As the nELAVL family has been linked to neuronal physiology and brain function (17,18,23,53), we examined whether any of the DEGs are linked with ASD. We identified a small but significant overlap of ELAVL2 DEGs- and ASD-related genes (Fig. 3C), finding well-characterized candidates such as AFF2, CD44, MCC, PRUNE2 and SEMA5A (Fig. 3D) (55–59). Others, such as DLAG1P1, GRIN2B and RELN are also implicated in FMRP-regulated pathways (31,60–62), suggesting ELAVL2 and FMRP regulate similar biological pathways implicated in ASD. Interestingly, we did not find significant overlap of ASD- or FMRP-related genes with either reported targets of ELAVL1 or nELAVLs (23) (data not shown), highlighting these as potential ELAVL2-specfic pathways.
ELAVL2 kd highlights modules involved in neuronal development and synaptic function
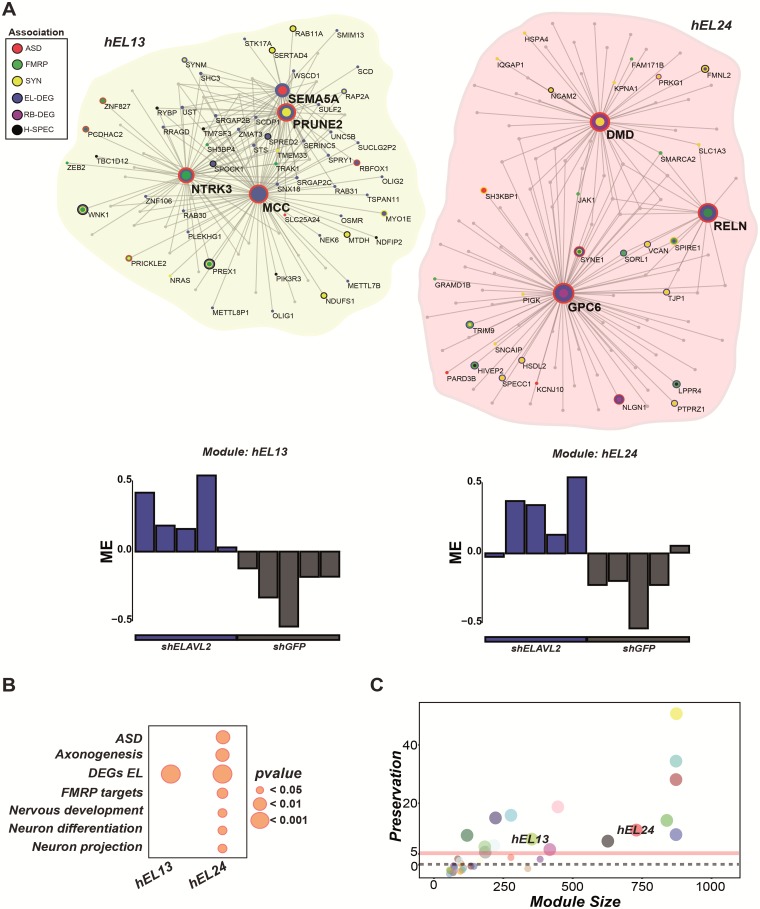
To better infer the genetic regulatory mechanisms altered by the ELAVL2 kd, we carried out weighted gene co-expression network analysis (WGCNA) (63,64) to identify the most highly co-expressed (hub) genes related to ELAVL2 expression and implicated in ASD, FMRP-regulated pathways or synaptic function. We found six modules that were directly correlated with ELAVL2 kd status. Among those, two modules showed strong association with DEGs, ASD and/or FMRP targets: hEL13 and hEL24, respectively (Fig. 4A and B, Supplementary Material, Table S3). We then asked whether the biological network modules dependent on ELAVL2 are conserved in RBFOX1 kd. We found a striking pattern of preservation, with the hEL13 and hEL24 key modules both strongly preserved within the RBFOX1 kd dataset (Fig. 4C). Taken together, these observations highlight the role of ELAVL2 in neurodevelopment and related disorders, by affecting the expression of multiple genes implicated in ASD. Moreover, we show how the gene co-expression networks identified with ELAVL2 kd are preserved within the RBFOX1 kd supporting a shared functional relationship in the regulation of these critical neurodevelopmental pathways.
Figure 4.
ELAVL2 kd alters genetic modules involved in neurodevelopmental networks. (A) ELAVL2 kd yielded two key modules (hEL13 and hEL24) enriched in synaptic genes, FMRP targets, and DEGs. Genes with higher connectivity are indicated by larger node size. ASD genes (red); FMRP targets (green); synaptic genes (yellow); ELAVL2 DEGs (blue); RBFOX1 DEGs (purple); Human frontal pole-specific genes derived by comparison of gene expression in human and non-human primate brains (black) (25). Barplots below each module show the relationship of each sample to the module eigengene. In both modules, ELAVL2 kd samples show strong positive correlation. (B) Gene ontology enrichment indicates modules strongly enriched for neurogenesis and neurodevelopmental pathways. (C) Preservation of the ELAVL2 modules in the RBFOX1 kd data set (significance Z score > 5). The two key modules of interest are indicated.
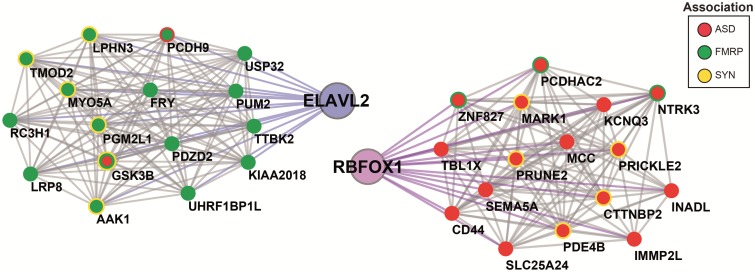
Since interacting proteins are more likely to be co-expressed compared with non-interacting proteins (65), we determined which proteins co-expressed with ELAVL2 and RBFOX1 showed differential expression driven by ELAVL2 in phNs. ELAVL2 itself is in the hEL19 module, which is also significantly enriched for FMRP targets (hypergeometric test, P = 0.012; FDR correction, perm = 0.0001) and includes other RNAbps such as PUM2 and RC3H1 implicated in FMRP-regulated pathways and found co-expressed with ELAVL2 in frontal pole development (Fig. 1D). RBFOX1 was found in the hEL24 module, which is enriched for ELAVL2 DEGs, as well as ASD and FMRP targets. RBFOX1 is also found co-expressed with several genes implicated in ASD, FMRP-regulation and synaptic function (e.g. NTRK3, PRUNE2 and SEMA5A) consistent with its involvement in neuronal development and related disorders (Fig. 5). Furthermore, we also identified additional RNAbps strongly co-expressed with RBFOX1 (Supplementary Material, Table S3). Among those was NOVA1, implicated in neuronal excitability and post-transcriptionally regulated by RBFOX1 (18,66), and CPEB3, implicated in synaptic plasticity and dendrite morphogenesis (67,68), indicating that RBFOX1, NOVA1 and CPEB3 are likely directing similar neurodevelopmental pathways in phNs. These data are consistent with previous results showing that RBFOX1 and NOVA1 synergistically regulate alternative splicing (69).
Figure 5.
ELAVL2 kd alters co-expression of genes involved in synaptic, ASD, and FMRP pathways. On the left, the co-expressed genes with ELAVL2; on the right, the co-expressed genes with RBFOX1. ASD genes (red); FMRP targets (green); Synaptic genes (yellow).
Because it has been previously shown that ELAVL2 is differentially expressed specifically in the human frontal pole compared with non-human primates (25), we asked whether the transcriptional circuitry of human frontal pole specific genes is altered by ELAVL2 kd. To test this hypothesis, we determined the intranetwork connectivity of genes with human frontal pole specific patterns of expression (25). Remarkably, the human-specific genes have significantly higher connectivity compared with all the pairs of the network (Wilcoxon rank sum test, P 0.01). Significant differences in connectivity were also found for ASD, FMRP, synaptic and alternatively spliced genes (Wilcoxon test, P < 0.01; Supplementary Material, Figure S5). These data suggest that dysregulation of ELAVL2 affects the normal circuitry of genes implicated in neurodevelopmental disorders and human-specific frontal pole expressed genes, highlighting ELAVL2 as a regulator of important neuronal pathways that may be involved in frontal lobe evolution.
Discussion
The genes and molecular pathways at risk in ASD and other neurodevelopmental disorders are still not fully delineated. As these disorders are thought to be a consequence of human brain evolution, the study of a gene with human-specific brain expression such as ELAVL2 provides direct insight into how brain disorders and brain evolution are connected. Furthermore, there is accumulating evidence that the pathways disrupted in ASD are highly convergent and post-transcriptional mechanisms are at play. Using a human cellular model and RNA-seq, we identified the transcriptional program regulated by ELAVL2. Although this cellular system cannot model several developmental properties associated with ASD pathophysiology such as cortical migration, axon guidance, and brain circuit formation, it can recapitulate in vivo gene expression patterns of the human brain. ELAVL2-associated co-expression networks are highly preserved with co-expression networks regulated by RBFOX1, also an ASD gene. We also found that these two splicing factors are co-expressed with FMR1 during the critical period of human fetal brain development, and all of these RNAbps regulate molecular pathways implicated in neurodevelopmental disorders and/or neuronal function. More importantly, we have uncovered relationships among these genes with other RNAbps suggesting the existence of an even larger network of RNAbps that underlie vulnerability to disorders of neurodevelopment. Among the additional ELAVL2-associated genes encoding RNAbps, we found well-characterized genes such as, HNRNPH2, RNPS1 and YTHDC2 that are associated with ASD (Fig. 1D) (52,61). The observation that several RNAbps implicated in ASD interact with ELAVL2 in frontal cortical development further supports the role of ELAVL2 in such disorders.
To better understand the functional role of ELAVL2 in human neuronal gene regulatory networks, we performed a genome-wide analysis examining transcriptional and splicing changes caused by loss of ELAVL2. We focused on genes implicated in ASD, FMRP-regulated pathways, and synaptic function. Although we identified a relatively small number of alternatively spliced or DEGs downstream of ELAVL2, these gene lists significantly overlapped with all of these functional categories. It is possible that our analysis is limited by the partial kd of ELAVL2 expression; however, we chose to recapitulate ELAVL2-haploinsufficiency that might most closely mimic a disease-state. It is unlikely that other nELAVL family members are compensating, as we did not observe changes in their expression (Supplementary Material, Table S2). The relatively low nuclear expression and enhanced perinuclear expression of ELAVL2 together with dispersed cytoplasmic expression (Fig. 1E) may suggest an additional role for ELAVL2 post-splicing although pre-mRNA splicing has been observed outside of the nucleus (70), particularly in the dendrites of neurons (71,72). However, while we did not observe an effect of ELAVL2 on RNA stability, future genome-wide binding studies can better ascertain whether ELAVL2 is involved in RNA trafficking or other aspects of RNA processing.
Among the ELAVL2 alternatively spliced events, we found CADM1, a synaptic cell adhesion molecule associated with ASD (73–76), CTNNB1, a neuronal development gene involved in ASD and FMRP-regulated pathways (31,61,77) and UBE3A, a gene important for dendritic patterning and associated with ASD (78–81) (Supplementary Material, Table S1). CADM1 and CTNNB1 have been previously described as targets of the nELAVL family (23,43), further supporting nELAVL regulation of ASD candidates genes. Moreover, CADM1 was also found as an alternatively spliced gene downstream of RBFOX1 and has previously been shown as an RBFOX1 target in a mouse brain HITS-CLIP study (17), suggesting co-regulation of CADM1 by ELAVL2 and RBFOX1.
Although the number of ELAVL2 and RBFOX1 overlapping targets is small (10 DEGs and 14 alternatively spliced), such an overlap is actually greater than previous studies examining nELAVL shared alternatively spliced targets (six genes; (23,43)). Moreover, by using a network approach, we uncovered conserved co-expression between ELAVL2 and RBFOX1 regulated genes that strengthen the involvement of ELAVL2 in neurodevelopmental disorders. The hEL24 module is strongly enriched for ASD genes, FMRP targets, and synaptic genes and the hEL24 module is enriched for DEGs implicated in ASD (Supplementary Material, Table S3). The hEL13 module is also significantly enriched for DEGs in ELAVL2 and RBFOX1 kd implicated in ASD and ID. In addition, the hEL24 module is remarkably enriched for genes involved in neurodevelopment, axonogenesis and neuronal projection (Fig. 4B).
Remarkably, we showed that the expression of ASD-associated genes is significantly affected by ELAVL2 kd. Among the most interesting candidates, we detected AFF2, CNTNAP5, GRIN2B, LRFN5, MCC, PRUNE2 and SEMA5A (Supplementary Material, Table S2). Such ASD-associated genes are also prioritized as module hubs genes. Taken together, these data demonstrate a strong connection between the ELAVL2-associated networks and neurodevelopmental disease. In addition, this data provide an important source for novel ELAVL2 candidate genes and potentially implicate ELAVL2 in etiologies such as ASD and/or ID.
Network analysis allowed us to determine alteration of the intranetwork connectivity by ELAVL2 kd. We found that ELAVL2 kd significantly affects the circuitry of genes associated with ASD, FMRP-targets, synaptic genes, and alternatively spliced genes, providing direct evidence for the role of ELAVL2 in the regulatory mechanisms of neurons. Also, of note, we identified a remarkable dysregulation at the circuitry level of genes with human specific expression patterns in frontal lobe compared with other non-human primates. These data lead us to speculate that ELAVL2 may have a key role in the regulatory networks of genes that might be implicated in human brain evolution.
The finding that ELAVL2 does not regulate transcript stability like RBFOX1 illustrates a key difference between RBFOX1 and ELAVL2 function and may explain why fewer differentially spliced transcripts were identified with reduction of ELAVL2 compared with RBFOX1. Recent work has shown that ELAVL1 (or HuR) kd affects protein localization rather than transcript level or splicing (54). However, another recent study has shown that ELAVL1 has a higher binding affinity to intronic regions compared with 3′UTRs supporting a role in splicing regulation (23,82). Consistent with this, our findings confirm a role for ELAVL2 in regulating splicing and transcript abundance; however, we cannot rule out the possibility that ELAVL2 also regulates protein localization through transcript trafficking. Such a role for nELAVL family members has been previously hypothesized (23,83). Future studies examining the interaction of ELAVL2 with its bound transcripts in neurons will provide insight into this possibility.
These data are the first report defining alternative splicing and differential gene expression downstream of ELAVL2 in human neurons; however, additional studies will be required to determine which changes are due to direct versus indirect regulation by ELAVL2. We have found that many of these targets are enriched for genes linked to ASD and neurodevelopment. Our data further supports a model where the function of co-expressed RNAbps may be important in the development of the human frontal cortex. Additionally, examining these novel and well-characterized RNAbps and their association with ELAVL2 predict pathways that may be important for neurodevelopmental disorders and suggests new targets for further study. We also find enrichment of genes involved in neuronal function, suggesting that ELAVL2 is an important factor in establishing normal neuronal and synaptic function in the brain, thus deepening our understanding of normal neurodevelopment and its relationship to ASD pathogenesis in humans.
Materials and Methods
Constructs
DNA constructs used in this study were as follows: pLenti6.4-ELAVL1-V5 (generated by Invitrogen Gateway system from pDONR223, Open Biosystems, OHS1770-202320774), pFRT-DestFLAGHA_HuB (ELAVL2) (Addgene, 65758), pFRT-TODestFLAGHA_HuC (ELAVL3) (Addgene, 65756) and pFRT-TODestFLAGHA_HuD (ELAVL4) (Addgene, 65757). All constructs were confirmed by sequencing.
Cell culture
phNs were cultured and differentiated as previously described in (18). phNs were transduced with lentiviruses containing either a specific hairpin against ELAVL2 or green fluorescent protein (GFP). Total RNA was extracted using a miRNeasy Mini kit (Qiagen, 217004). 293T cells (ATCC) were cultured in DMEM containing 10% fetal bovine serum (Invitrogen, 0437028) and antibiotic-antimycotic (Invitrogen, 15240-062) at 37 °C under 5% CO2. Forty-eight hours after transfection using FuGENE6 transfection reagent (Promega, E2691), cells were harvested. Cells were washed with PBS, lysed with a lysis buffer (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 0.1% (w/v) Triton X-100) with protease inhibitor cocktail (Sigma, P8340).
qRT-PCR
qRT-PCR was carried out as previously described in (18). Single-stranded cDNAs were made using DNaseI, Amplification grade (Invitrogen, 18068015) and SuperScript III First-Strand Synthesis SuperMix (Invitrogen, 18080400) and amplified by PCR according to the manufacturer’s instructions. qRT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, 172-5121) and a Touch Real-Time PCR Detection System (Bio-Rad Laboratories, CFX384). Data analysis was performed by the delta delta Cq method using a Touch Real-Time PCR Detection System and CFX Manager software (Bio-Rad Laboratories, CFX384). Each sample was normalized to beta-actin expression followed by normalization to an shRNA GFP control. Two to four biological replicates and three to four technical replicates were used for each experiment. Data analysis was performed by the delta delta Cq method. Primer sequences are available upon request.
Immunoblotting
Whole cell lysates were generated from transduced phNs as previously described in (18) and immunoblotted with the following antibodies: rabbit anti-HuB (ELAVL2; Hel-N1) (1:200; Sigma, H1538) or rabbit anti-ELAVL2 (1:15 000; ProteinTech, 14008-1-AP), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (1:5000; Millipore, MAB374) followed by goat anti-rabbit-poly-horseradish peroxidase (1:4000; Cell Signaling Technology), mouse anti-beta-actin-horseradish peroxidase (1:100 000; Sigma-Aldrich, A3854), IRDye 680RD donkey anti-mouse IgG (10 000; LI-COR Biosciences, 926-68072) or IRDye 800CW donkey anti-rabbit IgG, (10 000; LI-COR Biosciences, 926-32214). The images were collected using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunocytochemistry
Cultured phNs were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with a solution of 10% goat serum and 1% BSA in PBS. Cells were incubated with the following primary antibodies at 4 °C overnight: rabbit anti-HuB (ELAVL2; Hel-N1) (1:200; Sigma, H1538), mouse anti-FMRP (1:100; clone 2F5-1; 2F5-1 was deposited to the DSHB by Tartakoff, Alan M./Fallon, J.R. (DSHB Hybridoma Product 2F5-1) (84), anti-MAP2 (1:2000; Abcam, ab92434), anti-NeuN (1:100; Millipore, MAB377). Cells were washed and then incubated with species-specific secondary antibodies for 1 h at room temperature: Alexa Fluor 647 and/or Alexa Fluor 555 (1:2000; Invitrogen). For nuclear counterstaining, cells were coverslipped with ProLong Gold Antifade Mountant with DAPI (Invitrogen, P36934). TO-PRO-3 (Invitrogen, T3605) was also used to stain the nucleus. Confocal images were captured using Zeiss confocal laser scanning microscope (LSM 510 with META, Carl Zeiss).
RNA-seq analysis
RNA-seq was performed on an Illumina HiSeq 2500 using polyA-enriched RNA from five biological replicates from phNs: 5 shGFP and 5 shELAVL2. 100 bp single-end reads were generated and uniquely mapped to the human genome (hg19/GRCh37) using Segemehl (85) with the following parameters -S -A 95 -T. Uniquely mapped reads where extracted. Mapped reads were counted and RPKM were calculated using a custom script in R based on the GenomicFeatures library and Ensembl gene reference (86,87). In order to make a direct comparison to our previously published data, we performed the same protocol for shRBFOX1 and shGFP samples from (18). Alternative splicing, isoform levels, and bayes factors for splicing events were inferred using the MISO algorithm with default parameters using both isoform-centric and exon-centric analysis based on MISO annotation (88). RefSeq transcript annotation was used for exon junction annotation and exons with Ψ ≤ 0.1 were excluded from the analysis. Alternative splicing events and/or exon dosage were manually curated using the UCSC web-browser and validated using DEXseq (89–91)
We compared each shELAVL2 sample with each shGFP sample sequentially and the total alternatively spliced genes where then summarized into a single list from all the pairwise comparisons. The same approach was used for shRBFOX1 and shGFP samples. The DEGs were calculated using DESeq (92). FDR < 0.05 and |log2FC| > 0.3 thresholds were applied.
RNAbp site analysis
To perform RNAbp site enrichment, we downloaded intronic and 3′-UTR sequences from the UCSC web interface (91). We interrogated only flanking region of 500 bp upstream and 500 bp downstream of exons as well as the full 3′-UTR region. RNAbp site enrichment was performed using RBPmap (93) with high stringency using nELAVL and RBFOX1 core sequences. Predicted motif detections were manually confirmed using a custom Perl script.
Additional large-scale datasets
Human brain expression data were downloaded and subsetted from BrainSpan (www.brain-map.org, 2011). The protein–protein interaction database was built using Biogrid and inWeb human datasets (94,95). The RNAbp gene sets were downloaded from RBPDB (96), autism related genes were downloaded from SFARI according to the latest release (97), synaptic-related genes were downloaded from SynaptomeDB (98) and FMRP-target genes and human-specific frontal pole genes were used from independent publications (25,31). The proteome data was collected from the Human Proteome Map (44) and the Human Proteome Atlas (45).
Network construction
For the protein–protein interaction network, we considered a protein-protein interaction present if found in either one of the datasets used. We used the DEGs of shELAVL2 and shRBFOX1 and their known protein–protein interactions. To infer the dorsolateral frontal cortex RNAbp network, we applied the wTO approach (38) using the RNAbp as nodes for the network interaction. In contrast to other methods, we calculate the wTO by taking into account that an RNAbp can act either as an activator or repressor of splicing. Therefore, edges between RNAbps show the same or opposite predicted function according to their commonly correlated gene sets. WGCNA of the shELAVL2 and shGFP samples was performed using a custom R script (63). We defined the modules using mid-weight bicorrelation followed by unsigned network approach, minimum module size = 60, power = 12, deepSplit = 4 and cut height threshold = 0.1.
We calculated the module preservation between the ELAVL2 and RBFOX1 datasets using the module preservation function implemented in the WGCNA package applying 100 permutations (64). Data integration and statistical analysis were based on custom R and SQL scripts. The hypergeometric test was used to detect gene set enrichment in WGCNA modules. As background for population size, we used an independent cortical gene expression set data as described in Parikshak et al. 2013 (99) (15585 total genes), defining a more appropriate background set for phNs. To evaluate the enrichment, we performed permutation testing by randomizing each gene set 10000 times and assessing for ASD, FMRP and synaptic genes within the cortically expressed genes and assessing significance by the hypergeometric test. None of the randomized overlaps showed significant P-values (perm P = 0.0001).
Gene ontology analysis
Gene ontology enrichment was performed using DAVID (100) and confirmed using WebGestalt (101,102). As background, we used all the expressed genes. In Supplementary Material, Table S4, we indicate the FDR values and number of gene belonging to the specific gene ontology category from DAVID.
Code
All scripts are available upon request to the corresponding authors.
Accession numbers
The NCBI Gene Expression Omnibus accession number for the RNA-seq data reported in this manuscript is GSE69092.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
B.L.F. thanks Cameron Baker, Xizhe Wang and Andrew Curnow for technical support. We also thank Dr Kimberly Huber for the FMRP antibody, Dr Shin Yamazaki and Ms Stephanie Hickey for technical assistance.
Conflict of Interest statement. None declared.
Funding
This work was supported by Uehara Memorial Foundation (N.U.); NIH Grant R00MH090238, R01MH102603, R21MH107672, Research Grant No. 5-FY13-19 from the March of Dimes Foundation, Once Upon a Time Foundation, Commercial Real Estate Women (CREW) Dallas, Friends of the Alzheimer’s Disease Center at UT Southwestern, and a Jon Heighten Scholar in Autism Research at UT Southwestern Endowment (G.K.); and NIH Grants K08MH086297 and R01NS082094, and NIH/National Center for Advancing Translational Science (NCATS) UCLA Clinical and Translational Science Institute (CTSI) Grant Number UL1TR000124 (B.L.F).
References
- 1. De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S. et al. (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature, 515, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Butler M.G., Rafi S.K., Hossain W., Stephan D.A., Manzardo A.M. (2015) Whole exome sequencing in females with autism implicates novel and candidate genes. Int. J. Mol. Sci., 16, 1312–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature, 485, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu T.W., Chahrour M.H., Coulter M.E., Jiralerspong S., Okamura-Ikeda K., Ataman B., Schmitz-Abe K., Harmin D.A., Adli M., Malik A.N. et al. (2013) Using whole-exome sequencing to identify inherited causes of autism. Neuron, 77, 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Raj B., Blencowe B.J. (2015) Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron, 87, 14–27. [DOI] [PubMed] [Google Scholar]
- 6. Quesnel-Vallières M., Irimia M., Cordes S.P., Blencowe B.J. (2015) Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev., 29, 746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raj B., Irimia M., Braunschweig U., Sterne-Weiler T., O'Hanlon D., Lin Z.Y., Chen G.I., Easton L.E., Ule J., Gingras A.C. et al. (2014) A global regulatory mechanism for activating an exon network required for neurogenesis. Mol. Cell., 56, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grabowski P. (2011) Alternative splicing takes shape during neuronal development. Curr. Opin. Genet. Dev., 21, 388–394. [DOI] [PubMed] [Google Scholar]
- 9. Iijima T., Hidaka C., Iijima Y.H. (2016) Spatio-temporal regulations and functions of neuronal alternative RNA splicing in developing and adult brains. Neuroscience Res., 109, 1–8. [DOI] [PubMed] [Google Scholar]
- 10. Scotti M.M., Swanson M.S. (2016) RNA mis-splicing in disease. Nat. Rev. Genet., 17, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bryant C.D., Yazdani N. (2016) RNA‐binding proteins, neural development and the addictions. Genes Brain Behav., 15, 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Norris A.D., Gao S., Norris M.L., Ray D., Ramani A.K., Fraser A.G., Morris Q., Hughes T.R., Zhen M., Calarco J.A. (2014) A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol. Cell, 54, 946–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yano M., Okano H.J., Okano H. (2005) Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem., 280, 12690–12699. [DOI] [PubMed] [Google Scholar]
- 14. Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature, 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R. et al. (2015) RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science, 347, 1254806.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irimia M., Weatheritt R.J., Ellis J.D., Parikshak N.N., Gonatopoulos-Pournatzis T., Babor M., Quesnel-Vallieres M., Tapial J., Raj B., O’Hanlon D. et al. (2014) A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell, 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weyn-Vanhentenryck S.M., Mele A., Yan Q., Sun S., Farny N., Zhang Z., Xue C., Herre M., Silver P.A., Zhang M.Q. et al. (2014) HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep., 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fogel B.L., Wexler E., Wahnich A., Friedrich T., Vijayendran C., Gao F., Parikshak N., Konopka G., Geschwind D.H. (2012) RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum. Mol. Genet., 21, 4171–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bill B.R., Lowe J.K., Dybuncio C.T., Fogel B.L. (2013) Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int. Rev. Neurobiol., 113, 251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee J.A., Damianov A., Lin C.H., Fontes M., Parikshak N.N., Anderson E.S., Geschwind D.H., Black D.L., Martin K.C. (2016) Cytoplasmic Rbfox1 regulates the expression of synaptic and autism-related genes. Neuron, 89, 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta S., Ellis S.E., Ashar F.N., Moes A., Bader J.S., Zhan J., West A.B., Arking D.E. (2014) Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat. Commun., 5, 5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kraushar M.L., Thompson K., Wijeratne H.S., Viljetic B., Sakers K., Marson J.W., Kontoyiannis D.L., Buyske S., Hart R.P., Rasin M.R. (2014) Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc. Natl. Acad Sci. USA, 111, E3815–E3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ince-Dunn G., Okano H.J., Jensen K.B., Park W.Y., Zhong R., Ule J., Mele A., Fak J.J., Yang C., Zhang C. et al. (2012) Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron, 75, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohtsuka T., Yano M., Okano H. (2015) Acute reduction of neuronal RNA binding Elavl2 protein and Gap43 mRNA in mouse hippocampus after kainic acid treatment. Biochem. Biophys. Res. Commun., 466, 46–51. [DOI] [PubMed] [Google Scholar]
- 25. Konopka G., Friedrich T., Davis-Turak J., Winden K., Oldham M.C., Gao F., Chen L., Wang G.Z., Luo R., Preuss T.M. et al. (2012) Human-specific transcriptional networks in the brain. Neuron, 75, 601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamada K., Iwayama Y., Hattori E., Iwamoto K., Toyota T., Ohnishi T., Ohba H., Maekawa M., Kato T., Yoshikawa T. (2011) Genome-wide association study of schizophrenia in Japanese population. PloS One, 6, e20468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hormozdiari F., Penn O., Borenstein E., Eichler E.E. (2015) The discovery of integrated gene networks for autism and related disorders. Genome Res., 25, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tassone F., Hagerman R.J., Ikle D.N., Dyer P.N., Lampe M., Willemsen R., Oostra B.A., Taylor A.K. (1999) FMRP expression as a potential prognostic indicator in fragile X syndrome. Am. J. Med. Genet., 84, 250–261. [PubMed] [Google Scholar]
- 29. Stefani G., Fraser C.E., Darnell J.C., Darnell R.B. (2004) Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 24, 7272–7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown V., Jin P., Ceman S., Darnell J.C., O'Donnell W.T., Tenenbaum S.A., Jin X., Feng Y., Wilkinson K.D., Keene J.D. et al. (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 31. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. et al. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell, 146, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Delorme R., Ey E., Toro R., Leboyer M., Gillberg C., Bourgeron T. (2013) Progress toward treatments for synaptic defects in autism. Nat. Med., 19, 685–694. [DOI] [PubMed] [Google Scholar]
- 33. Ramani A.K., Li Z., Hart G.T., Carlson M.W., Boutz D.R., Marcotte E.M. (2008) A map of human protein interactions derived from co-expression of human mRNAs and their orthologs. Mol. Syst. Biol., 4, 180.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calarco J.A., Zhen M., Blencowe B.J. (2011) Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. Rna, 17, 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raj B., O’Hanlon D., Vessey J.P., Pan Q., Ray D., Buckley N.J., Miller F.D., Blencowe B.J. (2011) Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol. Cell, 43, 843–850. [DOI] [PubMed] [Google Scholar]
- 36. Okano H.J., Darnell R.B. (1997) A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci., 17, 3024–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ayoub A.E., Oh S., Xie Y., Leng J., Cotney J., Dominguez M.H., Noonan J.P., Rakic P. (2011) Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc. Natl. Acad. Sci. USA, 108, 14950–14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nowick K., Gernat T., Almaas E., Stubbs L. (2009) Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc. Natl. Acad. Sci. USA, 106, 22358–22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Y.H., Wu C.C., Chou C.K., Huang C.Y.F. (2011) A translational regulator, PUM2, promotes both protein stability and kinase activity of Aurora-A. PloS One, 6, e19718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H.H., Kuwano Y., Galban S., Becker K.G., Kamara D., de Cabo R. et al. (2009) Ubiquitin-mediated proteolysis of HuR by heat shock. Embo J., 28, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Villa C., Fenoglio C., De Riz M., Clerici F., Marcone A., Benussi L., Ghidoni R., Gallone S., Cortini F., Serpente M. et al. (2011) Role of hnRNP-A1 and miR-590-3p in neuronal death: genetics and expression analysis in patients with Alzheimer disease and frontotemporal lobar degeneration. Rejuvenation Res., 14, 275–281. [DOI] [PubMed] [Google Scholar]
- 42. Cao W., Razanau A., Feng D., Lobo V.G., Xie J. (2012) Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res., 40, 8059–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bolognani F., Contente-Cuomo T., Perrone-Bizzozero N.I. (2010) Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res., 38, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. (2014) A draft map of the human proteome. Nature, 509, 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. (2015) Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419.. [DOI] [PubMed] [Google Scholar]
- 46. Palmer T.D., Schwartz P.H., Taupin P., Kaspar B., Stein S.A., Gage F.H. (2001) Cell culture: Progenitor cells from human brain after death. Nature, 411, 42–43. [DOI] [PubMed] [Google Scholar]
- 47. Smith I., Silveirinha V., Stein J.L., de la Torre-Ubieta L., Farrimond J.A., Williamson E.M., Whalley B.J. (2015) Human neural stem cell-derived cultures in three-dimensional substrates form spontaneously functional neuronal networks. J. Tissue Eng. Regen. Med., doi:10.1002/term.2001. [DOI] [PubMed] [Google Scholar]
- 48. Stein J.L., de la Torre-Ubieta L., Tian Y., Parikshak N.N., Hernandez I.A., Marchetto M.C., Baker D.K., Lu D., Hinman C.R., Lowe J.K. et al. (2014) A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron, 83, 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Achuta V.S., Rezov V., Uutela M., Louhivuori V., Louhivuori L., Castrén M.L. (2014) Tissue plasminogen activator contributes to alterations of neuronal migration and activity-dependent responses in fragile X mice. J. Neurosci., 34, 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rao D.D., Vorhies J.S., Senzer N., Nemunaitis J. (2009) siRNA vs. shRNA: similarities and differences. Adv. Drug Deliv. Rev., 61, 746–759. [DOI] [PubMed] [Google Scholar]
- 51. Liu R., Loraine A.E., Dickerson J.A. (2014) Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics, 15, 364.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nguyen L.S., Kim H.G., Rosenfeld J.A., Shen Y., Gusella J.F., Lacassie Y., Layman L.C., Shaffer L.G., Gécz J. (2013) Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum. Mol. Genet., 22, 1816–1825. [DOI] [PubMed] [Google Scholar]
- 53. Lovci M.T., Ghanem D., Marr H., Arnold J., Gee S., Parra M., Liang T.Y., Stark T.J., Gehman L.T., Hoon S. et al. (2013) Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol., 20, 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berkovits B.D., Mayr C. (2015) Alternative 3 ′ UTRs act as scaffolds to regulate membrane protein localization. Nature, 522, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abrams M.T., Doheny K.F., Mazzocco M.M., Knight S.J., Baumgardner T.L., Freund L.S., Davies K.E., Reiss A.L. (1997) Cognitive, behavioral, and neuroanatomical assessment of two unrelated male children expressing FRAXE. Am. J. Med. Genet., 74, 73–81. [DOI] [PubMed] [Google Scholar]
- 56. Hu V.W., Frank B.C., Heine S., Lee N.H., Quackenbush J. (2006) Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics, 7, 118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barber J., Ellis K., Bowles L., Delhanty J., Ede R., Male B., Eccles D. (1994) Adenomatous polyposis coli and a cytogenetic deletion of chromosome 5 resulting from a maternal intrachromosomal insertion. J. Med. Genet., 31, 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vaags A.K., Lionel A.C., Sato D., Goodenberger M., Stein Q.P., Curran S., Ogilvie C., Ahn J.W., Drmic I., Senman L. et al. (2012) Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am. J. Hum. Genet., 90, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Melin M., Carlsson B., Anckarsater H., Rastam M., Betancur C., Isaksson A., Gillberg C., Dahl N. (2006) Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology, 54, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li J., Shi M., Ma Z., Zhao S., Euskirchen G., Ziskin J., Urban A., Hallmayer J., Snyder M. (2014) Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol. Syst. Biol. 10, 774.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature, 485, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Persico A.M., D’Agruma L., Maiorano N., Totaro A., Militerni R., Bravaccio C., Wassink T.H., Schneider C., Melmed R., Trillo S. et al. (2001) Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol. Psychiatry, 6, 150–159. [DOI] [PubMed] [Google Scholar]
- 63. Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Langfelder P., Luo R., Oldham M.C., Horvath S. (2011) Is my network module preserved and reproducible? PLoS Comput. Biol., 7, e1001057.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ge H., Liu Z., Church G.M., Vidal M. (2001) Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet., 29, 482–486. [DOI] [PubMed] [Google Scholar]
- 66. Gehman L.T., Stoilov P., Maguire J., Damianov A., Lin C.H., Shiue L., Ares M., Jr, Mody I., Black D.L. (2011) The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat. Genet. 43, 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pavlopoulos E., Trifilieff P., Chevaleyre V., Fioriti L., Zairis S., Pagano A., Malleret G., Kandel E.R. (2011) Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell, 147, 1369–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bestman J.E., Cline H.T. (2008) The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc. Natl. Acad. Sci. USA, 105, 20494–20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang C., Frias M.A., Mele A., Ruggiu M., Eom T., Marney C.B., Wang H., Licatalosi D.D., Fak J.J., Darnell R.B. (2010) Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science, 329, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. König H., Matter N., Bader R., Thiele W., Müller F. (2007) Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell, 131, 718–729. [DOI] [PubMed] [Google Scholar]
- 71. Glanzer J., Miyashiro K., Sul J.Y., Barrett L., Belt B., Haydon P., Eberwine J. (2005) RNA splicing capability of live neuronal dendrites. Proc. Natl. Acad. Sci. USA, 102, 16859–16864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khaladkar M., Buckley P.T., Lee M.T., Francis C., Eghbal M.M., Chuong T., Suresh S., Kuhn B., Eberwine J., Kim J. (2013) Subcellular RNA sequencing reveals broad presence of cytoplasmic intron-sequence retaining transcripts in mouse and rat neurons. PloS One, 8, e76194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fujita E., Dai H., Tanabe Y., Zhiling Y., Yamagata T., Miyakawa T., Tanokura M., Momoi M., Momoi T. (2010) Autism spectrum disorder is related to endoplasmic reticulum stress induced by mutations in the synaptic cell adhesion molecule, CADM1. Cell Death Dis., 1, e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhiling Y., Fujita E., Tanabe Y., Yamagata T., Momoi T., Momoi M.Y. (2008) Mutations in the gene encoding CADM1 are associated with autism spectrum disorder. Biochem. Biophys. Res. Commun., 377, 926–929. [DOI] [PubMed] [Google Scholar]
- 75. Takayanagi Y., Fujita E., Yu Z., Yamagata T., Momoi M.Y., Momoi T., Onaka T. (2010) Impairment of social and emotional behaviors in Cadm1-knockout mice. Biochem. Biophys. Res. Commun., 396, 703–708. [DOI] [PubMed] [Google Scholar]
- 76. Fujita E., Tanabe Y., Imhof B.A., Momoi M.Y., Momoi T. (2012) Cadm1-expressing synapses on Purkinje cell dendrites are involved in mouse ultrasonic vocalization activity. PloS One, 7, e30151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chenn A., Walsh C.A. (2002) Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science, 297, 365–369. [DOI] [PubMed] [Google Scholar]
- 78. Lu Y., Wang F., Li Y., Ferris J., Lee J.A., Gao F.B. (2009) The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum. Mol. Genet, 18, 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Samaco R.C., Hogart A., LaSalle J.M. (2005) Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet., 14, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nurmi E.L., Bradford Y., Chen Y., Hall J., Arnone B., Gardiner M.B., Hutcheson H.B., Gilbert J.R., Pericak-Vance M.A., Copeland-Yates S.A. et al. (2001) Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics, 77, 105–113. [DOI] [PubMed] [Google Scholar]
- 81. Noor A., Dupuis L., Mittal K., Lionel A.C., Marshall C.R., Scherer S.W., Stockley T., Vincent J.B., Mendoza-Londono R., Stavropoulos D.J. (2015) 15q11.2 duplication encompassing only the UBE3A gene is associated with developmental delay and neuropsychiatric phenotypes. Hum. Mutat., 36, 689–693. [DOI] [PubMed] [Google Scholar]
- 82. Li J.H., Liu S., Zheng L.L., Wu J., Sun W.J., Wang Z.L., Zhou H., Qu L.H., Yang J.H. (2015) Discovery of protein–lncRNA interactions by integrating large-scale CLIP-seq and RNA-seq datasets. Front. Bioeng. Biotechnol., 2, 88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lenzken S.C., Achsel T., Carrì M.T., Barabino S.M. (2014) Neuronal RNA‐binding proteins in health and disease. Wiley Interdiscip. Rev. RNA, 5, 565–576. [DOI] [PubMed] [Google Scholar]
- 84. Gabel L.A., Won S., Kawai H., McKinney M., Tartakoff A.M., Fallon J.R. (2004) Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J. Neurosci., 24, 10579–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hoffmann S., Otto C., Kurtz S., Sharma C.M., Khaitovich P., Vogel J., Stadler P.F., Hackermuller J. (2009) Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol., 5, e1000502.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol., 9, e1003118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Katz Y., Wang E.T., Airoldi E.M., Burge C.B. (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Anders S., Reyes A., Huber W. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res., 22, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hinrichs A.S., Raney B.J., Speir M.L., Rhead B., Casper J., Karolchik D., Kuhn R.M., Rosenbloom K.R., Zweig A.S., Haussler D. et al. (2016) UCSC Data Integrator and Variant Annotation Integrator. Bioinformatics (Oxford, England), 32, 1430–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M. et al. (2014) The UCSC genome browser database: 2014 update. Nucleic Acids Res., 42, D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Paz I., Kosti I., Ares M., Jr., Cline M., Mandel-Gutfreund Y. (2014) RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res., 42, W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lage K., Karlberg E.O., Storling Z.M., Olason P.I., Pedersen A.G., Rigina O., Hinsby A.M., Tumer Z., Pociot F., Tommerup N. et al. (2007) A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol., 25, 309–316. [DOI] [PubMed] [Google Scholar]
- 95. Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res., 34, D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cook K.B., Kazan H., Zuberi K., Morris Q., Hughes T.R. (2011) RBPDB: a database of RNA-binding specificities. Nucleic Acids Res., 39, D301–D308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Banerjee-Basu S., Packer A. (2010) SFARI Gene: an evolving database for the autism research community. Dis. Model Mech., 3, 133–135. [DOI] [PubMed] [Google Scholar]
- 98. Pirooznia M., Wang T., Avramopoulos D., Valle D., Thomas G., Huganir R.L., Goes F.S., Potash J.B., Zandi P.P. (2012) SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics, 28, 897–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Parikshak N.N., Luo R., Zhang A., Won H., Lowe J.K., Chandran V., Horvath S., Geschwind D.H. (2013) Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell, 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Huang D.W., Sherman B.T., Lempicki R.A. (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc., 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 101. Zhang B., Kirov S., Snoddy J. (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res., 33, W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wang J., Duncan D., Shi Z., Zhang B. (2013) Web-based gene set analysis toolkit (WebGestalt): Update 2013. Nucleic Acids Res., 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.