Abstract
Extracellular deposition of amyloid-beta (Aβ) peptide, a metabolite of sequential cleavage of amyloid precursor protein (APP), is a critical step in the pathogenesis of Alzheimer’s disease (AD). While death-associated protein kinase 1 (DAPK1) is highly expressed in AD brains and its genetic variants are linked to AD risk, little is known about the impact of DAPK1 on APP metabolism and Aβ generation. In this study, we demonstrated a novel effect of DAPK1 in the regulation of APP processing using cell culture and mouse models. DAPK1, but not its kinase deficient mutant (K42A), significantly increased human Aβ secretion in neuronal cell culture models. Moreover, knockdown of DAPK1 expression or inhibition of DAPK1 catalytic activity significantly decreased Aβ secretion. Furthermore, DAPK1, but not K42A, triggered Thr668 phosphorylation of APP, which may initiate and facilitate amyloidogenic APP processing leading to the generation of Aβ. In Tg2576 APPswe-overexpressing mice, knockout of DAPK1 shifted APP processing toward non-amyloidogenic pathway and decreased Aβ generation. Finally, in AD brains, elevated DAPK1 levels showed co-relation with the increase of APP phosphorylation. Combined together, these results suggest that DAPK1 promotes the phosphorylation and amyloidogenic processing of APP, and that may serve a potential therapeutic target for AD.
Introduction
One of the main hallmarks of Alzheimer’s disease (AD) is the formation of extracellular senile plaques, preferentially composed of amyloid-beta (Aβ) peptides (1–7). These Aβ polypeptides tend to aggregate and are believe to be neurotoxic. Aβ is a short peptide of 36–43 amino acids, of which the Aβ40 and Aβ42 peptides have been identified as the most common; these peptides are derived from the intracellular processing of amyloid precursor protein (APP). APP processing occurs via two pathways: an α-secretase (ADAM10)-mediated non-amyloidogenic pathway and a β-secretase (BACE1)-mediated amyloidogenic pathway (1–3,5,8–21). In the non-amyloidogenic pathway, cleavage occurs by α-secretase within the Aβ domain, generating a large, soluble N-terminal fragment (sAPPα) and an 83-amino acid C-terminal fragment (CTFα) (22–25). Further cleavage of CTFα by γ-secretase generates P3 peptide and APP intracellular domain (AICD), which are nontoxic (22,23). In the amyloidogenic pathway, BACE1 cleaves at the start of the Aβ domain, which generates a soluble N-terminal fragment (sAPPβ) and a 99-amino acid C-terminal fragment (CTFβ) (26–29). Further cleavage of CTFβ by γ-secretase generates AICD and cytotoxic Aβ (30).
APP is a transmembrane protein that can be phosphorylated at several residues, which may affect the proteolytic processing and secretion pathways of this protein (31–34). APP has potentially eight phosphorylation sites in its cytoplasmic domain, including Thr668 (31–34). Thr668 phosphorylation is induced during neurite differentiation and is elevated in AD brains (33). Furthermore, pThr668-APP and BACE1 co-localize in enlarged endosomes in AD and cultured primary neurons (33). Moreover, the constitutive phosphorylation of APP at Thr668 in the brain is thought to regulate the nuclear translocation of the AICD and induce neurodegeneration (31–38). In addition, the Thr668Ala mutation or Thr668 kinase inhibitors reduce Aβ production (33). Thr668 phosphorylation may also facilitate the BACE1-mediated cleavage of APP to increase Aβ generation (33). However, the mechanisms by which Thr668 phosphorylation is regulated are not fully understood.
Death-associated protein kinase 1 (DAPK1) is an actin filament-associated, calcium/calmodulin-dependent serine/threonine kinase that promotes apoptosis in response to a variety of stimuli, including Fas, γ-interferon and TNFα (39,40). DAPK1, a known tumor suppressor protein, is also a key player in several modes of neuronal death/injury and has been implicated in late-onset Alzheimer’s disease (LOAD) (41,42). For example, DAPK1 overexpression or activation increases neuronal cell death (43,44), and DAPK1 ablation in neurons is less sensitive to apoptotic stimuli in cell culture and knockout (KO) animal models (45,46). DAPK1 physically and functionally interacts with the N-methyl-d aspartate (NMDA) receptor NR2B subunit at extrasynaptic sites and this interaction mediates brain damage from strokes (46–49). Other studies also indicated that DAPK1 polymorphism is associated with the risk of developing LOAD and that DAPK1 kinase activity-deficient mice are more efficient learners and have better spatial memories than wild-type (WT) mice (39,50–53). Recently, we showed that DAPK1 expression is highly upregulated in the brains of AD patients compared with age-matched normal subjects (54). Moreover, DAPK1 promotes tau protein accumulation and phosphorylation (54), whereas DAPK1 ablation in mice reduces tau protein stability and phosphorylation at multiple AD-related sites (54). Furthermore, DAPK1 has been shown to regulate tau function by modulating microtubule assembly, neuronal differentiation and tau phosphorylation (54–56). Since Aβ has been found to exacerbate tangle formation in tau mutant mice and tau reduction blocks Aβ-mediated toxicity (57–59), DAPK1 might contribute to AD by regulating tau and APP. However, it is unknown whether DAPK1 regulates APP processing or Aβ secretion, or whether modulating DAPK1 affects the development of Aβ pathology in AD.
In this study, we examined the effect of DAPK1 on APP processing and Aβ generation in various neuronal cell culture models and an AD mouse model. We found that DAPK1 interacts with APP and triggers APP phosphorylation at Thr668. Interestingly, DAPK1 ablation in APP overexpressing AD mice dramatically suppressed Aβ production. Taken together, these results demonstrate that DAPK1 is a critical regulator of APP processing and that DAPK1 deregulation may contribute to the progression of AD.
Results
DAPK1 increases secretion of human and mouse Aβ40
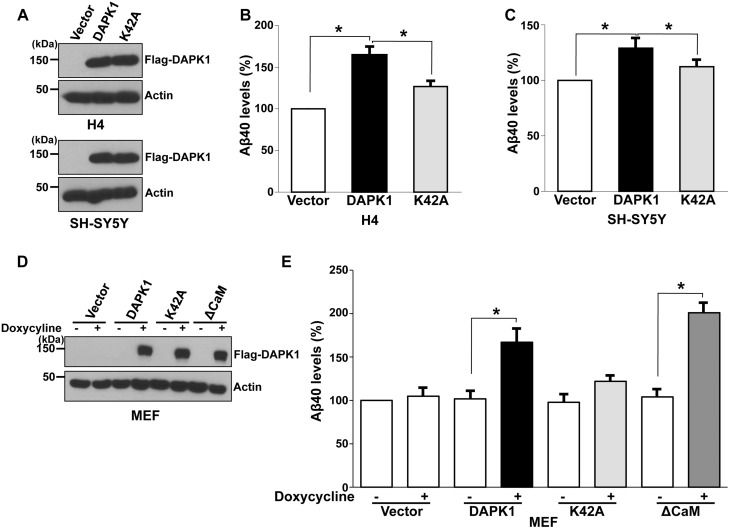
To determine whether DAPK1 plays a role in APP processing, we first determined the effects of DAPK1 overexpression on Aβ secretion in cultured cells by transfecting human neuroglioma H4 or human neuroblastoma SH-SY5Y cells with DAPK1 or the kinase activity-deficient mutant DAPK1K42A construct (Fig. 1A), followed by measuring total Aβ40 secretion from cells. Interestingly, in our enzyme-linked immunosorbent assay (ELISA), DAPK1 significantly increased human Aβ40 secretion compared with the empty vector (Fig. 1B and C). However, DAPK1K42A did not show significant effects compared with WT DAPK1 (Fig. 1B and C). Next, we used lentiviral expression systems to generate stable mouse embryonic fibroblast (MEF) cells expressing DAPK1 or its mutants in a tetracycline-inducible manner. The addition of doxycycline, a tetracycline analog, into the growth medium resulted in the production of DAPK1 (Fig. 1D). Again, doxycycline-induced expression of DAPK1 efficiently enhanced Aβ40 secretion (Fig. 1E). Moreover, its constitutively active mutant, DAPK1ΔCaM, also significantly increased Aβ40 secretion (Fig. 1E). However, Aβ42 were undetectable because these cell lines are not the pathological models. These results show that the overexpression of DAPK1 increases Aβ40 secretion and that its effects might depend on its kinase activity.
Figure 1.
DAPK1 stimulates the secretion of human- and mouse Aβ40. (A–C) Human H4 and SH-SY5Y cells were transfected with pRK5-Flag, pRK5-Flag-DAPK1 or pRK5-Flag-DAPK1K42A for 32 h. The cell lysates were subjected to western blot analysis with anti-Flag or anti-actin antibody and the levels of human Aβ40 in cell culture supernatants were determined by a solid phase sandwich ELISA assay. Each data point represents the mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). (D, E) MEF cells were transfected with pTRE-Tight vector, pTRE-Tight-DAPK1, pTRE-Tight-DAPK1K42A or pTRE-Tight-DAPK1ΔCaM for 8 h followed by treatment with or without 1 μg/ml doxycycline for 48 h. The cell lysates were subjected to western blot analysis with anti-Flag or anti-actin antibody and the levels of mouse Aβ40 in cell culture supernatants were determined by a solid phase sandwich ELISA assay. Each data point represents the mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test).
Inhibition of DAPK1 decreases secretion of Aβ40 and Aβ42
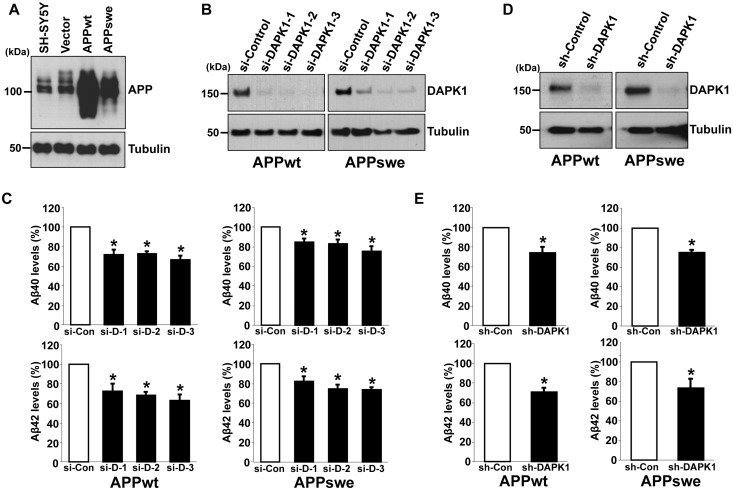
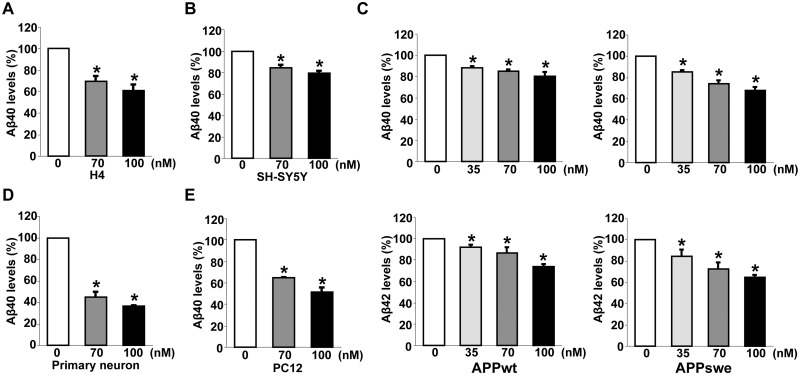
To examine whether DAPK1 also contributes to the secretion of Aβ42, we employed SH-SY5Y cells stably overexpressing either APP WT (SH-SY5Y APPwt) or Swedish mutant form (SH-SY5Y APPswe), which shifts APP toward β-secretase-mediated pathway (Fig. 2A). We efficiently knocked down the expression of endogenous DAPK1 using small interfering RNAs (si-RNAs) (Fig. 2B) or lentiviral-mediated small hairpin RNA (sh-RNA) (Fig. 2D) in APPwt and APPswe cells. In contrast to the control cells, the secretion of human Aβ40 and Aβ42 decreased markedly in DAPK1 knockdowned APPwt and APPswe cells (Fig. 2C and E). We next examined whether inhibition of DAPK1 catalytic activity affects Aβ secretion. For the efficient and selective pharmacological inhibition of DAPK1 activity, (4Z)-4-(3-pyridylmethylene)-2-styryl-oxazol-5-one was chosen as a DAPK1 inhibitor. This compound is known to inhibit DAPK1 activity effectively and selectively (IC50= 69 nm) (60) and, in previous studies, we have shown that this drug regulates DAPK1 activity-dependent neurite outgrowth and microtubule polymerization by affecting tau function (54). The DAPK1 inhibitor decreased Aβ40 secretion from both H4 and SH-SY5Y cells (Fig. 3A and B). The DAPK1 inhibitor also significantly reduced the secretion of both human Aβ40 and Aβ42 in SH-SY5Y APPwt and SH-SY5Y APPswe cells (Fig. 3C). Furthermore, inhibition of DAPK1 kinase activity using the DAPK1 inhibitor decreased mouse Aβ40 secretion from mouse primary cortical neurons and rat Aβ40 secretion from rat pheochromocytoma PC12 cells (Fig. 3D and E). Taken together, our data show that inhibition of DAPK1 expression or its activity suppresses the secretion of Aβ40 and Aβ42.
Figure 2.
DAPK1 inhibition reduces the secretion of human Aβ40 and Aβ42 from both SH-SY5Y APPwt and SH-SY5Y APPswe cells. (A) Baseline expression of APP in parent SH-SY5Y cells and SH-SY5Y cells stably expressing empty vector (pcDNA3), APP WT (pcDNA3-hAPP695) or APP Swedish mutant (pcDNA3-hAPP695swe). The cell lysates were subjected to western blot analysis with anti-APP or anti-tubulin antibody. The blots are representative of three independent experiments. (B, C) SH-SY5Y APPwt and SH-SY5Y APPswe cells were transfected with three different human DAPK1-sepcific siRNAs or scrambled control siRNA for 60 h. The cell lysates were subjected to western blot analysis with anti-DAPK1 or anti-tubulin antibody and the supernatants were subjected to human Aβ40 and Aβ42 ELISA analyses. The blots are representative of three independent experiments and ELISA data shown represent the mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). (D) SH-SY5Y APPwt and SH-SY5Y APPswe cells were infected with control lentiviral vector (pLKO.1) or vectors encoding human DAPK1-specific shRNA, and selected using puromycin (2 μg/ml) for 2 days. The obtained stable cell lines were maintained in low dose puromycin (0.25 μg/ml). The cell lysates were subjected to western blot analysis with anti-DAPK1 or anti-tubulin antibody. The blots are representative of three independent experiments. (E) SH-SY5Y APPwt and SH-SY5Y APPswe cells with stably silenced DAPK1 obtained from (D) were cultured for 30 h. The supernatants were subjected to Aβ40 and Aβ42 ELISA analyses as in (C). Each data point represents the mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test).
Figure 3.
The pharmacological inhibitor of DAPK1 reduces the secretion of Aβ40 and Aβ42. (A, B) Human H4 (A) and SH-SY5Y (B) cells were treated with the indicated concentrations of DAPK1 pharmacological inhibitor for 24 h. Levels of human Aβ40 in cell culture supernatants after 24 h incubation were determined by a solid phase sandwich ELISA assay as in (B) and (C). Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). (C) SH-SY5Y APPwt and SH-SY5Y APPswe cells were incubated with the indicated concentrations of DAPK1 pharmacological inhibitor for 24 h. The supernatants were subjected to human Aβ40 and Aβ42 ELISA analyses as in (C). Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). (D, E) Primary cortical neurons (D) at DIV 8–10 and the rat adrenal pheochromocytoma PC12 (E) cells were treated with the indicated concentrations of DAPK1 pharmacological inhibitor for 36 h. Levels of Aβ40 in cell culture supernatants after 36 h incubation were determined by a solid phase sandwich ELISA assay. Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test).
DAPK1 interacts with APP
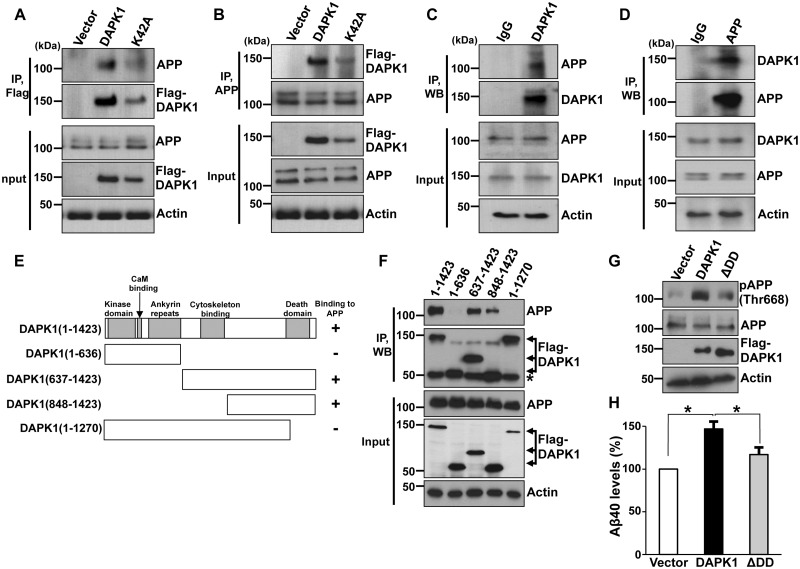
We next examined whether DAPK1 interacts with APP in cells. Chinese hamster ovary (CHO) cells expressing human APP WT (CHO APPwt) were transfected with empty Flag vector, Flag-DAPK1 or Flag-DAPK1K42A. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting analysis with human anti-APP antibody. Both DAPK1 and DAPK1K42A were associated with APP (Fig. 4A). Immunoprecipitation with human anti-APP antibody followed by immunoblotting with anti-Flag antibody also confirmed the interaction between DAPK1 and APP (Fig. 4B). Moreover, Flag-DAPK1 and Flag-DAPK1K42A were found to coimmunoprecipitate with anti-APP in CHO cells expressing the human APP Swedish mutant form (CHO APPswe) (Supplementary Material, Fig. S1A and B). Considering that the protein levels of DAPK1K42A were relatively low compared with WT DAPK1, these results show that the substitution of lysine 42 does not affect the interaction between DAPK1 and APP. Furthermore, endogenous DAPK1 and APP were reciprocally coimmunoprecipitated in human SH-SY5Y cells (Fig. 4C and D). Thus, both APP WT and the APP Swedish mutant form stable complexes in cells. Moreover, DAPK1 and APP were co-localized in CHO APPwt cells (Supplementary Material, Fig. S1C). Next, to map the domain in DAPK1 that is important for APP interaction, a series of truncated DAPK1 mutants was generated and used in a co-immunoprecipitation experiment (Fig. 4E). DAPK1 fragments from 1 to 636 and from 1 to 1270 failed to bind to APP (Fig. 4F). In contrast, the DAPK1 fragments from 1 to 1423, from 637 to 1423 and from 848 to 1423 efficiently bound to APP, suggesting that the death domain (1271–1423, DD) is likely bound to APP (Fig. 4F). To examine whether this interaction is important for Aβ secretion, SH-SY5Y cells were transfected with DAPK1 or DAPK1 ΔDD followed by measuring total Aβ40 secretion (Fig. 4G). While DAPK1 increased Aβ40 secretion, DAPK1 ΔDD did not show significant effects compared with vector control (Fig. 4H). These results show that DAPK1 and APP interaction is critical for the regulation of Aβ secretion.
Figure 4.
Interaction between DAPK1 and APP. (A, B) CHO cells stably expressing APP WT were transfected with pRK5-Flag, pRK5-Flag-DAPK1 or pRK5-Flag-DAPK1K42A for 12 h. Cell lysates were immunoprecipitated with rabbit anti-Flag or rabbit anti-APP antibody. The precipitated samples were analyzed by western blotting with mouse anti-Flag or mouse anti-APP antibody. Total lysates, corresponding to 1% input, were also analyzed by western blotting with same antibodies. Anti-actin antibody was used as a loading control. The blots are representative of three independent experiments. (C, D) Endogenous DAPK1 (C) or APP (D) in SH-SY5Y cells was immunoprecipitated with mouse anti-DAPK1 or rabbit anti-APP antibody, respectively. Immunoprecipitated DAPK1 and APP were subjected to western blot analysis with mouse anti-DAPK1 or mouse anti-APP antibody. Anti-actin antibody was used as a loading control and the blots are representative of three independent experiments. (E) Schematic representation of full-length DAPK1 and its truncated mutants. (F) CHO cells stably expressing APP WT were transfected with pRK5-Flag-DAPK1 or its truncated mutants. Cell lysates were immunoprecipitated with mouse anti-Flag antibody. The precipitated samples were analyzed by western blotting with rabbit anti-APP antibody. Total lysates, corresponding to 1% input, were also analyzed by western blotting with same antibodies. Anti-actin antibody was used as a loading control. The blots are representative of three independent experiments. Asterisks denote heavy chain. (G, H) SH-SY5Y cells were transfected with pRK5-Flag, pRK5-Flag-DAPK1 or pRK5-Flag-DAPK1ΔDD for 32 h. The cell lysates were subjected to western blot analysis to detect phosphorylated APP at Thr668 and total APP protein. Anti-actin antibody was used as a loading control. The levels of human Aβ40 in cell culture supernatants were determined by a solid phase sandwich ELISA assay. Each data point represents the mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test).
DAPK1 triggers APP phosphorylation at the Thr668 site
APP is known to contain potentially eight phosphorylation sites within its cytoplasmic domain (33). Among these sites, Thr668 is mechanistically important for the generation of Aβ by initiating and facilitating amyloidogenic APP cleavage. To determine whether DAPK1 may affect APP phosphorylation, SH-SY5Y or H4 cells were transfected with DAPK1 or K42A construct, followed by measuring phosphorylated APP levels. DAPK1, but not DAPK1K42A, significantly upregulated APP phosphorylation at Thr668 in SH-SY5Y and H4 cells (Fig. 5A and B, and Supplementary Material, Fig. S2A and B). However, APP phosphorylation at Tyr682 was not affected by either DAPK1 or DAPK1K42A (Fig. 5A and B and Supplementary Material, Fig. S2A and B). In order to confirm these results, we efficiently knocked down the expression of endogenous DAPK1 using lentiviral-mediated sh-RNA in SH-SY5Y APPwt or SH-SY5Y APPswe cells and evaluated APP phosphorylation. Consistent with the data shown in Fig. 5A and B, knockdown of DAPK1 significantly reduced APP phosphorylation at Thr668 but not Tyr682 (Fig. 5C and D, and Supplementary Material, Fig. S2C and D). Moreover, while transfection of APP WT with DAPK1 significantly promoted Aβ secretion, phosphorylation-deficient mutant APP T668A failed to increase Aβ level in the conditioned media (Supplementary Material, Fig. S3A and B). Furthermore, DAPK1-induced APP phosphorylation was suppressed when DAPK1 ΔDD was introduced (Fig. 4G), indicating that DAPK1 regulates Aβ secretion through APP Thr668 phosphorylation.
Figure 5.
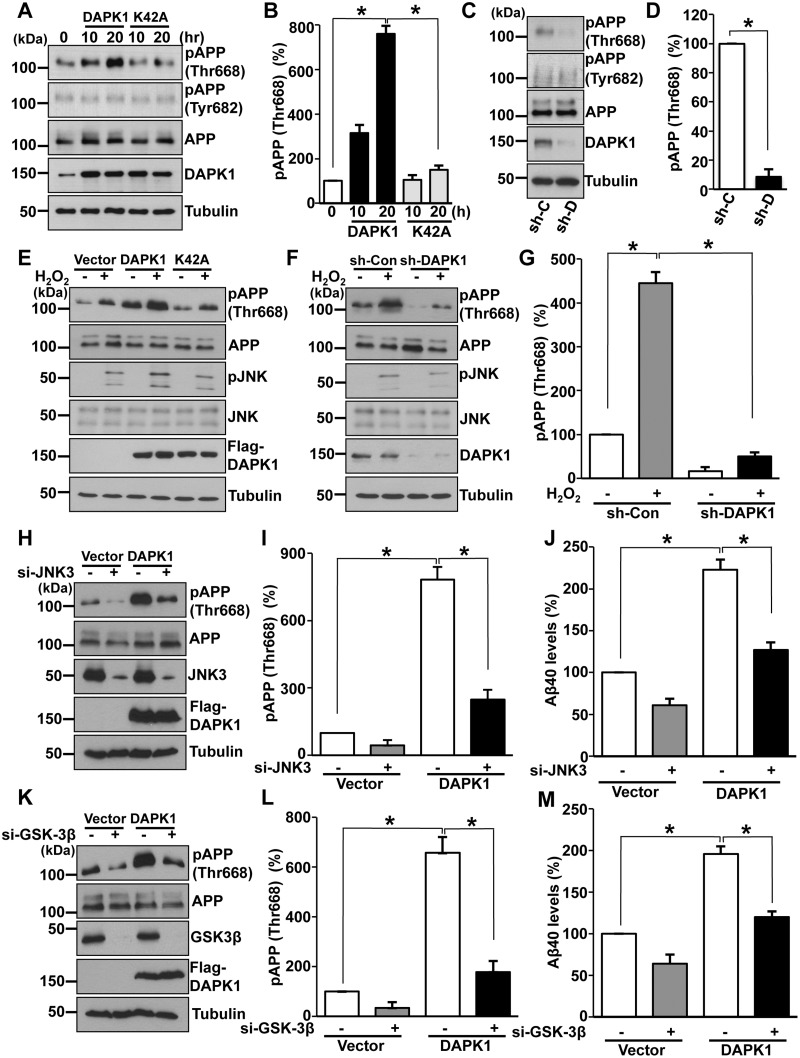
DAPK1 induces APP phosphorylation at Thr668. (A, B) SH-SY5Y cells were transfected with pRK5-Flag-DAPK1 or pRK5-Flag-DAPK1K42A for the indicated time periods. The cell lysates were subjected to western blot analysis to detect phosphorylated (Thr668 and Tyr682) and total APP protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. Phosphorylated APP intensities were quantified by computer-assisted densitometry (*P < 0.05; ANOVA/Dunnett’s test). (C, D) Lysates from SH-SY5Y APPwt cells with stably silenced DAPK1 were subjected to western blot analysis to detect phosphorylated (Thr668 and Tyr682) and total APP protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. Phosphorylated APP intensities were quantified by computer-assisted densitometry (*P < 0.05; ANOVA/Dunnett’s test). (E) SH-SY5Y cells were transfected with pRK5-Flag, pRK5-Flag-DAPK1 or pRK5-Flag-DAPK1K42A for 12 h and treated with 0.5 mm H2O2 for 2 h. The cell lysates were subjected to western blot analysis to detect phosphorylated (Thr668) and total APP protein and to detect phosphorylated (Thr183/Tyr185) and total JNK protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. (F, G) SH-SY5Y cells expressing scrambled control shRNA or human DAPK1-specific shRNA were treated with 0.5 mm H2O2 for 2 h. The cell lysates were subjected to western blot analysis to detect phosphorylated (Thr668) and total APP protein and to detect phosphorylated (Thr183/Tyr185) and total JNK protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. Phosphorylated APP intensities were quantified by computer-assisted densitometry (*P < 0.05; ANOVA/Dunnett’s test). (H–J) SH-SY5Y cells were transfected with human JNK3-specific siRNA or scrambled control siRNA in combination with pRK5-Flag or pRK5-Flag-DAPK1 for 60 h. The cell lysates were subjected to western blot analysis to detect phosphorylated (Thr668) and total APP protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. Phosphorylated APP intensities were quantified by computer-assisted densitometry and human Aβ40 levels in cell culture supernatants were determined by a solid phase sandwich ELISA assay (*P < 0.05; ANOVA/Dunnett’s test). (K–M) SH-SY5Y cells were transfected with human GSK-3β-specific siRNA or scrambled control siRNA in combination with pRK5-Flag or pRK5-Flag-DAPK1 for 60 h. The cell lysates were subjected to western blot analysis to detect phosphorylated (Thr668) and total APP protein. Anti-tubulin antibody was used as a loading control. The blots are representative of three independent experiments. Phosphorylated APP intensities were quantified by computer-assisted densitometry and human Aβ40 levels in cell culture supernatants were determined by a solid phase sandwich ELISA assay (*P < 0.05; ANOVA/Dunnett’s test).
Previous studies showed that APP is phosphorylated at Thr668 by many Ser/Thr kinases, including Cdc2, Cdk5, GSK-3β and JNK3/SAPK1β (31–38). Moreover, ERK increases APP Thr668 phosphorylation during staurosporine-induced apoptosis (61). Since DAPK1 activates JNK signaling under oxidative stress (62) and GSK-3β by inhibiting Pin1 activity (63,64), and DAPK1 catalytic activity is increased by ERK (65,66), we hypothesized that DAPK1 might regulate APP Thr668 phosphorylation through the regulation of multiple substrates. To examine this possibility, SH-SY5Y cells were transfected with DAPK1 or the kinase activity-deficient mutant DAPK1K42A expression construct and subjected to oxidative stress with hydrogen peroxide (H2O2); JNK activation and APP phosphorylation were then evaluated. DAPK1, but not the DAPK1K42A mutant, efficiently phosphorylated and activated JNK in response to H2O2-triggered oxidative stress (Fig. 5E). DAPK1 also dramatically increased APP phosphorylation at Thr668 after oxidative stress (Fig. 5E and Supplementary Material, Fig. S3C). Moreover, DAPK1 knockdown using shRNA significantly suppressed the oxidative stress-induced JNK phosphorylation at Thr183/Tyr185 and concomitant APP phosphorylation at Thr668 (Fig. 5F and G). Furthermore, inhibition of JNK using a pharmacological JNK inhibitor significantly prevented APP Thr668 phosphorylation in DAPK1 overexpressing cells (Supplementary Material, Fig. S3D and E). To investigate whether DAPK1 regulate specific JNK isoforms to promote APP phosphorylation, we knocked down expression of endogenous JNK1, JNK2 or JNK3 using si-RNAs in the SH-SY5Y cells. In contrast to the control cells, when JNK3 expression is suppressed, APP Thr668 phosphorylation and Aβ secretion was significantly decreased in DAPK1 overexpressing cells (Fig. 5H–J). However, DAPK1-mediated APP Thr668 phosphorylation was not inhibited after suppression of JNK1 or JNK2 expression (Supplementary Material, Fig. S3F and G). Since JNK3 activity is increased in human AD brains and significantly promotes Aβ secretion (67–69), DAPK1 might increase APP Thr668 phosphorylation and Aβ secretion through JNK3 activation. Moreover, APP Thr668 phosphorylation levels and Aβ levels were decreased when the expression of GSK-3β was suppressed in DAPK1 overexpressing SH-SY5Y cells (Fig. 5K–M). However, DAPK1-induced APP phosphorylation was not suppressed in ERK knockdown cells (Supplementary Material, Fig. S3H), indicating that JNK3 and GSK-3β activation is required for DAPK1-mediated APP phosphorylation at Thr668 and Aβ secretion.
We also examined whether DAPK1 exerts an effect on APP protein stability. Our cycloheximide (CHX) chase experiments showed that DAPK1 did not have any effect on the APP protein half-life in either H4 or SH-SY5Y cells (Supplementary Material, Fig. S4). Consistent with these results, overexpression or knockdown of DAPK1 did not have any effect on APP protein expression (Fig. 5A and C). Altogether, these results suggest that DAPK1 significantly enhanced APP Thr668 phosphorylation through JNK3 and GSK-3β activation and that this increase in phosphorylation was not due to increased protein levels.
KO of DAPK1 downregulates amyloidogenic processing of APP and Aβ secretion in the brain
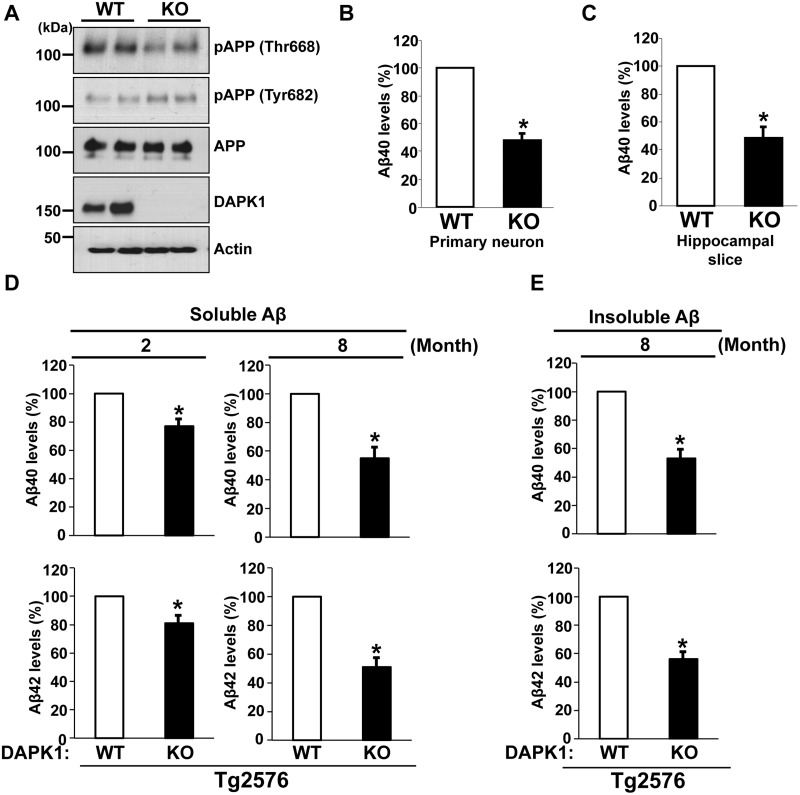
To test further the effects of DAPK1 on APP phosphorylation, we analyzed the brain lysates of DAPK1 KO and WT mice. Concordant with the results from cultured cell lines, depletion of DAPK1 resulted in the attenuation of APP phosphorylation at Thr668, while overall APP level and phosphorylation at Tyr682 were unchanged in the KO mice (Fig. 6A and Supplementary Material, Fig. S5A–C). To measure the effects of DAPK1 on endogenous mouse Aβ level, we employed primary cortical neuron and brain slice cultures. In conditioned media from both experimental settings, Aβ levels were significantly lower in cultures derived DAPK1 KO mice than WT controls (Fig. 6B and C).
Figure 6.
DAPK1 KO reduces the secretion of Aβ40 and Aβ42 in mouse models. (A) Whole brain lysates from WT and DAPK1 KO mice harvested at 6 month of age were analyzed for the levels of phosphorylated (Thr668 and Tyr682) and total APP protein as well as DAPK1. Anti-actin antibody was used as a loading control. The blots are representative of three independent experiments. Analyzed mice number: WT (n = 6, 3 male (M) and 3 female (F)), DAPK1 KO (3M, 3F). (B, C) Primary cortical neurons (B) at DIV 8-10 of WT and DAPK1 KO mice and the brain hippocampal tissue slices (C) prepared from WT and DAPK1 KO mice at 6 month of age were cultured for 36 h. Levels of mouse Aβ40 in cell culture supernatants after 36 h incubation were determined by a solid phase sandwich ELISA assay. Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). (D) TBS-buffer soluble hippocampal tissue fraction from Tg2576/WT or Tg2576/DAPK1 KO mice at 2 or 8 month of age was prepared. Levels of human Aβ40 and Aβ42 were determined by a solid phase sandwich ELISA assay. Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). Analyzed mice number, 2-month-old groups: Tg2576/WT (3M, 3F), Tg2576/DAPK1 KO (3M, 3F); 8-month-old groups: Tg2576/WT (3M, 3F), Tg2576/DAPK1 KO (3M, 3F). (E) TBS-buffer insoluble hippocampal tissue fraction from Tg2576/WT or Tg2576/DAPK1 KO mice at 8 month of age was prepared. Levels of human Aβ40 and Aβ42 were determined by a solid phase sandwich ELISA assay. Each data point represents mean ± standard error of three independent experiments (*P < 0.05; ANOVA/Dunnett’s test). Analyzed mice number: Tg2576/WT (3M, 3F), Tg2576/DAPK1 KO (3M, 3F).
To examine the effects of DAPK1 ablation on APP processing and Aβ generation in vivo, we crossed DAPK1 KO mice with Tg2576 mice overexpressing APP Swedish mutation (Supplementary Material, Fig. S5D). ELISA analysis of Tg2576/DAPK1 KO brain lysates revealed that the levels of TBS-soluble Aβ40 and Aβ42 were significantly lower than those of Tg2576 control, ∼20% at 2 months and 50% at 8 months of age (Fig. 6D). This DAPK1 KO-induced downregulation Aβ was more pronounced in the TBS-insoluble fraction at 8 months (Fig. 6E), while insoluble Aβ40 and Aβ42 were barely detectable in Tg2576 at 2 months.
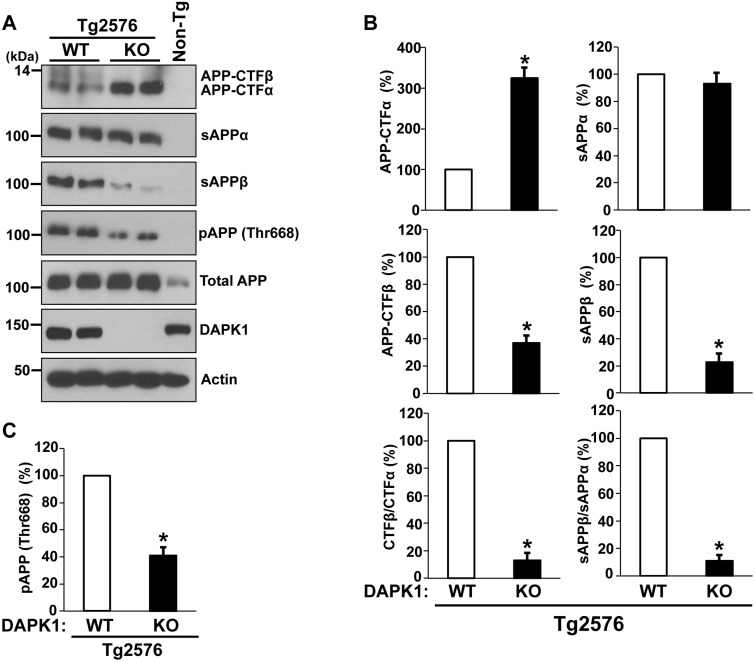
We next measured APP cleavage products in the brain by western blot analysis. In the Tg2576/DAPK1 KO mice, the level of APP-CTFβ, the direct substrate of γ-secretase for Aβ generation, was reduced to 35% of that in Tg2576/WT mice (Fig. 7A and B). Concordantly, sAPPβ, the other β-secretase cleavage product, was significantly decreased in the KO mice. However, while APP-CTFα, an α-secretase-cleaved membrane-bound fragment, increased more than 3-fold, sAPPα was not increased accordingly in the Tg2576/DAPK1 KO mouse brains (Fig. 7A and B). This discrepancy in the level of α-secretase cleavage product may be caused by the further degradation of excess sAPPα in the KO brain (21) or by another APP processing mechanism (70). The ratios of both CTFβ:CTFα and sAPPβ:sAPPα show that DAPK1 ablation leads to an 8-fold decrease in the amyloidogenic pathway. DAPK1 KO also results in the decrease of APP phosphorylation at Thr668 in the AD mouse brains (Fig. 7A and C). Combined together, these results suggest that DAPK1 KO downregulates APP phosphorylation, shifts APP processing toward non-amyloidogenic pathway and decreases Aβ generation in the AD brain.
Figure 7.
The effect of DAPK1 KO on APP processing in the Tg2576 APP transgenic mice. (A) APP processing in the brains of 8-month-old Tg2576/WT or Tg2576/DAPK1 KO mice was examined (3M, 3F). Total APP (G12A), sAPPα (6E10), sAPPβ-swedish (IBL), APP-CTFα (G12A), APP-CTFβ (G12A) and pThr668-APP were detected. (B) Densitometric analysis of APP-CTFα, APP-CTFβ, sAPPα, and sAPPβ and ratios of CTFβ:CTFα and sAPPβ:sAPPα (*P < 0.05; ANOVA/Dunnett’s test). (C) Densitometric analysis of pThr668-APP. (*P < 0.05; ANOVA/Dunnett’s test).
APP Thr668 phosphorylation correlates positively with DAPK1 levels in human AD hippocampus
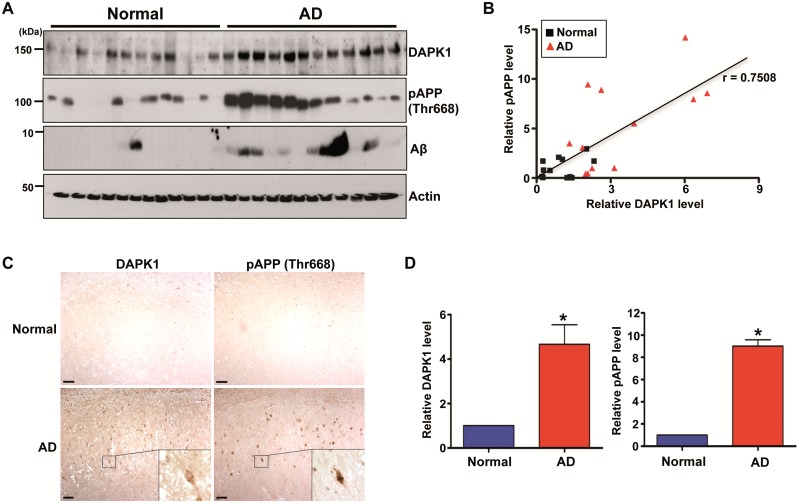
In order to measure the endogenous APP processing pattern in AD brains, we used the Aβ 6E10 antibody clone, which reacts to the processed isoforms, as well as precursor forms. As expected, Aβ peptides showed marked accumulation in AD brains compared with age-matched normal brains (Fig. 8A). Interestingly, we also found that APP phosphorylation at Thr668 and DAPK1 expression increased markedly in AD brains compared with age-matched normal brains (Fig. 8A). Moreover, there was a significant correlation between DAPK1 expression and APP Thr668 phosphorylation levels in AD brains, as indicated by the Pearson correlation coefficient (r = 0.7508) (Fig. 8A and B). Our immunohistochemical staining also confirmed that both DAPK1 and APP Thr668 phosphorylation levels increased markedly in the hippocampal neurons of Alzheimer’s patients compared to control brains (Fig. 8C and D). Collectively, these results indicate DAPK1 is upregulated and a positive correlation between DAPK1 expression and APP Thr668 phosphorylation levels in human AD brain tissues.
Figure 8.
The expression of DAPK1, APP phosphorylation at Thr668, and Aβ in the AD patient and normal brains. (A, B) Hippocampal tissues of 12 AD patients and 12 age-matched normal controls were harvested. (A) Proteins from normal and AD hippocampus were used for immunoblotting with anti-DAPK1, anti-pThr668-APP, anti-Aβ or anti-actin antibody. The blots are representative of three independent experiments. (B) Linear regression analysis was used to estimate the correlation between DAPK1 expression and APP phosphorylation at Thr668 (r = 0.7508; Pearson's correlation coefficient). (C) Immunohistochemistry using a monoclonal antibody specific for DAPK1 or a polyclonal antibody specific for phosphorylated APP at Thr668 was conducted on hippocampal regions from AD patient and normal control. ×100 (main photographs); ×400 (insets); scale bar = 50 μm. Representative images from five experiments are shown. (D) Quantitation of DAPK1 and pThr668 APP. Results shown are mean ± standard error (*P < 0.05; ANOVA/Dunnett’s test).
Discussion
Although the cumulative evidence indicates that DAPK1 may play a critical role in the development of AD, little is known about whether or how DAPK1 regulates APP processing and Aβ production. In this study, we showed for the first time that DAPK1 is a critical regulator of APP phosphorylation and processing in vitro and in vivo. First, DAPK1 increased both Aβ40 and Aβ42 secretion, but its kinase activity-deficient DAPK1K42A mutant failed to promote Aβ production. Second, inhibition of DAPK1 kinase activity using a pharmacological DAPK1 inhibitor significantly reduced the secretion of both human Aβ40 and Aβ42. Third, DAPK1 bound to APP and its binding was not dependent on DAPK1 kinase activity because the kinase-deficient DAPK1K42A mutant interacted with APP. Fourth, DAPK1 was found to increase the phosphorylation of APP at Thr668 through JNK or GSK-3β. Fifth, DAPK1 ablation decreased amyloidogenic APP processing and Aβ secretion in the APP overexpressing mouse brains. Finally, the significance of these findings is further substantiated by the demonstration that phosphorylated APP Thr668 and DAPK1 levels were highly correlated in the brain of AD patients. Taken together, these results demonstrate that DAPK1 stimulates APP phosphorylation at Thr668 and amyloidogenic APP processing pathway.
Aβ is derived from the cleavage of APP and varies in length from 39 to 43 amino acids. There are two predominant forms of the Aβ peptide in the brain, Aβ40 and Aβ42, which differ in length. While Aβ40 is the more common of the two in cerebrospinal fluid and plasma, Aβ42 is the most common form in plaques. Compared with Aβ40, Aβ42 tends to aggregate more rapidly and is more fibrillogenic, cytotoxic and pathogenic, and therefore, is associated with the AD state (71). Interestingly, our ELISA results demonstrated that DAPK1 stimulated the secretion of both Aβ40 and Aβ42 from neuronal cells expressing WT APP or APP bearing the Swedish 670/671 double mutation. It would be interesting to see whether DAPK1 exerts the same effects on other APP mutations.
APP, a type I integral membrane protein, has a large N-terminal extracellular domain and a short intracellular C-terminal domain that can be phosphorylated by various protein kinases. Phosphorylation of APP is a normal physiological process and affects the cellular function of APP linked to neurite extension of differentiating neurons and anterograde transport of vesicular cargo (69,72,73). However, the increased phosphorylation of APP can be a pathological cause of AD by regulating APP processing and Aβ production. To the best of our knowledge, there are at least eight potential phosphorylation sites in the cytoplasmic domain of APP (31–34). Among those sites, the Thr668 residue in the AICD is the most important for APP processing, Aβ generation and AD pathogenesis (31–34). First, it regulates APP localization to the growth cones and neurites. Second, phosphorylation of APP at this site contributes to Aβ generation induced by BACE1-mediated cleavage. Third, Thr668 phosphorylation regulates the nuclear translocation of the AICD, which then induces neurodegeneration. Fourth, Thr668 phosphorylation suppresses caspase-mediated APP cleavage and may increase susceptibility to neuronal cell death. Given that the expression of APP phosphorylated at Thr668 is also elevated in AD brains (33), Thr668 phosphorylation may be a target for AD therapy. In contrast, the Tyr682 site seems to be involved in APP regulation other than APP processing or Aβ generation. For example, phosphorylation at Tyr682 is required for Src homology 2 domain-containing binding (74). Fyn induces phosphorylation of APP at Tyr682 and increases cell surface expression of APP (74). Our data indicated that DAPK1, but not DAPK1K42A, significantly enhanced APP phosphorylation at the Thr668 site, but not the Tyr682 site in neuroblastoma cells. Concomitantly, knockdown or ablation of DAPK1 significantly reduced APP phosphorylation at Thr668, but not Tyr682. DAPK1 has been shown to increase JNK activity under oxidative conditions (62). Our data demonstrate that DAPK1 induces phosphorylation of APP at Thr668 and promotes Aβ secretion through JNK3. This phosphorylation might be facilitated by DAPK1 kinase activity-dependent JNK3 activation under oxidative conditions because JNK3 is a direct kinas of APP at Thr668 and increases Aβ secretion (67–69). Recent studies have also reported that a peptidyl-prolyl cis/trans isomerase, Pin1 inhibits GSK-3β activity (64) and that DAPK1 negatively regulates Pin1 activity through Ser71 phosphorylation (63,75). In addition, Pin1 directly binds to the phosphorylated Thr668-proline motif in APP and accelerates its cis to trans isomerization, helping APP maintain its proper form and thereby preventing AD pathology (75,76). Given the important role of APP phosphorylation at Thr668, the Ser/Thr kinase DAPK1 might exert its action on APP processing and Aβ generation through the upregulation of abnormal Thr668 phosphorylation by Pin1 inhibition in addition to JNK3 activation. ERK has been shown to activate DAPK1 kinase activity through its Ser735 phosphorylation and to increase its apoptotic ability (65). However, our results showed that DAPK1-induced APP phosphorylation was not suppressed when ERK expression was inhibited. Since ERK increased APP phosphorylation with staurosporine treatment (61), we would not rule out the possibility that ERK might promote APP Thr668 phosphorylation through DAPK1 activation under stresses such as staurosporine.
APP can also be regulated by protein interactions, as well as phosphorylation. Moreover, phosphorylation at Thr668 also results in significant conformational changes that may affect the interaction of APP with its binding partners. Our immunoprecipitation assay clearly demonstrated that both DAPK1 and its kinase-deficient mutant DAPK1K42A interact with APP, and that the Swedish mutation of APP did not inhibit this interaction. These results are in line with our findings that DAPK1 exerts its effects on Aβ secretion in both APPwt- and APPswe-overexpressing cells. These results suggest that DAPK1 might form a complex with APP, Pin1 and APP Thr668 kinases including JNK3 or GSK-3β. This complex might increase the activity or the local concentration of JNK3 or GSK-3β to promote APP Thr668 phosphorylation. The possible role of Thr668 phosphorylation in the interaction between APP and DAPK1 needs to be further investigated.
In order to validate our findings in vivo, we used Tg2576 mice, a well-established mouse model of an AD-like pathology (29). We crossed DAPK1 KO with Tg2576 mice and investigated the effects of DAPK1 KO on Aβ accumulation in the brains of AD mice. Our ELISA assay demonstrated that the production of Aβ40 and Aβ42 in Tg2576 mouse brains decreased significantly after DAPK1 KO in an age-dependent manner. Mechanistically, APP Thr668 phosphorylation and APP processing to generate sAPPβ and CTFβ were attenuated in Tg2576 mice with DAPK1 KO, suggesting that DAPK1 ablation exerts non-amyloidogenic effects in vivo and has potential therapeutic value in AD. Because increased Aβ secretion promotes Aβ pathology and leads to memory loss, further studies should include whether DAPK1 ablation would improve cognitive function and Aβ plaques.
Additionally, we introduced a selective DAPK1 inhibitor as a promising approach to suppress Aβ secretion. Given that the DAPK1 pharmacological inhibitor significantly inhibited the secretion of both Aβ40 and Aβ42 in various cell culture models in our results, a DAPK1 inhibitor might offer promising options for treatment of AD. To confirm this, more studies need to be conducted to determine whether a DAPK1 inhibitor penetrates the blood–brain barrier to inhibit Aβ levels in the brain and/or rescue age-related cognitive decline in transgenic AD mice. These trials may offer some encouragement for the clinical development of a disease-modifying drug for AD.
In conclusion, our data suggest a model in which DAPK1 regulates the amyloidogenic APP processing pathway and the resultant secretion of cytotoxic Aβ40 and Aβ42 via aberrant APP phosphorylation at Thr668 (Supplementary Material, Fig. S6). This study thus suggests that DAPK1 may be a therapeutic target for Aβ deposition in AD pathology.
Materials and Methods
Materials
CHX was purchased from Sigma and was used to inhibit protein synthesis and assess protein stability of APP. H2O2 was obtained from Sigma and was used as oxidant. Doxycycline and DAPI were obtained from Sigma. DAPK1 inhibitor (4Z)-4-(3-Pyridylmethylene)-2-styryl-oxazol-5-one and JNK inhibitor II were obtained from Calbiochem and were used to specifically inhibit the kinase activity of DAPK1 and JNK, respectively.
Brain samples
Whole brain tissues from WT and DAPK1 KO mice at 6 and 22 months of age and from WT/Tg2576 and DAPK1 KO/Tg2576 mice at 2 and 8 months of age were harvested. DAPK1 WT, KO or Tg2576 mice were previously described (77,78). DAPK1-WT or DAPK1-KO mice were maintained in pure C57B6 background. Tg2576 or Tg2576/DAPK1-KO mice were maintained in C57BL/6 × SJL hybrid background. Tg2576 mice in C57BL/6 × SJL F1 hybrid background were initially crossed with DAPK1 in KO mice in C57BL/6 background to obtain Tg2576:DAPK1+/− and control non-transgenic:DAPK1+/−. The mice analyzed for the study were derived from the cross of Tg2576:DAPK1+/− and non-transgenic: DAPK1+/− in order to maintain the hemizygosity of Tg2576. Dissected mouse brains were snap frozen in liquid nitrogen-cold isopentane and stored at −80 °C until use. Mouse brain hemispheres were homogenized in TBS buffer containing with protease inhibitor and phosphatase inhibitor cocktail. The half of lysates was resuspended in 3 volume of RIPA buffer (1% NP40, 0.1% SDS and 0.5% Sodium deoxycholate in TBS) for 10 min, followed by centrifugation at 13 000g for 10 min. The RIPA-soluble fractions were used as total protein lysates including both cytosolic and membrane proteins. The other half of lysates was centrifuged at 100 000g for 1 h. Supernatant was used for ‘soluble Aß’ measurements. Pellets were resuspended and further homogenized in 70% formic acid (equal volume of TBS), followed by centrifugation at 100 000g for 1 h. Formic acid supernatants were neutralized with 1 m Tris for ‘insoluble Aß’ analysis. Brain hippocampus tissues of human Alzheimer’s patients and age-matched normal controls were obtained from the Neuropathology Core of the Massachusetts Alzheimers Disease Research Center (MADRC), Massachusetts General Hospital (Boston, MA) and the Harvard Brain Tissue Resource Center (HBTRC), Mclean Hospital (Belmont, MA). Research was conducted in compliance with the policies and principles contained in the Federal Policy for the Protection for the Protection of Human subjects. Clinical information of 12 AD patients and 12 controls are reported in Supplementary Material, Table S1. Frozen brain samples were thawed on ice and homogenized into about 5-volume of sucrose lysis buffer by 10 up-and-down strokes in Potter-Elvehjem homogenizer.
Cell culture
The human neuroglioma cell line H4; the human neuroblastoma cell line SH-SY5Y; the Chinese hamster ovary cell line CHO; and rat pheochromocytoma cell line PC12 were obtained from the American Type Culture Collection (ATCC). MEF cells were isolated from mouse embryos between embryonic days 12.5 and 14.5. H4, SH-SY5Y, CHO and MEF cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) high-glucose supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin (all from Gibco). PC12 cells were cultured onto poly-l-lysine (Sigma)-coated plate in DMEM high-glucose supplemented with 10% horse serum and 5% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (all from Gibco). The cultures were maintained at 37 °C under 5% CO2.
Plasmid transfection
The cDNA fragment encoding the human DAPK1 or DAPK1K42A mutant (in which lysine 42 was replaced by alanine) or DAPK1ΔCaM mutant (in which a Ca2+/Calmodulin regulatory domain is deleted) was cloned into 5′ end-Flag tagged and CMV promoter-driven mammalian expression vector pRK5, as described (54). Cells were transiently transfected with different plasmids using Lipofectamine LTX and PLUS reagent (Invitrogen) according to the manufacturer’s instruction.
Si-RNA transfection and lentiviral infection
For generating transient DAPK1 knockdown in SH-SY5Y cells expressing WT human APP (SH-SY5Y APPwt) and APP with mutations at positions K670N and M671L (Swedish) (SH-SY5Y APPswe), three different DAPK1 si-RNAs were designed using the best-in-class design algorithm developed by Rosetta and were synthesized by Sigma. SH-SY5Y APPwt cells or SH-SY5Y APPswe cells were transiently transfected with three different si-RNAs against human DAPK1 or scrambled si-RNA using LipofectamineTM RNAiMAX Reagent (Invitrogen) according to the manufacturer’s instructions. For generating transient JNK1, JNK2, JNK3, GSK-3β or ERK knockdown, SH-SY5Y cells were transiently transfected with JNK1, JNK2, JNK3, GSK-3β, ERK1 or ERK2 si-RNAs or scrambled si-RNA using LipofectamineTM RNAiMAX Reagent. The si-RNA target sequences were as follows: DAPK1-#1, CAACTATGATGTTAACCAA; DAPK1-#2, GAGAATCGATGTCCAGGAT; DAPK1-#3, GACATGAAGGTACTTCGAA, JNK1, ATAACAAATCCCTTGCCTGACTGGC; JNK2, AGTTGAGTCTGCCACTTGTACACTG; JNK3, GATATATGGTCTGTGGGATGCATTA; GSK-3β, CCCAAATGTCAAACTACCAAA; ERK1, CGCTACACGC AGTTGCAGTACA; ERK2, TGTTCCCAAATGCTG ACTCCAA and scrambled si-RNA, GATCATACGTGCGATCAGA. A scrambled si-RNA does not match any human sequences in GeneBank search. For generating stable DAPK1 knockdown in SH-SY5Y, SH-SY5Y APPwt and SH-SY5Y APPswe cells, DAPK1 sh-RNA clone which is constructed within lentiviral pLKO.1-Puro vector were transfected together with VSV-G- and gag-pol-expressing plasmids into 293FT cells. Sixty hours after transfection, virus-containing supernatants were used to infect SH-SY5Y, SH-SY5Y APPwt and SH-SY5Y APPswe cells. A day after infection, the stable clones were selected using 2 μg/ml puromycin. The target sequence of DAPK1-specific sh-RNA used was GGTCAAGGATCCAAAGAAG.
Primary moue cortical neuronal cell culture
Primary mouse embryonic cortical neurons were prepared from the cerebral cortices of embryonic day E15 WT and DAPK1 KO anesthetized pregnant female C57BL/6 mouse according to the established procedures described previously with minor modifications (54). After removal of the meninges outside the cortex, freshly dissected cortical tissues were minced into ∼1–2 mm3 fragments and digested with 0.25% trypsin and 50 U/ml DNase1 (Sigma) for 15 min at 37 °C until tissue was thoroughly homogenized. The tissue was then dispersed mechanically with a fire-polished glass Pasteur pipette, and the cells were seeded at a density of 8 × 105 cells/ml on poly-d-lysine-coated dishes in neurobasal medium supplemented with 2% B27 supplement, 0.5 mm glutamine and 1% penicillin–streptomycin (all from Life Technologies) and cultured at 37 °C under 5% CO2. After 24 h of adaptation, the culture medium was replaced with a neurobasal-A medium containing 2% B27 supplement, 0.5 mm glutamine and 1% penicillin–streptomycin. All experiments were performed at days in vitro (DIV) 8–10.
Organotypic brain slice preparation
Organotypic mouse brain slices were prepared according to the method described previously with slight modifications (54). Brain tissue was obtained from 6-month-old WT and DAPK1 KO mice. Mice were anesthetized with isofluran (Baxter) and immediately decapitated under sterile conditions. Removed brains were immersed in ice-cold Krebs solution (126 mm NaCl, 2.5 mm KCl, 1.2 mm NaH2PO4, 1.2 mm MgCl2, 2.5 mm CaCl2, 11 mm glucose, 25 mm NaHCO3, 10 mm HEPES, pH 7.4). The cerebellum was trimmed off and the caudal end of the brain was glued onto the cutting table of vibratome (Leica VT1200). Brain tissues were cut into coronal slices of 260 μm with amplitude of 1.5 and an oscillation frequency of 90 Hz. Slices containing the hippocampus were cultured onto semi-porous (0.4-μm pore diameter) Millicell-CM membrane inserts (Millipore) in 6-well culture plates with culture media. The insert was positioned to orient the brain’s dorsal region the same way in each well.
Solid phase sandwich ELISA analysis of secreted Aβ40 and Aβ42
Aβ secretion into the medium was determined using mouse Aβ40, mouse Aβ42, human Aβ40 and human Aβ42 colorimetric ELISA Kits (all from Invitrogen), following the manufacturer’s instructions. The levels of mouse Aβ40 and Aβ42 in the TBS-insoluble fraction were measured using mouse Aβ40 and mouse Aβ42 ELISA Kits, respectively. The samples diluted in the provided diluent buffer or standards containing aggregated Aβ along with biotinylated rabbit detecting antibody specific for the C-terminus of Aβ were incubated on plate well that was pre-coated with a monoclonal capturing antibody specific for the N-terminus of Aβ. Samples were incubated for 3 h at room temperature allowing the Aβ to bind both capturing antibody and detecting antibody. After washing the wells four times, immobilized Aβ in samples or standards were incubated with horseradish peroxidase-labeled streptavidin for 30 min. Stabilized chromogen 3,3′,5,5′-tetramethylbenzidine (TMB) was added to produce a colorimetric solution. The TMB reaction was terminated using stop solution and the optical density was measured at 450 nm using a plate reader. The intensity of colored product is directly proportioned to the concentration of Aβ in the samples. The concentrations of Aβ40 or Aβ42 were calculated using standard curves for Aβ40 or Aβ42 peptide by comparing the sample’s absorbance with the absorbance of known concentrations of a standard solution. The results were expressed as percentage of control culture values for comparison.
Immunoprecipitation assay for DAPK1–APP interaction
Polyclonal anti-Flag (Cell Signaling) and anti-APP (Millipore) antibodies were used to immunoprecipitate transiently expressing Flag-DAPK1 (or Flag-DAPK1K42A) and stably expressing APPwt (or APPswe), respectively, in CHO cells. Monoclonal anti-DAPK1 (Sigma) and polyclonal anti-APP (Millipore) antibodies were used to immunoprecipitate endogenous DAPK1 and APP, respectively, in SH-SY5Y cells. Flags, APPs or DAPK1s were immunoprecipitated from 500 μg of whole cell lysate via the above antibodies. Cells were lysed in ice-cold lysis buffer (10 mm Tris–HCl [pH7.4], 150 mm NaCl, 1% Nonidet P-40) supplemented with protease inhibitor cocktail and lysates were incubated with primary antibodies at 4 °C for 12 h and then with 15 μl of protein A (for anti-Flag and anti-APP antibodies) or protein G (for anti-DAPK1 antibody) sepharose slurry for additional 2 h. Beads were washed four times with 1 ml of same lysis buffer.
Measurement of protein stability of APP
The half-life of APP protein was measured by incubating H4 and SH-SY5Y cells with 75 μg/ml of CHX for the indicated time. The relative protein levels were determined by immunoblotting and densitometry analysis.
Immunoblot analysis
Immunoblotting analyses were performed to monitor APP processing-related protein levels. Cellular lysates were prepared using ice-cold lysis buffer (50 mm Tris–HCl (pH 7.4), 50 mm NaCl, 1 mm EGTA, 1% Triton X-100, 1 mm DTT) supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail. Mouse and human brain extracts were prepared by homogenizing tissue in ice-cold lysis buffer [10 mm Tris–HCl (pH 7.4), 0.8 m NaCl, 1 mm EGTA, 10% sucrose, 1 mm DTT] supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail. The soluble protein concentration was determined by Bradford assay (Bio-Rad Laboratories). The Proteins (10 μg) were separated by SDS–PAGE and electrotransferred onto PVDF membranes (Perkin Elmer). The membranes were then blocked with 5% non-fat dry milk in TBS-T (0.1% Tween 20) and probed with antibodies against the following molecules: Flag, DAPK1 (Sigma); APP (G12A: rabbit polyclonal antibody targeting last 20 amino acids residues of APP. Custom-manufactured by Pierce Custom Antibodies); p-APP (Thr668), JNK1, JNK2, JNK2, p-JNK (Thr183/Tyr185), GSK-3β, ERK, β-actin, α-tubulin (Cell Signaling Technology); 6E10 (Covance); sAPPβ-swedish (IBL), APP-CTFα/β (G12A). Primary antibodies were detected using horseradish peroxidase (HRP)-conjugated secondary antibodies and the Western Lightning Plus-ECL (Perkin Elmer) or ECL Prime (GE Healthcare Life Sciences) chemiluminescence system.
Immunohistochemistry and immunostaining analyses
For immunohistochemical analysis of human brain tissues, immersion-fixed tissue (hippocampus) sections were embedded in paraffin. Coronal tissue sections were deparaffinized with xylene, rehydrated with descending grade of ethanol and incubated with 3% H2O2/PBS to quench the endogenous peroxidase activity. After antigen retrieval, sections were incubated in PBS blocking buffer with 5% horse serum, 5% bovine serum albumin and 0.1% v/v Tween 20. Primary antibodies were added in blocking buffer and incubated with sections overnight at 4 °C. Secondary antibody was biotinylated goat-anti-rabbit or -mouse IgG. Sections were processed with ABC reagents by using a Vector ABC kit (Vector Laboratories). After washing, the HRP reaction was detected with diaminobenzidine and H2O2. For immunofluorescent analysis, the slides were fixed in methanol and blocked with 7% serum in PBS for 1 h at room temperature, followed by incubation with anti-Flag and anti-APP at 1:1000 dilution overnight at 4 °C. After extensive wash with PBS, slides were incubated with Alexa488-labeled anti-mouse IgG and Alexa546-labeled anti-Rabbit IgG (Invitrogen) diluted 1:200 for 2 h at room temperature. After washing with PBS, the nuclei were stained with DAPI for 10 min at room temperature.
Statistical analysis
Statistical analyses were performed using the SAS 8.1 statistical analysis program for Windows (SAS Institute). One-way ANOVA followed by the Dunnett post-test was used to assess the significant differences between the groups. The Pearson correlation coefficient was used to assess the significant correlation between DAPK1 and APP Thr668 phosphorylation levels. The data are expressed as mean ± standard error and significance was set at P < 0.05.
Study approval
The biosafety guidelines set by Beth Israel Deaconess Medical Center’s Institutional Animal Care and Use Committee were applied to all research carried out in this study in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International regulations.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
Human brain tissues were provided by the Neuropathology Core of the Massachusetts Alzheimer Disease Research Center (P50AG05134) and the Harvard Brain Tissue Resources Center (R24MH068855).
Conflict of Interest statement. None declared.
Funding
This work was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health & Welfare Affairs, Republic of Korea (No. HI08C2149) and by a faculty research grant of Yonsei University College of Medicine for 2014 (No. 2014-32-0048) to B.M.K. and NIH grant (R00AG033104), the Alzheimer’s Association (NIRG-12-258863), the American Federation for Aging Research, and the Massachusetts Alzheimer’s Disease Research Center (P50AG005134) to T.H.L.
References
- 1. Goldgaber D., Lerman M.I., McBride O.W., Saffiotti U., Gajdusek D.C. (1987) Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer's disease. Science, 235, 877–880. [DOI] [PubMed] [Google Scholar]
- 2. Tanzi R.E., Gusella J.F., Watkins P.C., Bruns G.A., St George-Hyslop P., Van Keuren M.L., Patterson D., Pagan S., Kurnit D.M., Neve R.L. (1987) Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science, 235, 880–884. [DOI] [PubMed] [Google Scholar]
- 3. Kang J., Lemaire H.G., Unterbeck A., Salbaum J.M., Masters C.L., Grzeschik K.H., Multhaup G., Beyreuther K., Muller-Hill B. (1987) The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature, 325, 733–736. [DOI] [PubMed] [Google Scholar]
- 4. Tanzi R.E., Bertram L. (2005) Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell, 120, 545–555. [DOI] [PubMed] [Google Scholar]
- 5. Hardy J., Selkoe D.J. (2002) The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science, 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 6. Jack C.R, Jr., Holtzman D.M. (2013) Biomarker modeling of Alzheimer's disease. Neuron, 80, 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim D., Tsai L.H. (2009) Bridging physiology and pathology in AD. Cell, 137, 997–1000. [DOI] [PubMed] [Google Scholar]
- 8. Nunan J., Small D.H. (2002) Proteolytic processing of the amyloid-beta protein precursor of Alzheimer's disease. Essays Biochem., 38, 37–49. [DOI] [PubMed] [Google Scholar]
- 9. Selkoe D.J., Yamazaki T., Citron M., Podlisny M.B., Koo E.H., Teplow D.B., Haass C. (1996) The role of APP processing and trafficking pathways in the formation of amyloid beta-protein. Ann. NY Acad. Sci., 777, 57–64. [DOI] [PubMed] [Google Scholar]
- 10. Vetrivel K.S., Thinakaran G. (2006) Amyloidogenic processing of beta-amyloid precursor protein in intracellular compartments. Neurology, 66, S69–S73. [DOI] [PubMed] [Google Scholar]
- 11. De Strooper B., Annaert W. (2000) Proteolytic processing and cell biological functions of the amyloid precursor protein. J. Cell Sci., 113, 1857–1870. [DOI] [PubMed] [Google Scholar]
- 12. Estus S., Golde T.E., Kunishita T., Blades D., Lowery D., Eisen M., Usiak M., Qu X.M., Tabira T., Greenberg B.D. et al. (1992) Potentially amyloidogenic, carboxyl-terminal derivatives of the amyloid protein precursor. Science, 255, 726–728. [DOI] [PubMed] [Google Scholar]
- 13. Golde T.E., Estus S., Younkin L.H., Selkoe D.J., Younkin S.G. (1992) Processing of the amyloid protein precursor to potentially amyloidogenic derivatives. Science, 255, 728–730. [DOI] [PubMed] [Google Scholar]
- 14. Haass C., Koo E.H., Mellon A., Hung A.Y., Selkoe D.J. (1992) Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature, 357, 500–503. [DOI] [PubMed] [Google Scholar]
- 15. Haass C., Schlossmacher M.G., Hung A.Y., Vigo-Pelfrey C., Mellon A., Ostaszewski B.L., Lieberburg I., Koo E.H., Schenk D., Teplow D.B. et al. (1992) Amyloid beta-peptide is produced by cultured cells during normal metabolism. Nature, 359, 322–325. [DOI] [PubMed] [Google Scholar]
- 16. Shoji M., Golde T.E., Ghiso J., Cheung T.T., Estus S., Shaffer L.M., Cai X.D., McKay D.M., Tintner R., Frangione B. et al. (1992) Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Science, 258, 126–129. [DOI] [PubMed] [Google Scholar]
- 17. De Strooper B., Umans L., Van Leuven F., Van Den Berghe H. (1993) Study of the synthesis and secretion of normal and artificial mutants of murine amyloid precursor protein (APP): cleavage of APP occurs in a late compartment of the default secretion pathway. J. Cell Biol., 121, 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koo E.H., Squazzo S.L. (1994) Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J. Biol. Chem., 269, 17386–17389. [PubMed] [Google Scholar]
- 19. Koo E.H., Squazzo S.L., Selkoe D.J., Koo C.H. (1996) Trafficking of cell-surface amyloid beta-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J. Cell Sci., 109, 991–998. [DOI] [PubMed] [Google Scholar]
- 20. Perez R.G., Soriano S., Hayes J.D., Ostaszewski B., Xia W., Selkoe D.J., Chen X., Stokin G.B., Koo E.H. (1999) Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J. Biol. Chem., 274, 18851–18856. [DOI] [PubMed] [Google Scholar]
- 21. Suh J., Choi S.H., Romano D.M., Gannon M.A., Lesinski A.N., Kim D.Y., Tanzi R.E. (2013) ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron, 80, 385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Esch F.S., Keim P.S., Beattie E.C., Blacher R.W., Culwell A.R., Oltersdorf T., McClure D., Ward P.J. (1990) Cleavage of amyloid beta peptide during constitutive processing of its precursor. Science, 248, 1122–1124. [DOI] [PubMed] [Google Scholar]
- 23. Sisodia S.S., Koo E.H., Beyreuther K., Unterbeck A., Price D.L. (1990) Evidence that beta-amyloid protein in Alzheimer's disease is not derived by normal processing. Science, 248, 492–495. [DOI] [PubMed] [Google Scholar]
- 24. Parvathy S., Hussain I., Karran E.H., Turner A.J., Hooper N.M. (1999) Cleavage of Alzheimer's amyloid precursor protein by alpha-secretase occurs at the surface of neuronal cells. Biochemistry, 38, 9728–9734. [DOI] [PubMed] [Google Scholar]
- 25. Buxbaum J.D., Liu K.N., Luo Y., Slack J.L., Stocking K.L., Peschon J.J., Johnson R.S., Castner B.J., Cerretti D.P., Black R.A. (1998) Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem., 273, 27765–27767. [DOI] [PubMed] [Google Scholar]
- 26. Yan R., Bienkowski M.J., Shuck M.E., Miao H., Tory M.C., Pauley A.M., Brashier J.R., Stratman N.C., Mathews W.R., Buhl A.E. et al. (1999) Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature, 402, 533–537. [DOI] [PubMed] [Google Scholar]
- 27. Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., Loeloff R. et al. (1999) Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science, 286, 735–741. [DOI] [PubMed] [Google Scholar]
- 28. Hussain I., Powell D., Howlett D.R., Tew D.G., Meek T.D., Chapman C., Gloger I.S., Murphy K.E., Southan C.D., Ryan D.M. et al. (1999) Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol. Cell Neurosci., 14, 419–427. [DOI] [PubMed] [Google Scholar]
- 29. Cai H., Wang Y., McCarthy D., Wen H., Borchelt D.R., Price D.L., Wong P.C. (2001) BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat. Neurosci., 4, 233–234. [DOI] [PubMed] [Google Scholar]
- 30. Wolfe M.S., Xia W., Ostaszewski B.L., Diehl T.S., Kimberly W.T., Selkoe D.J. (1999) Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature, 398, 513–517. [DOI] [PubMed] [Google Scholar]
- 31. Pastorino L., Lu K.P. (2005) Phosphorylation of the amyloid precursor protein (APP): is this a mechanism in favor or against Alzheimer’s disease. Neurosci. Res. Commun., 35, 213–231. [Google Scholar]
- 32. Phiel C.J., Wilson C.A., Lee V.M., Klein P.S. (2003) GSK-3alpha regulates production of Alzheimer's disease amyloid-beta peptides. Nature, 423, 435–439. [DOI] [PubMed] [Google Scholar]
- 33. Lee M.S., Kao S.C., Lemere C.A., Xia W., Tseng H.C., Zhou Y., Neve R., Ahlijanian M.K., Tsai L.H. (2003) APP processing is regulated by cytoplasmic phosphorylation. J. Cell Biol., 163, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimberly W.T., Zheng J.B., Town T., Flavell R.A., Selkoe D.J. (2005) Physiological regulation of the beta-amyloid precursor protein signaling domain by c-Jun N-terminal kinase JNK3 during neuronal differentiation. J. Neurosci., 25, 5533–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki T., Oishi M., Marshak D.R., Czernik A.J., Nairn A.C., Greengard P. (1994) Cell cycle-dependent regulation of the phosphorylation and metabolism of the Alzheimer amyloid precursor protein. Embo J., 13, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aplin A.E., Gibb G.M., Jacobsen J.S., Gallo J.M., Anderton B.H. (1996) In vitro phosphorylation of the cytoplasmic domain of the amyloid precursor protein by glycogen synthase kinase-3beta. J. Neurochem., 67, 699–707. [DOI] [PubMed] [Google Scholar]
- 37. Iijima K., Ando K., Takeda S., Satoh Y., Seki T., Itohara S., Greengard P., Kirino Y., Nairn A.C., Suzuki T. (2000) Neuron-specific phosphorylation of Alzheimer's beta-amyloid precursor protein by cyclin-dependent kinase 5. J. Neurochem., 75, 1085–1091. [DOI] [PubMed] [Google Scholar]
- 38. Standen C.L., Brownlees J., Grierson A.J., Kesavapany S., Lau K.F., McLoughlin D.M., Miller C.C. (2001) Phosphorylation of thr(668) in the cytoplasmic domain of the Alzheimer's disease amyloid precursor protein by stress-activated protein kinase 1b (Jun N-terminal kinase-3). J. Neurochem, 76, 316–320. [DOI] [PubMed] [Google Scholar]
- 39. Bialik S., Kimchi A. (2006) The death-associated protein kinases: structure, function, and beyond. Annu. Rev. Biochem., 75, 189–210. [DOI] [PubMed] [Google Scholar]
- 40. Deiss L.P., Feinstein E., Berissi H., Cohen O., Kimchi A. (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes. Dev., 9, 15–30. [DOI] [PubMed] [Google Scholar]
- 41. Fujita Y., Yamashita T. (2014) Role of DAPK in neuronal cell death. Apoptosis, 19, 339–345. [DOI] [PubMed] [Google Scholar]
- 42. Li Y., Grupe A., Rowland C., Nowotny P., Kauwe J.S., Smemo S., Hinrichs A., Tacey K., Toombs T.A., Kwok S. et al. (2006) DAPK1 variants are associated with Alzheimer's disease and allele-specific expression. Hum. Mol. Genet., 15, 2560–2568. [DOI] [PubMed] [Google Scholar]
- 43. Yamamoto M., Hioki T., Ishii T., Nakajima-Iijima S., Uchino S. (2002) DAP kinase activity is critical for C(2)-ceramide-induced apoptosis in PC12 cells. Eur. J. Biochem., 269, 139–147. [DOI] [PubMed] [Google Scholar]
- 44. Fujita Y., Taniguchi J., Uchikawa M., Endo M., Hata K., Kubo T., Mueller B.K., Yamashita T. (2008) Neogenin regulates neuronal survival through DAP kinase. Cell Death Differ., 15, 1593–1608. [DOI] [PubMed] [Google Scholar]
- 45. Pelled D., Raveh T., Riebeling C., Fridkin M., Berissi H., Futerman A.H., Kimchi A. (2002) Death-associated protein (DAP) kinase plays a central role in ceramide-induced apoptosis in cultured hippocampal neurons. J. Biol. Chem., 277, 1957–1961. [DOI] [PubMed] [Google Scholar]
- 46. Tu W., Xu X., Peng L., Zhong X., Zhang W., Soundarapandian M.M., Balel C., Wang M., Jia N., Lew F. et al. (2010) DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell, 140, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shamloo M., Soriano L., Wieloch T., Nikolich K., Urfer R., Oksenberg D. (2005) Death-associated protein kinase is activated by dephosphorylation in response to cerebral ischemia. J. Biol. Chem., 280, 42290–42299. [DOI] [PubMed] [Google Scholar]
- 48. Velentza A.V., Wainwright M.S., Zasadzki M., Mirzoeva S., Schumacher A.M., Haiech J., Focia P.J., Egli M., Watterson D.M. (2003) An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury. Bioorg. Med. Chem. Lett., 13, 3465–3470. [DOI] [PubMed] [Google Scholar]
- 49. Liu S.B., Zhang N., Guo Y.Y., Zhao R., Shi T.Y., Feng S.F., Wang S.Q., Yang Q., Li X.Q., Wu Y.M. et al. (2012) G-protein-coupled receptor 30 mediates rapid neuroprotective effects of estrogen via depression of NR2B-containing NMDA receptors. J. Neurosci., 32, 4887–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li H., Wetten S., Li L., St Jean P.L., Upmanyu R., Surh L., Hosford D., Barnes M.R., Briley J.D., Borrie M. et al. (2008) Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol., 65, 45–53. [DOI] [PubMed] [Google Scholar]
- 51. Wu Z.C., Zhang W., Yu J.T., Zhang Q., Tian Y., Lu R.C., Yu N.N., Chi Z.F., Tan L. (2011) Association of DAPK1 genetic variations with Alzheimer's disease in Han Chinese. Brain Res., 1374, 129–133. [DOI] [PubMed] [Google Scholar]
- 52. Laumet G., Chouraki V., Grenier-Boley B., Legry V., Heath S., Zelenika D., Fievet N., Hannequin D., Delepine M., Pasquier F. et al. (2010) Systematic analysis of candidate genes for Alzheimer's disease in a French, genome-wide association study. J. Alzheimers Dis., 20, 1181–1188. [DOI] [PubMed] [Google Scholar]
- 53. Yukawa K., Tanaka T., Bai T., Li L., Tsubota Y., Owada-Makabe K., Maeda M., Hoshino K., Akira S., Iso H. (2006) Deletion of the kinase domain from death-associated protein kinase enhances spatial memory in mice. Int. J. Mol. Med., 17, 869–873. [PubMed] [Google Scholar]
- 54. Kim B.M., You M.H., Chen C.H., Lee S., Hong Y., Hong Y., Kimchi A., Zhou X.Z., Lee T.H. (2014) Death-associated protein kinase 1 plays a critical role in aberrant tau protein regulation and function. Cell Death. Dis., 5, e1237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu P.R., Tsai P.I., Chen G.C., Chou H.J., Huang Y.P., Chen Y.H., Lin M.Y., Kimchi A., Chien C.T., Chen R.H. (2011) DAPK activates MARK1/2 to regulate microtubule assembly, neuronal differentiation, and tau toxicity. Cell Death Differ., 18, 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Duan D.X., Chai G.S., Ni Z.F., Hu Y., Luo Y., Cheng X.S., Chen N.N., Wang J.Z., Liu G.P. (2013) Phosphorylation of tau by death-associated protein kinase 1 antagonizes the kinase-induced cell apoptosis. J. Alzheimers Dis., 37, 795–808. [DOI] [PubMed] [Google Scholar]
- 57. Lewis J., Dickson D.W., Lin W.L., Chisholm L., Corral A., Jones G., Yen S.H., Sahara N., Skipper L., Yager D. et al. (2001) Enhanced neurofibrillary degeneration in transgenic mice expressing mutant tau and APP. Science, 293, 1487–1491. [DOI] [PubMed] [Google Scholar]
- 58. Gotz J., Chen F., van Dorpe J., Nitsch R.M. (2001) Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science, 293, 1491–1495. [DOI] [PubMed] [Google Scholar]
- 59. Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.Q., Mucke L. (2007) Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science, 316, 750–754. [DOI] [PubMed] [Google Scholar]
- 60. Okamoto M., Takayama K., Shimizu T., Ishida K., Takahashi O., Furuya T. (2009) Identification of death-associated protein kinases inhibitors using structure-based virtual screening. J. Med. Chem., 52, 7323–7327. [DOI] [PubMed] [Google Scholar]
- 61. Sodhi C.P., Perez R.G., Gottardi-Littell N.R. (2008) Phosphorylation of beta-amyloid precursor protein (APP) cytoplasmic tail facilitates amyloidogenic processing during apoptosis. Brain Res., 1198, 204–212. [DOI] [PubMed] [Google Scholar]
- 62. Eisenberg-Lerner A., Kimchi A. (2007) DAP kinase regulates JNK signaling by binding and activating protein kinase D under oxidative stress. Cell Death Differ., 14, 1908–1915. [DOI] [PubMed] [Google Scholar]
- 63. Lee T.H., Chen C.H., Suizu F., Huang P., Schiene-Fischer C., Daum S., Zhang Y.J., Goate A., Chen R.H., Zhou X.Z. et al. (2011) Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol. Cell, 42, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma S.L., Pastorino L., Zhou X.Z., Lu K.P. (2012) Prolyl isomerase Pin1 promotes amyloid precursor protein (APP) turnover by inhibiting glycogen synthase kinase-3beta (GSK3beta) activity: novel mechanism for Pin1 to protect against Alzheimer disease. J. Biol. Chem., 287, 6969–6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen C.H., Wang W.J., Kuo J.C., Tsai H.C., Lin J.R., Chang Z.F., Chen R.H. (2005) Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. Embo J., 24, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Houle F., Poirier A., Dumaresq J., Huot J. (2007) DAP kinase mediates the phosphorylation of tropomyosin-1 downstream of the ERK pathway, which regulates the formation of stress fibers in response to oxidative stress. J. Cell Sci., 120, 3666–3677. [DOI] [PubMed] [Google Scholar]
- 67. Yoon S.O., Park D.J., Ryu J.C., Ozer H.G., Tep C., Shin Y.J., Lim T.H., Pastorino L., Kunwar A.J., Walton J.C. et al. (2012) JNK3 perpetuates metabolic stress induced by Abeta peptides. Neuron, 75, 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shen C., Chen Y., Liu H., Zhang K., Zhang T., Lin A., Jing N. (2008) Hydrogen peroxide promotes Abeta production through JNK-dependent activation of gamma-secretase. J. Biol. Chem., 283, 17721–17730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Muresan Z., Muresan V. (2005) c-Jun NH2-terminal kinase-interacting protein-3 facilitates phosphorylation and controls localization of amyloid-beta precursor protein. J. Neurosci., 25, 3741–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Willem M., Tahirovic S., Busche M.A., Ovsepian S.V., Chafai M., Kootar S., Hornburg D., Evans L.D., Moore S., Daria A. et al. (2015) eta-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature, 526, 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Citron M. (2010) Alzheimer's disease: strategies for disease modification. Nat. Rev. Drug. Discov., 9, 387–398. [DOI] [PubMed] [Google Scholar]
- 72. Muresan Z., Muresan V. (2005) Coordinated transport of phosphorylated amyloid-beta precursor protein and c-Jun NH2-terminal kinase-interacting protein-1. J. Cell Biol., 171, 615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ando K., Oishi M., Takeda S., Iijima K., Isohara T., Nairn A.C., Kirino Y., Greengard P., Suzuki T. (1999) Role of phosphorylation of Alzheimer's amyloid precursor protein during neuronal differentiation. J. Neurosci., 19, 4421–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hoe H.S., Minami S.S., Makarova A., Lee J., Hyman B.T., Matsuoka Y., Rebeck G.W. (2008) Fyn modulation of Dab1 effects on amyloid precursor protein and ApoE receptor 2 processing. J. Biol. Chem., 283, 6288–6299. [DOI] [PubMed] [Google Scholar]
- 75. Lee T.H., Pastorino L., Lu K.P. (2011) Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev. Mol. Med., 13, e21.. [DOI] [PubMed] [Google Scholar]
- 76. Pastorino L., Sun A., Lu P.J., Zhou X.Z., Balastik M., Finn G., Wulf G., Lim J., Li S.H., Li X. et al. (2006) The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature, 440, 528–534. [DOI] [PubMed] [Google Scholar]
- 77. Gozuacik D., Bialik S., Raveh T., Mitou G., Shohat G., Sabanay H., Mizushima N., Yoshimori T., Kimchi A. (2008) DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ., 15, 1875–1886. [DOI] [PubMed] [Google Scholar]
- 78. Hsiao K., Chapman P., Nilsen S., Eckman C., Harigaya Y., Younkin S., Yang F., Cole G. (1996) Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science, 274, 99–102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.