SUMMARY
RAD51 promotes homologous recombination repair (HR) of double-strand breaks and acts during DNA replication to facilitate fork reversal and protect nascent DNA strands from nuclease digestion. Several additional HR proteins regulate fork protection by promoting RAD51 filament formation. Here we show that RADX modulates stalled fork protection by antagonizing RAD51. Consequently, silencing RADX restores fork protection in cells deficient for BRCA1, BRCA2, FANCA, FANCD2, or BOD1L. Inactivating RADX prevents both MRE11- and DNA2-dependent fork degradation. Furthermore, RADX overexpression causes fork degradation that is dependent on these nucleases and fork reversal. The amount of RAD51 determines the fate of stalled replication forks, with more RAD51 required for fork protection than fork reversal. Finally, we find that RADX effectively competes with RAD51 for binding to single-stranded DNA, supporting a model in which RADX buffers RAD51 to ensure the right amount of reversal and protection to maintain genome stability.
INTRODUCTION
Single strand DNA binding proteins (SSBs) regulate DNA replication, recombination and repair. In eukaryotes, the major SSBs at replication forks include Replication Protein A (RPA) and RAD51. RAD51 is best known for its ability to form nucleoprotein filaments on resected double-strand breaks and catalyze strand invasion for HR (Kowalczykowski, 2015). RAD51 also has at least two functions at stalled replication forks. First, it cooperates with SNF2 family DNA translocases to promote fork reversal (Bétous et al., 2012; Ciccia et al., 2012; Kile et al., 2015; Vujanovic et al., 2017; Zellweger et al., 2015). Second, in cooperation with BRCA2, RAD51 inhibits degradation of the nascent DNA after fork reversal of persistently stalled forks (Hashimoto et al., 2010; Lemaçon et al., 2017; Mijic et al., 2017; Schlacher et al., 2011; Taglialatela et al., 2017). In addition to BRCA2-deficiency, loss of several other HR proteins cause nascent-strand degradation (Higgs et al., 2015; Schlacher et al., 2011, 2012). At least two nucleases, MRE11 and DNA2, are involved. How these pathways work together to maintain fork stability is still unclear. Small amounts of nuclease action could be beneficial to remove DNA lesions or end binding proteins, control the amount of ssDNA at a stalled fork, remodel the reversed fork and promote fork restart (Thangavel et al., 2015); however, unregulated degradation causes genome instability (Schlacher et al., 2011, 2012).
Fork reversal is independent of BRCA2, thus explaining how nascent strand degradation can proceed from reversed forks in BRCA2-deficient cells (Mijic et al., 2017). How RAD51 gains access to persistently stalled forks without BRCA2 to mediate an exchange with RPA is unknown. Nonetheless, the need for RAD51 to promote reversal explains why silencing RAD51 using RNA interference is reported to not cause degradation (Mijic et al., 2017; Thangavel et al., 2015; Zellweger et al., 2015). Paradoxically, some RAD51 mutations and inhibitors do yield fork degradation (Dungrawala et al., 2017; Kolinjivadi et al., 2017; Leuzzi et al., 2016; Mijic et al., 2017; Su et al., 2014; Taglialatela et al., 2017; Zadorozhny et al., 2017), raising the possibility that either fork reversal is not always required for nucleases to degrade the nascent strands or these ways of inhibiting RAD51 only interfere with some of its activities. Finally, fork degradation may be an important determinant of the viability of BRCA2-deficient cells and their sensitivity to PARP inhibitors (Chaudhuri et al., 2016; Ding et al., 2016; Dungrawala et al., 2017; Rondinelli et al., 2017).
We recently identified a new SSB with similarity to RPA named RADX (Dungrawala et al., 2017). RADX negatively regulates RAD51 accumulation at replication forks and we proposed that this regulation prevents inappropriate RAD51-dependent fork reversal in the absence of added replication stress. Presumably, at persistently stalled forks, the negative regulation of RAD51 by RADX is overcome by the positive regulation conferred by BRCA2 to sustain fork protection. Consistent with this hypothesis, knocking out RADX restores fork protection to BRCA2-deficient cells.
In this study, we sought to further test the hypothesis that RADX acts as a RAD51 antagonist and use RADX as a tool to investigate how fork protection pathways operate. Our findings support the model that RADX is a RAD51 antagonist that ensures the right amount of RAD51 fork reversal and protection activities to maintain genome stability. We also find a requirement for higher cellular levels of RAD51 to protect persistently stalled forks than to promote fork reversal.
RESULTS
RADX silencing suppresses MRE11-dependent fork degradation in cells with impaired RAD51 filament stability
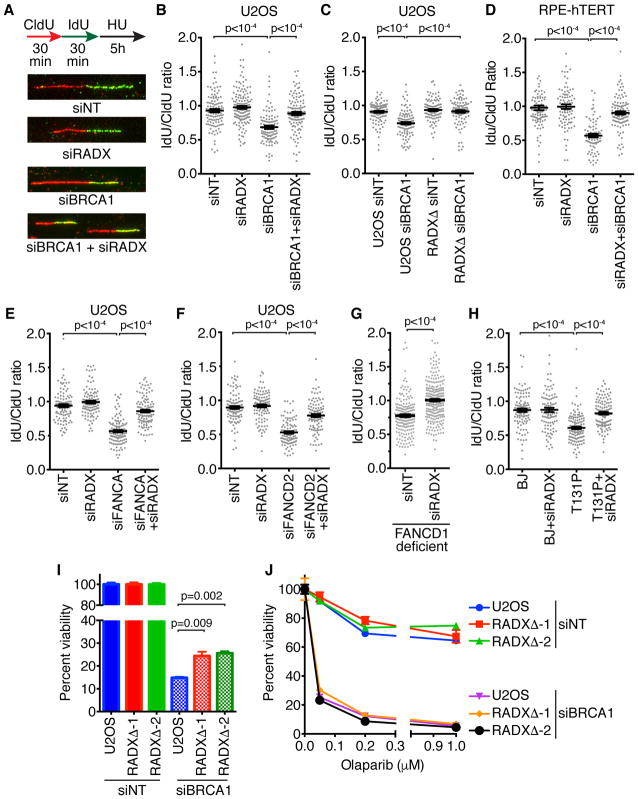
Multiple HR proteins including BRCA1 protect the nascent DNA at replication forks from MRE11 digestion by promoting RAD51 filament stability. To test the hypothesis that silencing RADX would restore fork protection in cells with decreased RAD51 activity, we utilized siRNA to deplete BRCA1 in U2OS cells and induce nascent strand degradation (Schlacher et al., 2012). Silencing RADX is sufficient to restore fork protection to BRCA1-depleted cells (Figures 1A, 1B, and Supplemental Figure 1). This is not an off-target effect of siRNA since deletion of RADX using CRISPR-CAS9 also prevents nascent strand degradation after BRCA1 silencing (Figure 1C). RADX-deficiency also restores fork protection to BRCA1-depleted RPE-hTERT cells (Figure 1D), indicating that this effect is not cell-type specific. We did not observe any defects in fork protection upon RADX depletion alone, either by siRNA or RADX deletion (Figure 1). These results differ from a previous report that suggested RADX deficiency causes nascent strand degradation (Schubert et al., 2017). The reason for this difference is unclear.
Figure 1. RADX silencing rescues the MRE11-dependent fork protection defects caused by loss of RAD51 stability.
(A) Graphical depiction of the fork protection assay with representative images. (A, B, C, E, F) U2OS or RADXΔ U2OS, (D) RPE-hTERT, (G) FANCD1/BRCA2-mutant fibroblasts, or (H) fibroblasts expressing the T131P RAD51 mutant were transfected with the indicated siRNAs then labeled sequentially with CldU and IdU before treatment with 3mM HU for 5 hours. The lengths of DNA fibers were measured and mean+/−SEM of the IdU/CldU ratio is depicted. P values were derived from Kruskal-Wallis ANOVA with a Dunn’s post-test. Each experiment was repeated at least twice and a representative result is depicted. (siNT = non-targeting siRNA). (I and J) siRNA transfected parental or RADXΔ U2OS were plated for clonogenic survival assays in the absence (I) or presence (J) of drug. P values were calculated from a two-way ANOVA with Tukey’s post-test. Mean+/−SEM from n=3 is depicted. See also Supplemental Figures S1 and S2.
In addition to their function in interstrand crosslink repair the FA pathway proteins FANCA and FANCD2 also prevent MRE11-dependent nascent strand degradation (Schlacher et al., 2012). The exact mechanism by which these proteins act is unknown, but since RAD51 overexpression rescues the fork degradation in FA cells, we predicted that RADX depletion should also suppress this phenotype. Indeed, silencing FANCA or FANCD2 in U2OS cells causes fork degradation and silencing RADX restores fork protection to these cells (Figures 1E, 1F and Figure S1). As expected, silencing RADX also restores fork protection to fibroblasts harboring a FA-patient derived mutation in FANCD1/BRCA2 (Figure 1G).
RAD51 T131P is an FA-associated, dominant-negative mutant (Wang et al., 2015). Patient derived fibroblasts harboring this mutation are defective in RAD51 fork protection, but are proficient in HR repair (Kolinjivadi et al., 2017; Mijic et al., 2017; Wang et al., 2015; Zadorozhny et al., 2017). We reasoned that since the cells harboring the T131P allele exhibit only a partial loss of RAD51 function, improving RAD51 function by RADX silencing should restore fork protection in these cells. As predicted, loss of RADX prevented the nascent strand degradation observed in the T131P cells (Figure 1H). These data further support the model that RADX loss facilitates increased RAD51 function at persistently stalled replication forks and prevents MRE11 nuclease action.
Restoration of fork protection to BRCA1-deficient U2OS cells does not cause PARP inhibitor resistance
Fork protection may be an important determinant of the chemosensitivity of BRCA-deficient cells to PARP inhibitors like Olaparib (Chaudhuri et al., 2016; Ding et al., 2016). Consistent with this idea, RADX silencing not only confers fork protection to BRCA2-mutant cells, but also improves their viability and resistance to Olaparib even though it does not alter HR (Dungrawala et al., 2017). Therefore, we tested if the restoration of fork protection in BRCA1-deficient RADXΔ cells is accompanied by an increase in cell viability and Olaparib resistance. BRCA1 knockdown reduces U2OS cell viability, and RADX deletion conferred a small, but significant, increase in viability to BRCA1-depleted cells in the absence of any drug (Figure 1I and Figure S1) consistent with what was observed in BRCA2-deficient cells (Dungrawala et al., 2017). However, unlike in BRCA2-deficient cells, RADX loss did not confer Olaparib-resistance to BRCA1-depleted U2OS cells (Figure 1H). RADX loss also did not confer hydroxyurea or camptothecin resistance to BRCA1-depleted cells (Figure S2A–C). In fact, the HU sensitivity caused by silencing RADX or BRCA1 by themselves is further increased in cells deficient for both proteins. Thus, despite restoring fork protection to BRCA1-deficient cells, RADX deficiency does not necessarily improve their sensitivity to replication stress inducing agents.
Loss of RADX protects against DNA2-dependent fork degradation
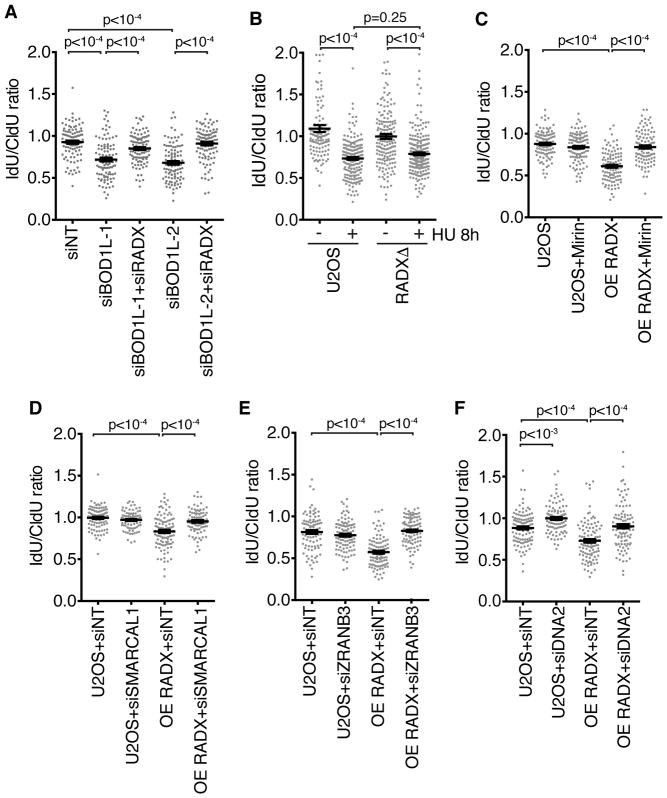
Thus far, our results indicate that silencing RADX can suppress fork degradation that is caused by RAD51 filament instability and MRE11 nuclease activity. Recently, BOD1L was shown to suppress fork degradation by promoting RAD51 filament stability, but the degradation in BOD1L-deficient cells is MRE11-independent. Instead, DNA2 degrades the nascent DNA in this setting (Higgs et al., 2015). To test if RADX also regulates fork protection in cases where the degradation is dependent on DNA2, we utilized siRNA against BOD1L and RADX. As reported previously, knocking down BOD1L causes DNA2-dependent nascent strand degradation (Figure S3). RADX silencing restored fork protection to the BOD1L-deficient cells (Figure 2A and Figure S1). Thus, RADX deficiency restores fork protection irrespective of the nuclease mediating the degradation.
Figure 2. RADX silencing rescues DNA2 dependent fork degradation and RADX overexpression causes degradation of reversed forks.
(A–F) Fork protection assays were completed in U2OS cells or RADX overexpressing (OE RADX) U2OS cells transfected with the indicated siRNAs or treated with Mirin. All cells were treated with HU for 5 hours except for an 8-hour treatment in (B). P values were derived from Kruskal-Wallis ANOVA with a Dunn’s post-test. See also Supplemental Figures S1 and S3.
DNA2 also degrades nascent DNA in U2OS cells without any genetic perturbation when these cancer cells are treated with HU for long times (Thangavel et al., 2015). In contrast to cells with compromised RAD51, RADX deletion does not restore fork protection in this circumstance (Figure 2B).
Overexpression of RADX causes nascent strand degradation that is rescued by inhibition of MRE11 or ZRANB3
While silencing RADX can restore fork protection to RAD51-compromised cells, RADX overexpression causes nascent strand degradation (Dungrawala et al., 2017). If the fork degradation is due to reduced RAD51 function, then it should be dependent genetically on the same factors that cause nascent strand degradation in BRCA2-deficient cells including the MRE11 nuclease and the fork reversal enzymes SMARCAL1 and ZRANB3 (Kolinjivadi et al., 2017; Mijic et al., 2017; Taglialatela et al., 2017). As predicted, inhibiting MRE11 (Figure 2C), depleting SMARCAL1 or ZRANB3 rescues the fork degradation caused by RADX overexpression (Figure 2D, 2E and Figure S1). We also observed a partial rescue of the RADX overexpression-induced fork degradation by silencing DNA2 (Figure 2F). The ability of either DNA2 or MRE11 inhibition to rescue fork degradation in RADX overexpression cells is consistent with the idea that RAD51 destabilization can lead to either MRE11-dependent degradation as in BRCA2-deficient cells, or DNA2-dependent degradation, as in BOD1L-deficient cells.
Differential requirements of RAD51 in fork reversal and protection
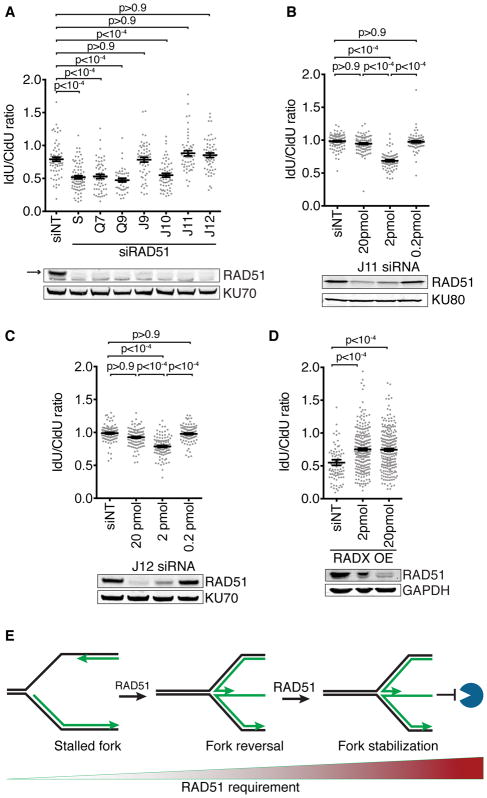
RADX deficiency in unstressed cells causes replication fork breakage that can be rescued by silencing fork reversal proteins including RAD51, ZRANB3, and SMARCAL1 (Dungrawala et al., 2017). Thus, we hypothesized that RADX prevents fork reversal by antagonizing RAD51 at unstressed forks; a model that is consistent with the reduced amount of RAD51 at forks in RADX overexpressing cells (Dungrawala et al., 2017). However, if RAD51 is required for fork reversal, which is in turn required for fork degradation, we might have expected that RADX overexpression in HU-treated cells would inhibit RAD51-dependent fork reversal yielding stable nascent strands instead of the fork degradation that we observed. A possible explanation is that different RAD51 functions could be needed for fork reversal and fork protection and RADX only antagonizes the fork protection function. Alternatively, the same RAD51 function could be required for both reversal and protection, but more of it may be needed for fork protection than fork reversal. Consistent with the second hypothesis, knocking down RAD51 with multiple different siRNAs yields different phenotypic outcomes – fork stability or degradation (Figure 3A). Importantly, titrating the amount of a potent RAD51 siRNA into cells to yield partial RAD51 knockdown initially yields fork degradation at low concentrations and fork protection at higher concentrations (Figure 3B). The same result is observed with a second potent RAD51 siRNA (Figure 3C). These results indicate that the different siRNA results are not due to off-target effects but rather that the amount of RAD51 in the cell determines whether forks reverse and are then protected from nucleases. Consistent with this interpretation and our model that RADX negatively regulates RAD51, even a modest knockdown of RAD51 with an siRNA concentration that caused fork degradation in wild-type U2OS cells is sufficient to prevent degradation in U2OS cells overexpressing RADX (Figure 3D). This is presumably because the combination of partial RAD51 knockdown by siRNA and RAD51 inhibition by overexpressing RADX reduces RAD51 activity below the threshold required to promote fork reversal.
Figure 3. More RAD51 is required for fork protection than fork reversal.
(A) Fork protection assays in U2OS cells transfected with seven different RAD51 siRNAs. (B–D) U2OS or RADX overexpressing (OE RADX) cells were transfected with the “J11” siRNA (B and D) or “J12” siRNA (C) to RAD51 at the indicated amounts prior to performing the fork protection assay. P values were derived from Kruskal-Wallis ANOVA with a Dunn’s post-test. Immunoblots from transfected cells corresponding to the same samples are shown below the graphs. (E) Model illustrating differential RAD51 requirements.
RADX outcompetes RAD51 for ssDNA but is expressed at lower levels
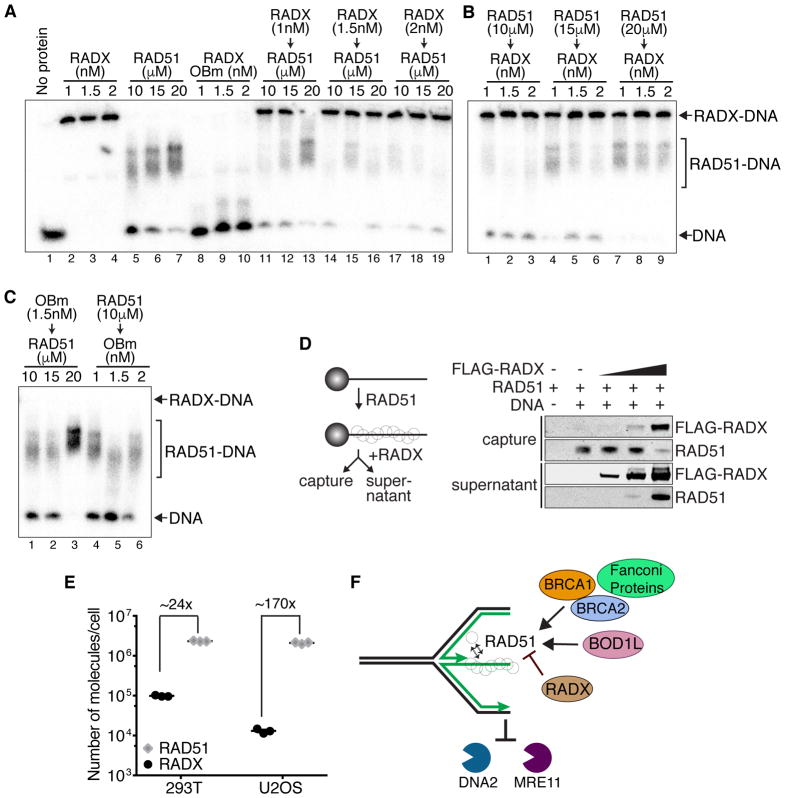
To investigate the mechanism by which RADX antagonizes RAD51 we considered whether they could compete for the same ssDNA ligand. We performed competition experiments with RADX pre-bound to the DNA followed by addition of increasing amounts of RAD51 (Figure 4A); or experiments with RAD51 pre-bound to the DNA followed by incubation with increasing RADX concentrations (Figure 4B). Low concentrations of RADX (1nM) are sufficient to fully bind the ssDNA ligand (Figure 4A, Lane 2); whereas, over 20μM RAD51 is needed (Figure 4A, lane 7), indicating a large difference in binding affinities. Even 1nM RADX pre-bound to the DNA is sufficient to block binding by 10μM RAD51 (Figure 4A, lane 11). At 20μM RAD51, the 1nM prebound RADX is partially displaced by RAD51 (Figure 4A, lane 13). At the highest concentration of RADX that we tested (2nM), the RADX-DNA complex resists displacement even with 20μM RAD51 (Figure 4A lanes 17–19). Conversely, when RAD51 is prebound to the DNA at micromolar concentrations, even 1nM RADX is sufficient to displace most of the RAD51 (Figure 4B). Thus, RADX can outcompete RAD51 for ssDNA binding even when RAD51 is at 10,000-fold molar excess. This competition is dependent on the ability of RADX to bind DNA, since a RADX mutant that abrogates most of its DNA binding activity (OB-m RADX, (Dungrawala et al., 2017)) cannot compete with RAD51 for ssDNA (Figure 4A, lanes 8–10 and Figure 4C). We also confirmed that RADX competes with RAD51 for ssDNA utilizing a biotin-ssDNA pull down assay in which pre-bound RAD51 is displaced by RADX (Figure 4D).
Figure 4. RADX can outcompete RAD51 for binding to ssDNA.
(A–C) Electrophoretic mobility shift assays for RADX and RAD51 binding to a dT-50 oligonucleotide. The experiment was performed by incubating 0.5 nM ssDNA with RADX, RAD51, or both proteins in the indicated order and concentrations in the presence of ATP. The Ob-m is a DNA binding mutant of RADX. (D) ssDNA pre-bound with RAD51 in the presence of ATP was incubated with increasing amounts of RADX prior to capture with magnetic beads and analysis of captured and supernatant proteins by immunoblotting. (E) Quantitation of RAD51 and RADX levels in 293T and U2OS cells (see also Supplemental Figure S4). The median is depicted. All experiments are representative of two repeats.
These results suggest that RADX and RAD51 compete for the same ssDNA ligand. RAD51 is abundantly expressed in both 293T and U2OS cells (~2X106 molecules/cell in both cell types) (Figures 4E and Figure S4). On the other hand, RADX is expressed at comparatively low levels (~1X105 molecules/cell in 293T and ~13000 molecules/cell in U2OS) (Figure 4D). Despite being 24x or 170x less abundant than RAD51 in 293T or U2OS cells respectively, the strong affinity of RADX for ssDNA suggests there is sufficient RADX in cells to inhibit RAD51 from binding ssDNA. Thus, additional regulatory mechanisms controlled by BRCA2 or other RAD51 mediator proteins are essential to overcome the antagonistic activities of RADX.
DISCUSSION
Our data indicates that loss of RADX mimics RAD51 overexpression and confers fork protection to cells lacking BRCA1/2, FANCA, FANCD2 or BOD1L—all situations where RAD51 filament stability is compromised. RADX loss prevents both DNA2- and MRE11-dependent fork degradation. Conversely, RADX overexpression mimics loss of BRCA1/2 and results in fork instability that is dependent on fork reversal. RADX can outcompete RAD51 for ssDNA even when present at concentrations that are 10,000-fold less than RAD51. There is less RADX per cell than RAD51, but even at these reduced concentrations, RADX would efficiently outcompete RAD51. Positive RAD51 regulators like BRCA2 are thus required to balance the antagonistic functions of RADX (Figure 4F).
The deleterious effects of both decreasing and increasing RADX expression levels and its relative stoichiometry with RAD51 is reminiscent of the relationship between the bacterial RecX and RecA proteins (Cox, 2007). In contrast to RecX and RecA, we have not observed evidence for a trimeric complex between RADX, RAD51, and ssDNA. Thus far, our data is most consistent with a competition mechanism to explain how RADX antagonizes RAD51. However, we cannot rule out the possibility that a trimeric complex could be detectable using other experimental conditions. Furthermore, the ability of RADX to easily outcompete RAD51 for ssDNA binding biochemically may be modulated by other proteins or regulatory mechanisms in cells.
An alternative model for how RADX depletion causes fork protection would be for RADX to activate or recruit the fork degradation nucleases. We do not favor this model for the following reasons: First, there is no difference in the amount of MRE11 or DNA2 at replication forks in RADXΔ cells (Dungrawala et al., 2017). Second, there is no difference in the amount of ssDNA or RPA S4/S8 phosphorylation upon RADX depletion (Dungrawala et al., 2017). Third, RADX silencing does not cause sensitivity to ionizing radiation (Dungrawala et al., 2017). Fourth, RADX silencing prevents both MRE11- and DNA2-dependent nascent strand degradation. Fifth, RADX silencing does not prevent the DNA2-dependent fork degradation caused by long HU treatments in U2OS cells with functional BRCA-RAD51. Finally, RADX overexpression causes decreased RAD51 accumulation in HU- or IR-treated cells (Dungrawala et al., 2017), which would be the opposite of what would be expected if RADX promoted the activities of MRE11 and DNA2. Thus, we favor the model that RADX functions either directly or indirectly by regulating RAD51.
Since RADX overexpression results in fork degradation, it must be insufficient to prevent the fork reversal function of RAD51 in the presence of persistent replication stress. Consistent with this idea, RADX overexpression results in only a partial decrease in RAD51 foci formation in HU-treated cells (Dungrawala et al., 2017). Thus, similar to the loss of BRCA2, the partial decrease in RAD51 function by RADX overexpression is sufficient to cause defects in fork protection, but not in fork reversal unless combined with partial silencing of RAD51 expression.
Finally, we found that the amount of RAD51 function is critical to determining the fate of persistently stalled forks. Wild-type levels allow stalled forks to be reversed which can serve as a way to accomplish template switching, repair DNA damage or otherwise promote fork restart. Moderately reduced levels of RAD51 can still facilitate fork reversal but are unable to stabilize the reversed fork leading to excessive nuclease mediated resection and genome instability if forks are persistently stalled. Very low levels of RAD51 prevent any fork reversal yielding stable nascent strands but defects in fork restart and challenges in completion of DNA replication. RADX helps to balance RAD51 activities ensuring its fork reversal and protection activities operate appropriately to maintain genome stability.
Replication fork protection as a determinant of chemosensitivity
Whether replication fork protection is an important determinant of PARP inhibitor and replication stress cell sensitivity appears to be dependent on genetic background and experimental model (Chaudhuri et al., 2016; Ding et al., 2016; Dungrawala et al., 2017; Feng and Jasin, 2017; Yazinski et al., 2017). For example, loss of some factors like EZH2 and MUS81 restore fork protection and chemoresistance only to BRCA2-mutant but not BRCA1-mutant cells (Lemaçon et al., 2017; Rondinelli et al., 2017). Our results suggest RADX also differentiates between BRCA2 and BRCA1 since deleting RADX confers partial chemoresistance to BRCA2-deficient U2OS cells and not BRCA1-deficient U2OS cells despite rescuing fork protection in both settings. The specific HR gene mutation and its severity in disrupting function may determine whether restoring fork protection would be sufficient to generate drug resistance. Identifying drug-resistance mechanisms in patients will be critical to testing this idea.
EXPERIMENTAL PROCEDURES
Cell culture
U2OS, RPE-hTERT, and HEK293T cells were cultured in DMEM with 7.5% fetal bovine serum (FBS). Fibroblasts were cultured in DMEM with 15% FBS and 1% non-essential amino acids. Cells are tested for mycoplasma and authenticated using short tandem repeat profiling. U2OS RADXΔ and RADX overexpressing cells were described previously and drug sensitivity assays were completed as described (Dungrawala et al., 2017). Plasmid and siRNA transfections were performed with polyethylenimine, Dharmafect1 (Dharmacon) for U2OS, or RNAiMax (Thermo Fisher) for RPE and fibroblast cell lines. siRNAs and antibodies are described in Supplemental Experimental Methods.
DNA-binding and DNA Fiber assays
RAD51 EMSA assay was performed largely as described (Wang et al., 2015). 0.5nM 32P-labeled oligo-dT50 ssDNA was incubated with RAD51 in 20μl reactions of binding buffer containing 50 mM Tris pH7.5, 100 ug/ml BSA, 2 mM CaCl2, 2 mM ATP, 100mM NaCl and 1 mM DTT. The reactions were incubated at 37 degrees for 30 minutes. The reactions were separated on 5% 0.5X TBE 37.5:1 gels at 50V for 180 minutes at 4 degrees.
The biotin-ssDNA pull-down assays utilized 4 μM RAD51 prebound to the DNA in the presence of 2mM ATP and 0.1% Tween-20. Increasing amounts of RADX was added and reactions were incubated for 30 minutes prior to separation of the DNA-bound and supernatant fractions and analysis by SDS-PAGE and immunoblotting.
DNA fiber analysis of DNA replication was carried out as described previously (Couch et al., 2013).
Statistical Methods
All statistical analyses are described in the figure legends and were completed with Prism. Investigators were blinded to sample identities and all experiments completed at least twice.
Supplementary Material
Acknowledgments
We thank Agata Smogorzewska for the gift of the RAD51 mutant and FANCD1 mutant cell lines and Grant Stewart for the BOD1L antibody. This work was supported by the National Institutes of Health [R01GM116616 to D.C. and F99CA212345 to K.B.]. RADX protein was purified with assistance from the SBDR EMB Core (P01CA092584). Additional funding was received from the Breast Cancer Research Foundation and the Vanderbilt-Ingram Cancer Center.
Footnotes
AUTHOR CONTRIBUTIONS
K.B., A.K., and H.D. performed the experiments. E.B.G. purified RAD51 with direction from M.M. K.B. and D.C. designed the study, interpreted the results and wrote the manuscript.
DECLARATION OF INTEREST
The authors declare no competing interests.
References
- Bétous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AR, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar J, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. The ZRANB3 translocase associates with poly-ubiquitinated PCNA to promote fork restart and limit recombination after replication stress. Mol Cell. 2012;47:396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Bétous R, Carroll CM, Jung SY, Qin J, Cimprich Ka, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Regulation of Bacterial RecA Protein Function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- Ding X, Chaudhuri AR, Callen E, Pang Y, Biswas K, Klarmann KD, Martin BK, Burkett S, Cleveland L, Stauffer S, et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat Commun. 2016;7:12425. doi: 10.1038/ncomms12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, Sharan SK, Wessel SR, Sathe AA, Zhao R, Cortez D. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol Cell. 2017;67:374–386e5. doi: 10.1016/j.molcel.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Jasin M. BRCA2 suppresses replication stress-induced mitotic and G1 abnormalities through homologous recombination. Nat Commun. 2017;8:525. doi: 10.1038/s41467-017-00634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, Zlatanou A, Nieminuszczy J, Ryan EL, Davies NJ, et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell. 2015;59:462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- Kile AC, Chavez DA, Bacal J, Eldirany S, Korzhnev DM, Bezsonova I, Eichman BF, Cimprich KA. HLTF’s Ancient HIRAN Domain Binds 3′ DNA Ends to Drive Replication Fork Reversal. Mol Cell. 2015;58:1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, Técher H, Baldi G, Shen R, Ciccia A, Pellegrini L, et al. Smarcal1-Mediated Fork Reversal Triggers Mre11-Dependent Degradation of Nascent DNA in the Absence of Brca2 and Stable Rad51 Nucleofilaments. Mol Cell. 2017;67:867–881e7. doi: 10.1016/j.molcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb Perspect Biol. 2015;7(11) doi: 10.1101/cshperspect.a016410. pii: a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon D, Jackson J, Quinet A, Brickner JR, Li S, Yazinski S, You Z, Ira G, Zou L, Mosammaparast N, et al. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzzi G, Marabitti V, Pichierri P, Franchitto A, Ammazzalorso F, Pirzio L, Bignami M, Franchitto A, Pichierri P, Basile G, et al. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 2016;35:1437–1451. doi: 10.15252/embj.201593265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijic S, Zellweger R, Chappidi N, Berti M, Jacobs K, Mutreja K, Ursich S, Ray Chaudhuri A, Nussenzweig A, Janscak P, et al. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat Commun. 2017;8:859. doi: 10.1038/s41467-017-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondinelli B, Gogola E, Yücel H, Duarte AA, Van De Ven M, Van Der Sluijs R, Konstantinopoulos PA, Jonkers J, Ceccaldi R, Rottenberg S, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert L, Ho T, Hoffmann S, Haahr P, Guérillon C, Mailand N. RADX interacts with single-stranded DNA to promote replication fork stability. EMBO Rep. 2017;18:1991–2003. doi: 10.15252/embr.201744877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Mukherjee S, Yang Y, Mori E, Bhattacharya S, Kobayashi J, Yannone SM, Chen DJ, Asaithamby A. Nonenzymatic Role for WRN in Preserving Nascent DNA Strands after Replication Stress. Cell Rep. 2014;9:1387–1401. doi: 10.1016/j.celrep.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, et al. Restoration of Replication Fork Stability in BRCA1- and BRCA2-Deficient Cells by Inactivation of SNF2-Family Fork Remodelers. Mol Cell. 2017;68:414–430e8. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, et al. DNA2 drives processing and restart of reversed replication forks in human cells. J Cell Biol. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic M, Krietsch J, Raso MC, Terraneo N, Zellweger R, Schmid JA, Taglialatela A, Huang JW, Holland CL, Zwicky K, et al. Replication Fork Slowing and Reversal upon DNA Damage Require PCNA Polyubiquitination and ZRANB3 DNA Translocase Activity. Mol Cell. 2017;67:882–890e5. doi: 10.1016/j.molcel.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, Kwan TT, Morris R, Lauffer S, Nussenzweig A, et al. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–332. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadorozhny K, Sannino V, Beláň O, Mlčoušková J, Špírek M, Costanzo V, Krejčí L. Fanconi-Anemia-Associated Mutations Destabilize RAD51 Filaments and Impair Replication Fork Protection. Cell Rep. 2017;21:333–340. doi: 10.1016/j.celrep.2017.09.062. [DOI] [PubMed] [Google Scholar]
- Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.