Summary
The Hippo signaling pathway is an evolutionarily conserved mechanism that controls organ size in animals. Yorkie is well known as a transcriptional co-activator that functions downstream of the Hippo pathway to positively regulate transcription of genes that promote tissue growth. Recent studies have shown that increased myosin activity activates both Yorkie and its vertebrate orthologue YAP, resulting in increased nuclear localization and tissue growth. Here we show that Yorkie also can accumulate at the cell cortex in the apical junctional region. Moreover, Yorkie functions at the cortex to promote activation of myosin through a myosin regulatory light chain kinase, Stretchin-Mlck. This Yorkie function is not dependent on its transcriptional activity and is required for larval and adult tissues to achieve appropriate size. Based on these results, we suggest that Yorkie functions in a feed-forward ‘amplifier’ loop that promotes myosin activation, and thereby greater Yorkie activity, in response to tension.
eTOC Blurb
Yorkie is well-established as a transcriptional co-activator mediating Hippo signaling to promote tissue growth. Xu et al. show that Yorkie additionally functions at the cell cortex, in a non-transcriptional manner, to promote myosin activation in a positive feedback loop that amplifies the effect of cortical tension on Hippo pathway activity.
Introduction
The Hippo signaling pathway is an important regulator of growth that controls cell proliferation, apoptosis and differentiation (Irvine and Harvey, 2015; Yu et al., 2015). The core components of the Hippo pathway consist of upstream regulators, a kinase cascade including Tao, Hippo (Hpo) and Warts (Wts), and a downstream effector, Yorkie (Yki). Despite intensive studies since its discovery more than a decade ago, the molecular mechanisms through which the Hippo pathway is regulated in vivo are yet to be fully understood.
Studies so far identify Yki as a transcriptional co-activator that functions in the nucleus to promote target gene expression. Current models propose that when the Hippo pathway is active Yki is phosphorylated and retained in the cytoplasm, while in the absence of pathway activity Yki remains unphosphorylated and accumulates in the nucleus. Thus, the activation state of the Hippo pathway regulates Yki’s subcellular localization and activity. This cytoplasmic versus nuclear localization of Yki has been the focus of past studies on Yki regulation, with an underlying assumption that cytoplasmic Yki is sequestered from the nucleus and therefore is transcriptionally inactive.
In epithelial cells, the Apical Junctional Region (AJR) is an important hub for regulation of Hippo pathway activity (Boggiano and Fehon, 2012; Meng et al., 2016; Sun and Irvine, 2016; Su et al., 2017). The AJR is a cortical region that extends from the lateral edge of the apical cortex through the adherens junctions (AJs). The upstream regulators of the Hippo pathway as well as the core kinases have been shown to localize to the AJR where they organize into signaling complexes (Boggiano and Fehon, 2012; Sun et al., 2015; Chung et al., 2016). The AJR also contains actomyosin networks that both generate and respond to cytoskeletal tension. Importantly, in both vertebrates and Drosophila cytoskeletal tension has been implicated as a negative regulator of the Hippo pathway. Increased cytoskeletal tension promotes Yki or YAP/TAZ (vertebrate orthologues of Drosophila yki) nuclear localization and transcriptional activity (Dupont et al., 2011; Aragona et al., 2013; Rauskolb et al., 2014).
In this study, we demonstrate a function of Yki at the cell cortex separate from its well characterized role as a transcriptional co-activator. We show that endogenously expressed Yki accumulates at the AJR, particularly under conditions where upstream pathway activity is reduced. Remarkably, experimentally tethering Yki to the cell cortex results in strong activation of cortical myosin in a manner that is independent of Yki’s transcriptional functions. Structure/function analysis indicates that the N-terminal domain of Yki is responsible for this function, and mutation of this domain using CRISPR/Cas9 genome editing results in tissue undergrowth. Furthermore, we show that Yki promotes myosin activation via the upstream regulatory kinase Stretchin-Mlck. We propose a model in which Yki at the cell cortex promotes myosin activation, which in turn activates Yki by suppressing the Hippo pathway, thereby forming a positive feedback loop. Such positive feedback may serve to amplify the effects of tension on Yki’s growth promoting activity independently of, and antagonistically to, the transcriptionally-based negative feedback previously proposed.
Results
Yki localizes to the apical junctional region (AJR) in addition to the nucleus
We recently described a YFP-tagged Yki transgene, expressed under its normal promoter, whose subcellular distribution responds to pathway activity (Su et al., 2017). To better characterize Yki subcellular localization at different states of pathway activity, we generated wts mutant clones and used confocal imaging to examine Yki-YFP localization in live wing imaginal tissues. While wild-type cells displayed a primarily diffuse cytoplasmic distribution of Yki-YFP, in wts clones we observed strong nuclear Yki-YFP localization (Fig. 1B–B′), as previously reported (Oh and Irvine, 2008; Su et al., 2017). We also found strong enrichment of Yki-YFP at the AJR in wts mutant cells (Fig. 1C–C′), as previously reported (Oh et al., 2 009). Furthermore, careful observation of apical sections through wild-type cells revealed less distinct but still clear junctional accumulation in normal tissues as well (Fig. 1D). We noted that while readily apparent in live tissues, this junctionally-localized Yki was much less obvious after fixation (data not shown), which likely explains why it has not been reported previously.
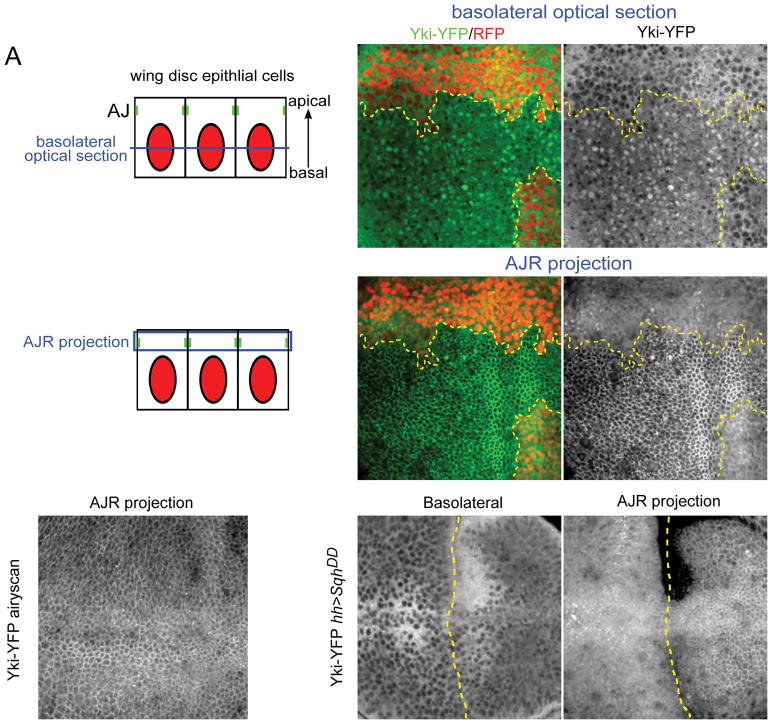
Figure 1. Yki localizes to the apical junctional region (AJR) in addition to the nucleus.
(A) Illustrations showing cross-sectional views of the imaginal epithelium and the approximate positions of basolateral and AJR images shown in this study. As indicated in blue, basolateral images are single sections while AJR views are maximal projections of a small number of apical sections to compensate for curvature of the epithelium.
(B–C′) The effect of Hippo pathway inactivation on Yki subcellular localization. In basal sections of live tissues containing wts null mitotic clones (clone marked by the absence of RFP and a yellow dashed line), Yki-YFP is primarily cytoplasmic in normal imaginal tissue, but is strongly nuclear in wts mutant cells (B–B′). Apically, Yki-YFP is slightly enriched at the apical cortex in normal tissues, but strongly recruited to the AJR in wts null clones (C–C′).
(D) Yki localizes to the AJR in normal tissues. A super-resolution image (using the Zeiss Airyscan that improves resolution and signal-to-noise) of a live wing disc carrying two copies of endogenously expressed Yki-YFP in a ykiB5 null background. A maximum projection of apical optical sections shows cortical localization of Yki-YFP apically.
(E–F) The effect of increasing cytoskeletal tension on Yki subcellular localization. Decreasing Hippo pathway activity by increasing cytoskeletal tension in cells that express a constitutively active form of the myosin regulatory light chain, SqhDD, caused increased nuclear and AJR localization of Yki-YFP. The boundary of SqhDD expression, under the control of hh>Gal4, is marked with a dashed yellow line.
One possible interpretation of these results is that when Hippo pathway activity is decreased, Yki not only accumulates in the nucleus, but also is recruited to the AJR. To further test this idea, we inactivated the Hippo pathway in an alternative fashion by inducing increased cytoskeletal tension. Recent studies in Drosophila have shown that cytoskeletal tension generated by activation of myosin represses pathway activity, resulting in increased Yki target gene expression (Rauskolb et al., 2014). We found that ectopic expression of activated Drosophila non-muscle myosin II regulatory light chain Spaghetti Squash (SqhDD), which constitutively activates myosin contractility (Mitonaka et al., 2007), resulted in a modest increase in nuclear Yki (Fig. 1E) and also resulted in marked Yki accumulation at the AJR (Fig. 1F). This result again indicates that Yki can accumulate both in the nucleus and at the AJR, and further that increased cytoskeletal tension results in greater cortical Yki accumulation.
Cortical Yki promotes activation of myosin independently of Yki transcriptional activity
We next asked if cortical Yki could have a previously unrecognized cellular function in developing tissues. As an initial test, we generated a form of Yki that is constitutively targeted to the cell cortex by fusing the myristoylation signal sequence from Drosophila Src kinase to the N-terminus of Yki (Myr-Yki, Fig. 2A). Myr-Yki expressed in the wing epithelium appeared tightly membrane associated and enriched in the apical domain of cells, as has previously been observed for other Myr-tagged proteins (Neisch et al., 2013; Fig. S1A–D″).
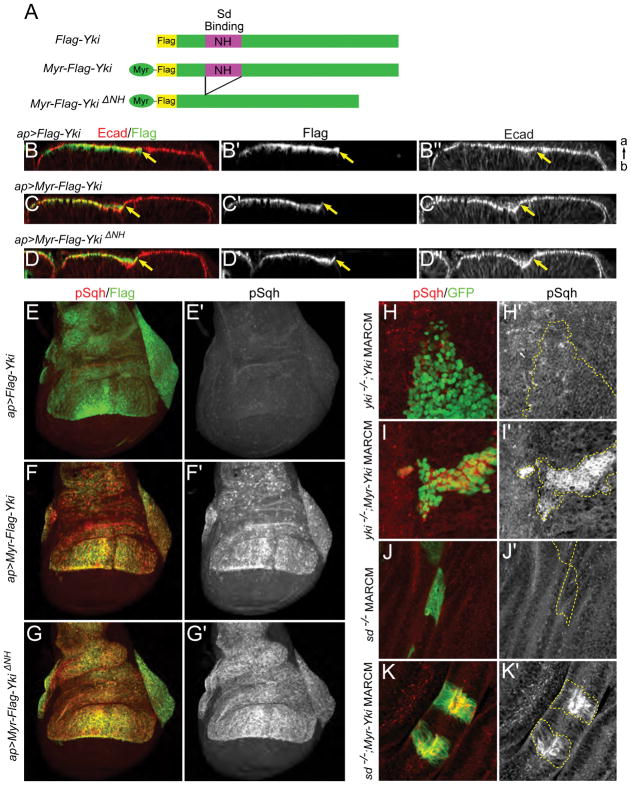
Figure 2. Cortical Yki promotes activation of myosin independently of Yki transcriptional activity.
(A) Cartoons showing UAS transgenes used in B–K. To provide a membrane tether, an N-terminal myristoylation signal sequence was added to Yki (Myr-Yki) or YkiΔNH (Myr-YkiΔNH). The NH domain binds Scalloped (Sd).
(B–D″) Ectopic expression of membrane-associated Yki induces indentation of the epithelium. apterous-Gal4 (ap-Gal4) was used to drive expression of transgenes in the dorsal compartment of wing discs. Optical cross-sections of the epithelium show that expression of Myr-Yki or Myr-YkiΔNH, but not wild-type Yki, induces indentation of wing disc epithelium at the expression boundary (marked by yellow arrows). Ecad staining marks adherens junctions. a, apical; b, basal.
(E–G′) Ectopic expression of membrane-associated Yki induces increased myosin activation. Anti-phospho-Sqh (pSqh), which specifically recognizes activated myosin regulatory light chain, was used to assay myosin activation. Expression of Myr-Yki or Myr-YkiΔNH, but not wild-type Yki, induces a dramatic increase in pSqh staining, indicating increased myosin activation. Images are maximal projections of apical optical sections.
(H–I′) Myosin activation caused by membrane-associated Yki is not dependent on endogenous Yki. Either wild-type Yki or Myr-Yki is expressed in yki null (ykiB5) mitotic clones (marked by GFP expression) using the MARCM technique. pSqh staining increases dramatically in Myr-Yki expressing cells but not in wild-type Yki expressing cells, suggesting that myosin activation is not downstream of Yki transcriptional activity.
(J–K′) Myosin activation caused by membrane-associated Yki is not dependent on Sd. When expressed in sd null (sd47M) mitotic clones (marked with GFP) using the MARCM technique, Myr-Yki induces increased pSqh, indicating that Yki/Sd-mediated transcription is not required for myosin activation.
See also Figure S1.
Observation of the wing imaginal disc revealed that expression of Myr-Yki using the apterous-Gal4 (ap-Gal4) driver caused an indentation or furrowing of the epithelium just inside the boundary of expression, while expression of wild-type Yki did not (Fig. 2B–C). We previously observed a similar epithelial indentation phenotype from mild Rho1 activation in the wing epithelium, which we inferred was caused by increased myosin-based contractility (Neisch et al., 2013). To test if this was the case in cells expressing Myr-Yki, we used a phospho-specific antibody against Sqh (pSqh), as an assay for myosin activity (Zhang and Ward, 2011). Remarkably, we observed a dramatic increase of pSqh staining that was specific to Myr-Yki expressing cells (Fig. 2E–F′). We also examined myosin levels because myosin is known to accumulate at the cell cortex in response to cytoskeleton tension (Fernandez-Gonzalez et al., 2009; Rauskolb et al., 2014; Kim et al., 2015; Pan et al., 2016). Sqh levels at the cortex assayed either by Sqh antibody staining or using the sqh:mCherry reporter (Martin et al., 2009) increased in cells expressing Myr-Yki. (Fig. S1E–H). Together these results indicate that cortically targeted Yki strongly induces myosin activation, and suggest that Yki, whose activity is regulated by actomyosin contractility, might in turn feedback on the actomyosin activation state in the disc epithelium.
Previous studies have shown that YAP positively regulates myosin activity by transcriptionally upregulating genes that activate the myosin regulatory light chain or inhibit myosin light chain phosphatases (Porazinski et al., 2015; Lin et al., 2017). However, in our experiments, Myr-Yki, which was targeted to the plasma membrane, should have been transcriptionally inactive. Consistent with the idea that Myr-Yki functions at the cell cortex rather than in the nucleus to promote myosin activation, ectopic expression of wild-type Yki caused neither indentation of the imaginal epithelium nor increased pSqh staining (Fig. 2B, E–E′).
An alternative possibility is that while Myr-Yki itself is sequestered at the cell cortex, it has a dominant-negative effect on the upstream Hippo pathway machinery. As a result, endogenously expressed Yki could promote greater expression of pathway targets that activate myosin. Therefore, we next tested if the Myr-Yki induced myosin activation depends on endogenous Yki. We examined myosin activation in yki null clones that expressed either wild-type Yki or Myr-Yki using mosaic analysis with a repressible cell marker (MARCM; Fig. 2H–I′). Expression of wild-type Yki did not cause upregulation of pSqh (Fig. 2H–H′). Conversely, in yki null clones that expressed Myr-Yki, pSqh staining was still dramatically increased (Fig. 2I–I′). These results indicate Myr-Yki induced myosin activation does not depend on increased expression of target genes driven by endogenous Yki.
Finally, to further rule out the possibility that Yki-mediated transcription plays a role, we tested the effect of removal of Scalloped (Sd), the main DNA-binding partner of Yki that is necessary for its transcriptional output (Wu et al., 2008). We generated sd null clones that simultaneously expressed Myr-Yki and found that pSqh was again dramatically increased (Fig. 2J–K′). In addition, we generated a Myr-ykiΔNH transgene that lacked the NH domain that mediates the binding between Yki and Sd (Wu et al., 2008). Expression of Myr-YkiΔNH also caused activation of myosin and indentation of the imaginal epithelium (Fig. 2D, G–G′). Together these results support the model that Yki has a non-transcriptional function at the cell cortex to promote myosin activation.
Yki promotes myosin activation under physiological conditions and in response to Hippo pathway inactivation
While myristoylated Yki is a useful tool for examining the role of Yki at the cell cortex, using ectopic expression of an artificially membrane-attached form of Yki could produce artifactual results. To test if endogenous Yki promotes myosin activation under normal physiological conditions, we depleted Yki with RNAi and assayed myosin activation using pSqh staining. Because Yki is required for tissue growth and strong knockdown causes severe tissue loss, we used a temperature sensitive allele of Gal80 (Gal80ts; McGuire et al., 2003) and shifted to restrictive temperature two days prior to dissection to reduce Yki expression without causing significant cell death. Consistent with our hypothesis, we found that depleting Yki resulted in reduced myosin activation (Fig. 3A–D). Together with the observations that endogenously expressed Yki-YFP localizes to the cortex (Fig. 1D) and ectopically expressed wild-type Yki did not appreciably affect pSqh levels (Fig. 2E–E′), this experiment suggests that Yki normally functions at the cortex to promote myosin activity.
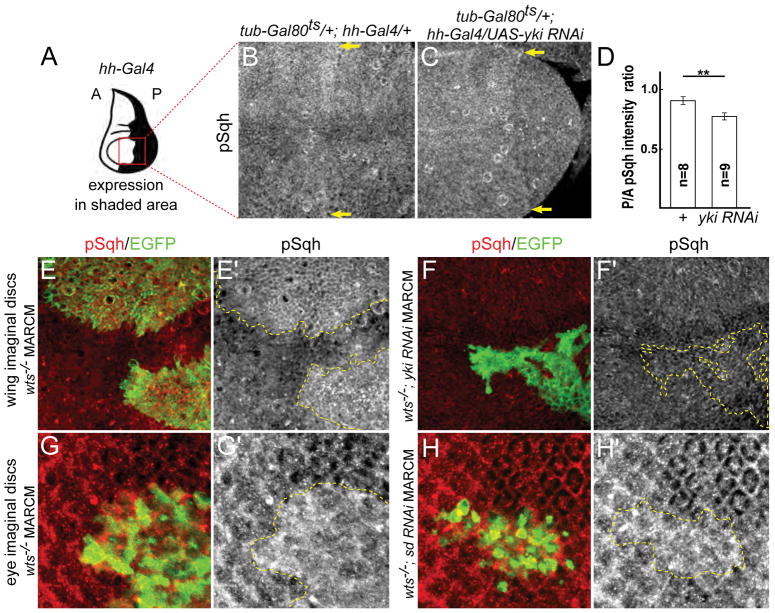
Figure 3. Yki promotes myosin activation in normal tissues and in response to Hippo pathway inactivation.
(A) Cartoon showing expression domain of hh-Gal4 in the wing disc. Red box indicates the approximate area of the wing shown in B–C. A: anterior, P: posterior.
(B–C) Depletion of Yki results in decreased Sqh activation. Expression of a yki-RNAi transgene for two days prior to dissection results in decreased pSqh staining, indicating Yki normally promotes Sqh activation. Images are maximal projections of apical optical sections. Yellow arrows indicate the hh-Gal4 expression boundary.
(D) Quantification of the ratio of pSqh staining fluorescence intensity between posterior (P) and anterior (A) compartments. There is a significant reduction of pSqh ratio in yki RNAi wing discs. Data are represented as mean ± SEM. Asterisks represent statistical significance of the difference between selected groups (** p<0.01, One-way ANOVA and Tukey’s HSD test, n = number of wing discs).
(E–F′) Yki promotes myosin activation in response to Hippo pathway inactivation. pSqh staining was increased in wts null (wtsX1) mitotic clones in wing discs (E–E′; clone marked with GFP and yellow dashed lines), suggesting Yki promotes myosin activation upon Hippo pathway inactivation. This increased pSqh staining is suppressed when yki is depleted in wts null clones (F–F′; clone marked with GFP and yellow dashed lines), suggesting endogenous Yki can promote myosin activation.
(G–H′) Yki promotes myosin activation independent of its transcriptional function. pSqh staining was increased in wtsX1 null mitotic clones in eye discs (G–G′; clone marked with GFP and yellow dashed lines). This increased pSqh staining is maintained when sd is depleted in wts null clones (H–H′; clone marked with GFP and yellow dashed lines), suggesting Yki promotes myosin activation independent of its transcriptional function.
See also Figure S2.
If the model that cortical Yki activates myosin is correct, then our observation that endogenous Yki strongly accumulates at the AJR in wts mutant cells (Fig. 1C) predicts that these cells should also display increased myosin activation. Indeed, we found that loss of wts caused increased pSqh staining (Fig. 3E–E′). We also examined myosin levels using Sqh:GFP and Zip:GFP (Zip=Zipper, Drosophila non-muscle myosin II heavy chain) reporters and found increased myosin levels at the cortex in wts clones (Fig. S2). Furthermore, depletion of Yki in wts clones prevented an increase in pSqh staining (Fig. 3F–F′), suggesting that endogenously expressed Yki can promote myosin activation in the wing epithelium.
While these results indicate that endogenous Yki promotes myosin activation in the imaginal epithelium, they do not distinguish between a transcriptional vs. non-transcriptional role in promoting myosin activation. Indeed, as noted earlier mammalian YAP, a Yki orthologue, is known to drive transcription of genes that promote myosin activation. To address this question further, we first asked if the increased pSqh staining observed in wts mutant cells is Sd-dependent by expressing a sd-RNAi transgene in wts mutant clones by MARCM. We performed these experiments in the eye imaginal disc where Sd is not required for cell viability (Koontz et al., 2013). Compared to wts control clones, these clones were irregular in shape, generally small (Fig. 3G–H′) and lethal in the wing (not shown), indicating that Sd was effectively depleted. In these clones, we observed increased pSqh staining relative to surrounding cells, consistent with the idea that Yki can promote myosin activation independent of its transcriptional activity. However, we note that pSqh staining in this experiment appeared somewhat weaker than in wts mutant control clones, suggesting the possibility that Yki has both non-transcriptional and transcriptional effects on myosin activity. The combined observations that Yki is required for pSqh activation in wts clones and that pSqh activation is retained in the absence of Sd indicate that endogenous Yki can activate myosin in a transcriptionally-independent manner.
As a further test of Yki’s ability to activate myosin at the cortex, we made use of the GrabFP system to recruit Yki-YFP to the junctional cortex by expressing the GrabFP-Aint trap, which uses the Bazooka minimal localization domain tethered to an anti-GFP nanobody (Harmansa et al., 2017). Expression of this transgene strongly recruited endogenously-expressed Yki-YFP to the cortex and promoted increased pSqh staining (Fig. S1I–L). Although qualitatively quite similar, the degree of Sqh activation was less than that observed from Myr-Yki (Supplemental Table 1), consistent with the much lower levels of Yki-YFP expressed from its endogenous promoter in this experiment compared to Myr-Yki expressed under the Gal4/UAS system.
Cortical Yki promotes growth via the Hippo pathway
Previous studies found that increased activation of myosin results in overgrowth due to inactivation of the Hippo pathway (Rauskolb et al., 2014; Deng et al., 2015). We therefore asked if myosin activation induced by Myr-Yki has a similar effect. Consistent with this idea, both Myr-Yki and Myr-YkiΔNH caused overgrowth when expressed in the wing, similar to expression of wild-type Yki (Fig. 4A–D,I). To test if Myr-Yki induced overgrowth is myosin-dependent, we genetically reduced myosin activity by removing one dose of sqh or zip and found that this partially suppressed the Myr-Yki overgrowth phenotype in the adult wing (Fig. 4E–I). In contrast, heterozygosity for either sqh or zip had no effect on the overgrowth induced by ectopic expression of wild-type, untethered Yki (Fig. 4I). Additionally, expression of Myr-Yki and Myr-YkiΔNH caused increased expression of the Hippo pathway reporter expanded-lacZ (ex-lacZ) (Fig. 4J–M), consistent with greater Yki activity.
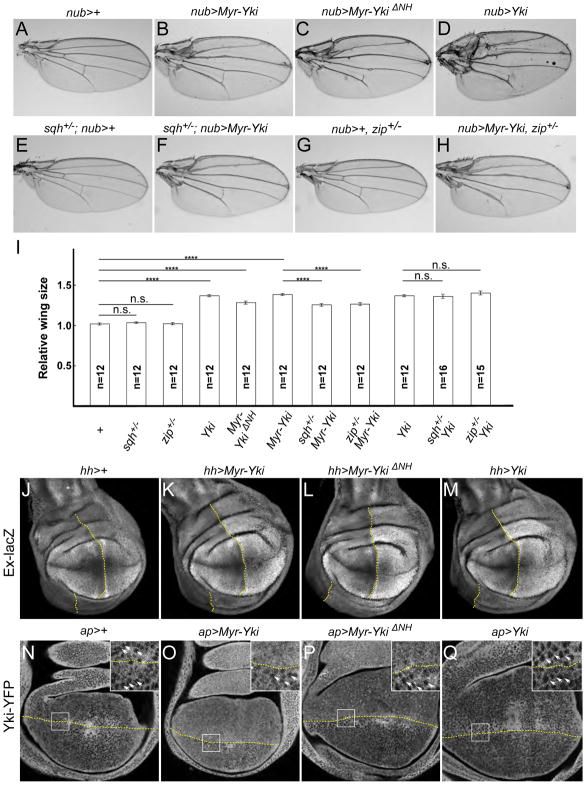
Figure 4. Cortical Yki promotes growth via the Hippo pathway.
(A–D) Expression of both wild-type and membrane tethered Yki in the wing causes overgrowth. nub-Gal4 was used to drive expression of the indicated Yki transgenes in the wing blade. Representative images of female adult wings (cultured at 18 °C) of indicated genotypes are shown. Myr-Yki, Myr-YkiΔNH, and wild-type Yki all caused overgrowth.
(E–H) Myr-Yki-mediated overgrowth depends on myosin activity. Removing one dose of the regulatory light chain sqh (sqhAX3) or the non-muscle myosin zip (zip1) did not affect wing size on its own and partially suppressed the Myr-Yki overgrowth phenotype.
(I) Quantification of wing sizes for the indicated genotypes. Heterozygosity for sqh or zip suppressed the overgrowth induced by cortical, Myr-tagged Yki but not wild-type Yki. Data are represented as mean ± SEM. Asterisks represent statistical significance of the difference between selected groups (**** p<0.0001; n.s.: not significant [p > 0.05], One-way ANOVA and Tukey’s HSD test, n = number of wings).
(J–M) Ectopic expression of Yki transgenes in wing discs causes up-regulation of the Hippo pathway reporter ex-lacZ. hh-Gal4 was used to drive expression of Yki transgenes in the posterior compartment of wing discs. The boundary of expression is marked with a dotted yellow line.
(N–Q) Ectopic expression of membrane-associated Yki in wing discs causes increased nuclear localization of Yki-YFP. At the position of these optical sections, the nuclei are seen as dark circles surrounded by diffuse, cytoplasmically localized Yki-YFP in normal tissue (below the dotted yellow lines). ap-Gal4 was used to drive expression of Yki transgenes in the dorsal compartment (above the dotted lines) of wing discs. Myr-Yki and Myr-YkiΔNH, but not wild-type Yki, caused increased nuclear localization of Yki-YFP. Boxes denote regions shown in high magnification insets; white arrows indicate nuclei.
See also Figures S3 and S4.
The model that cortical Yki promotes growth by repressing Hippo pathway activity predicts that the observed overgrowth should be dependent on endogenous Yki. To test this, first we examined the effect of Myr-Yki expression on nuclear localization of endogenously-expressed Yki-YFP. Both Myr-Yki and Myr-YkiΔNH caused increased nuclear accumulation of Yki-YFP relative to wild-type cells (Fig. 4N–P). Interestingly, ectopic expression of wild-type Yki, which caused similar overgrowth to Myr-Yki expression (Fig. 4A–D) actually resulted in slightly decreased nuclear localization of endogenously-expressed Yki-YFP (Fig. 4Q). This is expected if the ectopically expressed untethered Yki competes with endogenously-expressed Yki-YFP for nuclear entry or retention. Furthermore, we generated MARCM clones that expressed Myr-Yki, Myr-YkiΔNH, or wild-type Yki in the background of a yki null allele. Only wild-type Yki was able to rescue undergrowth of yki null clones (Fig. S3). Together our results indicate that ectopically expressed cortical Yki promotes growth by activating myosin and inactivating the Hippo pathway, thereby allowing endogenous Yki to enter the nucleus and promote growth.
Because Yki physically interacts with Sd and Wts, we considered the possibility that Myr-Yki might alter pathway activity by sequestering an interacting component at the cell cortex. To address this, we first stained wing imaginal discs with anti-Sd and found that while Myr-Yki expression caused dramatic cortical Sd accumulation, Myr-YkiΔNH did not (Fig. S4A–C‴). The observation that both the Myr-Yki and Myr-YkiΔNH transgenes caused myosin activation and overgrowth indicates that recruitment of Sd to the cell cortex is not required. Next, we asked if the effect of Myr-Yki on pathway output and growth might be attributable to the ability of Yki to bind Wts, which could sequester Wts cortically so that it cannot participate in Hippo pathway signaling. We detected no increased cortical accumulation of endogenously expressed Wts-YFP in response to Myr-Yki expression (Fig. S4D–E′), though we did notice a slight alteration in the appearance of Wts that we speculate might be related to increased cortical contractility induced by Myr-Yki. To further test if Myr-Yki interferes with Wts function, we compared the effect of ectopic Wts expression alone to co-expression with Myr-Yki and found that co-expression of Myr-Yki failed to suppress Wts induced undergrowth of the wing (Fig. S4F–I). Together these results argue against the hypothesis that Myr-Yki promotes growth by sequestering or otherwise inactivating Wts.
Yki activates Sqh via the Sqh kinase Stretchin-Mlck (Strn-Mlck)
To better understand the mechanism by which cortical Yki regulates myosin activity, we next screened known or predicted Drosophila myosin light chain kinases to determine if they were necessary for Yki-induced Sqh activation. Specifically, we simultaneously expressed Myr-Yki and one of at least two independent RNAi transgenes each for Rho kinase (Rok), Death-associated protein kinase related (Drak), bent (bt), spaghetti-squash activator (sqa), genghis khan (gek), Stretchin-Mlck (Strn-Mlck), sticky (sti), or AMP-activated protein kinase α subunit (AMPKα) in the wing epithelium (Maroto et al., 1992; Dong et al., 2002; D’Avino et al., 2004; Neubueser and Hipfner, 2010; Nie et al., 2014). RNAi lines for only two genes, AMPKα and Strn-Mlck, had any detectable effect on pSqh levels (Fig. S5). One out of six tested AMPKα RNAi lines (TRiP.JF01951) displayed weak suppression (Fig. S5U). Because this suppression was much weaker than that caused by Strn-Mlck RNAi and two validated AMPKα RNAi lines, VDRC1827 (Choi et al., 2011) and VCRC1060200 (Stenesen et al., 2013), did not suppress pSqh staining, we focused on Strn-Mlck and did not further pursue AMPKα.
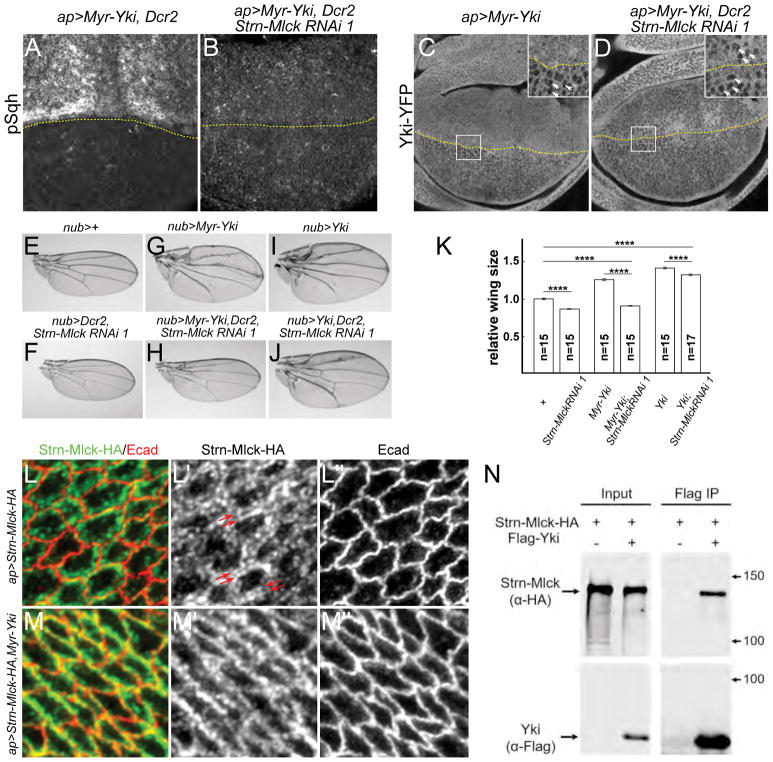
Two non-overlapping Strn-Mlck RNAi lines, TRiP.JF02278 (Strn-Mlck RNAi 1) and TRiP.JF021 (Strn-Mlck RNAi 2), suppressed the increased pSqh staining caused by Myr-Yki expression (Fig. S5N–O). Strn-Mlck is a member of the Titin/Myosin Light Chain Kinase family (Champagne et al., 2000). Adding expression of a Dicer-2 (Dcr-2) transgene to increase Strn-Mlck RNAi efficiency strongly increased this effect (Fig. 5A–B). No null Strn-Mlck alleles have been described, but the two RNAi lines described above recognize distinct domains and display indistinguishable phenotypes, suggesting they are specific for Strn-Mlck. We also found that Strn-Mlck RNAi 1 strongly repressed expression of a UAS-Strn-Mlck transgene (Fig. S6A–B).
Figure 5. Yki activates Sqh via the Sqh kinase Strn-Mlck.
(A–B) Strn-Mlck depletion by RNAi strongly suppresses Myr-Yki-mediated myosin activation. UAS-Dcr2 was used to enhance efficiency of the RNAi transgenes. The expression boundary is marked by dashed yellow lines.
(C–D) Strn-Mlck depletion suppresses Myr-Yki induced nuclear Yki-YFP localization. The expression boundary is marked by dashed yellow lines. Boxes denote regions shown in high magnification insets; white arrows indicate nuclei.
(E–K) Strn-Mlck depletion completely suppresses Myr-Yki mediated overgrowth. Representative images of adult male wings of the indicated genotypes are shown (E–J) together with quantification of the results (K). Expression of a Strn-Mlck RNAi transgene (line BL26736), either alone or in the background of Myr-Yki, caused undergrowth of the wing. In contrast, Strn-Mlck depletion did not completely suppress the overgrowth caused by expression of wild-type Yki. Data in (K) are represented as mean ± SEM. Asterisks represent statistical significance of the difference between selected groups (**** p<0.0001, One-way ANOVA and Tukey’s HSD test, n = number of wings).
(L–M″) Myr-Yki recruits Strn-Mlck to the junctional region. Transgenically-expressed, Strn-Mlck was localized to a sub-cortical band that is distinct from the junctional marker Ecad (L–L″). Co-expression of Myr-Yki resulted in greater junctional localization of Strn-Mlck. Strn-Mlck was tagged with HA and detected with HA antibody staining.
(N) Strn-Mlck and Yki form a complex in S2 cells. HA-tagged Strn-Mlck and Flag-tagged Yki were co-expressed in S2 cells. Lysates were immunoprecipitated with anti-Flag and immunoblotted. Strn-Mlck co-immunoprecipitated with Yki, suggesting they can form a complex.
See also Figure S5 and S6.
We next tested whether depleting Strn-Mlck also suppressed other Myr-Yki phenotypes. Strn-Mlck depletion strongly suppressed the increased nuclear Yki-YFP accumulation (Fig. 5C–D), increased expression of ex-lacZ (Fig. S6E–F), and overgrowth of adult wings (Fig. 5E–H, K) caused by Myr-Yki expression. Indeed, depletion of Strn-Mlck appeared epistatic to Myr-Yki with regard to overall wing growth. In contrast, Strn-Mlck depletion only weakly suppressed the overgrowth caused by expression of wild-type Yki (Fig. 5I–K). Altogether our results suggest that Strn-Mlck functions downstream of cortical Yki to activate Sqh but is not required for nuclear Yki to promote growth.
We confirmed that depletion of Strn-Mlck did not affect Myr-Yki localization (Fig. S6C–D), though we did note that the level of Myr-Yki expression was lower when Strn-Mlck was depleted. We do not understand the basis of this observation, though it does not appear to be due to transgene competition (data not shown) and we note that different Myr-Yki transgenes that express at different levels still promote overgrowth (data not shown) and Sqh activation to high levels (Supplemental Table 1).
To understand how Yki and Strn-Mlck cooperate to promote myosin activity, we next looked at the effect of Myr-Yki expression on localization of transgenically-expressed, tagged Strn-Mlck (isoform B; FlyBase) in the wing imaginal epithelium. When expressed alone, Strn-Mlck was localized to a sub-cortical band that is distinct from the junctional marker Ecad (Fig. 5L–L″), a localization that is strikingly similar to Sqh (called the apical cytocortex in Tsoumpekos et al., 2018). In contrast, co-expression of Myr-Yki and Strn-Mlck appeared to recruit Strn-Mlck to the junctional region (Fig. 5M–M″), suggesting that Yki and Strn-Mlck can form a complex. To address this possibility, we co-expressed these proteins in S2 cultured cells and found that they readily co-IP from cell lysates (Fig. 5N). Together these experiments suggest that Yki and Strn-Mlck form a complex and that in tissues cortical Yki may recruit Strn-Mlck, thereby promoting Sqh activation.
Since Strn-Mlck depletion alone caused undergrowth, we asked if Strn-Mlck regulates growth via the Hippo pathway during normal development. We found that strong depletion of Strn-Mlck caused reduced expression of the Hippo pathway reporters Diap1-lacZ and fj-lacZ in wing imaginal discs (Fig. S6G–K). Similar results have been found for other known negative regulators of the Hippo pathway (Das Thakur et al., 2010; Rauskolb et al., 2011), suggesting Strn-Mlck is a negative regulator of the Hippo pathway during normal development.
The N-terminal region of Yki is necessary and sufficient for myosin activation
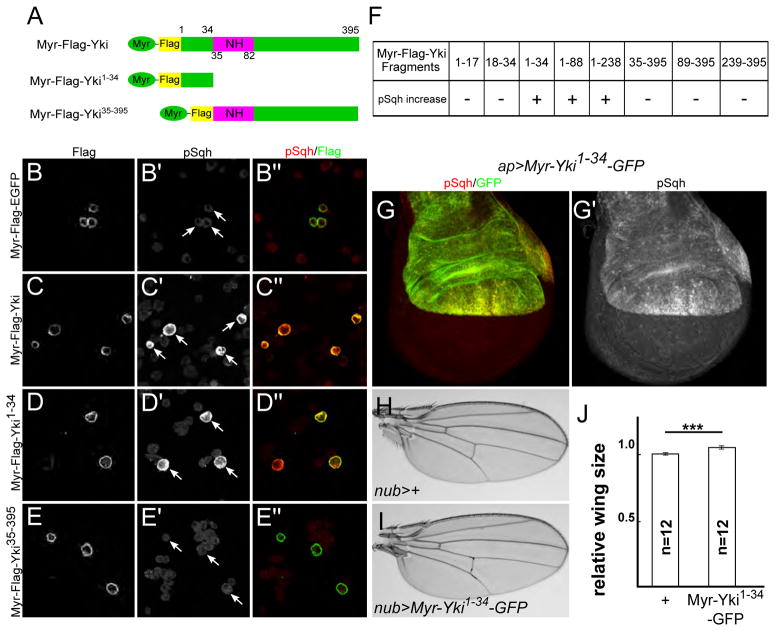
We next mapped the region within Yki that is responsible for activating myosin. We first tested if we could recapitulate the pSqh increase phenotype in Drosophila S2 cells. Expression of Myr-Yki caused a dramatic increase in pSqh staining. (Fig. 6A–C). To map the region of Yki responsible for this increase, we generated truncated Yki fragments with a myristoylation signal sequence, expressed them individually in S2 cells, and found that an N-terminal fragment encoding the first 34 amino acids was both necessary and sufficient to activate myosin (Fig. 6D–F).
Figure 6. The N-terminal region of Yki is necessary and sufficient for myosin activation.
(A) Cartoons showing Myr-Yki fragments used in (C–E″).
(B–F) Mapping of the myosin activation region using S2 cells transfected with the indicated constructs and stained for Flag and pSqh. Myr-Yki and Myr-Yki1-34 cause increased pSqh staining, while Myr-Yki35-395 and Myr-EGFP do not. White arrows indicate transfected cells. In (F), the full panel of tested deletion constructs is displayed. Fragments 1-34, 1-88, 1-238 each induce Sqh activation whereas their complementary C-terminal fragments 35-395, 89-395, 239-395 do not. Further subdivision of the 1-34 region (fragments 1-17 and 18-34) disrupted the ability to activate Sqh.
(G–G′) Expression of UAS-Myr-Yki1-34-GFP in the dorsal compartment of the wing is sufficient to induce myosin activation.
(H–J) Expression of UAS-Myr-Yki1-34-GFP causes overgrowth of the wing. Representative images of adult female wings of the indicated genotypes are shown (H–I), together with quantification of the results (J). Data are represented as mean ± SEM. Asterisks represent statistical significance of the difference between selected groups (*** p<0.001, One-way ANOVA, n = number of wings).
To test if this N-terminal fragment is sufficient to activate myosin in vivo, we generated UAS-Myr-yki1-34-GFP transgenic animals. When expressed in the wing imaginal disc, Myr-Yki1-34-GFP activated myosin (Fig. 6G–G′) and caused overgrowth of adult wings (Fig. 6H–J). These effects were similar to, but weaker than, the phenotypes observed for full-length Myr-Yki (Figs. 2F–F′, 4A–B, Supplemental Table 1), possibly because other regions of Yki also contribute to myosin activation.
Uncoupling the transcriptional and myosin-activating functions of Yki
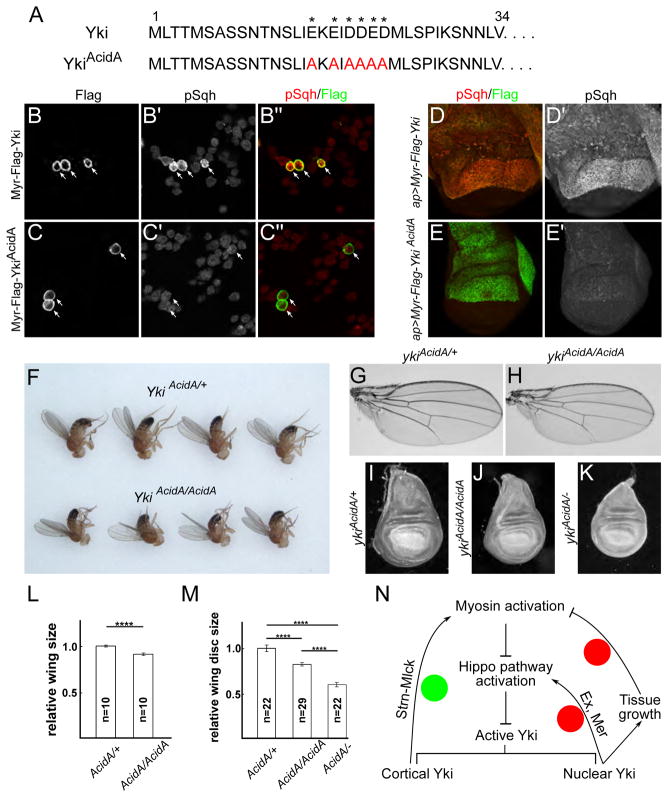
The data presented so far suggest that Yki has a non-transcriptional function at the cell cortex to promote myosin activation through its N-terminal domain. To understand the developmental significance of this function, we sought to disable it without affecting Yki’s transcriptional functions. An initial attempt—deleting aa1-34 from the yki locus via CRISPR/Cas9—resulted in strong reduction of Yki expression (data not shown). As an alternative approach, we generated a series of smaller deletions and point mutations in the aa1-34 region and tested their ability to activate Sqh when expressed in S2 cells (data not shown). These studies led us to focus on a short stretch of acidic residues in the first 34 amino acids of Yki protein sequence (Fig. 7A). Mutation of these residues to Alanine blocked Sqh activation in S2 cells (Fig. 7B–C″) and wing imaginal discs (Fig. 7D–E′). Using CRISPR/Cas9, we then generated the ykiAcidA allele at the yki locus and found that this mutation does not affect Yki expression (Fig. S7A–B′). To ask if this allele visibly affects Yki’s transcriptional activity, we examined expression levels of the Hippo pathway reporter Diap1-lacZ as well as the Hippo pathway transcriptional target expanded (ex) and found they were not detectably affected in ykiAcidA/AcidA clones (Fig. S7C–D′). Additionally, to test the ykiAcidA allele under conditions where it should be maximally transcriptionally active, we examined the expression of Yki targets in clones that were simultaneously mutant for ykiAcidA and wts. In these cells, Yki transcriptional activity should be uncoupled from any upstream pathway activity, as well as the tension mediated feedback we propose, because tension-mediated pathway regulation is thought to be carried out via sequestration of Wts (Rauskolb et al., 2014). Comparison of wts−/− (;yki+/AcidA) clones to wts−/−; ykiAcidA/AcidA double mutant clones showed that staining of Ex and Merlin (Mer), another Yki target, increased similarly in both genotypes (Fig. S7E–F‴). Taken together these results suggest that the ykiAcidA mutation affects Yki’s myosin activation function but not its transcriptional function.
Figure 7. Uncoupling the transcriptional and myosin-activating functions of Yki.
(A) Sequences of the first 34 amino acids of wild-type Yki and the YkiAcidA allele. Asterisks indicate acidic amino acids.
(B–E′) Mutation of acidic residues at the N-terminus of Yki blocks its ability to activate myosin. S2 cells expressing membrane-associated wild-type Myr-Yki (B–B‴) and Myr-YkiAcidA (C–C‴). Myr-YkiAcidA fails to activate Sqh. White arrows indicate transfected cells. Wing imaginal discs expressing Myr-YkiAcidA also fail to activate Sqh (D–E′). The two transgenes were integrated at the same landing site to ensure comparable expression levels.
(F–K) The myosin-activating function of Yki is required for proper growth. Homozygous ykiAcidA adults are smaller than controls (F). Representative images show that adult female wings from animals homozygous for the ykiAcidA allele are undergrown (G–H). Larval wing imaginal discs are similarly undergrown (I–K), with ykiAcidA/ykiB5 (ykiAcidA/−) animals displaying the strongest phenotype (the latter genotype does not survive to adults). Quantification of adult wing and larval wing disc sizes of the indicated genotypes confirms these differences (L–M). Data are represented as mean ± SEM. Asterisks represent statistical significance of the difference between selected groups (**** p<0.0001, One-way ANOVA and Tukey’s HSD test, n = number of wings [L]/wing discs [M]).
(N) Model of feedback regulation of the Hippo pathway. Two negative feedback mechanisms have been proposed in the regulation of the Hippo pathway (red circles). One is mediated through transcriptional upregulation of upstream components, including Mer and Ex, and the other is a consequence of morphogen-driven growth at the center of tissues and the resulting compression of those cells. We propose a positive feedback mechanism (green circle) in which cortical Yki at the AJR promotes myosin activation and increased tension via Strn-Mlck, which in turn further activates Yki by suppressing the Hippo pathway. See the Discussion for more details.
See also Figure S7.
We then analyzed the growth phenotypes of ykiAcidA animals. ykiAcidA homozygous adults were viable but smaller than heterozygotes, as were their imaginal discs and wings (Fig. 7F–M). Animals transheterozygous for ykiAcidA over ykiB5, a null allele, were pupal lethal and displayed even smaller wing imaginal discs (Fig. 7I–K, M). Interestingly, these wing imaginal discs not only were smaller, but also appeared more bilaterally symmetrical along anterior-posterior axis than normal wing discs (Fig. 7K). Together these results suggest that Yki’s function in promoting myosin activation is important for organ size control and perhaps organ morphogenesis. However, further studies will be required to fully understand how this domain and its ability to promote myosin activation function in tissue growth and morphogenesis.
Discussion
Collectively, our results reveal a previously unrecognized role for Yki at the cell cortex where it promotes myosin activation through the myosin regulatory light chain kinase Strn-Mlck. Two aspects of these findings are unexpected. First, Yki has traditionally been viewed as a transcriptional co-activator that functions in the nucleus to promote target gene expression. The results presented here indicate Yki also acts via a non-transcriptional mechanism to control non-muscle myosin II contractility. While surprising, several previous studies have suggested that Yki and its vertebrate orthologues YAP and TAZ, might have additional, non-nuclear functions. For example, cytoplasmic YAP has been shown to inhibit TGF-β/Smad signaling (Varelas et al., 2010a) and evidence suggests that TAZ and Yki can act in the cytoplasm to inhibit Wnt/β-catenin signaling (Varelas et al., 2010b). Interestingly, although YAP has been shown to localize to the cell-cell junctions in primary human keratinocytes (Schlegelmilch et al., 2011), and cortical localization of Yki in Drosophila has been reported in the eye (Badouel et al., 2009) and wing imaginal discs (Zhang et al., 2009), these observations have been interpreted as inhibition of Yki/YAP nuclear function by sequestration in the cytoplasm. The results presented here indicate that Yki also has an active role at the cell cortex, to promote myosin activity, and that this function is important for growth regulation.
A second unexpected aspect of our findings is that they suggest that Yki functions in a positive feedback loop that promotes myosin activation, and thereby greater Yki activity, in response to tension. We have shown that cortical Yki promotes myosin activation via the Sqh kinase Strn-Mlck, and that this increased myosin activity in turn can activate Yki, presumably by repressing the Hippo pathway. Further, our observations indicate that Hippo pathway downregulation and Yki activation caused by loss of wts result in 1) increased cortical accumulation of Yki and 2) increased myosin activation. Taken together, our results suggest the existence of a non-transcriptional, positive feedback loop between Yki and myosin activation (Fig. 7N). This positive feedback loop could potentially serve to amplify the effect of tension on Yki activity and therefore tissue growth.
A recent model suggests that Yki’s ability to sense tension and promote growth might be particularly important at the periphery of tissues, where morphogen levels, and therefore morphogen-driven proliferation, should be lower (Aegerter-Wilmsen et al., 2007, 2012; Hariharan, 2015). According to one version of this model, morphogen-driven growth at the center of tissues exerts tension at the periphery, which in turn might cause tension-mediated activation of Yki function. In agreement with this idea, recent empirical measurements in the wing imaginal disc indicate higher tension at the periphery of wing blade (Mao et al., 2013). The positive feedback mechanism described here could be particularly important for promoting growth at the periphery of developing tissues.
To further test the positive feedback model of Yki function in normal development, we identified a short stretch of amino acids in the N-terminus of Yki that is both necessary and sufficient for its ability to promote myosin activation. We mutated this sequence to generate the ykiAcidA allele that lacks myosin activation activity but at the same time retains normal transcriptional function. Strikingly, ykiAcidA homozygous animals display overall undergrowth, and smaller wing imaginal discs in particular, indicating that this region of Yki is important for its growth promoting function. In addition, ykiAcidA/− wing imaginal discs show morphological defects: they lost asymmetry along anterior-posterior axis outside of the pouch region (Fig. 7H). We do not know yet whether these defects are due to loss of the proposed feedback mechanism, but studies are currently underway to examine this question.
How is this positive feedback mechanism down-regulated to prevent overgrowth? Two sources of negative feedback have been proposed in Hippo pathway regulation in Drosophila (Fig. 7N, red circles). First, when Yki is activated, it drives expression of positive upstream regulators of the Hippo pathway including Ex and Mer. These proteins will in turn activate the Hippo pathway to suppress Yki activity (Boggiano and Fehon, 2012). Second, Yki activation causes growth, which can reduce tension at the center of tissues, leading to de-repression of the Hippo pathway and Yki inactivation (Moberg et al., 2005; Pan et al., 2016). These sources of negative feedback require target gene transcription (and growth) and therefore would be expected to act more slowly than the positive feedback we propose.
Another important question is how does Yki localize to the cell cortex to promote myosin activation? One possible mechanism is that Yki is recruited via a previously described interaction with Ex, which strongly localizes to the AJR (Badouel et al., 2009; Su et al., 2017). Additionally, Yki has been shown to interact with several other pathway components, including Kibra (Kib), Hpo, Wts, Ajuba (Jub) and 14-3-3 (Genevet and Tapon, 2011). Our preliminary experiments have not clearly implicated any of these proteins in localizing Yki to the cortex (data not shown), though it is possible either that partial depletion by RNAi is not sufficient to see an effect, or a multiprotein complex is responsible. Other factors could also recruit Yki to the AJR, and further experiments will be required to resolve this question.
Our results indicate that Strn-Mlck is an integral part of the cortical Yki-myosin positive feedback loop. The Strn-Mlck locus is proposed to generate three distinct sets of protein isoforms, with the largest isoform encoding a huge Titin-like 926 kDa protein (Champagne et al., 2000). Previous functional studies of Strn-Mlck have focused on its function in indirect flight muscles (Patel and Saide, 2005; Schönbauer et al., 2011) or actomyosin flow after wounding (Antunes et al., 2013). However, Strn-Mlck’s function in regulating myosin activity has not been thoroughly examined, nor has its expression in non-muscle tissues. Interestingly, although previous studies have shown that Rok driven cytoskeletal tension can regulate Yki activity, our results indicate that Yki acts through Strn-Mlck, but not Rok, to promote myosin activation. Thus, our data suggest that while all forms of cytoskeletal tension likely affect Hippo pathway activity, Yki itself regulates tension specifically via Strn-Mlck. Interestingly, Strn-Mlck has two flexible PEVK domains whose conformation changes when stretched (Champagne et al., 2000; Linke, 2000). Thus the functional interaction between Yki and Strn-Mlck could represent yet another means by which physical tension regulates pathway output in growing tissues.
Similar to Yki in Drosophila, vertebrate YAP also can promote myosin activation. However, the underlying mechanisms appear to be different. Our results uncover a role for Yki at the cell cortex to promote myosin activity whereas studies in mouse and in Medaka found that YAP does so by increasing transcription of genes encoding proteins that activate the myosin regulatory light chain (Porazinski et al., 2015; Lin et al., 2017). In addition, The N-terminal myosin activation domain of Drosophila Yki does not display clear sequence homology with vertebrate YAP. This could mean Drosophila has a distinctive, non-transcriptional mechanism to generate Yki-myosin positive feedback. Even though the specific mechanism through which Yki promotes myosin activation does not appear to be conserved in vertebrates, positive feedback between cytoskeletal tension and the activity of Yki or YAP seems to be a well-conserved but still poorly understood feature of the Hippo pathway. While it is clear that the non-transcriptional positive feedback we propose for Yki would be temporally distinct from the transcriptionally-based negative feedback mechanism, it is not as clear how YAP’s positive and negative feedback effects would be distinctly interpreted in developing tissues. Further studies in both Drosophila and vertebrate systems will be required to understand how these competing inputs are integrated into the process of tissue growth during development.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Please contact the Lead Contact, Richard G. Fehon (rfehon@uchicago.edu), for reagents and resources generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster was cultured using standard techniques at 25°C (unless otherwise noted). Both male and female animals were used.
METHOD DETAILS
Drosophila Genetics
Drosophila stocks used in this study are listed in Key Resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Guinea pig anti-Ex | Maitra et al., 2006 | N/A |
| Guinea pig anti-Sqh1P | Zhang and Ward, 2011 | N/A |
| Mouse anti Sqh | Zhang and Ward, 2011 | N/A |
| Mouse anti-Flag | Sigma | Cat#F1804 |
| Rat anti-Ecad | Developmental Studies Hybridoma Bank | Cat#AB_528120 |
| Guinea pig anti-Sd | Guss et al., 2013 | N/A |
| Rabbit anti-Yki | Oh and Irvine, 2008 | N/A |
| Rabbit anti-HA | Santa Cruz | Cat#sc-805 |
| Mouse anti-Flag agarose beads | Sigma | Cat#A2220 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Normal goat serum | Jackson ImmunoResearch | Cat# 005000001 |
| 16% formaldehyde | Electron Microscopy Sciences (EMS) | Cat#15710 |
| S2 medium | Sigma | Cat# S9895 |
| 10% Fetal Bovine Serum | Thermo Fisher Scientific | Cat#16000044 |
| Didodecyldimethylammonium bromide | Sigma | Cat# 359025 |
| Methyl salicylate | Thermo Fisher Scientific | Cat#119368 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: UAS-sqhDD | Mitonaka et al., 2007 | N/A |
| D. melanogaster: UAS-Flag-Yki | This study | N/A |
| D. melanogaster: UAS-Myr-Flag-Yki (Random insertion) | This study | N/A |
| D. melanogaster: UAS-Myr-Flag-Yki (Site specific integration) | This study | N/A |
| D. melanogaster: UAS-Myr-Flag-YkiΔNH | This study | N/A |
| D. melanogaster: UAS-Myr-Flag-Yki1-34-GFP(Site specific integration) | This study | N/A |
| D. melanogaster: UAS-Myr-Flag-YkiAcidA (Site specific integration) | This study | N/A |
| D. melanogaster: UAS-yki RNAi | Vienna Drosophila RNAi Center | 40497 |
| D. melanogaster: UAS-yki RNAi | Vienna Drosophila RNAi Center | 104523 |
| D. melanogaster: UAS-sd RNAi | Vienna Drosophila RNAi Center | 101497 |
| D. melanogaster: UAS-Dcr2 | Bloomington Drosophila Stock Center | 24650 |
| D. melanogaster: UAS-Dcr2 | Bloomington Drosophila Stock Center | 24651 |
| D. melanogaster: UAS-Strn-Mlck RNAi | Bloomington Drosophila Stock Center | JF02278 |
| D. melanogaster: UAS-Strn-Mlck RNAi | Bloomington Drosophila Stock Center | JF02170 |
| D. melanogaster: UAS-Strn-Mlck RNAi | Bloomington Drosophila Stock Center | HMS01665 |
| D. melanogaster: UAS-Strn-Mlck RNAi | Bloomington Drosophila Stock Center | HMS02663 |
| D. melanogaster: UAS-Rok RNAi | Vienna Drosophila RNAi Center | 104675 |
| D. melanogaster: UAS-Rok RNAi | Bloomington Drosophila Stock Center | HMS01311 |
| D. melanogaster: UAS-Drak RNAi | Bloomington Drosophila Stock Center | JF03385 |
| D. melanogaster: UAS-Drak RNAi | Bloomington Drosophila Stock Center | GL00087 |
| D. melanogaster: UAS-Drak RNAi | Bloomington Drosophila Stock Center | HMS02822 |
| D. melanogaster: UAS-sqa RNAi | Bloomington Drosophila Stock Center | JF02277 |
| D. melanogaster: UAS-sqa RNAi | Vienna Drosophila RNAi Center | 101640 |
| D. melanogaster: UAS-sqa RNAi | Vienna Drosophila RNAi Center | 107241 |
| D. melanogaster: UAS-bt RNAi | Bloomington Drosophila Stock Center | JF01107 |
| D. melanogaster: UAS-bt RNAi | Bloomington Drosophila Stock Center | JF01108 |
| D. melanogaster: UAS-gek RNAi | Bloomington Drosophila Stock Center | GL00299 |
| D. melanogaster: UAS-gek RNAi | Bloomington Drosophila Stock Center | HMS04488 |
| D. melanogaster: UAS-sti RNAi | Bloomington Drosophila Stock Center | GL00312 |
| D. melanogaster: UAS-sti RNAi | Bloomington Drosophila Stock Center | HMC03586 |
| D. melanogaster: UAS-sti RNAi | Bloomington Drosophila Stock Center | HMJ30325 |
| D. melanogaster: UAS-AMPKα RNAi | Bloomington Drosophila Stock Center | JF01951 |
| D. melanogaster: UAS-AMPKα RNAi | Bloomington Drosophila Stock Center | HMS00362 |
| D. melanogaster: UAS-AMPKα RNAi | Bloomington Drosophila Stock Center | GL00004 |
| D. melanogaster: UAS-AMPKα RNAi | Bloomington Drosophila Stock Center | HMC04979 |
| D. melanogaster: UAS-AMPKα RNAi | Vienna Drosophila RNAi Center | 1827 |
| D. melanogaster: UAS-AMPKα RNAi | Vienna Drosophila RNAi Center | 106200 |
| D. melanogaster: 82BFRT wtsX1 | Xu et al., 1995 | N/A |
| D. melanogaster:42DFRT ykiB5 | Huang et al., 2005 | N/A |
| D. melanogaster:19AFRT sd47M | Wu et al., 2008 | N/A |
| D. melanogaster: sqhAX3/FM | Jordan and Karess, 1997 | N/A |
| D. melanogaster: Diap1-lacZ | Hay et al., 1995 | N/A |
| D. melanogaster: ex-lacZ | Hamaratoglu et al., 2006 | N/A |
| D. melanogaster: fj-lacZ | Bloomington Drosophila Stock Center | 6370 |
| D. melanogaster: sqh:mCherry | Martin et al., 2009 | N/A |
| D. melanogaster: sqh:GFP | Vienna Drosophila RNAi Center | 318484 |
| D. melanogaster: zip:GFP | Rauskolb et al., 2014 | N/A |
| D. melanogaster: zip1/Cyo | Young et al., 1993 | N/A |
| D. melanogaster: w1118; ykiB5 {yki-YFP} VK37 | This study | N/A |
| D. melanogaster: nub-Gal4 | Bloomington Drosophila stock center | 38418 |
| D. melanogaster: hh-Gal4 | Tetsuya Tabata | N/A |
| D. melanogaster: ap-Gal4 | Bloomington Drosophila stock center | 3041 |
| D. melanogaster: w1118; {Wts-YFP} VK37 | This study | N/A |
| D. melanogaster: y M{vas-int.Dm}ZH-2A w; M{3xP3-RFP.attP}ZH-86Fb | Bloomington Drosophila stock center | 24749 |
| D. melanogaster: UAS-Strn-Mlck-HA | This study | N/A |
| D. melanogaster: Kr[If-1]/CyO; UAS-GrabFP-AInt/TM6 | Bloomington Drosophila stock center | 68178 |
| D. melanogaster: y w vas-Cas9 | Bloomington Drosophila stock center | 52669 |
| Oligonucleotides | ||
| Yki 5′ Homology arm
forward 5AGCACCTGCgatcTACTGAAAGTAGAGGTGATTAGTTGTTAACGAG3 |
Integrated DNA Technologies | N/A |
| Yki 5′ Homology arm
reverse 5TTTCTAGGGTTAACATATAAAAGGCACTTATTGTATACAAACGAAAAC3 |
Integrated DNA Technologies | N/A |
| Yki 3′ Homology arm forward including
AcidA
substitution 5TTCTAGGGTTAACGACGATGTCAGCCAGCAGCAATACAAACAGCCTGATCGccAAGGccATCGcCGcCGccGcCATGCTTTCGCCGATtAAGTCgAACAACCTG3 |
Integrated DNA Technologies | N/A |
| Yki 3′ Homology arm
reverse 5CGACGGCTCTTCCTTGTAGTATGACTGCAAGTAGAGAGAACAC3 |
Integrated DNA Technologies | N/A |
| Guide RNA
sequence 5ACCAGGTTGTTGGACTTGAT3 |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pU6-BbsI-chiRNA | Addgene | Cat#45946 |
| PAWH | Drosophila Genomics Resource Center | 1096 |
| pUAST-Myr | Neisch et al., 2013 | N/A |
| pUASTattB | Bischof et al., 2007 | N/A |
| Strn-Mlck EST | Drosophila Genomics Resource Center | RH61010 |
| pAWH-Strn-Mlck (Strn-Mlck-HA) | This study | N/A |
| pHD-ScarlessDsRed | Drosophila Genomics Resource Center | 1364 |
| Software and Algorithms | ||
| Image J | NIH | https://imagej.nih.gov/ij/ |
| R 3.5.0 | The R Foundation | https://www.r-project.org/ |
To generate mutant clones or MARCM clones, the following genotypes were used:
-
yki−/−; Yki MARCM clones:
hsFLP, tub-Gal4, UAS-GFP; 42DFRT ykiB5/42DFRT tub-Gal80; UAS-Flag-Yki/+
-
yki−/−; Myr-Yki MARCM clones:
hsFLP, tub-Gal4, UAS-GFP; 42DFRT ykiB5/42DFRT tub-Gal80; UAS-Myr-Flag-Yki/+
-
yki−/−; Myr-YkiΔNH MARCM clones:
hsFLP, tub-Gal4, UAS-GFP; 42DFRT ykiB5/42DFRT tub-Gal80; UAS-Myr-Flag-YkiΔNH/+
-
sd−/− MARCM clones:
hsFLP, tub-Gal80, 19AFRT/sd47M, 19AFRT; tub-Gal4, UAS-GFP/+
-
sd−/−; Myr-Yki MARCM clones:
hsFLP, tub-Gal80, 19AFRT/sd47M, 19AFRT; tub-Gal4, UAS-GFP/UAS-Myr-Flag-Yki
-
wts−/− clones with Yki-YFP:
hsFLP; ykiB5, Yki-YFP/ykiB5, Yki-YFP; 82BFRT wtsX1/82BFRT Ubi-RFP
-
wts−/−; yki RNAi clones:
hsFLP; UAS-yki RNAi/+; 82BFRT wtsX1/82BFRT Ubi-RFP
-
wts−/−; sd RNAi clones:
hsFLP; UAS-sd RNAi/+; 82BFRT wtsX1/82BFRT Ubi-RFP
-
wts−/− clones with sqh:GFP:
hsFLP; 82BFRT wtsX1, sqh:GFP/82BFRT Ubi-RFP
-
wts−/− clones with zip:GFP:
hsFLP; zip:GFP/+; 82BFRT wtsX1/82BFRT Ubi-RFP
The ykiAcidA allele was generated using the Scarless CRISPR/Cas9 genome engineering technique (http://flycrispr.molbio.wisc.edu/scarless). A 1.5kb left homology arm containing the 5′ UTR and first 3bp of exon1 of yki and a 1.0kb right homology arm containing 1kb of yki sequence from the 4th bp of exon1 were cloned by homologous recombination (Gibson et al., 2009) into pHD-ScarlessDsRed (Drosophila Genomics Resource Center) as the donor template. Two silent mutations were introduced at the 84th and 90th bp of exon1 (ATCAAGTCC was mutated to ATtAAGTCg) to destroy the guide recognition site in the donor template. This plasmid was co-injected with a pU6- Bbs1-chiRNA based plasmid expressing the guideRNA (5′-ACCAGGTTGTTGGACTTGAT-3′) into vas-Cas9 embryos. G0 flies were crossed to w1118 and individual DsRed positive F1s were selected to establish stocks. The DsRed marker cassette was then removed through a single cross to a source of the PBac transposase (w1118; CyO, P{Tub-PBac\T}2/wgSp-1; BL8285). The ykiAcidA allele was verified by sequencing.
All crosses were performed at 25°C unless otherwise noted. For clone induction, heat shocks were performed 60–84 hr after egg laying using the following program: 38°C for 1hr, 25°C for 1hr, 38°C for 1h, 25°C for 1hr in an EchoTherm IN35 incubator (Torrey Pines Scientific).
Expression Constructs and transgenic line generation
UAS-Myr-Flag-Yki was generated by cloning the Yki cDNA (based on transcript isoform F, NCBI RefSeqID NM_001043103) into a Gateway pTFW (Drosophila Genomics Resource Center) that had been modified to include a Drosophila Src kinase myristoylation consensus sequence at the N-terminus of any gene cloned in-frame (pUAST-Myr) (Neisch et al., 2013). UAS-Myr-Flag-YkiΔNH was generated by using site-directed mutagenesis primers designed to create an in-frame deletion of 47 amino acids from the coding sequence of Yki (deleting aa 35-82 based on transcript isoform F). UAS-Myr-Flag-Yki1-17, UAS-Myr-Flag-Yki18-34, UAS-Myr-Flag-Yki1-34, UAS-Myr-Flag-Yki35-395, UAS-Myr-Flag-Yki1-88, UAS-Myr-Flag-Yki89-395, UAS-Myr-Flag-Yki1-238, UAS-Myr-Flag-Yki239-395 were generated by cloning corresponding Yki fragments into pTFW-Myr construct.
Strn-Mlck-HA construct was generated by cloning Strn-Mlck isoform B CDS from BDGP ESTs collection clone RH61010 (Drosophila Genomics Resource Center) into a Gateway pAWH (Drosophila Genomics Resource Center) vector.
P-element transformation was used to generate transgenic animals carrying either UAS-Myr-Yki or UAS-Myr-YkiΔNH (Duke University Model Systems Genomics). Multiple lines were isolated and tested for each. Lines on the third chromosome were selected for further experimentation.
Site specific integration of UAS-Myr-Yki1-34-GFP, UAS-Myr-YkiAcidA, UAS-Myr-Yki were performed by cloning respective Yki constructs into a modified pUASTattB (Bischof et al., 2007) vector that adds a myristoylation consensus sequence and a 3X Flag at the N terminus, followed by injection into y M{vas-int.Dm}ZH-2A w; M{3xP3-RFP.attP}ZH-86Fb embryos.
Site specific integration of UAS-Strn-Mlck-HA was performed by cloning Strn-Mlck isoform B with C terminal HA tag into pUASTattB vector and injected into y M{vas-int.Dm}ZH-2A w; M{3xP3-RFP.attP}ZH-86Fb embryos.
Immunostaining and live imaging of wing imaginal discs
Wandering third instar wing discs were dissected in Schneider’s Drosophila Medium (Sigma) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific) and fixed in 2% paraformaldehyde/PBS solution for 20 min. The following primary antibodies were used: guinea pig anti-Sqh1P (1:300), mouse anti Sqh (1:1000) mouse anti-Flag (1:25000), rat anti-Ecad (1:2000), mouse anti-β-galactosidase (1:500), guinea pig anti-Sd (1:1000), rabbit anti-Yki (1:1000), guinea pig anti-Ex (1:5000). All secondary antibodies (Jackson ImmunoResearch Laboratories) were used at 1:500 or 1:1000. Immunostaining samples were imaged using either a Zeiss LSM 510, LSM800, or LSM 880 scanning confocal microscope and the images were manipulated with Image J.
Live imaging was done as previously described (Su et al., 2017). Briefly, wing imaginal discs from wandering third instar larvae were dissected and immersed in Schneider’s Drosophila Medium (Sigma) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific) inside a microwell dish (MatTeK Corporation, Part No.: P35G-1.5-14-C). Apical sides of wing imaginal discs were oriented toward the bottom cover glass of the microwell dish. Wing imaginal discs were covered by a ~8X8 mm No.1.5 coverslip fragment and supported with microspheres with a nominal diameter of 53μm (Bangs Laboratories). An inverted Zeiss LSM880 laser scanning confocal microscope equipped with a GaAsP spectral detector was used to detect YFP fluorescence excited using the 514 nm line from an argon ion laser.
Immunostaining and imaging of S2 cells
3.5×106 S2 cells were transfected with 250ng Ubi-Gal4 and 250ng of the indicated UAS constructs using DDAB (dimethyldioctadecyl-ammonium bromide, Sigma) (Han, 1996) at 250 μg/ml in six well plates. Cells were fixed 2–3 days after transfection in 2% paraformaldehyde/PBS solution for 10 min and stained and imaged as described above for imaginal tissues.
Co-Immunoprecipitation
3.5×106 S2 cells were transfected with 250ng Ubi-Gal4, 250ng of pAWH-Strn-Mlck (Strn-Mlck-HA under the actin5C promoter), and 250ng of pUASt constructs or 250ng Ubi-Gal4, 250ng of Strn-Mlck-HA, and 250ng of UAS-Flag-Yki constructs using DDAB (dimethyldioctadecyl-ammonium bromide, Sigma) (Han, 1996) at 250 μg/ml in six well plates. IPs were performed 2–3 days after transfection. Cells were harvested and lysed in buffer containing 25 mM Hepes, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.9 M glycerol, 0.1% Triton X-100, 0.05% deoxycholic acid, 0.5 mM DTT, and Complete protease inhibitor cocktail (Roche). Flag IPs were performed using anti-Flag M2 agarose beads (Sigma-Aldrich). IP reactions were performed at 4°C overnight. For immunoblotting, 8% SDS-PAGE gels were used and transferred onto nitrocellulose. Antibodies were used at the following concentrations: mouse anti-Flag M2 at 1:50,000 (Sigma-Aldrich), rabbit anti-HA at 1:5,000 (Rockland). Fluorescently labeled IRdye800 (Rockland) and Alexa Fluor 680 (Invitrogen) secondary antibodies were used at a concentration of 1:5,000. Images of the blots were obtained using Odyssey CLx with ImageStudio software (LI-COR Biosciences).
QUANTIFICATION AND STATISTICAL ANALYSIS
Image J was used to quantify mean fluorescence intensity in selected regions of the wing imaginal discs. To quantify adult wing sizes, wings were mounted in methyl salicylate and photographed with the same settings on a Zeiss Axioplan 2ie microscope using a Canon camera (EOS rebel T2i). Subsequent measurements of wing size and statistical analyses were processed using Image J and R, respectively.
All statistical comparisons between means were performed by One-way ANOVA with Tukey’s HSD test in R with the exception of Fig. S3 and Supplement Table 1, in which One-way ANOVA and Games-Howell test was used. Statistical significance of results between compared groups was indicated as follows: **** (p ≤ 0.0001), *** (p ≤ 0.001), ** (p≤0.01), n.s. (not significant, p>0.05).
Supplementary Material
Highlights.
Yorkie functions at the cell cortex to promote myosin activation.
Yorkie promotes myosin activation via the Stretchin-Myosin light chain kinase.
Mutation of Yorkie’s myosin activation domain results in growth defects.
Cortical Yorkie functions in a positive feedback loop that promotes growth.
Acknowledgments
We thank D. Pan, K. Irvine, R. Ward, the Developmental Studies Hybridoma Bank, TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947), the Bloomington stock center, and VDRC stock centers for fly stocks and other reagents. We thank Y. Wang for help testing RNAi lines. We thank C. Ferguson, S. Horne-Badovinac, M. Glotzer, and members of the Fehon lab for helpful discussions and comments on the manuscript. P.V., T.S., and J.X. were recipients of the Children’s Tumor Foundation Young Investigator Award (2012-01-033, 2013-01-020 and 2014-01-020, respectively). This work was supported by a grant from the National Institutes of Health to R.G.F (NS034783).
Footnotes
Author Contributions
J.X., P.V., and R.G.F. conceived the project. P.V. generated the UAS-Myr-Yki and UAS-Myr-YkiΔNH transgenes and performed initial characterization of UAS-Myr-Yki phenotypes. M.L. generated the Yki-YFP transgene. T.S. helped with experiments and performed live imaging of Yki-YFP in wts null clones. S.A.T. helped with mapping the region of Yki responsible for myosin activation and initiated the GrabFP experiments. J.X. generated all other reagents and performed all other experiments. J.X. and R.G.F. analyzed data and wrote the manuscript. R.G.F. supervised all aspects of the project.
Declaration of Interests
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aegerter-Wilmsen T, Aegerter CM, Hafen E, Basler K. Model for the regulation of size in the wing imaginal disc of Drosophila. Mech Dev. 2007;124:318–326. doi: 10.1016/j.mod.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Aegerter-Wilmsen T, Heimlicher MB, Smith AC, de Reuille PB, Smith RS, Aegerter CM, Basler K, Adler PN, Charlton J, Liu J, et al. Integrating force-sensing and signaling pathways in a model for the regulation of wing imaginal disc size. Development. 2012;139:3221–3231. doi: 10.1242/dev.082800. [DOI] [PubMed] [Google Scholar]
- Antunes M, Pereira T, Cordeiro JV, Almeida L, Jacinto A. Coordinated waves of actomyosin flow and apical cell constriction immediately after wounding. J Cell Biol. 2013;202:365–379. doi: 10.1083/jcb.201211039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1–13. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific C31 integrases. Proc Natl Acad Sci. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22:695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne MB, Edwards Ka Erickson HP, Kiehart DP. Drosophila stretchin-MLCK is a novel member of the Titin/Myosin light chain kinase family. J Mol Biol. 2000;300:759–777. doi: 10.1006/jmbi.2000.3802. [DOI] [PubMed] [Google Scholar]
- Choi S, Kim W, Chung J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC) J Biol Chem. 2011;286:2658–2664. doi: 10.1074/jbc.C110.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HLL, Augustine GJJ, Choi KWW. Drosophila Schip1 Links Expanded and Tao-1 to Regulate Hippo Signaling. Dev Cell. 2016;36:511–524. doi: 10.1016/j.devcel.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Feng Y, Jagannathan R, Seppa MJ, Skeath JB, Longmore GD. Ajuba LIM Proteins Are Negative Regulators of the Hippo Signaling Pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Savoian MS, Glover DM. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J Cell Biol. 2004;166:61–71. doi: 10.1083/jcb.200402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Wang W, Yu J, Zheng Y, Qing Y, Pan D. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. Elife. 2015;2015:1–17. doi: 10.7554/eLife.06567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J-M, Leung T, Manser E, Lim L. Cdc42 antagonizes inductive action of cAMP on cell shape, via effects of the myotonic dystrophy kinase-related Cdc42-binding kinase (MRCK) on myosin light chain phosphorylation. Eur J Cell Biol. 2002;81:231–242. doi: 10.1078/0171-9335-00238. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, de Simoes SM, Röper JCC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R, Venter JC, Hutchison Ca, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Guss Ka Benson M, Gubitosi N, Brondell K, Broadie K, Skeath JB. Expression and function of scalloped during Drosophila development. Dev Dyn. 2013;242:874–885. doi: 10.1002/dvdy.23942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Han K. An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res. 1996;24:4362–4363. doi: 10.1093/nar/24.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan IK. Organ Size Control: Lessons from Drosophila. Dev Cell. 2015;34:255–265. doi: 10.1016/j.devcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmansa S, Alborelli I, Bieli D, Caussinus E, Affolter M. A nanobody-based toolset to investigate the role of protein localization and dispersal in Drosophila. Elife. 2017;6:1–22. doi: 10.7554/eLife.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Harvey KF. Control of organ growth by patterning and hippo signaling in drosophila. Cold Spring Harb Perspect Biol. 2015;7:1–16. doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Karess R. Myosin light chain-activating phosphorylation sites are required for oogenesis in Drosophila. J Cell Biol. 1997;139:1805–1819. doi: 10.1083/jcb.139.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim T, Johnson RLL, Lim D-SS. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11:270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Koontz LM, Liu-chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, Pan D. The Hippo Effector Yorkie Controls Normal Tissue Growth by Antagonizing Scalloped-Mediated Default Repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yao E, Zhang K, Jiang X, Croll S, Thompson-Peer K, Chuang PT. YAP is essential for mechanical force production and epithelial cell proliferation during lung branching morphogenesis. Elife. 2017;6:1–25. doi: 10.7554/eLife.21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA. Stretching molecular springs: Elasticity of titin filaments in vertebrate striated muscle. Histol Histopathol. 2000;15:799–811. doi: 10.14670/HH-15.799. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The Tumor Suppressors Merlin and Expanded Function Cooperatively to Modulate Receptor Endocytosis and Signaling. Curr Biol. 2006;16:702–709. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- Mao Y, Tournier AL, Hoppe A, Kester L, Thompson BJ, Tapon N. Differential proliferation rates generate patterns of mechanical tension that orient tissue growth. EMBO J. 2013;32:2790–2803. doi: 10.1038/emboj.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Vinós J, Marco R, Cervera M. Autophosphorylating protein kinase activity in titin-like arthropod projectin. J Mol Biol. 1992;224:287–291. doi: 10.1016/0022-2836(92)90994-u. [DOI] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Guan K. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitonaka T, Muramatsu Y, Sugiyama S, Mizuno T, Nishida Y. Essential roles of myosin phosphatase in the maintenance of epithelial cell integrity of Drosophila imaginal disc cells. Dev Biol. 2007;309:78–86. doi: 10.1016/j.ydbio.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Neisch AL, Formstecher E, Fehon RG. Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with Moesin. Mol Biol Cell. 2013;24:1420–1433. doi: 10.1091/mbc.E12-11-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubueser D, Hipfner D. Overlapping roles of Drosophila Drak and Rok kinases in epithelial tissue morphogenesis. Mol Biol Cell. 2010;21:2869–2879. doi: 10.1091/mbc.E10-04-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J, Mahato S, Zelhof AC. The actomyosin machinery is required for Drosophila retinal lumen formation. PLoS Genet. 2014;10:e1004608. doi: 10.1371/journal.pgen.1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Reddy BVVG, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Heemskerk I, Ibar C, Shraiman BI, Irvine KD. Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc Natl Acad Sci U S A. 2016;113:E6974–E6983. doi: 10.1073/pnas.1615012113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Saide JD. Stretchin-klp, a novel Drosophila indirect flight muscle protein, has both myosin dependent and independent isoforms. J Muscle Res Cell Motil. 2005;26:213–224. doi: 10.1007/s10974-005-9012-y. [DOI] [PubMed] [Google Scholar]
- Porazinski S, Wang H, Asaoka Y, Behrndt M, Miyamoto T, Morita H, Hata S, Sasaki T, Krens SFG, Osada Y, et al. YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature. 2015:1–6. doi: 10.1038/nature14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Pan G, Reddy BVVG, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS Biol. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KDD. Cytoskeletal Tension Inhibits Hippo Signaling through an Ajuba-Warts Complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönbauer C, Distler J, Jährling N, Radolf M, Dodt HU, Frasch M, Schnorrer F. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature. 2011;479:406–409. doi: 10.1038/nature10559. [DOI] [PubMed] [Google Scholar]
- Stenesen D, Suh JM, Seo J, Yu K, Lee KS, Kim JS, Min KJ, Graff JM. Adenosine nucleotide biosynthesis and AMPK regulate adult life span and mediate the longevity benefit of caloric restriction in flies. Cell Metab. 2013;17:101–112. doi: 10.1016/j.cmet.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Ludwig MZ, Xu J, Fehon RG. Kibra and Merlin Activate the Hippo Pathway Spatially Distinct from and Independent of Expanded. Dev Cell. 2017;40:478–490 e3. doi: 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Irvine KD. Cellular Organization and Cytoskeletal Regulation of the Hippo Signaling Network. Trends Cell Biol. 2016;26:694–704. doi: 10.1016/j.tcb.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Reddy BVVG, Irvine KD. Localization of Hippo signalling complexes and Warts activation in vivo. Nat Commun. 2015;6:8402. doi: 10.1038/ncomms9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoumpekos G, Nemetschke L, Knust E. Drosophila Big bang regulates the apical cytocortex and wing growth through junctional tension. J Cell Biol. 2018;217:1033–1045. doi: 10.1083/jcb.201705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell. 2010a;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse Fa, Sakuma R, Pawson T, Hunziker W, McNeill H, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010b;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu YY, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart Ra, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Young PE, Richman AM, Ketchum AS, Kiehart DP. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993;7:29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- Yu FXX, Zhao B, Guan KLL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward RE. Distinct tissue distributions and subcellular localizations of differently phosphorylated forms of the myosin regulatory light chain in Drosophila. Gene Expr Patterns. 2011;11:93–104. doi: 10.1016/j.gep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.