Abstract
Recent studies have indicated that homeobox A3 (HOXA3) functions as a carcinogen in colon cancer and the methylation level of HOXA3 is significantly increased in lung adenocarcinoma (LUAD) tissues. However, at least to the best of our knowledge, few studies to date have been performed on HOXA3 in non-small cell lung cancer (NSCLC). Therefore, further studies on HOXA3 expression in NSCLC and the potential regulatory mechanisms are urgently required. In this study, HOXA3 expression in 55 tissues of cases of NSCLC and corresponding non-lung cancer tissues was detected by reverse transcription-quantitative PCR (RT-qPCR). In addition, the clinical significance of HOXA3 expression in NSCLC was evaluated using the Cancer Genome Atlas (TCGA) database. Bioinformatics analysis was then performed to elucidate the potential molecular mechanisms of action of HOXA3. Furthermore, the potential target microRNAs (miRNAs or miRs) of HOXA3 were predicted using miRWalk2.0. Based on Gene Expression Omnibus (GEO) and TGCA databases, standardized mean difference (SMD) and sROC methods were used for meta-analyses of the expression of potential target miRNAs of HOXA3 in NSCLC to evaluate their association with HOXA3. The results revealed that the HOXA3 expression levels in NSCLC, LUAD and lung squamous cell carcinoma (LUSC) were 0.1130±0.1398, 0.1295±0.16890 and 0.0906±0.0846, respectively. These values were all decreased compared with the normal tissues (0.1877±0.1975, 0.2337±0.2405 and 0.1249±0.0873, respectively, P<0.05). The TCGA database also revealed the low expression trend of HOXA3. The downregulation of HOXA3 may play an important role in the progression and the poor prognosis of LUAD. The TCGA database also suggested that HOXA3 in LUAD and LUSC tissues exhibited certain mutational levels. In addition, the methylation levels in the NSCLC, LUAD and LUSC tissues significantly increased [NSCLC: fold change (FC), 1.3226; P<0.001; LUAD: FC, 1.2712; P<0.001; and LUSC: FC, 1.3786; P<0.001]. According to the analyses using the Kyoto Encyclopedia of Genes and Genomes (KEGG), we found that the co-expression HOXA3 genes were mainly associated with the focal adhesion signalling pathway and the ECM-receptor interaction signalling pathway. Furthermore, the predicted miRNA, miR-372-3p, exhibited a high expression in both the NSCLC and LUAD tissues (P<0.05). On the whole, the findings of this study indicate that low HOXA3 expression may play a certain role in LUAD; however, its association with LUSC still requires further investigation. HOXA3 function may be achieved through different pathways or target miRNAs. However, the specific underlying mechanisms need to be confirmed through various functional studies.
Keywords: homeobox A3, lung adenocarcinoma, reverse transcription-quantitative PCR, co-expression, mechanism
Introduction
The most recent data indicate that lung cancer remains the type of cancer with the highest prevalence among men and women and is also one of the most common causes of cancer-related mortality (1). Approximately 1.82 million cases of lung cancer are newly diagnosed globally each year, and approximately 1.37 million individuals succumb to the disease (2–4). Based on the histological classification, lung cancers are divided into 2 types: Non-small cancer lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for 85% of all lung cancer cases (5). NSCLS includes a number of histological subtypes, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and large cell carcinoma (LCC). LUAD is one of the main subtypes of NSCLC (6). Despite in-depth studies and significant progress being made in lung cancer treatment in recent decades (7–10), comprehensive clinical evaluation and in-depth molecular mechanistic studies are still required. Therefore, further investigations of the pathological mechanisms of NSCLC are warranted. Thus, the screening of novel functional genes and targets in NSCLC may provide basic data and a preliminary theoretic basis for further studies on the pathogenesis of NSCLC.
The homeobox (HOX) gene family is composed of 39 genes. These genes play critical roles in normal embryonic development by encoding transcription factors. The HOX gene family is divided into 4 groups: HOXA, HOXB, HOXC and HOXD. These genes localize to different chromosomal regions. The human HOXA gene cluster localizes to chromosome 7. There are 12 genes, including HOXA1 to HOXA11 and HOXA13 (11–13). The HOXA gene cluster plays important roles in colorectal cancer, leukaemia, pancreatic cancer, oral squamous cell carcinoma and lung cancer (14–18). For HOXA3, only one study suggests that it nat be used as a biomarker of LUAD. This study was conducted by Daugaard et al (19). These researchers used a DNA methylation microarray to compare the genome-wide methylation patterns between tumour and tumour-adjacent lung tissues from 4 patients with LUAD and identified 74 differential methylation regions (DMRs). Through methylation sensitive-high resolution melting (MS-HRM) analyses, 18 DMRs were selected for validation in 52 LUAD tissues and 32 cancer-adjacent lung tissues. Finally, this study confirmed and validated 15 DMRs, including HOXA3, that could be used as biomarkers of LUAD (19). However, studies on the clinical significance and regulatory mechanisms of HOXA3 expression in NSCLC are still lacking. Therefore, further studies are urgently required to examine HOXA3 expression in NSCLC and to investigate the potential regulatory mechanisms in order to provide new insight into the pathogenic mechanisms of NSCLC.
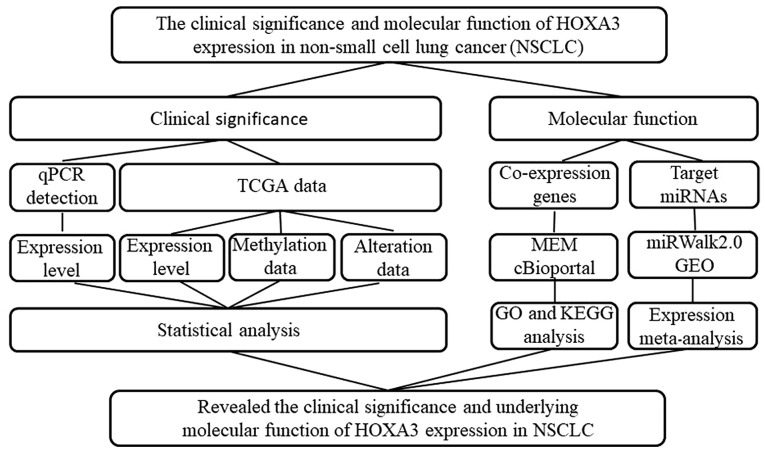
In this study, we used reverse transcription-quantitative PCR (RT-qPCR) to detect HOXA3 expression in NSCLC and normal lung tissues. HOXA3 expression in NSCLC was validated using the Gene Expression Profiling Interactive Analysis (GEPIA) database. In addition, informatics analysis methods, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI), were performed to investigate the potential molecular mechanisms of action of HOXA3. Furthermore, the potential target microRNAs (miRNAs or miRs) of HOXA3 were predicted using miRWalk2.0. The expression levels of miRNAs were validated using the Gene Expression Omnibus (GEO) and the Cancer Genome Atlas (TGCA) databases. The study design is presented in Fig. 1.
Figure 1.
Flow chart of the study design of this study.
Materials and methods
Clinical samples
A total of 55 tissues of cases of clinical NSCLC and cancer-adjacent lung tissues were collected (from January, 2012 to February, 2014) at the Department of Pathology of The First Affiliated Hospital of Guangxi Medical University (Nanning, China). Among these 55 patients, 32 cases were diagnosed with LUAD, and 23 cases were diagnosed with LUSC. There were 41 men and 14 women. Patients were 23–90 years of age with a mean age of 56.9 years. Among these 55 cases, tumours in 39 cases were larger than 3 cm. Table I summarizes the detailed clinicopathological data of all the patients. Samples were fixed in formalin and embedded in paraffin. This research programme was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University. All participants signed informed consent forms. All samples were diagnosed by 2 pathologists (G.C. and D.-M.W.) independently using the double-blind method.
Table I.
The association of between HOXA3 mRNA expression in NSCLC tissues and patient clinicopathological characteristics based on RT-qPCR detection.
| Clinicopathological characteristics | Case no. | HOXA3 expression
|
t-test of data
|
|||
|---|---|---|---|---|---|---|
| Mean | SD | FC | t-test | P-value | ||
| Group | ||||||
| Control | 55 | 0.19 | 0.20 | 1.00 | 2.86 | 0.006 |
| Cancer | 55 | 0.11 | 0.14 | 0.60 | ||
| Pathological type | ||||||
| Adenocarcinoma | 32 | 0.13 | 0.17 | 1.00 | 0.99 | 0.326 |
| Squamous cell carcinoma | 23 | 0.09 | 0.08 | 0.70 | ||
| LUAD | ||||||
| Control | 32 | 0.23 | 0.24 | 1.00 | −1.94 | 0.023 |
| Cancer | 32 | 0.13 | 0.17 | 0.55 | ||
| LUAD | ||||||
| Non-smoker | 18 | 0.18 | 0.20 | 1.00 | 2.42 | 0.025 |
| Smoker | 14 | 0.06 | 0.04 | 0.32 | ||
| LUSC | ||||||
| Control | 23 | 0.12 | 0.09 | 1.00 | −1.32 | 0.034 |
| Cancer | 23 | 0.09 | 0.08 | 0.73 | ||
| Sex | ||||||
| Male | 41 | 0.12 | 0.15 | 1.00 | 0.35 | 0.732 |
| Female | 14 | 0.10 | 0.12 | 0.87 | ||
| Age (years) | ||||||
| <60 | 35 | 0.12 | 0.15 | 1.00 | 0.25 | 0.801 |
| ≥60 | 20 | 0.11 | 0.12 | 0.91 | ||
| Smoking | ||||||
| No | 30 | 0.14 | 0.17 | 1.00 | 1.91 | 0.064 |
| Yes | 25 | 0.08 | 0.06 | 0.54 | ||
| Size | ||||||
| ≤3 cm | 16 | 0.12 | 0.12 | 1.00 | 0.09 | 0.931 |
| >3 cm | 39 | 0.11 | 0.15 | 0.97 | ||
| LNM | ||||||
| No | 29 | 0.08 | 0.10 | 1.00 | −1.66 | 0.103 |
| Yes | 26 | 0.14 | 0.17 | 1.78 | ||
| TNM | ||||||
| I–II | 29 | 0.09 | 0.11 | 1.00 | −1.26 | 0.212 |
| III–IV | 26 | 0.14 | 0.17 | 1.54 | ||
| Vascular invasion | ||||||
| No | 50 | 0.11 | 0.15 | 1.00 | 0.23 | 0.817 |
| Yes | 5 | 0.10 | 0.03 | 0.85 | ||
| Clinical grading | ||||||
| I | 5 | 0.09 | 0.08 | 0.664 | ||
| II | 40 | 0.12 | 0.16 | |||
| III | 10 | 0.08 | 0.07 | |||
HOXA3, homeobox A3, an mRNA of homeobox genes of clusters A; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SD, standard deviation; FC, fold change; LNM, lymph node metastasis. An independent sample t-test was adopted to test the data and single factor ANOVA was used to test the clinical grading data. The difference between the cancer and normal adjacent group was analyzed by a paired-samples t-test.
Detection of HOXA3 expression by RT-qPCR
HOXA3 expression in NSCLC samples was detected by RT-qPCR which was performed as previously described (20–22). Total RNA was extracted from the tumour and cancer-adjacent normal tissues using the RNeasy reagent (Qiagen, Shanghai, China). The RNA concentration was measured using NanoDrop2000 (Thermo Fisher Scientific, Wilmington, DE, USA). Total RNA was reverse transcribed (10 µl reaction system) using a reverse transcription kit (ABI, Life Technologies, Carlsbad, CA, USA) to obtain cDNA for RT-qPCR. SYBR-Green (Shanghai GeneCore BioTechnologies Co., Ltd., Shanghai, China) was used to perform RT-qPCR and the distinctiveness of the PCR product was differentiated based on melting curve (23–26). RT-qPCR was carried out using the following conditions: preheating for 10 min at temprature 95°C; and then repeating 40 cycles in temperature 95°C for 15 sec and 60 sec at 60°C. The primer sequences of HOXA3 were as follows: Upstream, TCATTTAAGAGCGCCTGGACA and downstream primer, GAGCTGTCGTAGTAGGTCGC. Using GAPDH as an internal reference gene and the primer sequence were as follows: Upstream, 5′-TGGTCCCTGCTCCTCTAAC-3′, downstream primer, 5′-GGCTCAATGGCGTACTCTC-3′. The relative expression level of HOXA3 in this study was calculated using the 2−ΔΔCq (27) formula (28, 29).
Retrieval of TCGA data
GEPIA (http://gepia.cancer-pku.cn/) is a visualization website based on TCGA data and contains differential gene expression between cancer and non-cancer, and analysis of the association between gene expression and overall survival (OS) or disease-free survival (DFS) and clinical stages (Stage). Survival analyses were performed using the log-rank test (also known as Mantel-Cox test) for hypothesis tests and calculation of the hazard ratio (HR) and 95% confidence interval (95% CI). Keywords, such as 'HOXA3', 'LUAD' and 'LUSC', were searched in GEPIA to retrieve data of the differential expression of HOXA3 in NSCLS, LUAD, LUSC and normal lung tissues, as well as information regarding the association between HOXA3 and clinical stages and prognostic survival.
The keywords 'lung' and 'HOXA3' were searched in the cBioPortal (http://www.cbioportal.org/) website to retrieve genetic alteration data of HOXA3 in LUAD and LUSC. Data were obtained from microarray detection and RNA sequencing. In addition, DNA methylation data in LUAD and LUSC were downloaded from the Cancer Genome Atlas (TCGA: https://cancergenome.nih.gov/abouttcga/overview) database. HOXA3 methylation data in LUAD and LUSC were screened.
HOXA3 co-expression genes
To investigate the intrinsic mechanisms of HOXA3 expression in NSCLC tissues, we acquired HOXA3-associated co-expression genes from the MEM (http://biit.cs.ut.ee/mem) and cBioPortal databases. In MEM, when the score/P-value was smaller, the association between that gene and HOXA3 was more significant. The results of 2 gene probes were extracted from MEM based on a P-value <0.0001, and the results were used for intersection. On the other hand, Pearson's correlation analysis was performed in cBioPortal to calculate the correlation between co-expression genes and HOXA3. When the absolute value of the Pearson's correlation coefficient was larger, the correlation was stronger. This study selected genes with an absolute value of the Pearsons' correlation coefficient >0.3 in NSCLC, LUAD and LUSC. Finally, the intersection results between MEM and cBioPortal were obtained and used for further analyses. The overlapping genes were confirmed using a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Bioinformatics analyses
In this study, we used bioinformatics analyses to preliminarily investigate the molecular mechanisms of action of HOXA3 in NSCLC. The final overlapping genes in MEM and cBioPortal were used for GO and KEGG analyses in David v6.7 (http://david.abcc.ncifcrf.gov/) to elucidate the enrichment of genes in biological processes and signalling pathways. PPI analyses were performed in STRING v10.5 (https://string-db.org/cgi/input.pl?input_page_show_search=on&UserId=HJJhhNv8hYyf&sessionId=Ce1Dx9pYDluc) to validate the protein interaction association to hypothesize the pathways through which HOXA3 carries out its functions in NSCLC. The construction of the functional network was performed using Cytoscape 3.5.0 software and ClueGO plugin.
Prediction and preliminary validation of target miRNAs of HOXA3
The prediction of target miRNAs of HOXA3 was performed in miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). miRWalk2.0 contains 12 on-line target gene prediction tools: Pictar2, miRWalk, miRMap, DIANA microT v4, PITA, miRanda, RNA22, mirBridge, miRDB, RNAhybrid, miRNAMap and TargetScan. miRNAs that were present in >8 software results were used as candidate miRNAs for further analyses.
The microarray results of differentially expressed miRNAs in lung cancer were searched in the GEO database and downloaded. The following search formula was employed: (lung OR pulmonary OR respiratory OR bronchi OR bronchioles OR alveoli OR pneumocytes OR 'air way') AND (cancer OR carcinoma OR tumor OR neoplas* OR malignan* OR adenocarcinoma) AND (microRNA OR miRNA OR 'micro RNA' OR 'small temporal RNA' OR 'noncoding RNA' OR ncRNA OR 'small RNA'). We included microarray data containing expression data of miRNAs in lung cancer tissues and normal lung tissues and a sample number >3 into this study. Microarray data that did not conform to this condition were excluded. In addition, miRNA expression data for the LUAD, LUSC and normal lung tissues were extracted from the TCGA database. The mean value ± standard deviation (means ± SD) of the expression of potential target miRNAs in tumour and non-tumour tissues in each microarray was calculated. The random effects model was used to calculate the standardized mean difference (SMD) in STATA 2.0 software to evaluate the expression trend of miRNAs in NSCLC and plot the forest plot. In addition, analyses using the sROC method were performed to further validate the expression trend of miRNAs in NSCLC.
Statistical analyses
Statistical analyses of the experimental data were performed using SPSS 22.0 software (IBM, New York, NY, USA). The HOXA3 expression levels detected by RT-qPCR were expressed as the means ± SD and the Mann-Whitney test was used to determine significance. The normal distribution was determined using the single-sample K-S test. The comparison of the association between the HOXA3 expression level and the patient clinicopathological characteristics, and the comparison of mean values of 2 factors were performed using the independent sample t-test when variances were homogeneous. When variances were not homogeneous, rank sum statistical analysis was performed. The comparison of the mean values of multiple factors was performed using one-way analysis of variance (ANOVA) followed by the Bonferroni post hoc test, with a default P-value of 0.05. The differences in HOXA3 expression levels between the NSCLC and normal lung tissues were further validated using the calculation of the area under the ROC curve. The Kaplan-Meier curve method was performed to analyse the association between the HOXA3 expression level and NSCLC patient survival. Pearson's correlation analysis was used to calculate the correlation between two genes. GraphPad Prism was used to generate the scatter plot. Meta-analysis of target miRNAs in NSCLC and normal lung tissues was performed using the STATA 12.0 software. P<0.05 in all analytic results indicated that the differences exhibited statistical significance.
Results
HOXA3 expression in NSCLC
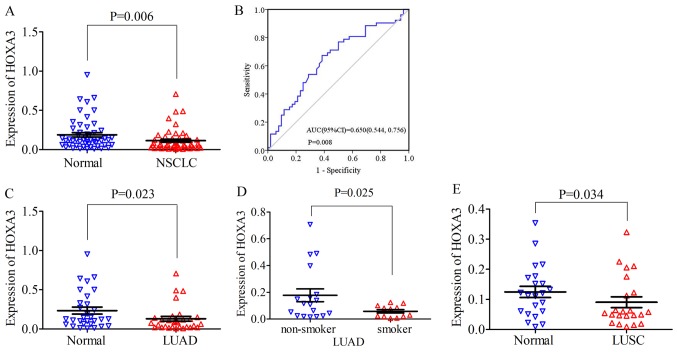
For NSCLC, the HOXA3 expression level in the cancer tissues was 0.11±0.14 and the fold change (FC), 0.60 (FC is the fold change of HOXA3 expression in tumour and non-tumour adjacent tissue). Compared with the normal lung tissues (0.19±0.20, P=0.006), HOXA3 expression was significantly decreased (P=0.006; Fig. 2A). The median of the HOXA3 level was 0.06 and the FC, 0.5 (control, 0.12; P=0.008). The area under the ROC curve (AUC) of low HOXA3 expression in NSCLC was 0.65 (95% CI: 0.54, 0.76; P=0.008, Fig. 2B). Therefore, a low HOXA3 expression may have some value in the occurrence of NSCLC. A low HOXA3 expression was not significantly associated with the clinicopathological characteristics (Table I). In addition, in LUAD, the mean expression level of HOXA3 was 0.13±0.17 and the FC, 0.55. HOXA3 expression was significantly decreased in LUAD compared with the cancer-adjacent normal tissues (0.24±0.24, P=0.023) (Fig. 2C and Table I). The median of the HOXA3 level was 0.08 and the FC, 0.62 (control, 0.13; P=0.034). The AUC of downregulated HOXA3 expression in LUAD was 0.66 (95% CI: 0.52, 0.80; P=0.035) (data not shown). Further analyses revealed that the HOXA3 level in smokers (0.06±0.04) was reduced compared with that in non-smokers (0.186±0.20, P=0.025) (Fig. 2D and Table I). The AUC of downregulated HOXA3 in the smoking group was 0.70 (95% CI: 0.51, 0.89; P=0.069) (data not shown). The combination of the above-mentioned data analyses demonstrated that the downregulation of HOXA3 expression in LUAD was significant. In addition, HOXA3 expression in LUAD may be associated with smoking, but did not exhibit a significant association with other clinicopathological characteristics. The analyses of the LUSC group alone revealed that HOXA3 expression differed significantly between the cancer tissues and non-cancer tissues (cancer, 0.09±0.08 vs. control, 0.12±0.09, P=0.034, FC, 0.72) (Fig. 2E and Table I). The median of the HOXA3 level was 0.06 and the FC, 0.5 (control, 0.12; P=0.113). The AUC of the low HOXA3 level in the LUSC group was 0.64 (95% CI: 0.47, 0.81; P=0.113) (data not shown). The low expression of HOX3A in LUSC was not significantly associated with disease progression.
Figure 2.
(A) Scatter diagram of HOXA3 expression between NSCLC tissue and normal lung tissue detected by RT-qPCR. (B) ROC curve of HOXA3 expression in NSCLC tissue. (C and D) Scatter plots of HOXA3 level in LUAD detected by RT-qPCR. (C) Differences between cancer and non-cancer. (D) Differences between smoker and non-smoker in LUAD. (E) HOXA3 expression between LUSC tissue and normal lung tissue detected by RT-qPCR. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
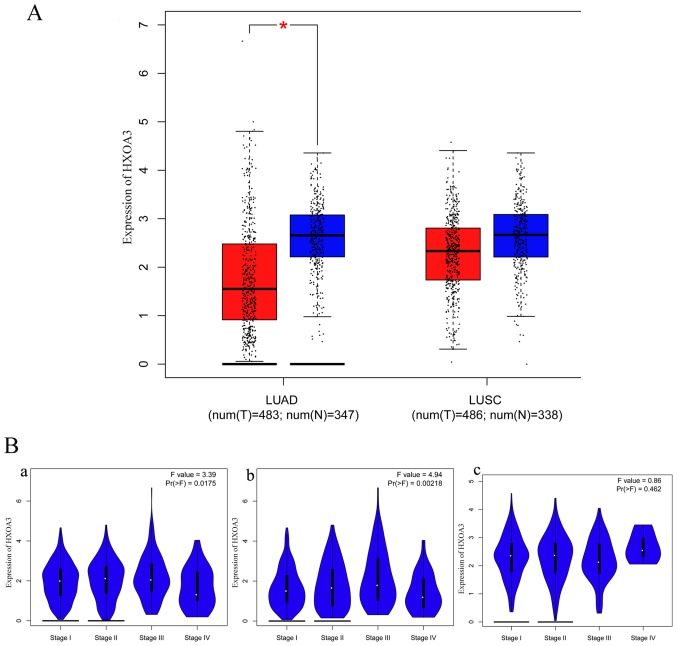
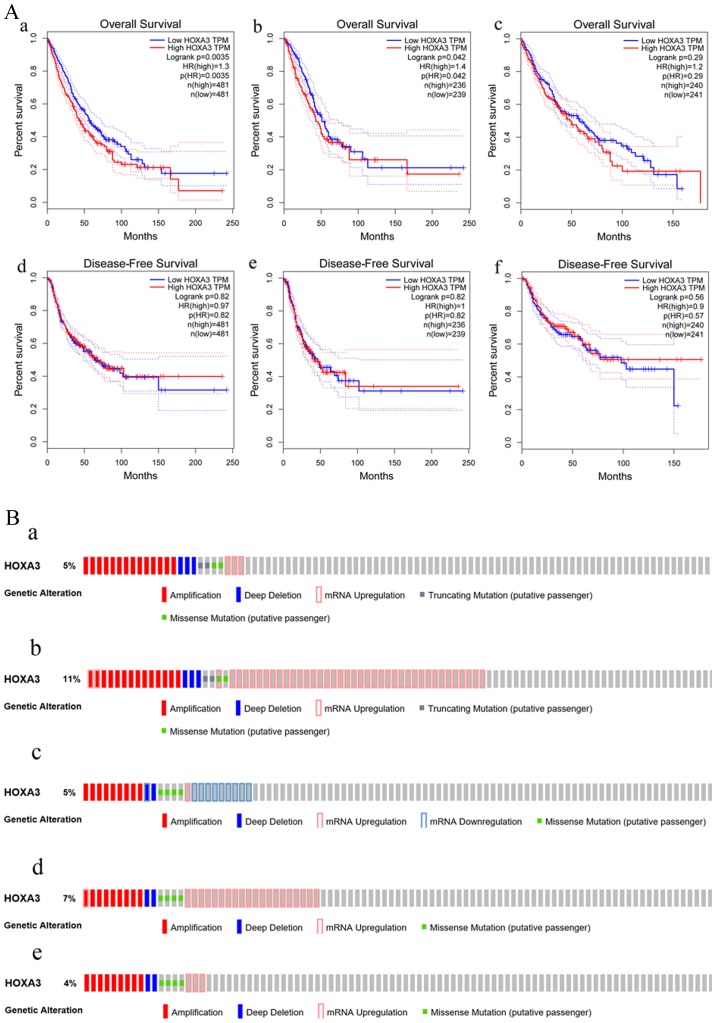
In addition, the GEPIA data revealed that HOXA3 expression in LUAD (483 cases) was also significantly decreased compared with the normal lung tissues (347 cases) and exhibited a low expression trend in LUSC (LUSC, 486 cases; normal, 338 cases) (Fig. 3A). These results were consistent with the results of RT-qPCR. However, the association between the HOXA3 expression level and the pathological stage did not exhibit an obvious pattern (Fig. 3B). We also acquired information regarding the association between the HOXA3 expression level and the survival of patients with NSCLC, LUAD and LUSC. Patients with NSCLC with a low HOXA3 expression exhibited a better OS [HR (high), 1.3; P=0.0035] (Fig. 4A, panel a). Thus, a low HOXA3 expression may be a protective factor of OS in patients with NSCLC. However, further analysis demonstrated that HOXA3 was not significantly associated with the DFS of patients with NSCLC (Fig. 4A, panel d). The analyses of OS in the LUAD group alone demonstrated that patients with a high HOXA3 expression exhibited an HR of 1.4 (P=0.042) (Fig. 4A, panel b), suggesting that a high HOXA3 level may be a risk factor for the prognosis of LUAD. However, the HOXA3 expression levels did not exhibit obvious predictive values as regards the DFS of patients with LUAD (Fig. 4A, panel e). In addition, the association between HOXA3 and the prognosis of patients LUSC was not statistically significant (Fig. 4A, panels c and f), and this may require further investigation.
Figure 3.
(A) Boxplot of HOXA3 expression in LUAD tissues vs. non-cancerous tissues and LUSC tissues vs. non-cancerous tissues; *P<0.05. (B) Pathological stage plot of HOXA3 levels in NSCLC. Panel a, NSCLC; panel b, LUAD; panel c, LUSC. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Figure 4.
(A) Survival curve of HOXA3 expression in NSCLC. Overall survival (OS: panel a, NSCLC; panel b, LUAD; panel c, LUSC) and disease-free survival (DFS: panel d, NSCLC; panel e, LUAD; panel f, LUSC). (B) The atlas of genetic alteration of HOXA3 mRNA in NSCLC. Panels a and b show HOXA3 mRNA expression z-scores by microarray and RNA Seq V2 RSEM in LUAD. Altered were 24 (5%) and 59 (11%) of 520 sequenced cases/patients (520 total) respectively; panels c, d and e show HOXA3 mRNA expression z-scores by U133 microarray only, microarray and RNA Seq V2 RSEM in LUSC. Altered were 18 (4%), 25 (5%) and 35 (7%) of 504 sequenced cases/patients (504 total). HOXA3, homeobox A3; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Furthermore, based on the microarray and RNA sequencing technologies, the sequencing results of 520 patients with LUAD revealed that the genetic alteration rates of HOXA3 were 5% (24/520) and 11% (59/520). In addition, the sequencing results of 504 patients LUSC suggested that the genetic alteration rates of HOXA3 were 5% (25/504), 7% (35/504) and 4% (18/504). Although the genetic alternation rates obtained using different methods differed, the final results all confirmed that HOXA3 harboured genetic alterations in the LUAD and LUSC populations (Fig. 4B).
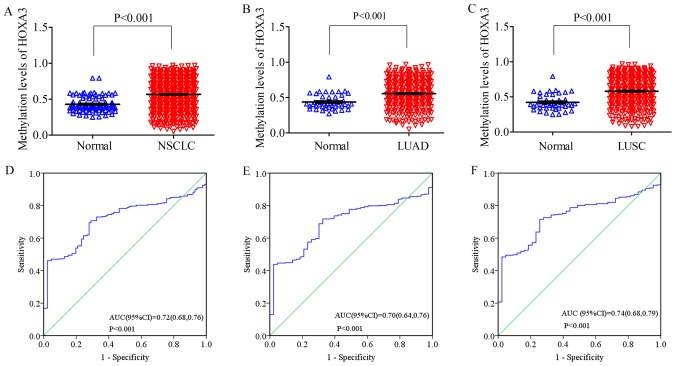
On the other hand, the methylation level of HOXA3 in the NSCLC tissues was 0.57±0.21 (FC, 1.32; P<0.001; Fig. 5A), with an AUC value of 0.72 (95% CI: 0.68, 0.76; P<0.001, Fig. 5D). The methylation level in the LUAD tissues was 0.56±0.20 (FC, 1.27; P<0.001; Fig. 5B), and an AUC value of 0.70 (95% CI: 0.64, 0.76; P<0.001, Fig. 5E). The methylation level in the LUSC tissues was 0.58±0.21 (FC, 1.38; P<0.001; Fig. 5C), and the AUC value was 0.74 (95% CI: 0.68, 0.79; P<0.001, Fig. 5F) (Table II). Compared with the normal tissues (NSCLC, 0.43±0.11; LUAD, 0.44±0.11; LUSC, 0.42±0.11), these levels were all significantly upregulated.
Figure 5.
Scatter plot of HOXA3 methylation levels between lung cancer tissues and normal lung tissues (A) NSCLC; (B) LUAD; (C) LUSC. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; ROC curve of HOXA3 methylation levels between lung cancer tissues and normal lung tissues (D) NSCLC; (E) LUAD; (F) LUSC.
Table II.
The association of between the HOXA3 mRNA methylation level in NSCLC tissues and normal adjacent tissues base on the TCGA data.
| Histological type | Case no. | HOXA3 methylation level
|
t-test of data
|
|||
|---|---|---|---|---|---|---|
| Mean | SD | FC | t-test | P-value | ||
| NSCLC | ||||||
| Normal adjacent | 86 | 0.43 | 0.11 | 1.00 | 9.89 | <0.001 |
| Cancer | 744 | 0.57 | 0.21 | 1.32 | ||
| LUAD | ||||||
| Normal adjacent | 43 | 0.44 | 0.11 | 1.00 | 6.19 | <0.001 |
| Cancer | 372 | 0.56 | 0.20 | 1.27 | ||
| LUSC | ||||||
| Normal adjacent | 43 | 0.42 | 0.11 | 1.00 | 7.72 | <0.001 |
| Cancer | 372 | 0.58 | 0.21 | 1.38 | ||
HOXA3, homeobox A3, an mRNA of homeobox genes of clusters A; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
HOXA3 co-expression genes
Based on MEM, 11,488 and 4,730 HOXA3 co-expression mRNA genes detected by 2 independent gene probes were obtained. The intersection between the results of 2 gene probes was collected, and a total of 1,709 more convincing co-expression mRNA genes were extracted. The data from cBioPortal revealed 3,600 and 2,676 co-expression mRNA genes in the NSCLC group and the LUAD group, respectively. Finally, the search results of the NSCLC group and the LUAD group in MEM and cBioPortal were processed using the crossing method (data not shown). In total, 293 and 213 genes were obtained, respectively, for further analysis.
Bioinformatics analyses
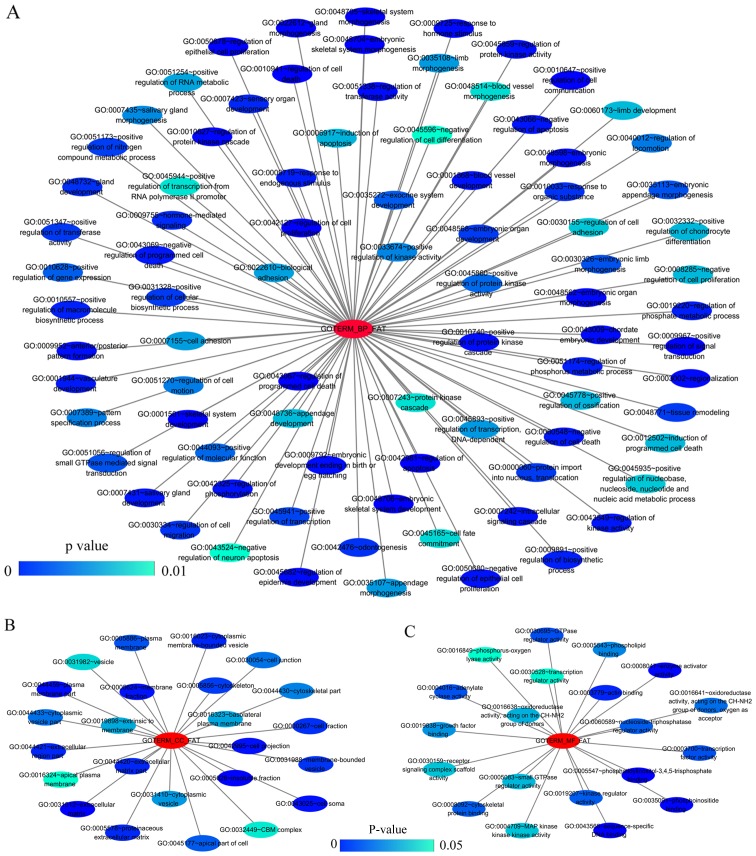
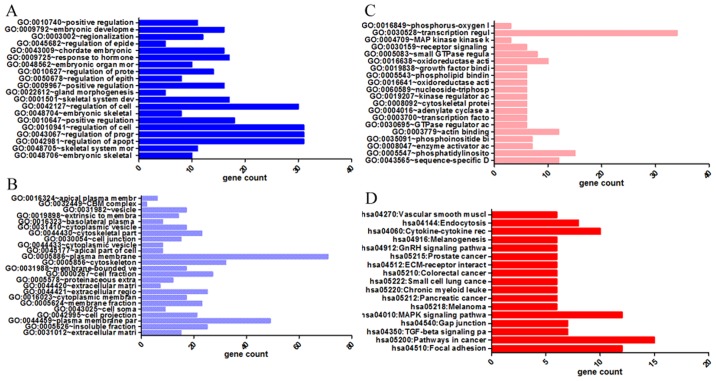
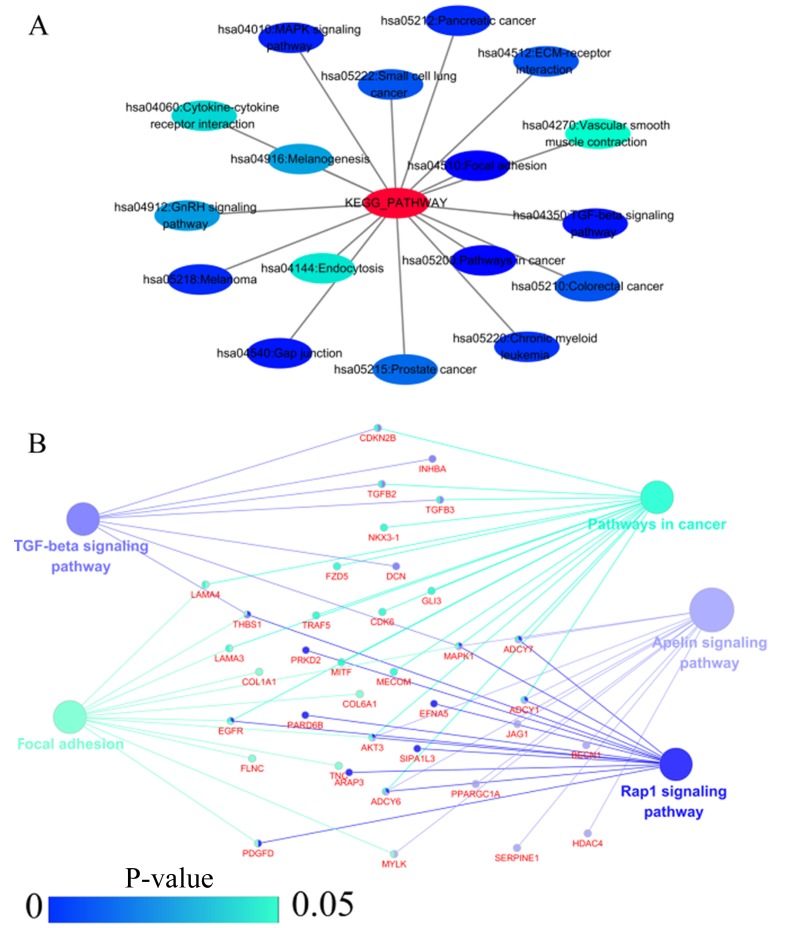
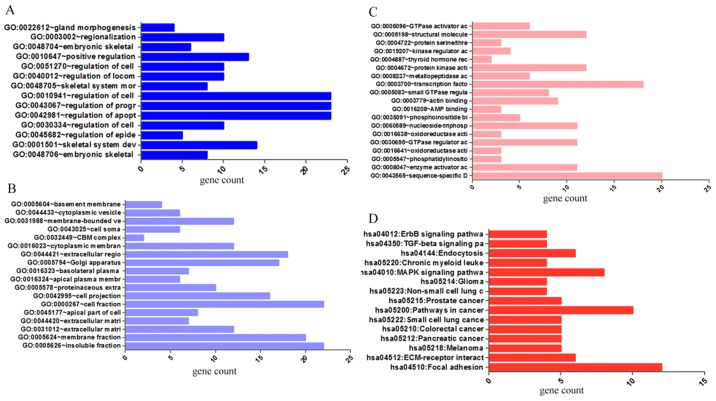
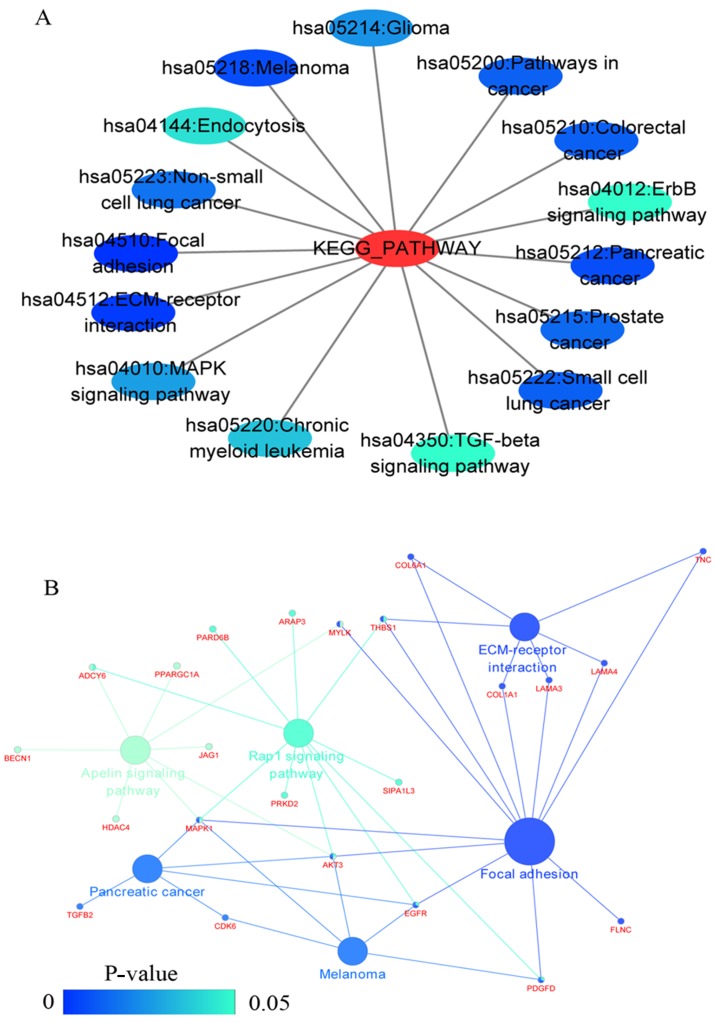
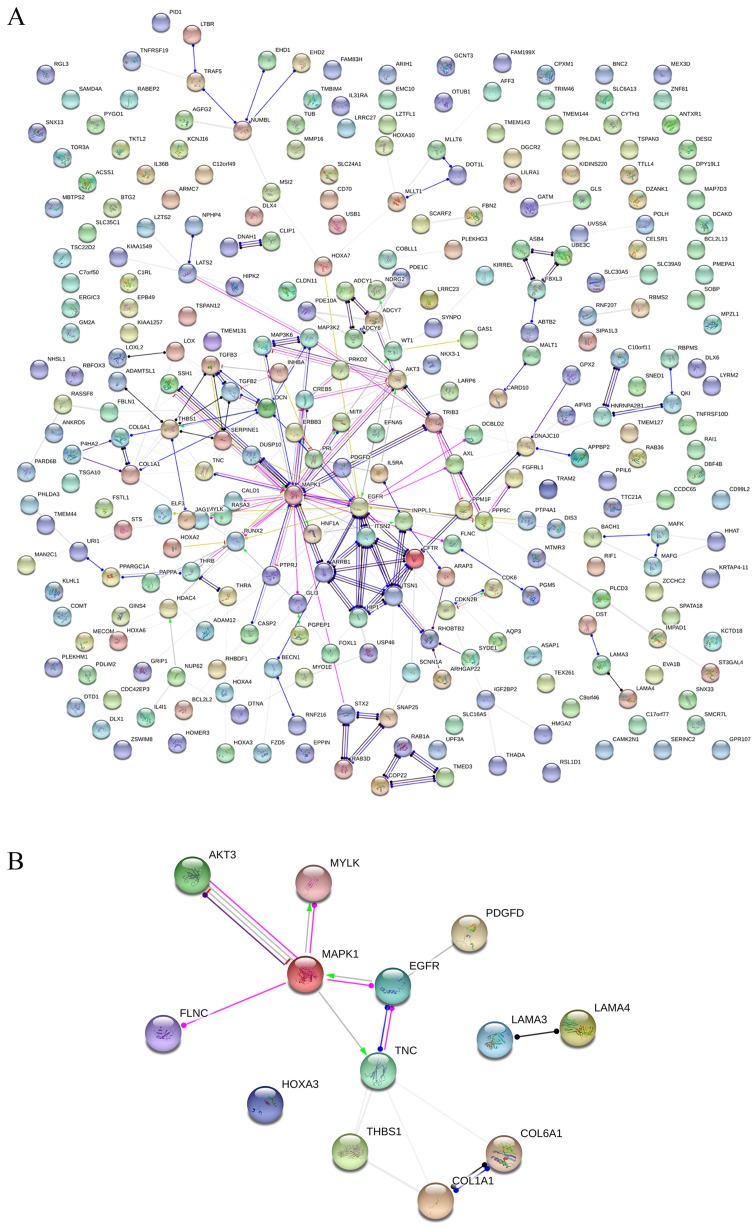
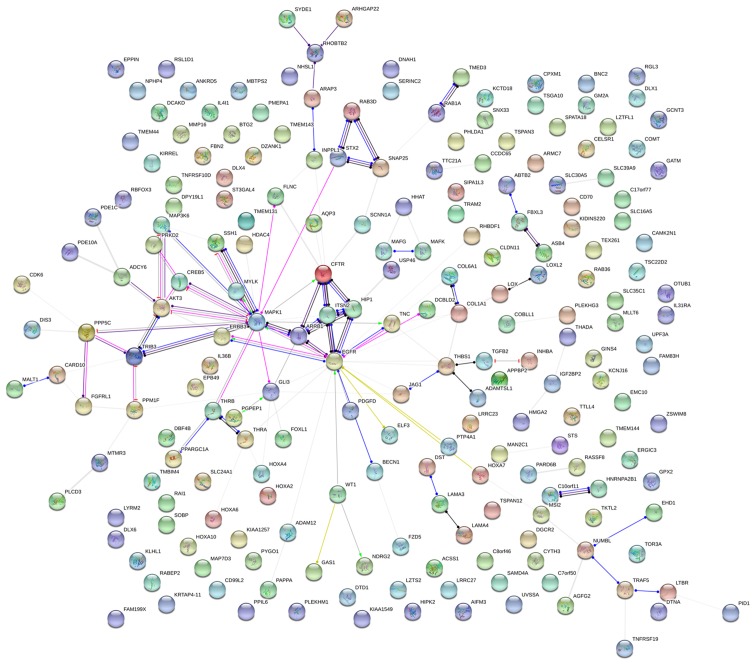
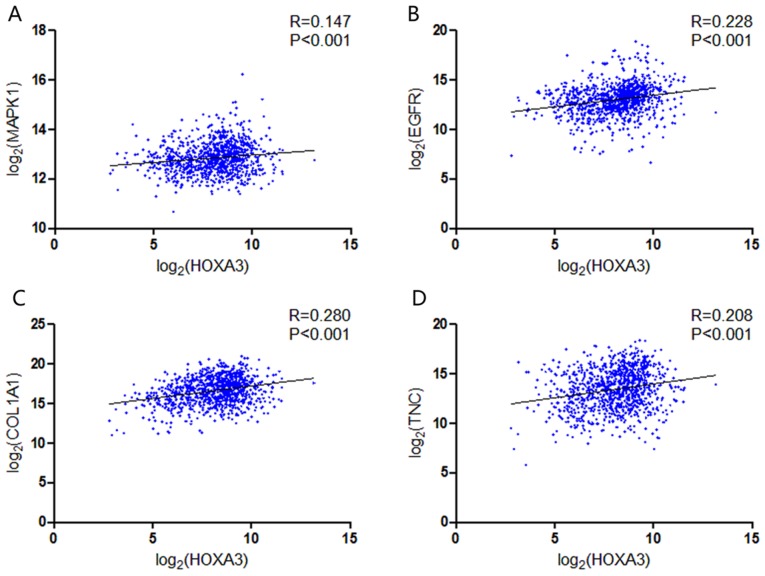
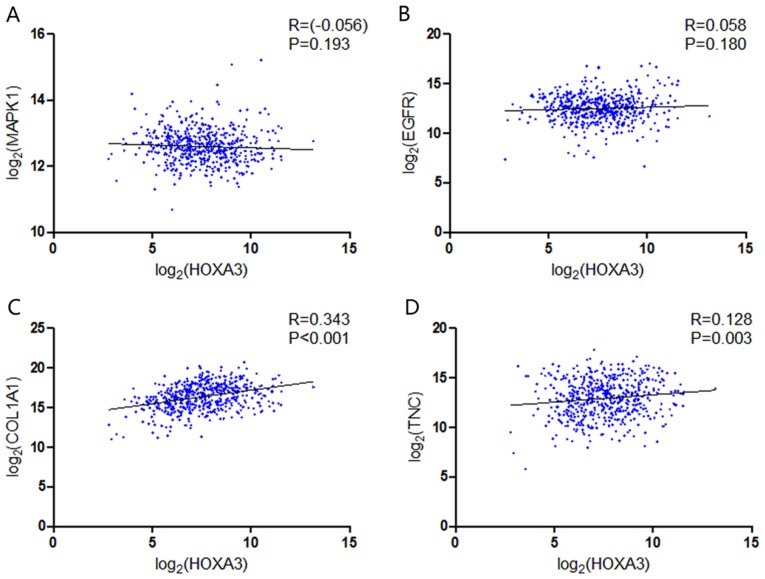
GO and KEGG analyses were performed on 293 and 213 genes in DAVID, and PPI network analyses were performed in STRING. The results from GO analysis revealed that genes in the NSCLC group mainly participated in biological processes, such as the regulation of cell migration; the regulation of apoptosis; and the regulation of programmed cell death; cellular components, such as insoluble fraction, membrane fraction, and extracellular matrix; and molecular functions, such as sequence-specific DNA binding, enzyme activator activity, and phosphatidylinositol-3,4,5-tri-sphosphate binding (Figs. 6 and 8A–C, and Table III). The results of KEGG pathway analyses suggested that HOXA3 may be involved in the functional regulation of 22 signalling pathways in the NSCLC group. The most important pathways were focal adhesion, pathways in cancer, and the TGF-β signalling pathway (Figs. 7 and 8D, and Table IV). In addition, genes in the LUAD group mainly participated in biological processes, such as the regulation of cell migration, regulation of apoptosis and the regulation of programmed cell death. Significant enrichment also occurred in cellular components and molecular functions; for example, insoluble fraction and membrane fraction in cellular components and sequence-specific DNA binding and enzyme activator activity in molecular functions (Figs. 9 and 11A–C, and Table V). The analytical results in the LUAD group suggested that HOXA3 may be involved in the biological regulation of 15 signalling pathways. Of these pathways, the most significant pathways were focal adhesion and ECM-receptor interaction (Figs. 10 and 11D, and Table VI). In PPI analyses, 278 interaction nodes were obtained in the NSCLC group (Fig. 12A). The genes with the top 10 overall scores are presented in Table VII. The results of interaction network analyses demonstrated that MAPK1, EGFR, DCN and CFTR represented key genes in these networks. In addition, genes enriched in 'Focal adhesion' were used for PPI analyses, and a total of 13 interaction nodes were obtained (Fig. 12B). The genes with the top 10 overall score are presented in Table VIII. The results suggested that MAPK1, EGFR, TNC and COL1A1 were key genes in this pathway. PPI analyses were also performed on genes in the LUAD group, and the results generated 129 protein interaction nodes (Fig. 13). The nodes with the top 10 overall scores are presented in Table IX. The protein interaction network suggested that MAPK1, EGFR, CFTR and GLI3 were key nodes in these interaction networks. Genes in the LUAD group that were enriched in 'Focal adhesion' were used for PPI analyses, and 14 protein interaction network nodes were also obtained. The result was the same as that for the NSCLC group. MAPK1, EGFR, TNC and COLIA1 were key genes in these pathways (refer to Fig. 12B and Table X). Analytic results revealed that genes in the NSCLC and LUAD groups all participated in the regulation of the 'Focal adhesion' pathway. MAPK1 and EGFR played key roles in NSCLC and LUAD, whereas MAPK1, EGFR, CFTR and GLI3 represented key genes in the 'Focal adhesion' pathway. Therefore, these 4 genes were analysed further. Based on TCGA data, the results of Pearson's correlation analysis revealed that the 4 genes, EGFR, MAPK1, COL1A1 and TNC, all exhibited a potential positive correlation with HOXA3 expression in NSCLC (Fig. 14). In the LUAD group, only COL1A1 and TNC exhibited a possible positive correlation with HOXA3 (Fig. 15C and D).
Figure 6.
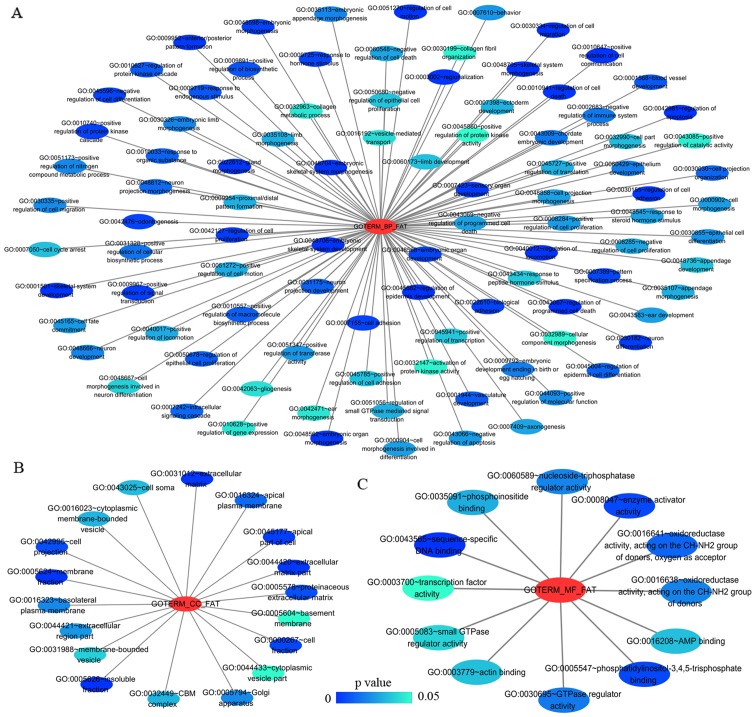
Network of gene ontology (GO) terms of HOXA3 co-expression genes in NSCLC constructed by cytoscape v3.5.0. (A-C) Show biological processes (BPs), cellular components (CCs) and molecular functions (MFs), respectively. In this network, only terms with statistically significant [P<0.01 (BP) or P<0.05 (CC and MF)] are displayed. Each node represents a GO term, and the node color indicates the P-value of a GO term. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer.
Figure 8.
Significantly enriched annotation of GO categories and KEGG pathways of HOXA3 co-expression genes in NSCLC. (A) Biological processes (BPs); (B) Cellular components (CCs); (C) Molecular factors (MFs); (D) KEGG pathways. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; HOXA3, homeobox A3; NSCLC, non-small cell lung cancer.
Table III.
The top ten most significant items of Gene Ontology (GO) terms of the co-expression genes of homeobox A3 (HOXA3) in non-small cell lung cancer.
| Category | Term | Count | P-value | FDR |
|---|---|---|---|---|
| Biological processes | ||||
| GOTERM_BP_FAT | GO:0048706~embryonic skeletal system development | 8 | 3.41E-05 | 0.06 |
| GOTERM_BP_FAT | GO:0001501~skeletal system development | 14 | 1.07E-04 | 0.18 |
| GOTERM_BP_FAT | GO:0045682~regulation of epidermis development | 5 | 1.17E-04 | 0.19 |
| GOTERM_BP_FAT | GO:0030334~regulation of cell migration | 10 | 1.80E-04 | 0.30 |
| GOTERM_BP_FAT | GO:0042981~regulation of apoptosis | 23 | 1.97E-04 | 0.33 |
| GOTERM_BP_FAT | GO:0043067~regulation of programmed cell death | 23 | 2.27E-04 | 0.38 |
| GOTERM_BP_FAT | GO:0010941~regulation of cell death | 23 | 2.39E-04 | 0.40 |
| GOTERM_BP_FAT | GO:0048705~skeletal system morphogenesis | 8 | 3.64E-04 | 0.60 |
| GOTERM_BP_FAT | GO:0040012~regulation of locomotion | 10 | 4.64E-04 | 0.77 |
| GOTERM_BP_FAT | GO:0051270~regulation of cell motion | 10 | 4.81E-04 | 0.80 |
| Cellular components | ||||
| GOTERM_CC_FAT | GO:0005626~insoluble fraction | 22 | 0.000184 | 0.24 |
| GOTERM_CC_FAT | GO:0005624~membrane fraction | 20 | 0.000837 | 1.07 |
| GOTERM_CC_FAT | GO:0031012~extracellular matrix | 12 | 0.001071 | 1.37 |
| GOTERM_CC_FAT | GO:0044420~extracellular matrix part | 7 | 0.001490 | 1.90 |
| GOTERM_CC_FAT | GO:0045177~apical part of cell | 8 | 0.002892 | 3.66 |
| GOTERM_CC_FAT | GO:0000267~cell fraction | 22 | 0.004675 | 5.86 |
| GOTERM_CC_FAT | GO:0042995~cell projection | 16 | 0.006747 | 8.35 |
| GOTERM_CC_FAT | GO:0005578~proteinaceous extracellular matrix | 10 | 0.006859 | 8.48 |
| GOTERM_CC_FAT | GO:0016324~apical plasma membrane | 6 | 0.013372 | 15.92 |
| GOTERM_CC_FAT | GO:0016323~basolateral plasma membrane | 7 | 0.020688 | 23.61 |
| Molecular functional | ||||
| GOTERM_MF_FAT | GO:0043565~sequence-specific DNA binding | 20 | 0.000041 | 0.06 |
| GOTERM_MF_FAT | GO:0008047~enzyme activator activity | 11 | 0.004149 | 5.55 |
| GOTERM_MF_FAT | GO:0005547~phosphatidylinositol-3,4,5-trisphosphate binding | 3 | 0.004179 | 5.59 |
| GOTERM_MF_FAT | GO:0016641~oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 3 | 0.010187 | 13.11 |
| GOTERM_MF_FAT | GO:0030695~GTPase regulator activity | 11 | 0.014540 | 18.21 |
| GOTERM_MF_FAT | GO:0016638~oxidoreductase activity, acting on the CH-NH2 group of donors | 3 | 0.016640 | 20.58 |
| GOTERM_MF_FAT | GO:0060589~nucleoside-triphosphatase regulator activity | 11 | 0.016712 | 20.66 |
| GOTERM_MF_FAT | GO:0035091~phosphoinositide binding | 5 | 0.025094 | 29.45 |
| GOTERM_MF_FAT | GO:0016208~AMP binding | 3 | 0.028747 | 33.00 |
| GOTERM_MF_FAT | GO:0003779~actin binding | 9 | 0.028844 | 33.09 |
Figure 7.
(A) Network mappings were Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of HOXA3 co-expression genes in NSCLC. (B) The significant signalling pathways (P<0.05) enriched in HOXA3 co-expression genes in NSCLC constructed by ClueGO plugin. The color of the circles represents the degree of enrichment of the pathway (P-value). HOXA3, homeobox A3; NSCLC, non-small cell lung cancer.
Table IV.
KEGG pathways enriched by co-expression genes of homeobox A3 (HOXA3) mRNA in non-small cell lung cancer.
| Category | Term | Count | P-value | FDR |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa04510:Focal adhesion | 12 | 0.001 | 0.76 |
| KEGG_PATHWAY | hsa05200:Pathways in cancer | 15 | 0.001 | 1.67 |
| KEGG_PATHWAY | hsa04350:TGF-beta signaling pathway | 7 | 0.004 | 4.34 |
| KEGG_PATHWAY | hsa04540:Gap junction | 7 | 0.004 | 4.84 |
| KEGG_PATHWAY | hsa04010:MAPK signaling pathway | 12 | 0.006 | 7.01 |
| KEGG_PATHWAY | hsa05218:Melanoma | 6 | 0.008 | 8.33 |
| KEGG_PATHWAY | hsa05212:Pancreatic cancer | 6 | 0.008 | 8.81 |
| KEGG_PATHWAY | hsa05220:Chronic myeloid leukemia | 6 | 0.010 | 10.36 |
| KEGG_PATHWAY | hsa05222:Small cell lung cancer | 6 | 0.015 | 15.93 |
| KEGG_PATHWAY | hsa05210:Colorectal cancer | 6 | 0.015 | 15.93 |
| KEGG_PATHWAY | hsa04512:ECM-receptor interaction | 6 | 0.015 | 15.93 |
| KEGG_PATHWAY | hsa05215:Prostate cancer | 6 | 0.019 | 19.63 |
| KEGG_PATHWAY | hsa04912:GnRH signaling pathway | 6 | 0.028 | 27.23 |
| KEGG_PATHWAY | hsa04916:Melanogenesis | 6 | 0.029 | 28.15 |
| KEGG_PATHWAY | hsa04060:Cytokine-cytokine receptor interaction | 10 | 0.039 | 35.71 |
Figure 9.
The network of GO terms of HOXA3 co-expression genes in LUAD were constructed by cytoscape v3.5.0. (A-C) Biological processes (BPs), cellular components (CCs) and molecular functions (MFs), respectively. In this network, only terms with statistically significant (P<0.05) are displayed. Each node represents a GO term, and the node color indicates the P-value of a GO term. GO, Gene Ontology; HOXA3, homeobox A3; LUAD, lung adenocarcinoma.
Figure 11.
Significantly enriched annotation of GO categories and KEGG pathways of HOXA3 co-expression genes in LUAD. (A) Biological processes (BPs); (B) Cellular components (CCs); (C) Molecular factors (MFs); (D) KEGG pathway. HOXA3, homeobox A3; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUAD, lung adenocarcinoma.
Table V.
The top 10 most significant items of Gene Ontology (GO) terms of the co-expression genes of homeobox A3 (HOXA3) mRNA in lung adenocarcinoma.
| Category | Term | Count | P-value | FDR |
|---|---|---|---|---|
| Biological Processes | ||||
| GOTERM_BP_FAT | GO:0048706~embryonic skeletal system development | 8 | 3.41E-05 | 0.056837 |
| GOTERM_BP_FAT | GO:0001501~skeletal system development | 14 | 1.07E-04 | 0.178201 |
| GOTERM_BP_FAT | GO:0045682~regulation of epidermis development | 5 | 1.17E-04 | 0.194494 |
| GOTERM_BP_FAT | GO:0030334~regulation of cell migration | 10 | 1.80E-04 | 0.299680 |
| GOTERM_BP_FAT | GO:0042981~regulation of apoptosis | 23 | 1.97E-04 | 0.328453 |
| GOTERM_BP_FAT | GO:0043067~regulation of programmed cell death | 23 | 2.27E-04 | 0.377505 |
| GOTERM_BP_FAT | GO:0010941~regulation of cell death | 23 | 2.39E-04 | 0.397502 |
| GOTERM_BP_FAT | GO:0048705~skeletal system morphogenesis | 8 | 3.64E-04 | 0.604848 |
| GOTERM_BP_FAT | GO:0040012~regulation of locomotion | 10 | 4.64E-04 | 0.770756 |
| GOTERM_BP_FAT | GO:0051270~regulation of cell motion | 10 | 4.81E-04 | 0.798787 |
| Cellular ψomponents | ||||
| GOTERM_CC_FAT | GO:0005626~insoluble fraction | 22 | 0.000184 | 0.237025 |
| GOTERM_CC_FAT | GO:0005624~membrane fraction | 20 | 0.000837 | 1.072817 |
| GOTERM_CC_FAT | GO:0031012~extracellular matrix | 12 | 0.001071 | 1.370868 |
| GOTERM_CC_FAT | GO:0044420~extracellular matrix part | 7 | 0.001490 | 1.902184 |
| GOTERM_CC_FAT | GO:0045177~apical part of cell | 8 | 0.002892 | 3.662636 |
| GOTERM_CC_FAT | GO:0000267~cell fraction | 22 | 0.004675 | 5.857488 |
| GOTERM_CC_FAT | GO:0042995~cell projection | 16 | 0.006747 | 8.351548 |
| GOTERM_CC_FAT | GO:0005578~proteinaceous extracellular matrix | 10 | 0.006859 | 8.483923 |
| GOTERM_CC_FAT | GO:0016324~apical plasma membrane | 6 | 0.013372 | 15.921503 |
| GOTERM_CC_FAT | GO:0016323~basolateral plasma membrane | 7 | 0.020688 | 23.607821 |
| Molecular ϕunctional | ||||
| GOTERM_MF_FAT | GO:0043565~sequence-specific DNA binding | 20 | 0.000041 | 0.056109 |
| GOTERM_MF_FAT | GO:0008047~enzyme activator activity | 11 | 0.004149 | 5.548392 |
| GOTERM_MF_FAT | GO:0005547~phosphatidylinositol-3,4,5-trisphosphate binding | 3 | 0.004179 | 5.587211 |
| GOTERM_MF_FAT | GO:0016641~oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 3 | 0.010187 | 13.113973 |
| GOTERM_MF_FAT | GO:0030695~GTPase regulator activity | 11 | 0.014540 | 18.214548 |
| GOTERM_MF_FAT | GO:0016638~oxidoreductase activity, acting on the CH-NH2 group of donors | 3 | 0.016640 | 20.575093 |
| GOTERM_MF_FAT | GO:0060589~nucleoside-triphosphatase regulator activity | 11 | 0.016712 | 20.655661 |
| GOTERM_MF_FAT | GO:0035091~phosphoinositide binding | 5 | 0.025094 | 29.453780 |
| GOTERM_MF_FAT | GO:0016208~AMP binding | 3 | 0.028747 | 32.997529 |
| GOTERM_MF_FAT | GO:0003779~actin binding | 9 | 0.028844 | 33.089357 |
Figure 10.
(A) Network mappings of KEGG pathway anlaysis of HOXA3 co-expression genes in LUAD. (B) The significantly signalling pathways (P<0.05) enriched HOXA3 co-expression genes in LUAD constructed by the ClueGO plugin. The color of circles represents the P-value of the signalling pathways. HOXA3, homeobox A3; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUAD, lung adenocarcinoma.
Table VI.
KEGG pathway enriched by co-expression genes of homeobox A3 (HOXA3) mRNA in lung adenocarcinoma.
| Category | Term | Count | P-value | FDR |
|---|---|---|---|---|
| KEGG_PATHWAY | hsa04510:Focal adhesion | 12 | 3.84E-05 | 0.042706 |
| KEGG_PATHWAY | hsa04512:ECM-receptor interaction | 6 | 0.004145 | 4.514261 |
| KEGG_PATHWAY | hsa05218:Melanoma | 5 | 0.012451 | 13.009202 |
| KEGG_PATHWAY | hsa05212:Pancreatic cancer | 5 | 0.013063 | 13.603250 |
| KEGG_PATHWAY | hsa05210:Colorectal cancer | 5 | 0.021874 | 21.802852 |
| KEGG_PATHWAY | hsa05222:Small cell lung cancer | 5 | 0.021874 | 21.802852 |
| KEGG_PATHWAY | hsa05200:Pathways in cancer | 10 | 0.022490 | 22.349251 |
| KEGG_PATHWAY | hsa05215:Prostate cancer | 5 | 0.026406 | 25.738703 |
| KEGG_PATHWAY | hsa05223:Non-small cell lung cancer | 4 | 0.031134 | 29.652037 |
| KEGG_PATHWAY | hsa05214:Glioma | 4 | 0.045993 | 40.760927 |
| KEGG_PATHWAY | hsa04010:MAPK signaling pathway | 8 | 0.052597 | 45.164053 |
| KEGG_PATHWAY | hsa05220:Chronic myeloid leukemia | 4 | 0.070302 | 55.541161 |
| KEGG_PATHWAY | hsa04144:Endocytosis | 6 | 0.084942 | 62.734617 |
| KEGG_PATHWAY | hsa04350:TGF-beta signaling pathway | 4 | 0.099275 | 68.735075 |
| KEGG_PATHWAY | hsa04012:ErbB signaling pathway | 4 | 0.099275 | 68.735075 |
Figure 12.
(A) PPI network analysis of genes co-expressed in HOXA3 in NSCLC and (B) the genes most implicated by the signalling pathways. PPI, protein-protein interaction; HOXA3, homeobox A3; NSCLC, non-small cell lung cancer.
Table VII.
Top 10 with combined co-expression score that the protein-protein interaction (PPI) of homeobox A3 (HOXA3) co-expression genes in non-small cell lung cancer.
| #Node1 | Node2 | Node1 STRING internal ID | Homology | Co-expression | Experimentally determined interaction | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| STX2 | SNAP25 | 1858001 | 0 | 0.093 | 0.939 | 0.905 | 0.998 |
| CDKN2B | CDK6 | 1846519 | 0 | 0.054 | 0.432 | 0.763 | 0.985 |
| DUSP10 | MAPK1 | 1854467 | 0 | 0.053 | 0.498 | 0.705 | 0.984 |
| CALD1 | MYLK | 1854225 | 0 | 0.248 | 0 | 0.791 | 0.982 |
| MITF | MAPK1 | 1847592 | 0 | 0.048 | 0.395 | 0.733 | 0.982 |
| ARRB1 | MAPK1 | 1860799 | 0 | 0 | 0.43 | 0.658 | 0.978 |
| ITSN1 | ITSN2 | 1857363 | 0.965 | 0 | 0.702 | 0.726 | 0.969 |
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| MALT1 | CARD10 | 1850132 | 0 | 0 | 0.115 | 0.63 | 0.964 |
| RUNX2 | MAPK1 | 1855482 | 0 | 0 | 0 | 0.639 | 0.962 |
Table VIII.
Top 10 with combined co-expression score that the protein-protein interaction (PPI) of homeobox A3 (HOXA3) co-expression gene intersection in non-small cell lung cancer enriched in significant signalling pathways.
| #Node1 | Node2 | Node1 STRING internal ID | Homology | Co-expression | Experimentally determined interaction | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| COL6A1 | COL1A1 | 1854318 | 0.617 | 0.504 | 0 | 0.49 | 0.957 |
| MYLK | MAPK1 | 1853981 | 0.636 | 0 | 0 | 0.711 | 0.925 |
| LAMA3 | LAMA4 | 1850649 | 0.804 | 0 | 0 | 0.115 | 0.901 |
| PDGFD | EGFR | 1858075 | 0 | 0 | 0 | 0.49 | 0.893 |
| EGFR | TNC | 1846445 | 0 | 0 | 0.36 | 0.72 | 0.813 |
| THBS1 | COL1A1 | 1844779 | 0 | 0.1 | 0.324 | 0.463 | 0.645 |
| TNC | MAPK1 | 1845696 | 0 | 0 | 0 | 0.576 | 0.576 |
| TNC | THBS1 | 1845696 | 0 | 0.052 | 0 | 0.533 | 0.538 |
| FLNC | MAPK1 | 1850897 | 0 | 0 | 0.066 | 0.475 | 0.49 |
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| COL6A1 | COL1A1 | 1854318 | 0.617 | 0.504 | 0 | 0.49 | 0.957 |
| MYLK | MAPK1 | 1853981 | 0.636 | 0 | 0 | 0.711 | 0.925 |
| LAMA3 | LAMA4 | 1850649 | 0.804 | 0 | 0 | 0.115 | 0.901 |
| PDGFD | EGFR | 1858075 | 0 | 0 | 0 | 0.49 | 0.893 |
Figure 13.
PPI network analysis of genes co-expressed in HOXA3 in LUAD. PPI, protein-protein interaction; HOXA3, homeobox A3; LUAD, lung adenocarcinoma.
Table IX.
Top 10 with combined score co-expression correlation by STRING 10.5 of homeobox A3 (HOXA3) co-expression genes in lung adenocarcinoma.
| #Node1 | Node2 | Node1 STRING internal ID | Homology | Co-expression | Experimentally determined interaction | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| STX2 | SNAP25 | 1858001 | 0 | 0.093 | 0.939 | 0.905 | 0.998 |
| ARRB1 | MAPK1 | 1860799 | 0 | 0 | 0.43 | 0.658 | 0.978 |
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| MALT1 | CARD10 | 1850132 | 0 | 0 | 0.115 | 0.63 | 0.964 |
| COL6A1 | COL1A1 | 1854318 | 0.617 | 0.504 | 0 | 0.49 | 0.957 |
| PDE10A | ADCY6 | 1862089 | 0 | 0.207 | 0.169 | 0.384 | 0.954 |
| ARRB1 | EGFR | 1860799 | 0 | 0 | 0.165 | 0.416 | 0.947 |
| EGFR | CFTR | 1846445 | 0 | 0 | 0 | 0.445 | 0.942 |
| EGFR | ERBB3 | 1846445 | 0.921 | 0.05 | 0.36 | 0.956 | 0.938 |
| TGFB2 | THBS1 | 1854475 | 0 | 0.046 | 0 | 0.364 | 0.934 |
| STX2 | SNAP25 | 1858001 | 0 | 0.093 | 0.939 | 0.905 | 0.998 |
| ARRB1 | MAPK1 | 1860799 | 0 | 0 | 0.43 | 0.658 | 0.978 |
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| MALT1 | CARD10 | 1850132 | 0 | 0 | 0.115 | 0.63 | 0.964 |
| COL6A1 | COL1A1 | 1854318 | 0.617 | 0.504 | 0 | 0.49 | 0.957 |
Table X.
Top 10 with combined co-expression score that the protein-protein interaction (PPI) of homeobox A3 (HOXA3) co-expression gene intersection in lung adenocarcinoma enriched in significant signalling pathways.
| #Node1 | Node2 | Node1 STRING internal ID | Homology | Co-expression | Experimentally determined interaction | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
| COL6A1 | COL1A1 | 1854318 | 0.617 | 0.504 | 0 | 0.49 | 0.957 |
| MYLK | MAPK1 | 1853981 | 0.636 | 0 | 0 | 0.711 | 0.925 |
| LAMA3 | LAMA4 | 1850649 | 0.804 | 0 | 0 | 0.115 | 0.901 |
| PDGFD | EGFR | 1858075 | 0 | 0 | 0 | 0.49 | 0.893 |
| EGFR | TNC | 1846445 | 0 | 0 | 0.36 | 0.72 | 0.813 |
| THBS1 | COL1A1 | 1844779 | 0 | 0.1 | 0.324 | 0.463 | 0.645 |
| TNC | MAPK1 | 1845696 | 0 | 0 | 0 | 0.576 | 0.576 |
| TNC | THBS1 | 1845696 | 0 | 0.052 | 0 | 0.533 | 0.538 |
| FLNC | MAPK1 | 1850897 | 0 | 0 | 0.066 | 0.475 | 0.49 |
| AKT3 | MAPK1 | 1845421 | 0.65 | 0.134 | 0.292 | 0.576 | 0.486 |
| EGFR | THBS1 | 1846445 | 0 | 0 | 0 | 0.457 | 0.457 |
| COL6A1 | TNC | 1854318 | 0 | 0.055 | 0 | 0.424 | 0.432 |
| TNC | COL1A1 | 1845696 | 0 | 0.1 | 0 | 0.369 | 0.408 |
| EGFR | MAPK1 | 1846445 | 0.632 | 0 | 0.503 | 0.932 | 0.965 |
Figure 14.
Correlation between the hub genes and the expression of HOXA3 in NSCLC. (A) MAPK1; (B) EGFR; (C) COL1A1; (D) TNC. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer.
Figure 15.
Correlation between the hub genes and the expression of HOXA3 in LUAD. (A) MAPK1; (B) EGFR; (C) COL1A1; (D) TNC. HOXA3, homeobox A3; LUAD, lung adenocarcinoma.
Prediction and preliminary validation of HOXA3 target miRNAs
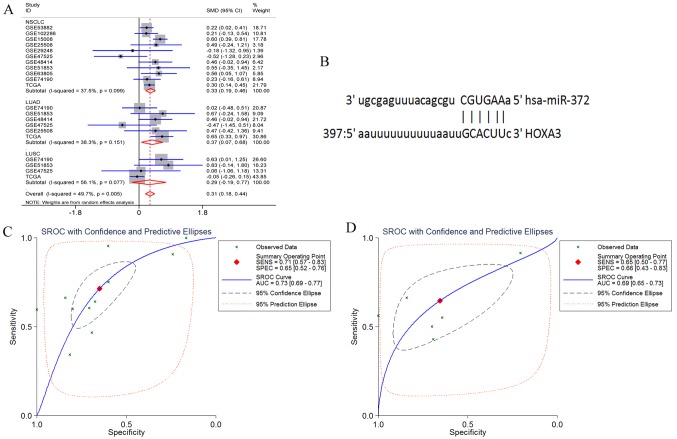
HOXA3 had 3 target miRNAs that passed the target prediction. Based on GEO microarray search and TCGA database, the random effects model was used for meta-analyses of the expression of these 3 miRNAs in NSCLC tissues. The results revealed that the expression of hsa-miR-372-3p was significantly increased in the NSCLC and LUAD tissues, but only exhibited an increasing trend in the LUSC tissues (NSCLC P<0.001, LUAD P=0.017, and LUSC P=0.243) (Fig. 16A). In addition, hsa-miR-372-3p and HOX3A had 1 fragment of complementary sequences of ′3′CGUGAA5′-3′ GCACUU-5″ (Fig. 16B). Furthermore, the sensitivity and specificity of miR-372-3p expression in NSCLC were examined using the sROC method. The results suggested that the optimal sensitivity and specificity of a high miR-372-3p expression in the NSCLC group were SENS, 0.71 (0.57, 0.83) and SPES, 0.65 (0.52, 0.76), respectively, and the AUC was 0.73 (95% CI: 0.69, 0.77). The optimal sensitivity and specificity of the LUAD group were SENS, 0.65 (0.5, 0.77) and SPES, 0.66 (0.43, 0.83), respectively, and the AUC was 0.69 (95% CI: 0.65, 0.73) (Fig. 16C and D).
Figure 16.
(A) Forest profile of SMD of miR-372-3p expression between NSCLC and normal lung tissue. (B) The complementary sequences existing between miR-372-3p and HOXA3. (C and D) The combined sROC curve for miR-372-3P expression between NSCLC and normal lung tissue; (C) NSCLC and (D) LUAD. HOXA3, homeobox A3; NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma.
Discussion
This study used qPCR detection and TCGA RNA-seq data to confirm that HOXA3 expression was significantly down-regulated in LUAD tissues. The downregulation of HOXA3 may be closely associated with a more favourable prognosis of LUAD. In addition, the TCGA data indicated that HOXA3 harboured genetic alterations in 11–55% of LUAD tissues, and the methylation level of HOXA3 in LUAD tissues was significantly upregulated. Combined with the TCGA data, the upregulation of the HOXA3 level in LUSC was not significant. Therefore, we paid more attention to the clinical significance and molecular mechanisms of action of HOXA3 in LUAD. Through the collection of co-expression genes and GO and KEGG analyses, the preliminary results revealed that HOXA3 may play important roles in LUAD through the regulation of focal adhesion and ECM-receptor interaction signalling pathways. Furthermore, through the prediction of HOXA3 target miRNAs and meta-analyses of target miRNA expression based on the GEO microarray and TCGA data, the results confirmed that the expression of miR-372-3p that had complementary sequences with HOXA3 was significantly increased in LUAD tissues.
There have been some studies of the association between the HOXA3 level and tumours. For example, Zhang et al (30) demonstrated that HOXA3 exhibited a high expression in both colorectal cancer tissues and cell lines. Shen et al (31) assessed the association between HOX family genes and nasopharyngeal carcinoma and demonstrated that HOXA3 expression was also upregulated. Kuasne et al (32) suggested that HOXA3 exhibited a low expression in penile carcinoma. At least to the best of our knowledge, only 1 methylation-related study on the association between HOXA3 and LUAD has been reported (19). This study, to the best of our knowledge, was the first to detect HOXA3 mRNA expression in LUAD. Combined with qPCR detection and TCGA data, we confirmed that HOXA3 expression was downregulated in LUAD tissues, and a low HOXA3 expression may play a important role in LUAD.
The investigation of the association between HOXA3 and tumour progression has demonstrated that a high HOXA3 expression is associated with low survival rates in colon cancer (33). In penile carcinoma, a low HOXA3 expression was found to be associated with a poor prognosis (32). However, the association between HOXA3 and the progression of nasopharyngeal carcinoma has not been studied in depth (31). Our analyses of HOXA3 expression in all pathological stages of LUAD did not exhibit an obvious pattern. The survival analyses demonstrated that patients LUAD with a low HOXA3 expression had a better OS. Therefore, we hypothesized that the downregulation of HOXA3 expression may be an independent protective factor in LUAD.
Furthermore, based on the microarray and RNA Seq data in TCGA, we preliminarily revealed that HOXA3 harboured a certain level of genetic alterations in LUAD tumour tissues. Thus, we would like to acquire more information for further validation. However, relevant studies are currently lacking, and literature reports are not available, at least to the best of our knowledge. As regards gene methylation, Kuasne et al performed genome-wide methylation studies on penile carcinoma and confirmed that 8 genes, including HOXA3, exhibited high methylation levels (32). In a validation study of epigenetic biomarkers in LUAD, Daugaard et al (19) performed genome-wide methylation microarray sequencing on 4 cases of LUAD tissues and normal cancer-adjacent lung tissues to screen differentially methylated genes in LUAD. They selected the 18 most significantly differentially methylated genes, including HOXA3, for validation in 52 samples. The methylation level of HOXA3 significantly increased (19). This study acquired HOXA3 methylation data from the TCGA database to perform analyses. The results suggested that the HOXA3 methylation level was significantly increased in LUAD tissues. This finding was consistent with the results in literature reports. Combined with HOXA3 mRNA expression levels in LUAD, the function of HOXA3 in LUAD may be associated with the increase in the methylation level. In addition, the study by Daugaard also demonstrated that the increase of HOXA3 methylation was even more evident in patients with tumour metastasis; however, its association with the prognostic survival rates in LUAD patients has not been elucidated (19). Despite the results of this study, sufficient evidence is not yet available to confirm the association between the methylation level of HOXA3 and LUAD progression. Thus, further studies are required for confirmation. As regards studies on the association between DNA methylation and mRNA expression, Kuasne et al (32) performed qPCR detection to demonstrate that genes with high methylation levels exhibited a low expression in penile carcinoma. These results confirmed that the methylation level and mRNA expression exhibited a negative correlation (32). Zhang et al performed cell experiments to confirm that a group of genes (S100P, GDA, WISP2, LOXL1, TIMP4, ICAM1, CLMP, HSP8, GAS1 and BMP2) exhibited a downregulated mRNA expression through high methylation levels to induce drug resistance in NSCLC (34). Jin et al demonstrated that 19 genes mediated the downregulation of mRNA expression through the increase in upstream methylation levels in LUAD (35). This study confirmed the low expression of HOXA3 in LUAD and the increase in the methylation level. Based on these findings, we hypothesized that HOXA3 methylation has a negative regulatory association with mRNA expression and plays critical roles in tumour development and progression.
To investigate the underlying molecular mechanisms, GO and KEGG analyses were performed to compare the enrichment of HOXA3 co-expression genes between the NSCLC group and the LUAD group. The results demonstrated that the enrichment conditions in biological processes, cellular components, and molecular functions were basically the same. Some differences in the KEGG signalling pathways were noted. The LUAD group was more concentrated on the focal adhesion and ECM-receptor interaction pathways. HOXA3 may carry out its regulatory functions in LUAD through participation in these 2 signalling pathways. The focal adhesion signalling pathway is a critical pathway to activate focal adhesion kinase (FAK). Kinases activated by this pathway mainly mediate a variety of cellular metabolic processes, including cell metastasis, growth factor signal transduction, cell survival, cell cycle progression, and cell movement, and are closely associated with the development of malignant tumours (36–38). Currently, there are literature reports on using FAK as a novel potential biological target to investigate targeted therapy of NSCLC (39,40). The ECM-receptor interaction signalling pathway mediates the expression of enriched genes and regulates functions of these genes to promote tumour cell proliferation and migration and participate in tumour metastasis and infiltration (41,42). In NSCLC unrelated to smoking, the ECM-receptor interaction signalling pathway plays a critical role (43). The above-mentioned 2 signalling pathways significantly influence the development and progression of human malignant tumours and act as the bridge between abnormal gene expression and malignant tumour development.
Therefore, in this study, we performed PPI analyses on genes enriched in the focal adhesion and ECM-receptor interaction signalling pathways and demonstrated that MAPK1, EGFR, TNC and COL1A1 served as core genes in these 2 pathways. In addition, based on the TCGA data, Pearson's correlation analyses demonstrated that only TNC and COL1A1 exhibited significantly positive correlations with HOXA3 expression in LUAD. One relevant study detected 72 cases of NSCLC tissues and normal lung tissues (44). TNC expression was upregulated in NSCLC and was closely associated with TNM stage, lymph node metastasis and tumour pleural invasion. Other studies have also demonstrated that TNC upregulation is associated with a poor prognosis in LUAD (45,46). The study by Oleksiewicz et al (47) on 156 cases of NSCLC and normal lung tissues revealed that COL1A1 was significantly overexpressed in tumour tissues. That study also performed cell experiments to confirm that COL1A1 was closely associated with hypoxia responses in NSCLC (47). Combined with bioinformatics analysis results, HOXA3 may collaborate with co-expression genes, such as TNC and COL1A1, to regulate focal adhesion and ECM-receptor interaction signalling pathways, thus carrying out is functions in LUAD. However, more functional studies are required for further validation.
In this study, we also performed meta-analyses to confirm that the expression of the predicted miR-372-3p was upregulated in LUAD. Complementary sequences were noted between miR-372-3p and HOXA3. Combined with qPCR detection, HOXA3 and miR-372-3p expression exhibited a negative correlation trend in LUAD. The results of a literature search demonstrated that miR-372-3p was highly expressed in both testicular cancer and LUSC. miR-372-3p promotes cell growth and migration in LUSC through the downregulation of the target gene FGF9 (48,49). Therefore, based on results of this study and those of studies in the literature, we hypothesized that the downregulation of HOXA3 expression in LUAD tissues may be associated with the increase in miR-372-3p expression and HOXA3 may carry out its function through the targeted upregulation of miR-372-3p. However, this hypothesis needs to be validated using various experiments.
In conclusion, combined with the RT-qPCR detection results and data from the TCGA and GEO databases, we confirmed that HOXA3 expression was downregulated in LUAD tissues and exhibited a certain level of genetic alteration. The functions of downregulated HOXA3 in tumour progression and a poor prognosis of patients with LUAD may be associated with increases in the methylation level. Furthermore, HOXA3 may collaborate with co-expression genes, such as TNC and COL1A1, to regulate the focal adhesion and ECM-receptor interaction signalling pathways together or upregulate miR-372-3p to promote its carcinogenesis function. These study results provide true and reliable experimental data and rigorous molecular theories for further studies on the molecular mechanisms of HOXA3 in LUAD.
Acknowledgments
The authors would like to thank all members of the Molecular Oncology Group of the First Affiliated Hospital of Guangxi Medical University (Nanning, Guangxi Zhuang Autonomous Region 530021, China) for their professional suggestions.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Authors' contributions
BLG collected data from public datasets and analyzed the data and performed the statistical analysis. RQH and YZ collected the clinical samples and performed in-house RT-qPCR experiments. RQH and GC participated in the conception and design of the study and in language modification. BLG and RQH drafted the manuscript and analyzed the GO and KEGG terms. DMW and XHH conceived and designed the study and assisted in the drafting of the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This research programme was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University. All participants signed informed consent forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Cancer UK: Lung cancer mortality statistics. 2014. [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 5.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien TD, Jia P, Caporaso NE, Landi MT, Zhao Z. Weak sharing of genetic association signals in three lung cancer subtypes: Evidence at the SNP, gene, regulation, and pathway levels. Genome Med. 2018;10:16. doi: 10.1186/s13073-018-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham C, Garsa A, Badiyan SN, Drzymala R, Yang D, DeWees T, Tsien C, Dowling JL, Rich KM, Chicoine MR, et al. Internal dose escalation is associated with increased local control for non-small cell lung cancer (NSCLC) brain metastases treated with stereotactic radiosurgery (SRS) Adv Radiat Oncol. 2017;3:146–153. doi: 10.1016/j.adro.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes P, Lareiro S, Vouga L, Guerra M, Miranda J. Uniportal video-assisted thorascoscopic surgery - The new paradigm in the surgical treatment of lung cancer. Rev Port Cir Cardiotorac Vasc. 2017;24:127. [PubMed] [Google Scholar]
- 9.Jean F, Tomasini P, Barlesi F. Atezolizumab: Feasible second-line therapy for patients with non-small cell lung cancer? A review of efficacy, safety and place in therapy. Ther Adv Med Oncol. 2017;9:769–779. doi: 10.1177/1758834017741074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Q, Xie M, He M, Yan F, Zhang X, Yu S. Anti-PD-1/PD-L1 antibodies versus docetaxel in patients with previously treated non-small-cell lung cancer. Oncotarget. 2017;9:7672–7683. doi: 10.18632/oncotarget.23584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne M, Martinez P, Morris V. Evolution of a pentameral body plan was not linked to translocation of anterior Hox genes: The echinoderm HOX cluster revisited. Evol Dev. 2016;18:137–143. doi: 10.1111/ede.12172. [DOI] [PubMed] [Google Scholar]
- 12.Kim HS, Kim BM, Lee BY, Souissi S, Park HG, Lee JS. Identification of Hox genes and rearrangements within the single homeobox (Hox) cluster (192.8 kb) of the cyclopoid copepod (Paracyclopina nana) J Exp Zoolog B Mol Dev Evol. 2016;326:105–109. doi: 10.1002/jez.b.22668. [DOI] [PubMed] [Google Scholar]
- 13.Schiemann SM, Martín-Durán JM, Børve A, Vellutini BC, Passamaneck YJ, Hejnol A. Clustered brachiopod Hox genes are not expressed collinearly and are associated with lophotrochozoan novelties. Proc Natl Acad Sci USA. 2017;114:E1913–E1922. doi: 10.1073/pnas.1614501114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Bu J, Liu X, Wang W, Mai W, Lv B, Zou J, Mo X, Li X, Wang J, et al. miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget. 2017;8:63935–63948. doi: 10.18632/oncotarget.19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. 2017;21:3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, Li W, Zheng S, Ye H, Wang L, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Yu J, Lv X, Zhang L. HOXA9 is critical in the proliferation, differentiation, and malignancy of leukaemia cells both in vitro and in vivo. Cell Biochem Funct. 2017;35:433–440. doi: 10.1002/cbf.3293. [DOI] [PubMed] [Google Scholar]
- 18.Sang Y, Zhou F, Wang D, Bi X, Liu X, Hao Z, Li Q, Zhang W. Up-regulation of long non-coding HOTTIP functions as an oncogene by regulating HOXA13 in non-small cell lung cancer. Am J Transl Res. 2016;8:2022–2032. [PMC free article] [PubMed] [Google Scholar]
- 19.Daugaard I, Dominguez D, Kjeldsen TE, Kristensen LS, Hager H, Wojdacz TK, Hansen LL. Identification and validation of candidate epigenetic biomarkers in lung adenocarcinoma. Sci Rep. 2016;6:35807. doi: 10.1038/srep35807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, et al. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: A study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. doi: 10.1038/s41598-017-05856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang RX, Chen WJ, He RQ, Zeng JH, Liang L, Li SK, Ma J, Luo DZ, Chen G. Identification of a RNA-Seq based prognostic signature with five lncRNAs for lung squamous cell carcinoma. Oncotarget. 2017;8:50761–50773. doi: 10.18632/oncotarget.17098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Xie Y, Zhong T, Zhang X, Dang Y, Gan T, Chen G. Expression and clinical contribution of MRGD mRNA in non-small cell lung cancers. J BUON. 2015;20:1101–1106. [PubMed] [Google Scholar]
- 23.Muraoka T, Soh J, Toyooka S, Aoe K, Fujimoto N, Hashida S, Maki Y, Tanaka N, Shien K, Furukawa M, et al. The degree of microRNA-34b/c methylation in serum-circulating DNA is associated with malignant pleural mesothelioma. Lung Cancer. 2013;82:485–490. doi: 10.1016/j.lungcan.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Singh J, Batish VK, Grover S. Simultaneous detection of Listeria monocytogenes and Salmonella spp. in dairy products using real time PCR-melt curve analysis. J Food Sci Technol. 2012;49:234–239. doi: 10.1007/s13197-011-0278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Yang Z, Zhang D, Wu W, Guo Y, Chen Q, Xu D, Cui W. Rapid detection of immunoglobulin heavy chain gene rearrangement by PCR and melting curve analysis using combined FR2 and FR3 primers. Diagn Pathol. 2015;10:140. doi: 10.1186/s13000-015-0370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaghloul H, El Morsi AA, Soweha HE, Elsayed A, Seif S, El-Sharawy H. A simple real-time polymerase chain reaction assay using SYBR Green for hepatitis C virus genotyping. Arch Virol. 2017;162:57–61. doi: 10.1007/s00705-016-3059-5. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Tang R, Liang L, Luo D, Feng Z, Huang Q, He R, Gan T, Yang L, Chen G. Downregulation of miR-30a is associated with poor prognosis in lung cancer. Med Sci Monit. 2015;21:2514–2520. doi: 10.12659/MSM.894372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren F, Ding H, Huang S, Wang H, Wu M, Luo D, Dang Y, Yang L, Chen G. Expression and clinicopathological significance of miR-193a-3p and its potential target astrocyte elevated gene-1 in non-small lung cancer tissues. Cancer Cell Int. 2015;15:80. doi: 10.1186/s12935-015-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Liu G, Ding L, Jiang T, Shao S, Gao Y, Lu Y. HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell Biochem. 2018;119:2864–2874. doi: 10.1002/jcb.26461. [DOI] [PubMed] [Google Scholar]
- 31.Shen ZH, Zhao KM, Du T. HOXA10 promotes nasopharyngeal carcinoma cell proliferation and invasion via inducing the expression of ZIC2. Eur Rev Med Pharmacol Sci. 2017;21:945–952. [PubMed] [Google Scholar]
- 32.Kuasne H, Cólus IM, Busso AF, Hernandez-Vargas H, Barros-Filho MC, Marchi FA, Scapulatempo-Neto C, Faria EF, Lopes A, Guimarães GC, et al. Genome-wide methylation and transcriptome analysis in penile carcinoma: Uncovering new molecular markers. Clin Epigenetics. 2015;7:46. doi: 10.1186/s13148-015-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Liu G, Ding L, Jiang T, Shao S, Gao Y, Lu Y. HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell Biochem. 2018;119:2864–2874. doi: 10.1002/jcb.26461. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YW, Zheng Y, Wang JZ, Lu XX, Wang Z, Chen LB, Guan XX, Tong JD. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9:896–909. doi: 10.4161/epi.28601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin X, Liu X, Li X, Guan Y. Integrated analysis of DNA methylation and mRNA expression profiles data to identify key Genes in lung adenocarcinoma. Biomed Res Int. 2016;2016:4369431. doi: 10.1155/2016/4369431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai YL, Chen LC, Shen TL. Emerging roles of focal adhesion kinase in cancer. Biomed Res Int. 2015;2015:690690. doi: 10.1155/2015/690690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min A, Zhu C, Wang J, Peng S, Shuai C, Gao S, Tang Z, Su T. Focal adhesion kinase knockdown in carcinoma-associated fibroblasts inhibits oral squamous cell carcinoma metastasis via downregulating MCP-1/CCL2 expression. J Biochem Mol Toxicol. 2015;29:70–76. doi: 10.1002/jbt.21669. [DOI] [PubMed] [Google Scholar]
- 38.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 39.Zhou B, Wang GZ, Wen ZS, Zhou YC, Huang YC, Chen Y, Zhou GB. Somatic mutations and splicing variants of focal adhesion kinase in non-small cell lung cancer. J Natl Cancer Inst. 2017;110:195–204. doi: 10.1093/jnci/djx157. [DOI] [PubMed] [Google Scholar]
- 40.Tang KJ, Constanzo JD, Venkateswaran N, Melegari M, Ilcheva M, Morales JC, Skoulidis F, Heymach JV, Boothman DA, Scaglioni PP. Focal adhesion kinase regulates the DNA damage response and its inhibition radiosensitizes mutant KRAS lung cancer. Clin Cancer Res. 2016;22:5851–5863. doi: 10.1158/1078-0432.CCR-15-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang HJ, Tao J, Sheng L, Hu X, Rong RM, Xu M, Zhu TY. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–1812. doi: 10.2147/OTT.S96535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Ye J, Weng X, Liu F, He L, Zhou D, Liu Y. Comparative transcriptome analysis reveals that the extracellular matrix receptor interaction contributes to the venous metastases of hepatocellular carcinoma. Cancer Genet. 2015;208:482–491. doi: 10.1016/j.cancergen.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Lv M, Wang L. Comprehensive analysis of genes, pathways, and TFs in nonsmoking Taiwan females with lung cancer. Exp Lung Res. 2015;41:74–83. doi: 10.3109/01902148.2014.971472. [DOI] [PubMed] [Google Scholar]
- 44.GE ZH, Cheng QS, LI WM, Feng Z, Wen MM, Wang WC, Zhang ZP. Expression of TNC in NSCLC and its clinical significance. Xiandai Shengwu Yixue Jinzhan. 2014;31:6103–6106. In Chinese. [Google Scholar]
- 45.Gocheva V, Naba A, Bhutkar A, Guardia T, Miller KM, Li CM, Dayton TL, Sanchez-Rivera FJ, Kim-Kiselak C, Jailkhani N, et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc Natl Acad Sci USA. 2017;114:E5625–E5634. doi: 10.1073/pnas.1707054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edlund K, Lindskog C, Saito A, Berglund A, Pontén F, Göransson-Kultima H, Isaksson A, Jirström K, Planck M, Johansson L, et al. CD99 is a novel prognostic stromal marker in non-small cell lung cancer. Int J Cancer. 2012;131:2264–2273. doi: 10.1002/ijc.27518. [DOI] [PubMed] [Google Scholar]
- 47.Oleksiewicz U, Liloglou T, Tasopoulou KM, Daskoulidou N, Gosney JR, Field JK, Xinarianos G. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143:1133–1141. doi: 10.1007/s00432-017-2381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Liu S, Zhao X, Wang Y, Tian D, Jiang W. MiR-372-3p promotes cell growth and metastasis by targeting FGF9 in lung squamous cell carcinoma. Cancer Med. 2017;6:1323–1330. doi: 10.1002/cam4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syring I, Bartels J, Holdenrieder S, Kristiansen G, Müller SC, Ellinger J. Circulating serum miRNA (miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as biomarkers in patients with testicular germ cell cancer. J Urol. 2015;193:331–337. doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.