Abstract
Many human tumors harbor mutations that result in deregulation of Cdk4 activity. Most of these mutations involve overexpression of D-type cyclins and inactivation of INK4 inhibitors. In addition, a mutation in the Cdk4 protein has been described in patients with familial melanoma (Wolfel, T., Hauer, M., Schneider, J., Serrano, M., Wolfel, C., et al. (1995) Science 269, 1281–1284; Zuo, L., Weger, J., Yang, Q., Goldstein, A. M., Tucker, M. A., et al. (1996) Nat. Genet. 12, 97–99). This mutation, R24C, renders the Cdk4 protein insensitive to inhibition by INK4 proteins including p16INK4a, a major candidate for the melanoma susceptibility locus. Here we show that knock-in mice expressing a Cdk4 R24C allele are highly susceptible to melanoma development after specific carcinogenic treatments. These tumors do not have mutations in the p19ARF/p53 pathway, suggesting a specific involvement of the p16INK4a/Cdk4/Rb pathway in melanoma development. Moreover, by using targeted mice deficient for other INK4 inhibitors, we show that deletion of p18INK4c but not of p15INK4b confers proliferative advantage to melanocytic tumor growth. These results provide an experimental scenario to study the role of Cdk4 regulation in melanoma and to develop novel therapeutic approaches to control melanoma progression.
Increasing evidence indicates that regulators of the cell cycle including cyclins, cyclin-dependent kinases (Cdks), and Cdk inhibitors either are themselves targets for genetic modification in cancer or are disrupted secondarily by other oncogenic events (1). This evidence has been obtained from studies in human patients or cultured cell lines (2–4) and from the characterization of mouse models carrying genetic alterations in the genes encoding these cell cycle regulators (5, 6). Among them, the INK4a locus, encoding p16INK4a, was identified initially in a screening for melanoma suppressor regions (7), and its alteration is among the most frequent genetic changes found in human tumors (4). Subsequent studies revealed that this locus encodes two different tumor suppressor proteins, p16INK4a and p19ARF (8, 9). p16INK4a is a member of the INK4 family of Cdk4/6 kinase inhibitors that regulate the retinoblastoma protein suppressor pathway (1). In contrast, p19ARF acts on the p53 pathway (1, 10). The fact that tumor-associated mutations frequently affect both INK4a-encoded suppressor proteins as well as the adjacent and highly related INK4b locus, encoding p15INK4b, has led to controversial issues regarding the role of these tumor suppressor pathways in melanoma development (11, 12).
INK4 proteins inhibit complexes formed by the cell cycle kinases Cdk4 and Cdk6 and the D-type cyclins. When active, these complexes phosphorylate the retinoblastoma protein, thereby helping to cancel its growth-repressive functions. Retinoblastoma protein inactivation by Cdks enables E2F proteins to function as transcriptional activators triggering progression of the cell cycle toward the synthesis of DNA (1). Both tumor-associated alterations, cyclin D overexpression and INK4 inactivation, therefore result in Cdk4/6 up-regulation. Cdk4 and Cdk6 also are overexpressed themselves in some human tumors such as sarcomas or leukemias because of amplification or translocation of their respective loci (13, 14). In addition, a specific point mutation in the Cdk4 gene itself has been found in melanoma patients. This point mutation, resulting in the substitution R24C, was found initially in human patients with spontaneous melanoma (15) and was confirmed later in human familiar melanoma (16). Arg-24 is involved in binding to INK4 inhibitors, and biochemical analysis of this interaction showed that the Cdk4 R24C mutant is not able to bind p16INK4a (15). The Cdk4 R24C mutation therefore is presumed to be functionally similar to inactivation of all members of the INK4 family, p16INK4a, p15INK4b, p18INK4c, and p19INK4d, all of which (except for p19INK4d) have been implicated in tumor development (4, 6).
To investigate the contribution of this mutation and the consequential lack of response to INK4 inhibitors to tumor development, we have generated knock-in mice carrying a Cdk4 R24C allele (17). Expression of this mutant allele does not cause major developmental defects in the knock-in mice. Cdk4 R24C mice are, however, 5–20% larger than wild-type controls and develop hyperplasia in some of the endocrine cells such as pancreatic β cells and Leydig cells (17). In addition, Cdk4 R24C mice develop a wide spectrum of tumors within a year (18). However, we have not observed the appearance of spontaneous melanoma even after 18 months of age, when most mutant mice are dead because of other tumors. To analyze the involvement of the Cdk4/INK4 regulatory pathway in melanoma we have subjected Cdk4- and INK4-targeted mice to specific carcinogenic treatments. Here we report that Cdk4 R24C mice are highly susceptible to develop melanoma after carcinogenic insults. In addition, a lack of p18INK4c confers proliferative advantages to melanocytes, whereas deficiency in p15INK4b seems not to affect melanocyte proliferation or transformation in vivo.
Materials and Methods
Mice and Carcinogenic Treatment.
Knock-in mice carrying the Cdk4 R24C allele (17) were maintained in a mixed 129/SvJ × CD-1 background. Mice carrying the mutant allele in heterozygosity (Cdk4+/R24C) or homozygosity (Cdk4R24C/R24C) were used in this study. p15INK4b or p18INK4c null mice (19) as well as INK4aΔ2,3 mutants (20) were maintained in a pure C57BL/6J genetic background. The corresponding 129/SvJ × CD-1 or C57BL/6J control mice were used in all the assays. Seven-day-old mice were painted with a single dose of 0.5 mg of 7,12-dimethylbenz[a]anthracene (DMBA). Two weeks later, tumor growth was promoted by treating with 5 μg of 12-O-tetradecanoylphorbol-13-acetate (TPA) three times a week for 12 weeks. Mice were shaved before each treatment to allow a better distribution of the chemicals. Animals were observed on a daily basis, and the size and characteristics of the skin lesions were annotated. Tumors were measured weekly in two bisecting diameters by using a caliper. Most mice developed multiple lesions. The position and number of each tumor were recorded, and each lesion was followed individually. Tumor incidence is expressed in the text as the percentage of animals with one or more tumors.
Pathological and Molecular Analysis of Tumors.
The status of the INK4a/ARF and INK4b loci and the methylation of the p16INK4a, p19ARF, and p15INK4b promoters were analyzed by Southern blot hybridization as described previously (21, 22). Amplification of Myc was determined by Southern blot hybridization with a probe specific for the murine gene. p53 mutations were analyzed by amplification of exons 4–9 and direct sequencing (18). The presence of mutations in codons 12, 13, and 61 of the H-ras, K-ras, and N-ras genes was analyzed by a PCR-restriction fragment length polymorphism strategy as described (23). Activation of Erk proteins was measured by immunological detection of protein lysates with the anti-active MAPK antibody (Promega) that specifically recognizes the dually phosphorylated forms of Erk1 and Erk2. The level of phosphorylated Erk was compared with the total amount of Erk proteins detected with an antibody (Santa Cruz Biotechnology, clone C-16) that recognizes the phosphorylated and nonphosphorylated forms of Erk1. Cell proliferation was quantified in paraffin sections by using a polyclonal antibody against the Ki67 antigen (NovoCastra, Newcastle, U.K.). Expression of p53, p21Cip1, and p16INK4a was detected by Western blot or immunohistochemistry using antibodies from NovoCastra (p53, clone CM5) and Santa Cruz Biotechnology (p21Cip1, clone C-19, and p16INK4a, clone M-156). The neuroectodermic origin of melanomas was determined by immunostaining with antibodies against the S100 antigen (Dako).
Results and Discussion
Susceptibility to Melanoma of Cdk4 R24C Mice.
Cdk4R24C/R24C mice do not develop visible melanocytic tumors spontaneously (18). To determine whether Cdk4R24C/R24C mice harbor abnormal melanocytes indicative of preneoplastic stages, we measured the abundance of these cells in the skin of adult mice (14 months), a time when tumors are developing readily in other tissues (18). Whereas wild-type mice had 3.6 ± 1.3 melanocytes (cells positive for S100 staining) per high power field (×40), Cdk4R24C/R24C mice had 3.9 ± 2.3, thus indicating that melanocytes do not hyperproliferate in these mutant mice. Accordingly, no significant differences were observed when cells were stained with antibodies against Ki67, because most melanocytes are not dividing either in wild-type or Cdk4R24C/R24C mice (data not shown).
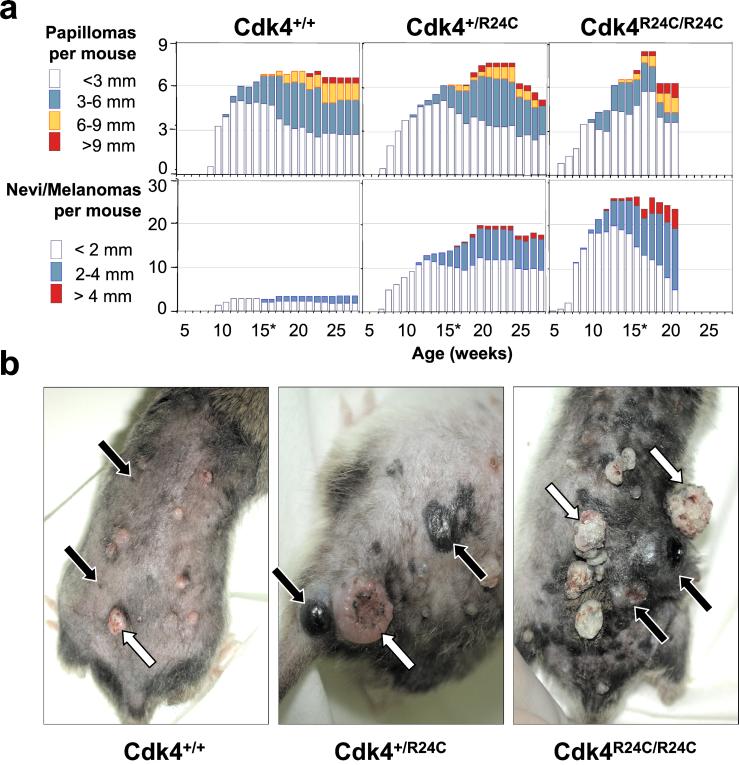
To examine whether the Cdk4 R24C mutation may predispose these mutant mice to melanoma, we used the classical DMBA + TPA two-stage carcinogenic protocol, a treatment known to induce the rapid development of skin papillomas. These tumors are hyperplastic, well differentiated, benign lesions that presumably originate from a single initiated cell. When compared with wild-type littermates, Cdk4 R24C mice develop larger papillomas that frequently progress to skin-invasive carcinomas with shorter latency (Fig. 1). For example, by 23 weeks of age, 15% of Cdk4R24C/R24C mice develop squamous cell carcinomas, whereas no carcinomas are found in wild-type littermates by that age. In addition to skin papillomas, DMBA + TPA treatment also induces the appearance of a few small nevi in the skin of wild-type mice. However, these nevi do not progress to malignant melanoma. In contrast, Cdk4 R24C mice insulted with the same DMBA + TPA carcinogenic treatment develop a high number of nevi that rapidly grow in size, leading to the formation of tumors of more than 60 mm2 by 20 weeks of age (Fig. 1). These lesions are usually dark and have a radial phase of growth, from which they often spread to neighbor papillomas. After the initial radial phase, these dark lesions show a vertical phase of growth frequently accompanied with necrotic images in the center of the lesion (Fig. 1). Alternative treatments with DMBA plus UVB light resulted in the development of similar tumors, albeit with reduced efficiency (data not shown). DMBA treatment alone also resulted in the appearance of nevi in the skin of Cdk4R24C/R24C mice. However, these lesions do not progress frequently to the melanoma stage. No melanocytic tumors were observed after treatment with UVB light alone.
Figure 1.
Development of skin lesions in mice treated with DMBA + TPA. (a) The average number of papillomas (top panels) or macroscopic nevi/melanomas (lower panels) per mouse was scored, grouped by size (diameter of the lesions in mm), and represented for each genotype: Cdk4+/+ (n =13), Cdk4+/R24C (n =13), and Cdk4R24C/R24C (n =20). Because a significant fraction of Cdk4R24C/R24C mice are dead by week 21, the statistical representation of their skin lesions was stopped at week 20. (b) Representative pictures taken at week 20. White arrows indicate papillomas, and black arrows show the position of small nevi (Cdk4+/+) or melanomas (Cdk4+/R24C and Cdk4R24C/R24C).
Histological examination of the skin of Cdk4R24C/R24C mice treated with the DMBA + TPA protocol also revealed increased proliferation of melanocytes even in areas with no visible lesions. Thus, whereas treated Cdk4+/+ mice maintain a density of melanocytes similar to that of untreated animals [≈5.4 ± 2.8 melanocytes per high power field (hpf, ×40)], Cdk4R24C/R24C mice display frequent areas with increased density (12 ± 25.2 melanocytes per hpf) including regions with as many as 60–80 melanocytes per hpf (Fig. 2 a and b). Pathological examination of the skin lesions confirmed the presence of melanomas in 70% of treated Cdk4R24C/R24C mice. Cdk4R24C/R24C animals that do not develop melanomas usually die earlier in the treatment because of the development of lymphomas (20% of cases) or other pathologies. Lymphoma was found also in treated wild-type littermates with an incidence of 13%, thus suggesting that development of this tumor type is not influenced significantly by the Cdk4 R24C mutation.
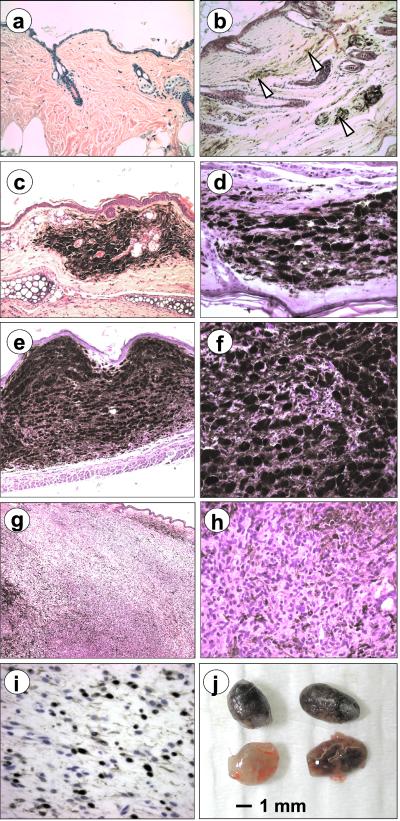
Figure 2.
Representative sections of nevi and melanomas in Cdk4 R24C mice. Hematoxylin and eosin images of Cdk4+/+ (a) and Cdk4R24C/R24C (b) mice after treatment with the DMBA + TPA protocol are shown. At this magnification (×200), melanocytes are barely visible in wild-type mice but are abundant in Cdk4R24C/R24C (arrow heads). (c and d) Small benign lesions similar to human blue nevus. (e and f) Cellular lesions with compacted groups of spindle-shaped melanocytes with a small load of melanin. (g and h) Invasive melanomas with evident loss of melanin content and infiltration to nearby tissues. (i) Melanocytic origin of advanced lesions was determined by positive immunostaining for the S100 antigen. Original magnifications: a and c, ×100; b, d, and f, ×400; e, ×45; g, ×630. (j) Enlarged lymph nodes with high melanin content commonly found in Cdk4R24C/R24C mice with primary melanomas.
The small nevi observed in Cdk4 R24C mice are typically 1–3 mm in diameter and are localized dermal lesions composed of heavily pigmented melanocytes admixed with numerous large polyhedral macrophages fully loaded with melanin (Fig. 2). After a few weeks, cell density increases because the appearance of groups of spindle-shaped epithelioid cells closely packed with a minor load of cytoplasmic melanin or even amelanotic cells. These nevic cells show occasional large atypical nuclei, but mitotic figures are uncommon. In the more advanced melanomas, the tumor is highly cellular, composed mainly of proliferating atypical melanocytes frequently spindle-shaped with low or no load of melanin (Fig. 2). Mitotic figures and areas of necrosis can be found easily. In these cases, melanocyte proliferation is usually invasive to neighbor tissues with infiltrating margins. The neuroectodermal origin of the melanomas was confirmed by positive immunological staining for the S100 antigen (Fig. 2). The highly melanotic nature of these multiple tumors makes them highly reminiscent of human familial atypical mole melanoma syndrome. Moreover, as observed in human melanomas, the ratio of melanotic cells decreases in the more advanced malignancies, because active proliferation and melanocyte dedifferentiation often results in decreased production of melanin. After necropsy, the draining and regional lymph nodes were found commonly to be enlarged and pigmented. However, we have been unable to detect melanocytic metastases in distal organs. Thus, it is possible that melanomas in Cdk4 R24C mice, albeit invasive to neighboring tissues, may not have distal metastatic potential. Alternatively, the early death of these mice, likely caused by the concomitant development of sarcomas and carcinomas (data not shown), may prevent distal progression of the invasive melanoma cells. To overcome this problem, we currently are obtaining mice that express the Cdk4 R24C knock-in allele specifically in melanocytes.
Cooperative Alterations in Melanocytic Lesions of Cdk4 R24C Mice.
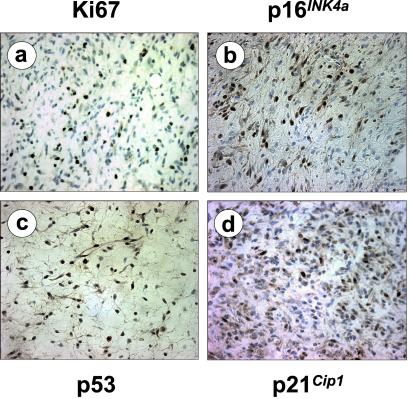
The development of melanoma in Cdk4 R24C mice provides an opportunity to assess whether a loss of the INK4a locus is essential for melanoma genesis in mice. Southern blot analysis of Cdk4 R24C mouse melanomas failed to detect deletion, rearrangement, or promoter methylation in p16INK4a, p15INK4b, and p19ARF genes. Similarly, immunological analysis of p16INK4a expression showed positive staining in all tumors analyzed, suggesting that loss of this inhibitor is not needed for melanoma development in Cdk4 R24C mice (Fig. 3). The presence of p19ARF in these tumors suggests that alteration of the p53 pathway is not needed for induction and/or progression of these melanomas. Although we did not find any mutations in the p53 locus (exons 4–9) in these tumors, we examined the expression pattern of p53 as well as of one of its main targets, p21Cip1. As illustrated in Fig. 3, immunohistochemistry analysis showed a normal pattern of expression for p53 and p21Cip1. These observations correlate well with the low incidence of p53 alterations in primary or metastatic human melanomas (24).
Figure 3.
Representative staining of Cdk4R24C/R24C melanomas against Ki67 proliferative antigen (a), p16INK4a (b), p53 (c), and p21Cip1 (d). Original magnification is ×400.
We have searched for other genetic alterations that could cooperate with the Cdk4 R24C mutation in inducing invasive melanoma. Among other molecular markers frequently involved in melanoma development, we observed the presence of Ras mutations (CAA/CTA and CAA/AAA transversions in codon 61 of H-ras and N-ras, respectively) in 2 of 20 melanomas (10% incidence) analyzed for mutations in codons 12, 13, or 61 of the H-ras, K-ras, and N-ras genes. In contrast, we observed a 70% incidence of the same CAA/CTA transversion in codon 61 of H-ras in papillomas of both wild-type and Cdk4 mutant mice, a frequency described often for this carcinogenic protocol. The point mutation observed in the N-ras allele has not been linked previously to the DMBA + TPA treatment. However, it corresponds to the Ras mutation found most frequently in human melanoma. Analysis of the level of Erk activation using phosphorylated-specific antibodies revealed additional samples with increased Erk phosphorylation (2 of 6 tumors), suggesting that the Ras/Raf/Mek/Erk pathway could be altered by mechanisms that do not involve mutations in ras loci.
Although Myc amplification is a commonly found alteration in melanoma development, we have not observed c-Myc amplification in a panel of 12 Cdk4 R24C melanomas. These results contrast with those obtained in Ras-induced melanomas in p53 mutant mice, in which Myc amplification is observed in ≈50% of samples (25). We currently are pursuing the analysis of other candidate molecules in the p53 tumor suppressor pathway such as Apaf-1, recently implicated in the induction of apoptosis in melanoma cells and in their response to chemotherapy (26).
Involvement of INK4 Inhibitors in Melanoma Susceptibility.
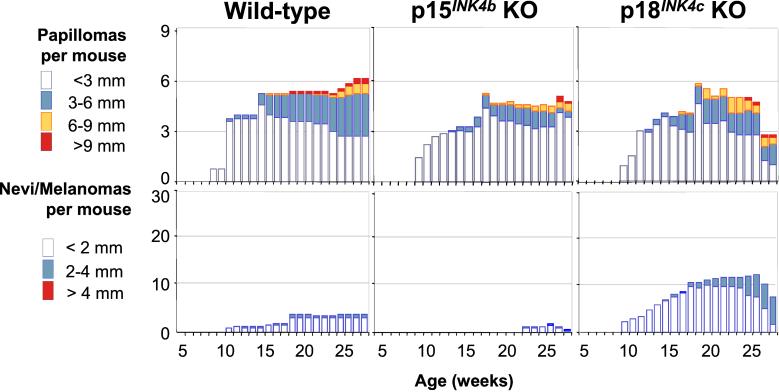
Because Cdk4 R24C is insensitive to inhibition by all members of the INK4 family, our results do not define which INK4 molecule(s) confers susceptibility to melanoma. On the basis of results obtained with human tumors, the obvious candidate is p16INK4a (4, 7, 11, 24). However, these results do not rule out that other INK4 inhibitors also might contribute to melanoma development. To address this question, we applied the same DMBA + TPA carcinogenic treatment to mice deficient in p15INK4b or p18INK4c (19). INK4aΔ2,3 mice, deficient in both p16INK4a and p19ARF (20), were used also. However, DMBA + TPA treatment of INK4aΔ2,3 mice resulted in increased susceptibility to lymphoma, and all treated animals died by 10–12 weeks of age (data not shown). Because of their early death, no skin lesions could be evaluated in these mice. The p15INK4b tumor suppressor gene maps very closely to the INK4a-ARF locus, and in fact 80% of deletions of the INK4a locus also affect p15INK4b sequences (4, 7). When insulted with the DMBA + TPA protocol, p15INK4b null mice do not show increased susceptibility to papilloma or melanoma development (Fig. 4). In contrast, p18INK4c-deficient mice submitted to the same carcinogenic treatment displayed an increased number of melanocytic lesions that reach larger size than wild-type control mice. Twenty percent of these lesions could be classified as early stage melanomas based on histological characteristics. In general, these tumors were similar to those observed in Cdk4 R24C but with a less aggressive phenotype and slower growth rate (Fig. 5). These results do not seem to be caused by an overall tumor susceptibility, because p18INK4c-deficient mice do not show increased numbers of papillomas after DMBA + TPA treatment (Fig. 4). Most treated p18INK4c null mice died by 50 weeks of age, mainly of lymphomas (70% incidence) and sarcomas (17% incidence). These malignancies correspond to tumor types known to develop spontaneously in p18INK4c null mice albeit with longer latency (19). The early death of these mice prevented us from studying the putative progression of these melanocytic lesions into more aggressive or metastatic tumors.
Figure 4.
Development of papillomas and melanocytic lesions in INK4-deficient mice. The average number of skin lesions per mouse is represented, and the same conventions described for Fig. 1 are maintained. The following genotypes were analyzed: wild type (n = 9), p15INK4b null (p15INK4b KO, n = 8), and p18INK4c null (p18INK4c KO, n =15). INK4a-ARF null mice (n =14) were treated also, but these animals died before 12 weeks without obvious skin lesions and are not represented in the figure.
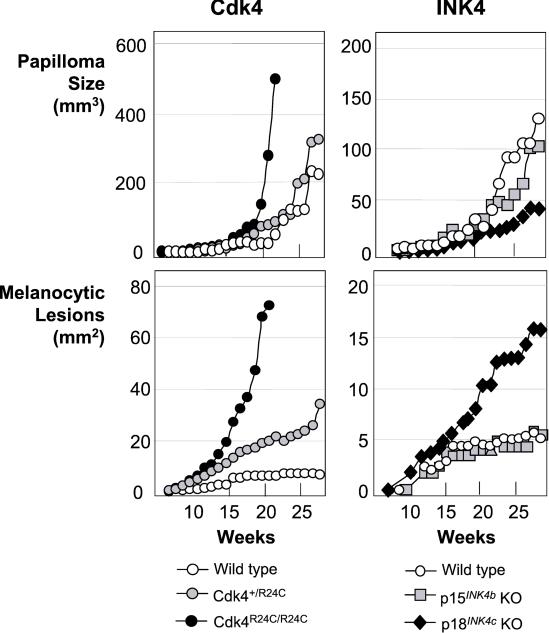
Figure 5.
Growth kinetics of papillomas or melanocytic lesions in Cdk4- (Left) or INK4- (Right) targeted mice. Tumor volume (mm3) or area (mm2) was scored and represented against time, because the lesion appears in the skin (0 weeks). Because melanocytic lesions are flat in the early stages, the area is represented, and the vertical phase of growth therefore is not considered. Please note that the size of lesions is represented at a different scale for Cdk4- and INK4-targeted mice.
Conclusions
The different levels of melanoma susceptibility in Cdk4R24C/R24C and p18INK4b null mice (Figs. 1 and 4) suggest that other members of the INK4 family also must play a role in preventing melanoma development. As illustrated in this (Fig. 4) and a previous study (19), a loss of p15INK4b, which frequently occurs in human melanoma as a consequence of deletions that target the INK4a locus, does not contribute to melanoma. Likewise, a loss of p19INK4d, the fourth member of the INK4 family, does not result in tumor development (27). Therefore, it is likely that, as illustrated previously in human tumors, p16INK4a plays an important role in preventing melanoma in mice.
While this article was being revised, the phenotype of p16INK4a null mice that retain p19ARF expression has been published (28, 29). These mice show very low susceptibility to spontaneous tumor development including a single melanoma in 39 p16INK4a−/− mice (29). DMBA treatment of p16INK4a null mice results in a low incidence of melanocytic tumors (7% in ref. 29). Thus, it would be interesting to determine whether melanoma development in Cdk4 R24C mice is mediated only by a lack of p16INK4a activity or whether there is partial compensation by other members of the INK4 family. Our observations indicating that p18INK4c null mice have a certain predisposition to melanoma development makes this protein a likely candidate to play a role in suppressing melanoma. Heterozygosity of p19ARF considerably increases melanoma incidence in p16INK4a mutant mice (28). Cdk4 R24C mice carrying either homozygous or heterozygous deletion of the p53 locus do not develop melanomas spontaneously (18). Whether this result is because of the short lifespan of these animals (3.3 months for Cdk4R24C/R24C;p53−/− and 9.5 months for Cdk4R24C/R24C;p53+/− mice; ref. 18 and unpublished observations) or to a specific role of p19ARF in melanoma suppression needs to be examined.
A loss of Cdk4 regulation is not sufficient to elicit uncontrolled melanocyte proliferation (ref. 18 and this study). Thus, DMBA + TPA treatment must contribute to the generation of additional mutations that cooperate with Cdk4 R24C in eliciting melanoma. This carcinogenic protocol is known to induce the frequent activation of H-ras oncogenes in skin tumors (30). Yet in melanocytes, ras loci do not seem to be frequent targets of DMBA. Thus, it will be very important to determine the full spectrum of these DMBA + TPA-induced mutations for both initiation as well as progression of the melanocytic tumors observed in these animals. In any event, Cdk4 R24C knock-in mice should provide an excellent model system to study the molecular biology of melanoma progression. Moreover, these mice should be of value to those interested in the development of melanoma vaccine protocols and in evaluating the activity of cell cycle inhibitors, two of the most promising venues for the discovery of novel antitumor agents.
Acknowledgments
We thank Ernesto de la Cueva, Maribel Muñoz, and Ignacio Segovia for technical expertise, M. Serrano for the INK4aΔ2,3 mice, and L. Sánchez for help with immunostaining. R.S. and J.M. were supported by fellowships from the Fondo de Investigaciones Sanitarias (Madrid). This work was supported by grants from the Association pour la Recherche contre le Cancer (to P.D.), Fundación Pzifer, V Framework Program of the European Union (to M.B.), and Comunidad Autónoma de Madrid (to M.M.).
Abbreviations
- Cdk
cyclin-dependent kinase
- DMBA
7,12-dimethylbenz[a]anthracene
- TPA
12-O-tetradecanoylphorbol-13-acetate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sherr C J. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- 2.Hirama T, Koeffler H P. Blood. 1995;86:841–854. [PubMed] [Google Scholar]
- 3.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 4.Ruas M, Peters G. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 5.Roussel M F. Oncogene. 1999;18:5311–5317. doi: 10.1038/sj.onc.1202998. [DOI] [PubMed] [Google Scholar]
- 6.Malumbres M, Ortega S, Barbacid M. Biol Chem. 2000;381:827–838. doi: 10.1515/BC.2000.105. [DOI] [PubMed] [Google Scholar]
- 7.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian S V, Stockert E, Day R S, III, Johnson B E, Skolmick M H. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 8.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 9.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 10.Sharpless N E, DePinho R A. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 11.Chin L, Merlino G, DePinho R A. Genes Dev. 1998;12:3467–3481. doi: 10.1101/gad.12.22.3467. [DOI] [PubMed] [Google Scholar]
- 12.Bardeesy N, Wong K K, DePinho R A, Chin L. Adv Cancer Res. 2000;79:123–156. doi: 10.1016/s0065-230x(00)79004-x. [DOI] [PubMed] [Google Scholar]
- 13.Khatib Z A, Matsushime H, Valentine M, Shapiro D N, Sherr C J, Look A T. Cancer Res. 1993;53:5535–5541. [PubMed] [Google Scholar]
- 14.Corcoran M M, Mould S J, Orchard J A, Ibbotson R E, Chapman R M, Boright A P, Platt C, Tsui L-C, Scherer S W, Oscier D G. Oncogene. 1999;18:675–684. doi: 10.1038/sj.onc.1203033. [DOI] [PubMed] [Google Scholar]
- 15.Wolfel T, Hauer M, Schneider J, Serrano M, Wolfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Buschenfelde K H, Beach D. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 16.Zuo L, Weger J, Yang Q, Goldstein A M, Tucker M A, Walker G J, Hayward N, Dracopoli N C. Nat Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 17.Rane S G, Dubus P, Mettus R V, Galbreath E J, Boden G, Reddy E P, Barbacid M. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 18.Sotillo, R., Dubus, P., Martín, J., de la Cueva, E., Ortega, S., Malumbres, M. & Barbacid, M. (2001) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 19.Latres E, Malumbres M, Sotillo R, Martín J, Ortega S, Martín-Caballero J, Flores J M, Cordón-Cardo C, Barbacid M. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano M, Lee H-W, Chin L, Cordón-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 21.Malumbres M, Pérez de Castro I, Santos J, Meléndez B, Mangues R, Serrano M, Pellicer A, Fernández-Piqueras J. Oncogene. 1997;14:1361–1370. doi: 10.1038/sj.onc.1200969. [DOI] [PubMed] [Google Scholar]
- 22.Meléndez B, Malumbres M, Pérez de Castro I, Santos J, Pellicer A, Fernández-Piqueras J. Carcinogenesis. 2000;21:817–821. doi: 10.1093/carcin/21.4.817. [DOI] [PubMed] [Google Scholar]
- 23.Mangues R, Kahn J M, Seidman I, Pellicer A. Cancer Res. 1994;54:6395–6401. [PubMed] [Google Scholar]
- 24.Meier F, Satyamoorthy K, Nesbit M, Hsu M Y, Schttek B, Garbe C, Herlyn M. Front Biosci. 1998;3:d1005–d1010. doi: 10.2741/a341. [DOI] [PubMed] [Google Scholar]
- 25.Bardeesy N, Bastian B C, Hezel A, Pinkel D, Depinho R A, Chin L. Mol Cell Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soengas M S, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman J G, Gerald W L, Lazebnik Y A, et al. Nature (London) 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 27.Zindy F, van Deursen J, Grosveld G, Sherr C J, Roussel M F. Mol Cell Biol. 2000;20:372–378. doi: 10.1128/mcb.20.1.372-378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krimpenfort P, Quon K C, Mooi W J, Loonstra A, Berns A. Nature (London) 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 29.Sharpless N E, Bardeesy N, Lee K-H, Carrasco D, Castrillon D H, Aguirre A J, Wu E A, Horner J W, DePinho R A. Nature (London) 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 30.Balmain A, Pragnell I B. Nature (London) 1983;303:72–74. doi: 10.1038/303072a0. [DOI] [PubMed] [Google Scholar]