Summary
The mitochondrial unfolded protein response (UPRmt) can be triggered in a cell-non-autonomous fashion across multiple tissues in response to mitochondrial dysfunction. The ability to communicate information about the presence of mitochondrial stress enables a global response that can ultimately better protect an organism from local mitochondrial challenges. We find that animals use retromer-dependent Wnt signaling to propagate mitochondrial stress signals from the nervous system to peripheral tissues. Specifically, the polyQ40-triggered activation of mitochondrial stress or reduction of cco-1 (complex IV subunit) in neurons of C. elegans results in the Wnt-dependent induction of cell-non-autonomous UPRmt in peripheral cells. Loss-of-function mutations of retromer complex components responsible for recycling the Wnt secretion-factor/MIG-14 prevent Wnt secretion and thereby suppress cell-non-autonomous UPRmt. Neuronal expression of the Wnt ligand/EGL-20 is sufficient to induce cell-non-autonomous UPRmt in a retromer complex-, Wnt signaling-, and serotonin-dependent manner, clearly implicating Wnt signaling as a strong candidate for the ‘mitokine’ signal.
Keywords: Mitochondrial unfolded protein response (UPRmt), Retromer complex, VPS-35, Wnt signaling, Mitokine
TOC image
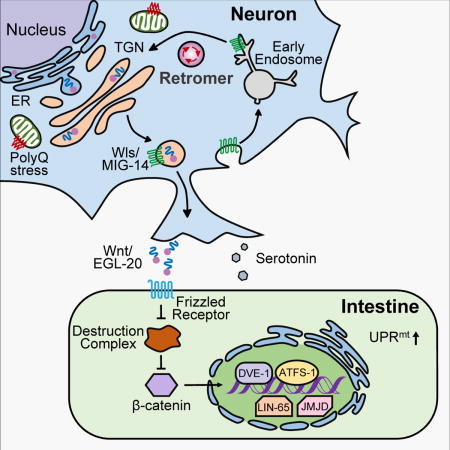
The Wnt/EGL-20 relays mitochondrial stress signals from neurons to peripheral tissues.
Introduction
Metazoan evolution required functional monitoring and communication of cellular stress across cells and tissues. The development of the nervous system plays a vital role in organismal homeostasis and coordinates diverse processes ranging from metabolism to circadian control of sleep. However, neurons appear to be one of the cell types most susceptible to the challenges of proteotoxic stress. How and why neurons succumb to the adverse effects of neurodegenerative diseases caused by the imbalance of mal-folded proteins such as Aβ, α-synuclein, and polyglutamine repeats found in the huntingtin protein, while other non-neuronal cell types do not, is unknown. Even less well-understood is the mechanism for the age onset decline in processes responsible for the proper handling of the proteome and the proteostasis network, allowing the imbalance of misfolded and toxic proteins that leads to neurodegeneration (Labbadia and Morimoto, 2015). However, many age onset neurodegenerative diseases are accompanied by peripheral maladies that present as metabolic abnormalities and deficiencies in non-neuronal tissues (Cai et al., 2012; Duarte et al., 2013).
Studies in C. elegans have established that expression of the Huntington’s disease-causing polyglutamine expansion protein (Q40) in neurons results not only in neuronal detriment for the animal, but also peripheral decline in metabolic homeostasis, muscle function, and lifespan (Berendzen et al., 2016; Morley et al., 2002). Surprisingly, it is also associated with the induction of a protective stress response, the mitochondrial unfolded protein response (UPRmt), which causes global alteration of transcription networks to maintain a functional mitochondrial proteome during challenges (Berendzen et al., 2016). Neuronal expressed Q40 associates with mitochondria, causing a local activation of the UPRmt that is further communicated extracellularly across the animal to promote UPRmt activation in non-neuronal, non-innervated cells and tissues. This process requires intact UPRmt machinery, the active participation of serotonin, and the release of dense core vesicles (Berendzen et al., 2016). While serotonin is necessary for the cell-non-autonomous communication of UPRmt stress response, it is not sufficient, suggesting other secreted factors might also be responsible for this signaling event.
Within C. elegans, the UPRmt is activated when the transcription factor ATFS-1 translocates from mitochondria to the nucleus in response to mitochondrial perturbations (Nargund et al., 2012). A number of other proteins work together with ATFS-1 during UPRmt activation, including the mitochondrial matrix protease ClpP, ubiquitin-like protein UBL-5, and the transcription factor DVE-1 (Benedetti et al., 2006; Haynes et al., 2007). ATFS-1 and DVE-1/UBL-5 induce the expression of genes involved in mitochondrial quality control and cellular metabolism to restore proteostasis within mitochondria (Nargund et al., 2015). More recent studies have demonstrated the role of epigenetic regulation in UPRmt induction. Two histone lysine demethylases (JMJD-1.2 and JMJD-3.1) that modify H3K27me2/me3 sites are necessary and sufficient for UPRmt induction (Merkwirth et al., 2016). Chromatin remodeling is mediated by the MET-2/LIN-65 histone methyltransferases, which regulate UPRmt-associated transcriptional networks (Tian et al., 2016).
Within metazoan, the induction of the UPRmt can be coordinated across multiple tissues, preparing the entire organism to better cope with a locally sensed stress (Higuchi-Sanabria et al., 2018). For example, UPRmt induction resulting from the knockdown of the mitochondrial electron transport chain (ETC) subunit cco-1 (Cytochrome c oxidase-1) in neurons is sensed and then reacted to by mitochondria located in the intestine, a physically distinct, non-innervated tissue, resulting in beneficial effects for the whole organism, including increased lifespan (Durieux et al., 2011). Further, the neuron-specific expression of the Q40 protein in C. elegans causes cell-non-autonomous induction of UPRmt in the intestine (Berendzen et al., 2016).
Collectively, these findings have led to the mechanistic hypothesis that a factor, termed a mitokine, is generated in neurons experiencing mitochondrial stress, and is secreted, propagated, and subsequently perceived by cells of peripheral tissues to regulate organismal mitochondrial homeostasis. Humans with mitochondrial diseases, such as IOSCA (Infantile onset spinocerebellar ataxia), suffer from muscle debilitation and produce excess levels of FGF21, a cytokine that enters and circulates in the blood (Suomalainen et al., 2011). Flies with disrupted muscle mitochondrial function have elevated levels of ImpL2 (an ortholog to human insulin-like growth factor binding protein 7), which systemically antagonizes insulin signaling, and prolongs lifespan (Owusu-Ansah et al., 2013). However, the molecular nature of a mitokine first perceived in the nervous system and sufficient to induce the mitochondrial stress response in distal cells has remained unknown.
Here, to identify a putative mitokine, we characterized C. elegans EMS-mutants that are defective in cell-non-autonomous UPRmt signaling but retain the ability for cell-autonomous UPRmt induction. Intriguingly, within the 16 mutants identified, we found VPS-35, a retromer component, is required for the cell-non-autonomous UPRmt induction through retrieval of the Wnt secretion factor, MIG-14. We further identified that the Wnt ligand, EGL-20, is not only essential, but also sufficient for cell-non-autonomous UPRmt induction. Collectively, EGL-20 presents itself as a strong candidate to be a mitokine.
Results
The retromer complex is required for cell-non-autonomous UPRmt induction
Neuronal expression of a polyglutamine repeat protein of 40 repeats (Q40) is sufficient to induce cell-non-autonomous UPRmt signaling in peripheral, non-neuronal cells of C. elegans (Berendzen et al., 2016). UPRmt activation can be monitored by measuring the expression of the transcriptional UPRmt reporter hsp-6p::gfp (Brignull et al., 2006). We performed an EMS mutagenesis screen using the hsp-6p∷gfp reporter to identify mutants that suppress UPRmt in peripheral cells of animals expressing neuronal Q40. Subsequently, we evaluated whether the mutants identified in this screen retained their ability to induce UPRmt in response to cell-autonomous mitochondrial stress, such as cco-1 RNAi delivered by bacterial feeding (Durieux et al., 2011; Houtkooper et al., 2013). Screening of 2,400 mutagenized genomes with these criteria yielded 16 mutant strains and uth13, the strain with the strongest suppression phenotype, was further characterized (Figure S1A).
Single nucleotide polymorphism (SNP) mapping and whole genome deep sequencing indicated that the uth13 carries a mutation in the vps-35 gene at the codon for amino acid 523 (CAA-TAA [Gln-Stop]) (Figure S1C). Using the deletion allele vps-35(ok1880), we confirmed that vps-35 was required for the induction of the hsp-6p∷gfp reporter in the peripheral cells of animals expressing Q40 in neurons (Figures 1A and 1B). Western blot analysis confirmed that both vps-35(uth13) and vps-35(ok1880) mutant animals lost hsp-6p∷gfp expression without any alteration of neuronal Q40 expression levels (Figures 1C and S1B). Furthermore, the induction of cell-autonomous UPRmt in vps-35 mutant animals was not affected when animals were treated with the cell autonomous stressor cco-1 RNAi via bacterial feeding (Figures 1D and 1E). While multiple challenges to mitochondrial proteostasis in neurons can activate cell-non-autonomous UPRmt, knockdown of the mitochondrial ETC subunit cco-1 in neurons also activates UPRmt in the peripheral tissue (Durieux et al., 2011). Intriguingly, we found that the vps-35 mutation also suppressed the induction of hsp-6p∷gfp expression in animals with neuronal cco-1 knockdown (Figures 1F and 1G).
Figure 1. The retromer complex is required for cell-non-autonomous UPRmt induction in animals expressing Q40∷YFP in neurons.
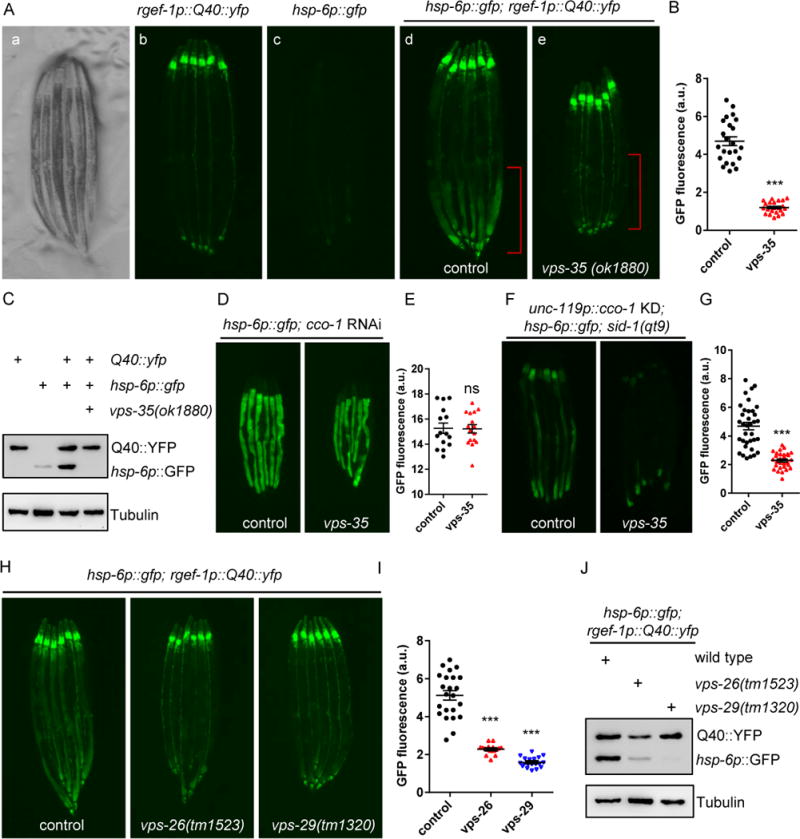
(A) Representative photomicrographs demonstrating: a) bright field images of aligned, WT animals; b) Q40∷yfp expression in neurons; c) hsp-6p∷gfp expression in WT animals; d) hsp-6p∷gfp was up-regulated in the intestine in day2 adult animals expressing neuronal Q40∷YFP; e) hsp-6p∷gfp expression was suppressed in vps-35 mutants. The posterior region of the intestine where hsp-6p∷gfp is induced or suppressed is highlighted in (d) and (e). Scale bar: 250μm.
(B) Quantification of hsp-6p∷gfp expression of the entire intestine in animals expressing Q40∷YFP in neurons with the presence (d) or absence of the vps-35 mutation (e) as shown in (A).
(C) Immunoblots of GFP expression in animals as indicated.
(D) Representative photomicrographs of hsp-6p∷gfp animals with the presence or absence of the vps-35 mutation grown on empty vector (EV) or with cco-1 RNAi from hatching.
(E) Quantification of hsp-6p∷gfp expression. The genotypes are as in (D).
(F) Representative photomicrographs of neuronal cco-1 knockdown; sid-1(qt9); hsp-6p∷gfp animals with the presence or absence of vps-35 mutation.
(G) Quantification of hsp-6p∷gfp expression. The genotypes are as in (F).
(H) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, vps-26, and vps-29 animals.
(I) Quantification of hsp-6p∷gfp expression. The genotypes are as in (H).
(J) Immunoblots of GFP expression in animals as indicated.
YFP can be recognized by the GFP antibody. Anti-tubulin serves as a loading control. *** p < 0.0001, ns denotes p > 0.05 via t-test. Error bars, SEM. n ≥15 worms.
See also Figure S1.
The non-autonomous nature of the unfolded protein response is not confined to the UPRmt; the UPRER and UPRCyt also function in a cell-non-autonomous manner to coordinate stress responses across different tissues. For example, ectopic activation of the UPRER in neurons, by overexpression of XBP-1s, activates the expression of hsp-4p∷gfp reporter, a marker of UPRER expression, in the intestine of C. elegans (Ron and Walter, 2007; Taylor and Dillin, 2013). Similarly, overexpression of the heat shock transcription factor, HSF-1, in the nervous system induces the expression of the sod-3p∷gfp reporter, a target gene of DAF-16, in the intestine (Douglas et al., 2015; Link et al., 1999). However, neither neuronal induction of the UPRER or UPRCyt activates the UPRmt.
We found that vps-35 was not involved in either the cell-autonomous or the cell-non-autonomous induction of the UPRER or UPRCyt (Figures S1D-S1G). Therefore, vps-35 plays an essential and specific role in cell-non-autonomous UPRmt communication. vps-35 encodes an essential subunit of the retromer complex, a highly conserved multi-subunit complex that mediates the retrograde transport of cargo between endosomes and the trans-Golgi network (Bonifacino and Rojas, 2006). We additionally found that animals carrying mutations in two additional core retromer components (vps-26 and vps-29) (Hierro et al., 2007) exhibited suppression phenotypes in animals expressing neuronal Q40 similar to those of the vps-35 mutants (Figures 1H–1J). Thus, the retromer complex, and not solely vps-35, functions in the regulation of cell-non-autonomous UPRmt signaling from the nervous system to the periphery.
Retrieval of the Wnt secretion factor, MIG-14, by the retromer complex is required for cell-non-autonomous UPRmt signaling
The retromer complex mediates at least two essential aspects of cellular biology: the regulation of mitochondrial dynamics and the retrieval of various cargo proteins from endosomes to the trans-Golgi network (e.g., glutamate, TGF-beta, Wnt, and phagocytic secretion factors) (Belenkaya et al., 2008; Chen et al., 2010; Gleason et al., 2014; Zhang et al., 2012). We first tested the role of the retromer complex in mitochondrial dynamics. Here, VPS35 facilitates the degradation of the dynamin-like protein, DLP1, via the mitochondrial-derived vesicle pathway (Wang et al., 2016). We examined the effects of a loss-of-function mutation and of overexpression of drp-1, the C. elegans DLP1 homolog (Labrousse et al., 1999). The drp-1(tm1108) mutant and the drp-1 overexpression strain both induced the expression of hsp-6p∷gfp reporter and did not suppress the induction of cell-non-autonomous UPRmt signaling in Q40 animals (Figures S2A and S2C). Likewise, the removal of mitochondrial fusion proteins eat-3 and fzo-1 also induced UPRmt and did not suppress the induction of cell-non-autonomous UPRmt of neuronal expressed Q40 animals (Figure S2B). Therefore, the role of the retromer complex in mitokine signaling does not appear to be linked to mitochondrial dynamics.
We next tested the retrieval function of the retromer complex, which is critical for efficient recycling of signaling receptors to allow proper transcellular signal transduction. We hypothesized that the observed loss of cell-non-autonomous UPRmt signaling in the vps-35 mutant was due to a defect in the retromer-mediated recycling of receptors that function in the cell-non-autonomous mitochondrial stress signaling pathway. We initially characterized receptors known to be retrieved by the retromer complex and found that the Wnt secretion factor, MIG-14, was required for induction of the hsp-6p∷gfp reporter in animals expressing Q40 in neurons (Figures 2A and 2B). In contrast, the glutamate receptor GLR-1, the phagocytic receptor CED-1, and the TGF-beta receptor SMA-6, which are also retrieved by the retromer complex, are not involved in cell-non-autonomous UPRmt signaling (Figure S2C).
Figure 2. Retromer-dependent Wnt secretion is required for cell-non-autonomous UPRmt induction in animals expressing Q40∷YFP in neurons.
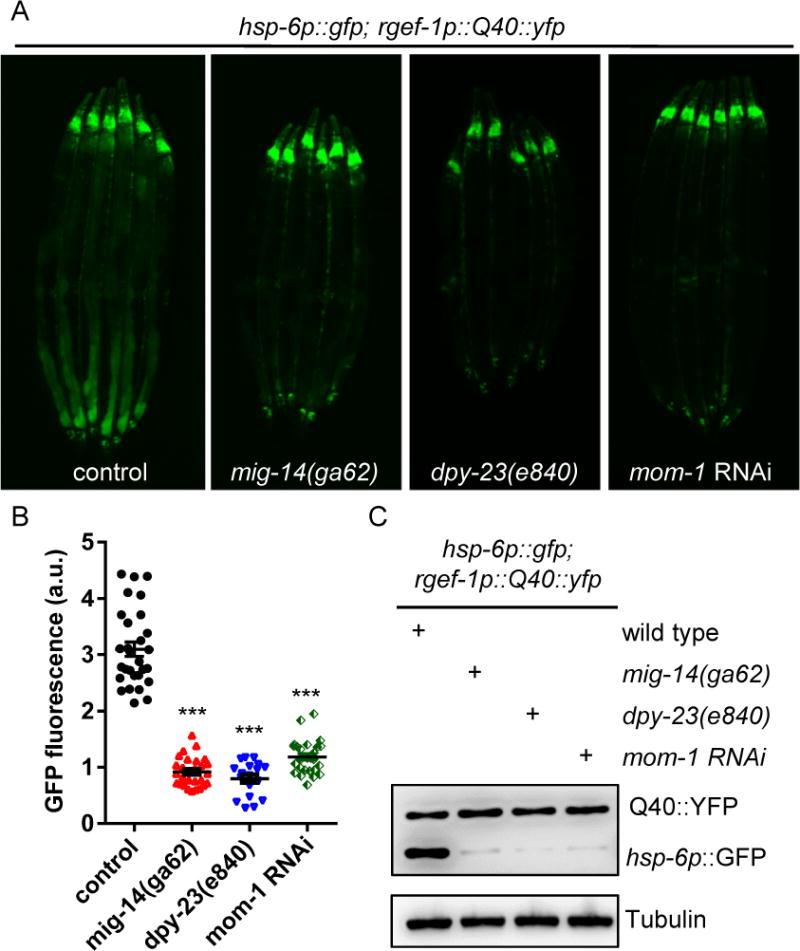
(A) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, mig-14, dpy-23, or mom-1 RNAi animals.
(B) Quantifications of the hsp-6p∷gfp expression as shown in (A). *** p < 0.0001 via t test. Error bars, SEM. n ≥20 worms.
(C) Immunoblots of GFP expression in animals as indicated.
See also Figure S2.
Studies across multiple organisms have demonstrated that Wnt ligands are post-translationally modified with lipids (Clevers and Nusse, 2012). Lipidation of Wnt ligands occurs in the ER and is catalyzed by the enzyme porcupine (mom-1 in C. elegans) (Rocheleau et al., 1997). The Wnt secretion factor, MIG-14, can then bind these modified Wnt ligands and transport them from the Golgi to the cell surface (Bänziger et al., 2006). After Wnt ligands have been released to the extracellular space, MIG-14 becomes internalized via the clathrin adaptor DPY-23/AP2-mediated endocytosis pathway (Pan et al., 2008). Subsequently, the retromer-dependent retrieval pathway delivers MIG-14 back to the Golgi, where it becomes available for further Wnt secretion (Yang et al., 2008). We found that loss of the DPY-23/AP2 adaptor in dpy-23 mutants resulted in suppression of the mitokine signaling that phenocopied the mig-14 mutant animals (Figures 2A and 2B). We also found that lipid modification of Wnt is essential for the mitokine signaling induction: mom-1 RNAi strongly suppressed induction of the hsp-6p∷gfp reporter in neurons of Q40 animals (Figures 2A and 2B). Western blot analysis verified that hsp-6p∷GFP levels were dramatically reduced in mig-14, dpy-23, and mom-1 mutants as compared to wild type (WT) animals expressing neuronal Q40 (Figure 2C). Moreover, we observed no differences in the induction of cell-autonomous UPRmt when we fed cco-1 RNAi bacteria to mig-14, dpy-23, and mom-1 mutants compared to control animals (Figure S2D). Furthermore, we overexpressed mig-14 in vps-35 mutant animals expressing neuronal Q40 and found that mig-14 overexpression partially rescued the hsp-6p∷gfp suppression phenotypes in vps-35 mutants. Our observation that excess MIG-14 ameliorated vps-35 suppression on UPRmt induction is consistent with the prediction that retromer-dependent MIG-14 recycling is essential for mitokine signaling (Figure S2E).
The Wnt ligand, EGL-20, is required for cell-non-autonomous UPRmt signaling
We performed a candidate screen against all known Wnt ligands in C. elegans (egl-20, lin-44, cwn-1, cwn-2, and mom-2) (Gm et al., 1993; Herman et al., 1995; Maloof et al., 1999; Rocheleau et al., 1997; Thorpe et al., 1997) for their role in UPRmt signaling. egl-20(n585) was the only Wnt ligand mutation able to suppress the induction of cell-non-autonomous UPRmt signaling of animals expressing neuronal Q40 (Figures 3A and 3B). The other Wnt ligand mutants did not affect either the cell autonomous or cell-non-autonomous UPRmt induction (Figures S3A-S3C). Western blot analysis verified that hsp-6p∷GFP levels were dramatically reduced in egl-20 mutants compared to the control animals expressing neuronal Q40 (Figure 3C). Furthermore, we measured the endogenous hsp-6 mRNA levels and found that it was also dramatically reduced by the loss of egl-20 in the neuronal expressed Q40 animals (Figure S3D).
Figure 3. Wnt ligand/EGL-20 is required for cell-non-autonomous UPRmt induction in animals with Q40∷YFP expression in neurons.
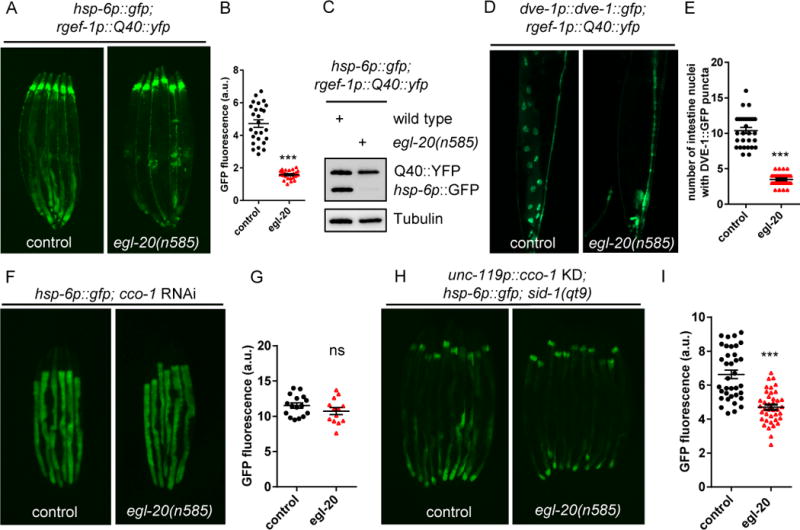
(A) Representative photomicrographs of D2 adult animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT or egl-20 animals.
(B) Quantification of hsp-6p∷gfp expression. The genotypes are as in (A).
(C) Immunoblots of GFP expression. The genotypes are as in (A).
(D) Representative photomicrographs of D2 adult animals expressing neuronal Q40∷YFP; DVE-1∷GFP in WT and egl-20 animals. Arrows highlight the DVE-1∷GFP signal localized in the intestinal nuclei.
(E) Quantification of the number of intestinal nuclei with DVE-1 puncta. The genotypes are as in (D).
(F) Representative photomicrographs of D1 animals expressing hsp-6p∷gfp in WT or egl-20 animals grown on EV or cco-1 RNAi from hatching.
(G) Quantification of hsp-6p∷gfp expression. The genotypes are as in (F).
(H) Representative photomicrographs of hsp-6p∷gfp expression in neuronal specific cco-1 RNAi animals in WT or egl-20 background.
(I) Quantification of hsp-6p∷gfp expression. The genotypes are as in (H).
*** p< 0.0001, ns denotes p > 0.05 via t-test. Error bars, SEM. n ≥ 15 worms.
See also Figure S3.
To further measure UPRmt induction, we followed the nuclear localization of the DVE-1 transcription factor using the DVE-1∷GFP fusion protein reporter. DVE-1 translocates to the nucleus in response to mitochondrial stress to induce the UPRmt (Haynes et al., 2007). We observed robust nuclear accumulation of DVE-1∷GFP in the intestinal cells of neuronal Q40 animals, but only moderate DVE-1∷GFP nuclear accumulation in egl-20 mutants (Figures 3D and 3E). In contrast, when cco-1 RNAi producing bacteria were fed to the egl-20 mutants, the hsp-6p∷gfp induction was comparable to that of wild type animals (Figures 3F and 3G). Therefore, egl-20 is not required for cell autonomous UPRmt signaling, much like the retromer complex, DPY-23/AP2, and MIG-14. Of the five Wnt ligands found in C. elegans, EGL-20 is the only Wnt ligand that can act across the entire length of the animal, whereas the other four act more proximal to their site of secretion (Hardin and King, 2008). Furthermore, we found that the egl-20 mutant also strongly suppressed the induction of cell-non-autonomous UPRmt upon neuronal cco-1 RNAi (Figures 3H and 3I). Taken together, EGL-20 is specifically required for the cell-non-autonomous UPRmt signaling from the nervous system to the periphery.
Neuronal expression of egl-20 is sufficient for the induction of mitokine signaling
Because egl-20 is required for cell-non-autonomous communication of the UPRmt, we tested whether tissue-specific expression of egl-20 was sufficient to induce the UPRmt cell-non-autonomously. To this end, we generated transgenic strains expressing egl-20 with pan-neuronal promoter (rgef-1p, two independent lines) or in the intestine (gly-19p, two independent lines). Intriguingly, expression of egl-20 in either neurons or the intestine was sufficient for the induction of hsp-6p∷gfp reporter and for the nuclear translocation of DVE-1∷GFP reporter (Figures 4A–4F, S4A, S4B, S4C, and S4E). Likewise, expression of egl-20 in the Wnt-producing cells was sufficient to induce cell-non-autonomous UPRmt signaling (Figures 4G, 4H, and 4I). However, expression of egl-20 in the body wall muscle (myo-3p), pharyngeal muscle (myo-2p), or hypodermal cells (lin-26p) failed to activate the expression of the hsp-6p∷gfp reporter (Figure S4D). Furthermore, neither the hsp-4p∷gfp reporter (UPRER) nor the hsp-16.2p∷gfp reporter (UPRCyt) were induced by neuronal or intestinal Wnt/EGL-20 expression, indicating that expression of Wnt/EGL-20 had a specific effect on mitokine signaling (Figures S4F and S4G).
Figure 4. Expression of Wnt ligand/EGL-20 is sufficient to induce UPRmt in both cell-autonomous and cell-non-autonomous manners.
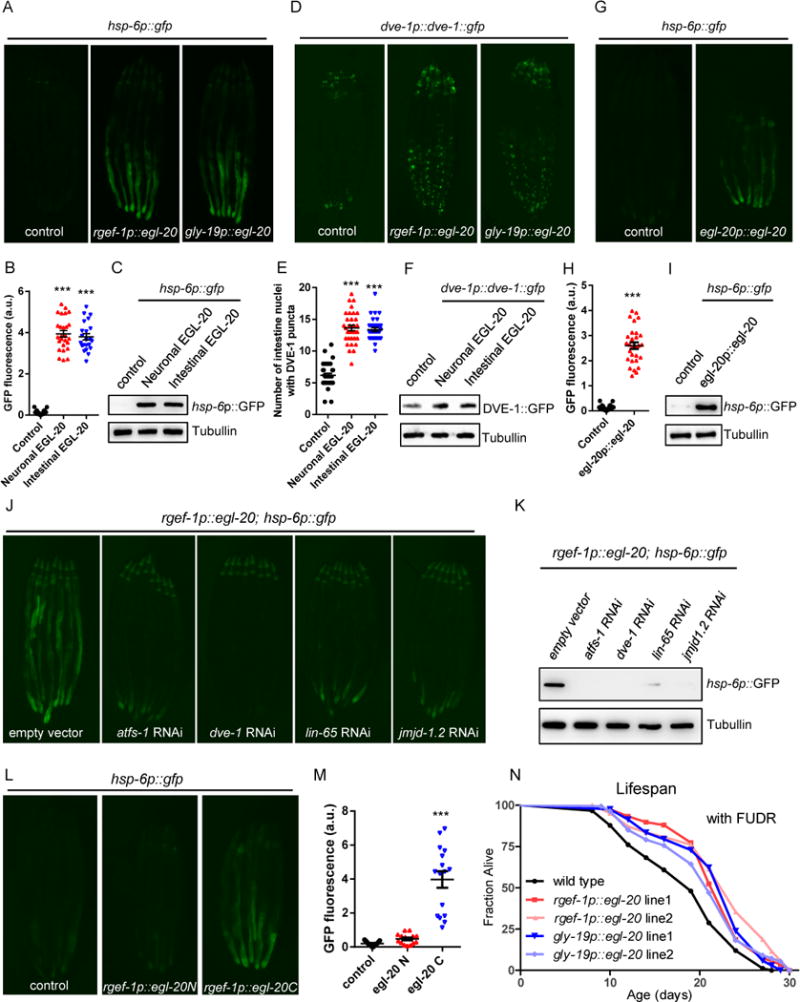
(A) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT, rgef-1p∷egl-20 (neuronal), and gly-19p∷egl-20(intestinal) background.
(B) Quantification of hsp-6p∷gfp expression. The genotypes are as in (A).
(C) Immunoblot of hsp-6p∷gfp expression. The genotypes are as in (A).
(D) Representative photomicrographs of DVE-1∷GFP reporter animals in WT, rgef-1p∷egl-20, and gly-19p∷egl-20 background.
(E) Quantification of the number of intestinal nuclei with DVE-1 puncta. The genotypes are as in (D).
(F) Immunoblot of DVE-1∷GFP expression. The genotypes are as in (D).
(G) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT or egl-20p∷egl-20∷mCherry background.
(H) Quantification of hsp-6p∷gfp expression. The genotypes are as in (G).
(I) Immunoblot of hsp-6p∷gfp expression. The genotypes are as in (G).
(J) Representative photomicrographs of rgef-1p∷egl-20; hsp6p∷gfp reporter animals grown on EV, atfs-1, dve-1, lin-65, or jmjd-1.2 RNAi from hatching.
(K) Immunoblot of hsp-6p∷gfp expression. The genotypes are as in (J).
(L) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT, rgef-1p∷egl-20N(1-720)∷mCherry, or rgef-1p∷egl-20C(721-1182)∷mCherry background.
(M) Quantification of hsp-6p∷gfp expression. The genotypes are as in (L).
(N) Overexpression of egl-20 in neurons or in the intestine extends C. elegans lifespan. Lifespan analysis of two independent transgenic lines of rgef-1p∷egl-20 or gly-19p∷egl-20 expressing animals compared to WT animals. See also Table S1.
*** p< 0.0001 via t-test. Error bars, SEM. n ≥ 20 worms.
See also Figure S4.
Ectopic induction of mitokine signaling by egl-20 expression required core components of the UPRmt response. RNAi against atfs-1 or dve-1 strongly suppressed hsp-6p∷gfp induction in both neuronal and intestinal egl-20 overexpression animals (Figures 4J, 4K, S4I, and S4J). Additionally, RNAi against the epigenetic factors lin-65 and the demethylase jmjd-1.2, both of which specifically function in UPRmt signaling (Merkwirth et al., 2016; Tian et al., 2016), also strongly suppressed hsp-6p∷gfp induction in both neuronal and intestinal egl-20 expression animals (Figures 4J, 4K, S4I, and S4J).
The crystal structure of the Xenopus Wnt8 (XWnt8) in complex with mouse Frizzled-8 (Fz8) cysteine-rich domain (CRD) reveals a two-domain Wnt structure which comprises an N-terminal α-helical domain (NTD) that contains the lipid-modified sites and a C-terminal cysteine-rich domain (CTD, also termed “mini-Wnt”) that autonomously engages the Fz8-CRD in a receptor-specific manner (Janda et al., 2012). Interestingly, neuronal expression of the C-terminal end of EGL-20 (mini-Wnt) was sufficient for hsp-6p∷gfp induction, whereas N-terminal EGL-20 failed to induce hsp-6p∷gfp expression (Figures 4L and 4M).
To characterize the physiological function of egl-20 overexpression, we performed mitochondrial morphology analyses. Mitochondria formed more fragmented structures in muscle cells of the posterior region of animals with egl-20 overexpression than in the WT control animals using the myo-3p∷GFP(mit) reporter (Figure S4H). Furthermore, both neuronal and intestinal egl-20 expressing animals induced greater lifespan extension than did the WT animals (Figure 4N and Table S1).
β-Catenin and the TCF transcription factor are required for cell-non-autonomous UPRmt signaling
Wnt ligands engage with various receptors to activate multiple downstream signaling pathways. Wnt signaling pathways can be classified as either canonical (β-catenin-dependent) or non-canonical (β-catenin-independent) (Komiya and Habas, 2008). In the absence of a Wnt ligand, the destruction complex (known as APC/Axin/CK1/GSK3β) phosphorylates cytosolic β-catenin, ultimately resulting in the degradation of β-catenin by the proteasome. Binding of a Wnt ligand to a Frizzled/LRP-5/6 receptor causes the recruitment of the destruction complex leading to the stabilization of β-catenin. Stabilized β-catenin can then enter the nucleus and interact with the TCF/LEF transcription factor to activate the expression of Wnt target genes (Clevers, 2006).
To determine which downstream signaling pathways of Wnt activation are involved in mitokine signaling, we analyzed the requirement for each of the Wnt receptors. The C. elegans genome encodes multiple genes for Wnt-Frizzled receptors, including lin-17, mom-5, mig-1, and cfz-2 (Rocheleau et al., 1997; Ruvkun and Hobert, 1998; Sawa et al., 1996). Using the mig-1(e1787) mutation, we found that loss of mig-1, though not of the other Wnt receptor mutants, resulted in significantly lower expression of the hsp-6p∷gfp reporter in neuronal Q40 animals (Figures 5A, 5B, 5C, and S5A). Furthermore, RNAi-mediated knockdown of bar-1(β-catenin) or pop-1(TCF/LEF transcription factor) resulted in strong suppression of hsp-6p∷gfp induction in the animals expressing neuronal Q40 (Figures 5A, 5B, and 5C). The roles of mig-1, bar-1, and pop-1 appear specific to mitokine signaling because mutations in any of the three genes did not block induction of hsp-6p∷gfp induction when animals were fed cco-1 RNAi bacteria (Figure S5B). To test whether stabilization of β-catenin was sufficient to induce UPRmt, we disrupted the activity of pry-1, a component of the destruction complex that targets β-catenin for degradation in the absence of Wnt signaling (Korswagen et al., 2002). As expected, we observed nuclear accumulation of the DVE-1∷GFP fusion protein in pry-1 RNAi animals (Figures S5C and S5D). Furthermore, disruption of the E3 ligase lin-23, which is required for β-catenin degradation (Dreier et al., 2005), resulted in robust ectopic induction of the hsp-6p∷gfp reporter (Figures S5E and S5F).
Figure 5. The canonical Wnt/β-catenin signaling pathway is required for cell-non-autonomous UPRmt induction in animals expressing Q40∷YFP in neurons.
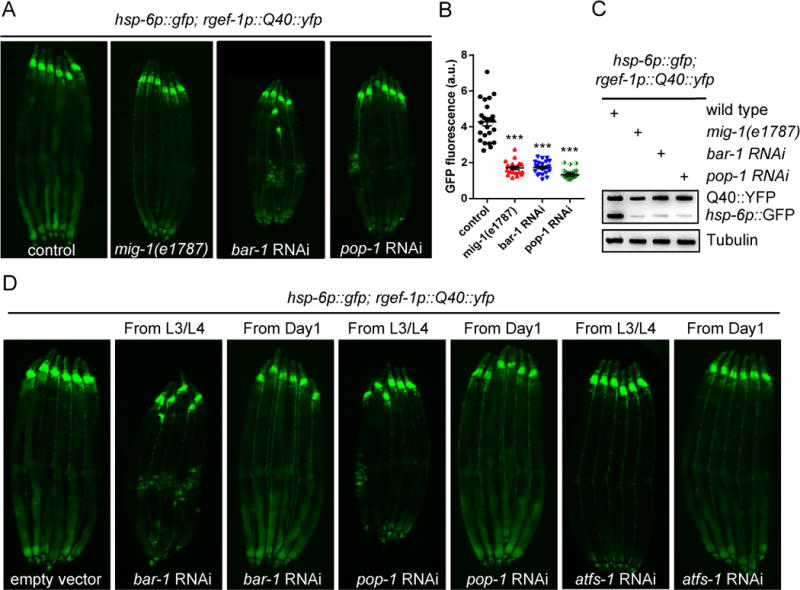
(A) Representative photomicrographs of neuronal Q40∷YFP; hsp-6p∷gfp expressing animals in WT, mig-1, bar-1 RNAi, or pop-1 RNAi background.
(B) Quantification of hsp-6p∷gfp expression. The genotypes are as in (A). *** p< 0.0001 via t-test. Error bars, SEM. n ≥ 20 worms.
(C) Immunoblots of GFP expression. The genotypes are as in (A).
(D) Representative photomicrographs of neuronal Q40∷YFP; hsp-6p∷gfp expressing animals transferred to RNAi plates either from L3/L4 stages or D1 adulthood.
See also Figure S5.
To understand whether Wnt signaling is required during development to mediate cell-non-autonomous UPRmt induction, we tested the timing requirement of bar-1, pop-1, and atfs-1 in animals expressing Q40 in neurons. Animals in which RNAi was induced from the L3/L4 stages during late development were able to strongly suppress cell-non-autonomous UPRmt induction. In contrast, animals treated with RNAi during adulthood failed to suppress UPRmt induction. These results indicate that a critical timing window during the L3/L4 stages of early development is essential for cell-non-autonomous UPRmt induction (Figure 5D). This observation is very consistent with previous timing requirements found for mitokine signaling (Dillin et al., 2002; Durieux et al., 2011; Merkwirth et al., 2016).
Induction of cell-non-autonomous UPRmt signaling upon neuronal Wnt/EGL-20 expression requires retromer and canonical Wnt signaling
The retromer complex enables Wnt molecules to be secreted for long range transduction of Wnt signals (Coudreuse et al., 2006). We tested whether VPS-35 is also required for cell-non-autonomous UPRmt activation when EGL-20 is expressed in neurons. Intriguingly, in the vps-35 mutant, the induction of UPRmt was strongly suppressed in animals expressing egl-20 in neurons but was unaffected in animals expressing egl-20 in the intestine (Figures 6A–6F). Notably, bar-1 and pop-1, two downstream regulators of Wnt signaling, are required for the induction of UPRmt signaling in animals with neuronal or intestinal expression of Wnt/EGL-20 (Figures 6A–6F). Collectively, these results indicate that canonical Wnt/β-catenin signaling is required for both local and long range induction of EGL-20-induced mitokine signaling, while retromer activity is only essential for long range UPRmt signaling originating from the nervous system.
Figure 6. The induction of cell-non-autonomous UPRmt signaling upon neuronal Wnt/EGL-20 expression is dependent on the retromer complex and canonical Wnt signaling.
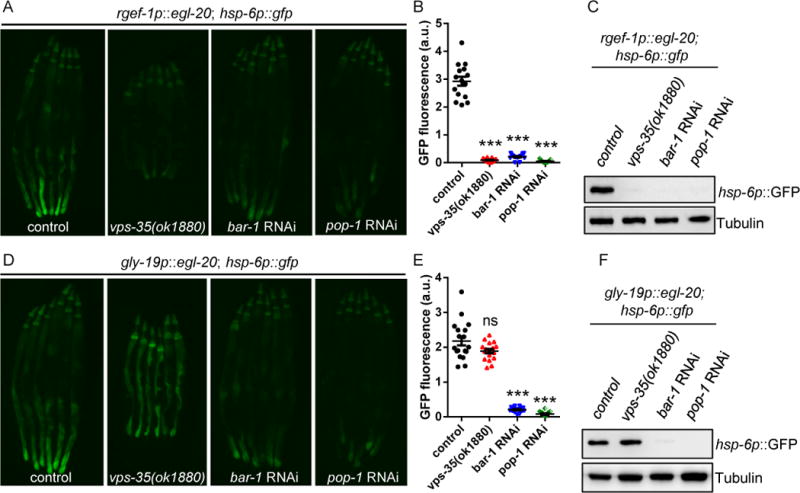
(A) Representative photomicrographs of rgef-1p∷egl-20; hsp-6p∷gfp reporter animals in WT, vps-35, bar-1 RNAi, or pop-1 RNAi background.
(B) Quantification of hsp-6p∷gfp expression. The genotypes are as in (A).
(C) Immunoblot of GFP expression. The genotypes are as in (A).
(D) Representative photomicrographs of gly-19p∷egl-20; hsp-6p∷gfp reporter animals in WT, vps-35, bar-1 RNAi, or pop-1 RNAi background.
(E) Quantification of hsp-6p∷gfp expression. The genotypes are as in (D).
(F) Immunoblot of GFP expression. The genotypes are as in (D).
*** p< 0.0001 via t-test. Error bars, SEM. n ≥ 15 worms.
See also Figure S6.
To identify which tissue is essential for mediation of cell-non-autonomous UPRmt by the retromer complex, we performed rescue experiments to induce vps-35 expression using tissue-specific promoters in the vps-35 mutant animals. Expression of vps-35 in Wnt/EGL-20 producing cells strongly rescued the suppression of the hsp-6p∷gfp reporter in the vps-35 mutants with neuronal Q40 expression (Figures S6A and S6B). However, expression of vps-35 in neurons or in the intestine failed to restore cell-non-autonomous UPRmt signaling in the vps-35 mutants (Figures S6A and S6B). Collectively, these results suggest that the retromer complex is required in Wnt-producing cells to mediate mitokine signaling.
Serotonin is essential for EGL-20 mitokine signaling
Serotonin can define non-autonomous from autonomous UPRmt induction. Consistent with the role of serotonin for cell-non-autonomous UPRmt induction in animals expressing Q40 in neurons (Berendzen et al., 2016), mutations in tryptophan hydroxylase (tph-1), a key enzyme for serotonin synthesis, completely suppressed the induction of the hsp-6p∷gfp reporter in the animals expressing egl-20 in neurons (Figures 7A, 7B, and 7C), whereas it only slightly suppressed the induction of the hsp-6p∷gfp reporter in the animals expressing egl-20 in the intestine (Figures 7D, 7E, and 7F). Importantly, the addition of serotonin rescued the suppression caused by the tph-1 mutation in animals expressing egl-20 in neurons (Figures 7G, 7H, 7I and 7J). Serotonin released from neurons may exert its function by targeting a serotonin receptor. We therefore tested mutations within the four serotonin receptors (ser-1, ser-4, ser-7, and mod-1) found in C. elegans (Chase, 2007) and they had no effect on cell-non-autonomous UPRmt signaling (Figures S7A and S7B). Moreover, expression of egl-20 solely in serotonergic neurons was sufficient to induce mitokine signaling in the periphery (Figures 7K and 7L).
Figure 7. Serotonin is required for the cell-non-autonomous UPRmt induction upon neuronal Wnt/EGL-20 expression.
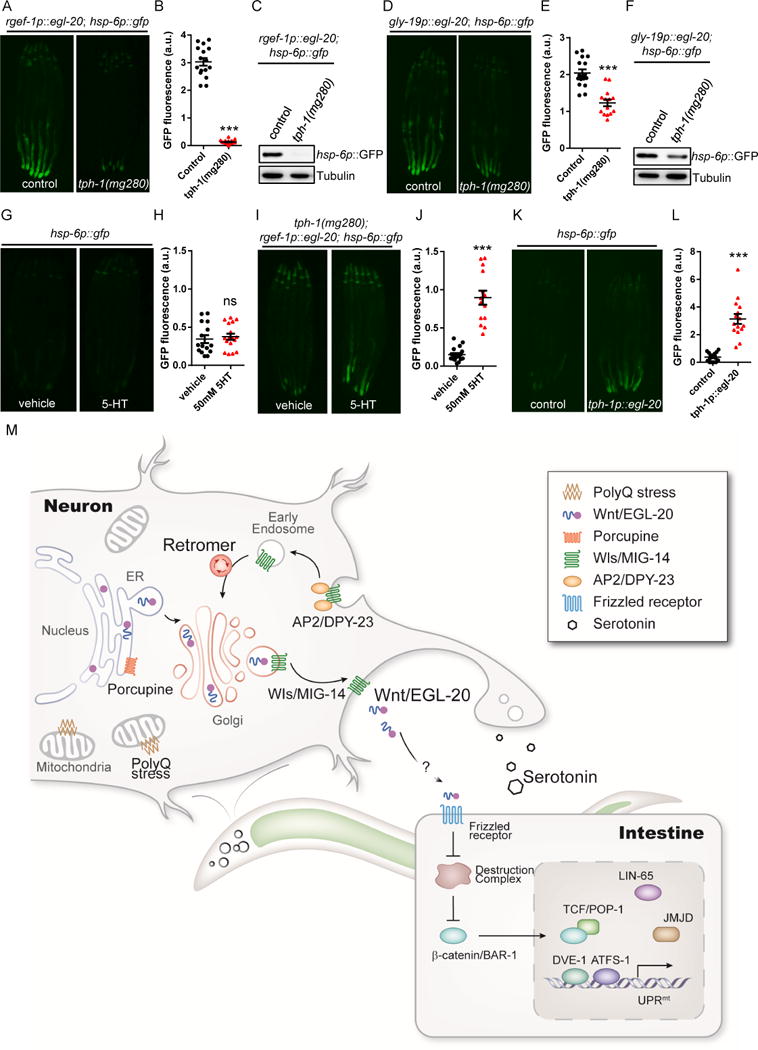
(A) Representative photomicrographs of rgef-1p∷egl-20; hsp-6p∷gfp animals in WT or tph-1 background.
(B) Quantification of hsp-6p∷gfp expression. The genotypes are as in (A).
(C) Immunoblot of hsp-6p∷gfp expression. The genotypes are as in (A).
(D) Representative photomicrographs of gly-19p∷egl-20; hsp-6p∷gfp transgenic animals in WT or tph-1 background.
(E) Quantification of hsp-6p∷gfp expression. The genotypes are as in (D).
(F) Immunoblot of hsp-6p∷gfp expression. The genotypes are as in (D).
(G) Representative photomicrographs of hsp-6p∷gfp expression in animals treated with vehicle control or 50mM serotonin (5-HT).
(H) Quantification of hsp-6p∷gfp expression. The genotypes are as in (G).
(I) Representative photomicrographs of hsp-6p∷gfp expression in neuronal Q40; tph-1 expressing animals treated with vehicle control or 50mM 5-HT.
(J) Quantification of hsp-6p∷gfp expression. The genotypes are as in (I).
(K) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT and tph-1p∷egl-20 background.
(L) Quantification of hsp-6p∷gfp expression. The genotypes are as in (K).
(M) Model of the mitokine signaling pathway. Q40 specifically binds to mitochondria in neurons, initiating a signaling cascade across tissues that requires retromer-dependent Wnt secretion, canonical Wnt signaling, serotonin, and functional components of the UPRmt to ensure cell-non-autonomous UPRmt induction in peripheral tissues.
*** p< 0.0001 via t-test. Error bars, SEM. n ≥ 15 worms.
See also Figure S7.
Discussion
The development and survival of multicellular organisms relies to a large extent on the ability of cells to communicate via extracellular signals, such as the well-characterized Wnt signaling pathway (Nusse and Clevers, 2017). This pathway directs processes as diverse as embryonic cell division, limb development, and neuronal patterning and migration (Wodarz and Nusse, 1998), as well as the regulation of stem cell maintenance and differentiation (Reya and Clevers, 2005). Aberrant Wnt activation has been implicated in several types of cancer (Logan and Nusse, 2004). Additionally, it has been proposed that Wnt signaling functions in a protective role against neurodegenerative diseases (Inestrosa and Arenas, 2010). However, very few reports have linked Wnt signaling to mitochondrial function (Yoon et al., 2010). We found that retromer-dependent Wnt signaling is required for the induction of cell-non-autonomous UPRmt from the nervous system to the periphery.
Given the large number of proteins known to depend upon the retromer complex for endosomal sorting, it is not surprising that retromer dysfunction has been implicated in a number of human diseases, including forms of Alzheimer’s and Parkinson’s diseases (Wang and Bellen, 2015). The present study broadens our understanding of the functional roles of the retromer complex in transducing intercellular signals by demonstrating that the retromer complex is directly involved in a cell-non-autonomous mitochondrial stress response—UPRmt.
EGL-20 is the only Wnt in C. elegans that has been demonstrated to signal over a long distance (Hardin and King, 2008; Whangbo and Kenyon, 1999). During early larval development, EGL-20 is expressed by a group of cells located at the posterior end of the animal (Gleason et al., 2006). The retromer complex and MIG-14 function in EGL-20-producing cells to allow the formation of an EGL-20 gradient along the anteroposterior axis during the larval stages (Coudreuse et al., 2006) (Figure S7C). However, it remains unclear whether neuronal mitochondrial stress can activate Wnt signaling and induce UPRmt over a distance. Due to the very low expression level of egl-20 in C. elegans, no significant differences were observed in expression levels of either the endogenous egl-20 mRNA level or the expression of an egl-20p∷egl-20∷mCherry translational reporter in animals expressing Q40 in neurons as compared to WT animals (Figures S7D, S7E and S7H). Additionally, we found that some Wnt target genes (Gorrepati et al., 2015; Jackson et al., 2014) are strongly up-regulated in animals expressing neuronal Q40 (Figure S7F). Notably, our results appear to suggest that the overall level of Wnt/EGL-20 might not be up-regulated upon mitochondrial stress; however, either transient or undetectable local up-regulation of Wnt signaling could still be occurring in cells experiencing mitochondrial stress or the localization/secretion of EGL-20 could also be affected during neuronal mitochondrial stress.
While EGL-20/Wnt expression is limited in early larval stages, its potential expression in neurons has not been extensively explored. Here, we carefully examined the distribution of EGL-20. The egl-20p∷mCherry reporter showed the co-localization of the signal with the ventral nerve cord in the posterior region of C. elegans (Figure S7G). Furthermore, the EGL-20∷mCherry fusion protein formed aggregates along or co-localized with the ventral and dorsal nerve cord (Figure S7G), indicating a potential function for Wnt/EGL-20 in neurons.
Wnt-receptor interactions can elicit a variety of intracellular responses, the best-known of which results in the activation of β-catenin, which was demonstrated to regulate mitokine signaling in this study. β-catenin is frequently found to be hyperactivated in most intestinal cancers and in a variety of other maladies. Further studies of the interactions between β-catenin and the key regulators of the UPRmt pathway may elucidate new therapeutic interventions to cancers.
The mitokine hypothesis was put forth to explain how RNAi of mitochondrial ETC complex in the nervous system could coordinate whole body aging and induction of the UPRmt. Further studies on this potentially unique signaling system determined parameters for such a mitokine molecule. The mitokine should depend upon serotonin and the downstream UPRmt factors as well as the newly identified LIN-65 chromatin remodeling complex (Tian et al., 2016). Furthermore, the mitokine should be sufficient to induce UPRmt even in the absence of mitochondrial stress, be able to originate from nerve tissues, and able to act at a distance. Our analysis and mutant screen show that egl-20, known as Wnt16b in humans, follows these mandates. One of the most difficult rules to comply with is that of sufficiency. Few molecules responsible for stress induction are sufficient. Such molecules include HSF1, controlling the heat shock response, xbp-1s for the ER stress response, and PHF-8 dictating the UPRmt response, all of which are transcription factors or chromatin remodelers. Therefore, the description of a Wnt ligand which can play a necessary, specific, and sufficient role in mitokine signaling provides important knowledge in the biology of transcellular stress responsiveness.
In summary, we discovered that the retromer complex is essential for cell-non-autonomous UPRmt, and our subsequent identification and characterization of the proteins recycled by the retromer complex demonstrated that the Wnt signaling pathway is also required for this cellular response to mitochondrial stress. Further, we found that Wnt expression is sufficient to induce the UPRmt in both a cell-autonomous and a cell-non-autonomous manner, thereby identifying Wnt as a strong candidate for the proposed “mitokine” signal. In keeping with the recently-discovered role of the neurotransmitter serotonin in mitochondrial function, we also found that serotonin is essential for the induction of cell-non-autonomous UPRmt in response to neuronal Wnt signaling (Figure 7M). Ultimately, our study provides strong evidence that retromer-dependent Wnt signaling regulates the induction of the UPRmt in the peripheral tissues, suggests that Wnt signaling regulates proteostasis, and indicates that Wnt pathway components should thus be viewed as therapeutic targets for age-related neurodegenerative diseases.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Carenorhabdities elegans Maintenance and Transgenic Lines
The following strains used in this study were obtained from the Caenorhabditis Genome Center (Minneapolis, MN): Bristol (N2) strain as wild-type (WT) strain, SJ4100 (zcIs13[hsp-6p∷gfp] V), AM101 (rmIs110[rgef-1p∷Q40∷yfp]), SJ4005 (zcIs4[hsp-4p∷gfp] V), CL2070 (dvIs70[hsp-16.2p∷gfp + rol-6(su1006)]), SJ4197 (zcIs39[dve-1p∷dve-1∷gfp]), VC1390 (vps-35(ok1880) II), EW12 (mig-14(ga62) II), CB40 (dpy-23(e840) X), KP4 (glr-1(n2461) III), CB3203 (ced-1(e1735) I), MT1215 (egl-20(n585) IV), RB763 (cwn-1(ok546) II), VC636 (cwn-2(ok895) IV), MT5383 (lin-44(n1792) I), CB3303 (mig-1(e1787) I), PS1403 (lin-17(sy277) I), MT15434 (tph-1(mg280) II).
The following strains used in this study were obtained from the the National BioResource Project (Tokyo, Japan): vps-26(tm1523) IV, vps-29(tm1320) III, drp-1(tm1108) IV.
The following strains used in this study were generated in our lab: ythIs6[egl-20p∷mCherry + rol-6], ythIs8[egl-20p∷egl-20∷mCherry + rol-6], ythIs1[gly-19p∷egl-20 + myo-2p∷tdTomato], ythIs2[gly-19p∷egl-20 + myo-2p∷tdTomato], ythIs3[rgef-1p∷egl-20 + myo-2p∷tdTomato], ythIs4[rgef-1p∷egl-20 + myo-2p∷tdTomato], ythEx4[egl-20p∷egl-20∷mCherry + rol-6], ythEx47[rgef-1p∷yfp], LTY109(ythEx22[egl-20p∷vps-35 + myo-2p∷tdTomato]; vps-35(ok1880) II; rmIs110[rgef-1p∷Q40∷yfp]; zcIs13[hsp-6p∷gfp]), LTY183 (ythEx43[gly-19p∷vps-35 + myo-2p∷tdTomato]; vps-35(ok1880) II; rmIs110[rgef-1p∷Q40∷yfp]; zcIs13[hsp-6p∷gfp]), LTY238 (ythEx51[rgef-1p∷vps-35 + myo-2p∷tdTomato]; vps-35(ok1880) II; rmIs110[rgef-1p∷Q40∷yfp]; zcIs13[hsp-6p∷gfp]), LTY181(ythEx43[myo-2p∷egl-20∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY177(ythEx39[myo-3p∷egl-20∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY120(ythEx33[tph-1p∷egl-20∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY114(ythEx27[tph-1p∷egl-20∷mCherry + rol-6]; zcIs39[dve-1p∷dve-1∷gfp]), LTY195(ythEx44[lin-26p∷egl-20∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY169(ythEx35[rgef-1p∷egl-20N∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY170(ythEx36[rgef-1p∷egl-20C∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]), LTY110(ythEx23[rgef-1p∷egl-20∷mCherry + rol-6]; zcIs39[dve-1p∷dve-1∷gfp]), LTY118(ythEx31[gly-19p∷egl-20∷mCherry + rol-6]; zcIs39[dve-1p∷dve-1∷gfp]), LTY135(ythEx34[rgef-1p∷egl-20∷mCherry + rol-6]; zcIs13[hsp-6p∷gfp]; egl-20(n585) IV), LTY245 (ythEx56[drp-1p∷mCherry∷drp-1 + myo-2p∷tdTomato]; rmIs110[rgef-1p∷Q40∷yfp]; zcIs13[hsp-6p∷gfp]), LTY248 (ythEx60[mig-14p∷mig-14∷mCherry + myo-2p∷tdTomato]; zcIs13[hsp-6p∷gfp]; vps-35(ok1880) II).
Nematodes were maintained and experimentally examined at 20℃ on standard nematode growth medium agar plates seeded with Escherichia coli OP50.
For generation of egl-20p∷mCherry strain, the egl-20 1.8 kb promoter was PCR amplified from genomic DNA and cloned into pNB23(sur-5p∷mCherry) using SphI and XmaI. For the egl-20p∷egl-20∷mCherry strain, the egl-20 cDNA (1179 bp) without stop codon was inserted into egl-20p∷mCherry construct with XmaI and XbaI. To replace egl-20 promoter, the rgef-1, gly-19, lin-26, myo-2, myo-3 and tph-1 promoter was cloned in place of the egl-20 promoter in the egl-20p∷egl-20∷mCherry plasmid. To replace egl-20 cDNA with half of egl-20, we use KOD-plus-Mutagenesis Kit to generate egl-20N (720bp) and egl-20C (462bp) plasmid.
Transgenic strains were generated by microinjecting target constructs (50ng/ul) mixed with a pRF4(rol-6) (50ng/ul) or a myo-2p∷tdTomato (25ng/ul) co-injection maker. Integrated lines were generated using UV irradiation and backcrossed six times.
METHOD DETAILS
RNAi Feeding
Age synchronized worms were bleached and grown from hatch on Escherichia coli HT115 strains containing an empty vector control or double-stranded RNA. RNAi strains were from the Vidal library if present, or the Ahringer library if absent from the Vidal library.
Heat shock assay
Synchronized Day 1 adult worms of different genetic backgrounds grown on solid NGM plates were incubated in 34℃ for 20 minutes.
ER stress assay
Synchronized L4 stage worms of different genetic backgrounds were incubate in M9 buffer containing 25 ng/μl tunicamycin or equivalent DMSO for 4 hours.
Analysis of the fluorescence intensity in whole worm
For whole-animal fluorescence image, worms were anesthetized with 50mM sodium azide, and imaged using a Leica M165 FC dissecting microscope. To quantify GFP fluorescence, the entire intestine regions were outlined and quantified using ImageJ software. For quantifying the localization of DVE-1∷GFP fusion reporter, worms were mounted on 2% agarose pads with 50Mm and photographs were taken using a Zeiss Imager M2 microscope. Confocal images were taken using Zeiss LSM700.
Western Blot Analysis
Worms were synchronized and collected for western blot analysis. 100-150 worms were picked into 16μl M9 buffer and frozen in liquid nitrogen and kept at −80 until all the samples are ready for analysis. Before running the western blot gels, 5 × SDS loading buffer were added to each sample, mixed well and boiled for 15 min and resolved by Bio-Rad gels.
Antibodies
Antibodies used for western blot analysis were as follows: anti-GFP antibody (Santa Cruz Biotechnology sc-9996); anti-Tubulin antibody (Sigma T6074); anti-mCherry (GTX128508), anti-mouse secondary antibody (EarthOx E030110); anti-rabbit secondary antibody (EASYBIO BE1010).
RNA Isolation and quantitative PCR analyses
Total RNA was isolated using TRIzol (Invitrogen). Worms were synchronized and washed off the plates using M9 buffer, and 500 ul TRIzol were added to the samples and homogenized by repeated freezing and thawing using liquid nitrogen. RNA was isolated according to manufacturer’s instructions. DNA was wiped off using RQ1 RNase-Free DNase (Promega). cDNA was synthesized using the M-MLV Reverse Transcriptase (Invitrogen). Gene expression levels were determined by real-time PCR using iTaq Universal SYBR Green Supermix (Biorad) and Biorad CFX96 Real-Time PCR Detection Systems. Relative gene expression was normalized to act-1(T04C12.6) mRNA levels (Chun et al., 2015). In each experiment at least three biological samples were analyzed. The primer sequences used in the quantitative PCR are shown in Table S2.
Feed serotonin (5-HT)
5-HT hydrochloride powder (Sigma) was dissolved in water to a concentration of 0.1M as a stock concentration. Plates were seeded with E. coli strain OP50 and 100uL drug was added at a final concentration of 50 mM. Plates were allowed to equilibrate overnight and used the next day (Berendzen et al., 2016).
Lifespan Analysis
Lifespan experiments were performed on NGM plates at 20℃ as previously described (Dillin et al., 2002). To prevent progeny production, which resulted in unnatural death of the EGL-20 overexpressed strains, 100uL 10mg/mL 5-fluoro-2′-deoxyuridine (FUdR) was added to seeded plates. Worms were synchronized by egg bleach and were grown on OP50 from hatch, and transit to FUdR plates from L4 to early adulthood. Worms were treated a second time at day 5 of adulthood. Worms were scored every second day. Prism6 software was used for statistical analysis. Log-rank (Mantel-Cox) method was used to determine the significance difference.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters, including the exact value of n and descriptive statistics (mean ± SEM) and statistical significance are reported in the Figures and the Figure Legends. Data are judged to be statistically significant when p < 0.05 by two-tailed Student’s t test. In figures, asterisks denote statistical significance as calculated by Student’s t test (*, p < 0.05, **, p < 0.001, ***, p < 0.0001) as compared to appropriate controls. Lifespans were analyzed using PRISM6 software to determine median survival.
Supplementary Material
Figure S1. vps-35(uth13) is required for the induction of cell-non-autonomous UPRmt in animals expressing Q40 in neurons. Related to Figure 1.
(A) Representative photomicrographs of vps-35(uth13) suppressed hsp-6p∷gfp induction in the animals expressing Q40∷YFP in neurons.
(B) Immunoblots of GFP expression in neuronal Q40∷YFP; hsp-6p∷gfp animals in the presence or absence of vps-35(uth13). Anti-tubulin serves as a loading control.
(C) A schematic of the two mutant alleles uth13 and ok1880 of vps-35 in C. elegans.
(D) vps-35(ok1880) was not required for the induction of UPRER reporter hsp-4p∷gfp when animals were treated with tunicamycin. Synchronized L4 animals were treated with 25ng/μl tunicamycin (Tm) or DMSO and rotated in an incubator at 20°C for 4h prior to imaging.
(E) Representative photomicrographs of vps-35(ok1880) which had no influence on UPRER reporter hsp-4p∷gfp induction in animals expressing spliced XBP-1 in neurons.
(F) vps-35(ok1880) is not required for the induction of the cytosolic heat shock stress response reporter hsp-16.2p∷gfp. Synchronized Day 1 adult animals were placed at 34°C for 20min and allowed to recover for 6h prior to imaging.
(G) Representative photomicrographs of vps-35(ok1880) which had no influence on UPRcyt reporter sod-3p∷gfp in animals expressing HSF-1 in neurons.
Figure S2. Other cargo proteins retrieved by the retromer complex are not required for the induction of cell-non-autonomous UPRmt. Related to Figure 2.
(A) Representative photomicrographs of Day 2 adult animals overexpressing DRP-1 in neuronal Q40∷YFP; hsp-6p∷gfp background.
(B) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp grown on empty vector, eat-3, or fzo-1 RNAi background from hatching.
(C) Representative photomicrographs of Day 2 adult animals expressing neuronal Q40∷YFP; hsp-6p∷GFP in WT, glr-1(n2461), ced-1(e1735), sma-6(wk7), and drp-1(tm1108) animals.
(D) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, mig-14(ga62), dpy-23(e840), or mom-1 RNAi background grown on cco-1 RNAi from hatching.
(E) Representative photomicrographs of overexpressing MIG-14 in vps-35(ok1880); hsp-6p∷gfp; rgef-1p∷Q40∷yfp background animals. The posterior region of the intestine where hsp-6p∷gfp is suppressed or induced is shown.
Figure S3. Other Wnt ligands are not required for the induction of cell-non-autonomous UPRmt in animals expressing Q40 in neurons. Related to Figure 3
(A) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, lin-44(n1792), cwn-1(ok546), cwn-2(ok895), or mom-2 RNAi background.
(B) Quantification of hsp-6p∷gfp expression in neuronal Q40∷YFP animals in the WT, lin-44(n1792), cwn-1(ok546), or cwn-2(ok895) background as shown in (A). (*** denotes p<0.0001 via t-test. Error bars indicate SEM, n ≥ 20 worms,).
(C) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷GFP in WT, lin-44(n1792), cwn-1(ok546), cwn-1(ok546), or mom-2 RNAi background grown on cco-1 RNAi from hatching.
(D) Quantitative PCR of hsp-6 mRNA level. Synchronized Day 1 adult animals of WT, rgef-1p∷Q40∷yfp, egl-20(n585), or rgef-1p∷Q40∷yfp;egl-20(n585) were collected for qPCR.
*p < 0.05, **p < 0.01 via t-test. Error bars indicate the SEM from four biological replicates.
Figure S4. Expression of Wnt ligand/EGL-20 had a specific effect on UPRmt. Related to Figure 4.
(A) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT and two additional egl-20 transgenic animals. Strains used in this figure are ythIs4[rgef-1p∷egl-20; myo-2p∷tdTomato] and ythIs2[gly-19p∷egl-20; myo-2p∷tdTomato].
(B) Representative photomicrographs of DVE-1∷GFP reporter animals in WT, ythIs4[rgef-1p∷egl-20; myo-2p∷tdTomato], or ythIs2[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(C) Representative photomicrographs of rgef-1p∷egl-20∷mCherry animals and gly-19p∷egl-20∷mCherry animals in a WT background. Images were taken at Day 1 of adulthood. EGL-20 was specifically expressed in the neurons with rgef-1 promoter and expressed in the intestine with gly-19 promoter.
(D) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT, ythEx43[myo-2p∷egl-20∷mCherry], ythEx39[myo-3p∷egl-20∷mCherry] or ythEx44[lin-26p∷egl-20∷mCherry] background. EGL-20 expression in the pharynx (myo-2 promoter), in the muscle cells (myo-3 promoter), or in the hypodermal cells (lin-26 promoter) are not able to induce the expression of the hsp-6p∷gfp reporter.
(E) Transcript levels of egl-20 in animals expressing egl-20 in neurons (ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato]), in the intestine (ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato]), or in the Wnt-producing cells (ythIs8[egl-20p∷egl-20∷mCherry; pRF6(rol-6)]) at Day 1 of adulthood were measured by QPCR. Results are shown relative to transcript levels in WT animals with error bars indicate SEM from three biological replicates. (** denotes p < 0.01, * denotes p <0.05 via-t test)
(F) Representative photomicrographs of hsp-4p∷gfp UPRER reporter animals in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(G) Representative photomicrographs of hsp-16.2p∷gfp UPRcyt reporter animals in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(H) Representative photomicrographs of the mitochondrial morphology of muscle cells in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background, which was marked by myo-3p∷GFP(mit). Scale bar represents 10 m.
(I) Representative photomicrographs of ythIs3[gly-19p∷egl-20; myo-2p∷tdTomato]; hsp-6p∷gfp reporter animals grown on EV, atfs-1 RNAi, dve-1 RNAi, lin-65 RNAi, or jmjd-1.2 RNAi from hatching. Images were taken of Day 1 adult animals.
(J) Immunoblot of hsp-6p∷GFP expression in animals grown on EV, atfs-1 RNAi, dve-1 RNAi, lin-65 RNAi, or jmjd-1.2 RNAi from hatching.
Figure S5. Other Wnt receptors are not required for the induction of cell-non-autonomous UPRmt in animals expressing Q40∷YFP in neurons. Related to Figure 5.
(A) Representative photomicrographs of neuronal Q40∷YFP; hsp-6p∷gfp animals in WT, lin-17(sy277), or mom-5 RNAi background.
(B) Representative photomicrographs of hsp-6p∷gfp animals in WT, mig-1(e1787), bar-1 RNAi, or pop-1 RNAi background grown on cco-1 RNAi from hatching as indicated. hsp-6p∷gfp expression was not affected in mig-1(e1787), bar-1 RNAi, and pop-1 RNAi animals.
(C) Representative photomicrographs of animals expressing DVE-1∷GFP reporter grown on EV or pry-1 RNAi from hatching as indicated. DVE-1∷GFP is accumulated in the pry-1 RNAi animals.
(D) Quantification of the number of intestinal nuclei with DVE-1 expression. The genotypes are as in (C). (*** denotes p< 0.0001 via t-test. Error bars indicate SEM, n ≥ 30 worms).
(E) Representative photomicrographs of animals expressing hsp-6p∷GFP grown on EV or lin-23 RNAi from hatching. hsp-6p∷GFP expression was induced in lin-23 RNAi animals.
(F) Immunoblot of GFP expression in hsp-6p∷gfp animals grown on EV or lin-23 RNAi from hatching. The genotypes are as in (E). Anti-tubulin serves as a loading control.
Figure S6. Expression of vps-35 in Wnt/EGL-20 producing cells strongly rescued the suppression of the hsp-6p∷gfp reporter in the vps-35 mutants with neuronal Q40 expression. Related to Figure 5.
(A) Representative photomicrographs of vps-35(ok1880); neuronal Q40∷YFP; hsp-6p∷gfp animals with or without egl-20p∷vps-35, rgef-1p∷vps-35, gly-19p∷vps-35 rescue as indicated.
(B) Quantification of hsp-6p∷gfp expression in vps-35(ok1880); neuronal Q40∷YFP; hsp-6p∷gfp animals with or without egl-20p∷vps-35, rgef-1p∷vps-35, gly-19p∷vps-35 rescue. The genotypes are as in (A). (*** denotes p< 0.0001 via t-test, ns denotes p > 0.05 via t-test. Error bars indicate SEM, n ≥ 30 worms)
Figure S7. EGL-20 is expressed in neurons in C. elegans. Related to Figure 7.
(A) Representative photomicrographs of animals expressing neuronal EGL-20; hsp-6p∷gfp in WT, ser-4(ok152), ser-7(tm1325), ser-1(ok345), or ser-1(ok345); ser-7(tm1325) double mutants background.
(B) Representative photomicrographs of animals expressing neuronal EGL-20; dve-1p∷dve-1∷gfp in WT or mod-1(ok103) background.
(C) Representative photomicrographs of egl-20p∷egl-20∷mcherry in WT, vps-35(ok1880), or mig-14(ga62) background.
(D) Quantitative PCR of egl-20 mRNA level. Synchronized Day 1 adult animals of WT, rgef-1p∷Q40∷yfp were collected for qPCR. ns denotes p > 0.05 via t-test. Error bar indicates the SEM from six biological replicates.
(E) Immunoblots of mCherry and YFP expression in egl-20p∷egl-20∷mcherry animals in the presence or absence of neuronal Q40. Anti-tubulin serves as a loading control.
(F) Quantitative PCR of Wnt target genes mRNA level. Synchronized Day 1 adult animals of WT and rgef-1p∷Q40∷yfp were collected for qPCR. *p < 0.05, **p < 0.01, ns denotes p > 0.05 via t-test. Error bars indicates the SEM from six biological replicates.
(G) Representative of three center images of confocal photomicrographs of animals expressing egl-20p∷mcherry or egl-20p∷egl-20p∷mCherry in combination with rgef-1p∷Q40∷yfp. Scale bar represents 50 m.
(H) Representative photomicrographs of animals expressing egl-20p∷egl-20p∷mCherry in combination with rgef-1p∷yfp or rgef-1p∷Q40∷yfp. Scale bar represents 50 m.
Highlights.
The retromer complex is required for the induction of cell-non-autonomous UPRmt
Retromer retrieval of the Wntless/MIG-14, is essential for the induction of UPRmt
The Wnt/EGL-20 is essential, specific, and sufficient for the induction of UPRmt
Neuronal EGL-20-induced cell-non-autonomous UPRmt requires serotonin
Acknowledgments
We thank Tian lab members for assistance with strain maintenance and reporter crosses. Several C. elegans strains used in this work were provided by CGC, which is supported by the NIH-Officer of Research Infrastructure Programs (P40 OD010440) and the Japanese National BioResource Project. The DRP-1 construct was kindly provided by Dr. Chonglin Yang’s lab. Y.T. was supported by the National Key R&D Program of China (2017YFA0506400), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13000000), and the National Natural Science Foundation of China (31771333). A.D. is supported by the Glenn Foundation for Medical Research, HHMI, NIH(R01 ES021667) and NIH(R37 AG024365).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Y.T. and A.D. conceived the study and wrote the manuscript. Y.T. performed the EMS screen and isolated the mutants in Dr. Andrew Dillin’s lab. Q.Z., X.W., and P.C. performed the C. elegans crosses, strain generation, RNAi experiments, and the fluorescence microscopy. Q.Z. and X.W. performed the qPCR experiments and the western blotting of C. elegans. Q.Z. performed the lifespan experiments. X.W. and N.X. made the transgene constructs. L.L. performed the microinjection experiments.
Declaration of Interests
A.D. is a cofounder of Proteostasis Therapeutics, Inc. and Mitobridge, Inc. and declares no financial interest related to this work.
References
- Bänziger C, Soldini D, Schütt C, Zipperlen P, Hausmann G, Basler K. Wntless, a Conserved Membrane Protein Dedicated to the Secretion of Wnt Proteins from Signaling Cells. Cell. 2006;125:509–522. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- Belenkaya TY, Wu Y, Tang X, Zhou B, Cheng L, Sharma YV, Yan D, Selva EM, Lin X. The Retromer Complex Influences Wnt Secretion by Recycling Wntless from Endosomes to the Trans-Golgi Network. Dev Cell. 2008;14:120–131. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen KM, Durieux J, Shao LW, Tian Y, Kim H eui, Wolff S, Liu Y, Dillin A. Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell. 2016;166:1553–1563.e10. doi: 10.1016/j.cell.2016.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol. 2006;7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- Brignull HR, Moore FE, Tang SJ, Morimoto RI. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J Neurosci Off J Soc Neurosci. 2006;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Cong W, Ji S, Rothman S, Maudsley S, Martin B. Metabolic Dysfunction in Alzheimer’s Disease and Related Neurodegenerative Disorders. 2012 doi: 10.2174/156720512799015064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D. Biogenic amine neurotransmitters in C. elegans. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xiao H, Zhang K, Wang B, Gao Z, Jian Y, Qi X, Sun J, Miao L, Yang C. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- Chun L, Gong J, Yuan F, Zhang B, Liu H, Zheng T, Yu T, Xu XZS, Liu J. Metabotropic GABA signalling modulates longevity in C. elegans. Nat Commun. 2015;6:1–10. doi: 10.1038/ncomms9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Coudreuse DYM, Roël G, Betist MC, Destrée O, Korswagen HC. Wnt gradient formation requires retromer function in Wnt-producing cells. Science (80−) 2006;312:921–924. doi: 10.1126/science.1124856. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science (80−) 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Douglas PM, Baird NA, Simic MS, Wolff SC, Kennedy BK, Dillin A, Douglas PM, Baird NA, Simic MS, Uhlein S, et al. Heterotypic Signals from Neural HSF-1 Separate Thermotolerance from Longevity Article Heterotypic Signals from Neural HSF-1 Separate Thermotolerance from Longevity. CellReports. 2015;12:1196–1204. doi: 10.1016/j.celrep.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier L, Burbea M, Kaplan JM. LIN-23-mediated degradation of beta-catenin regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Neuron. 2005;46:51–64. doi: 10.1016/j.neuron.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Duarte JMN, Schuck PF, Wenk GL, Ferreira GC. Metabolic Disturbances in Diseases with Neurological Involvement. Aging Dis. 2013;5:238–255. doi: 10.14336/AD.2014.0500238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JE, Szyleyko EA, Eisenmann DM. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev Biol. 2006;298:442–457. doi: 10.1016/j.ydbio.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Gleason RJ, Akintobi AM, Grant BD, Padgett RW. BMP signaling requires retromer-dependent recycling of the type I receptor. Proc Natl Acad Sci U S A. 2014;111:2578–2583. doi: 10.1073/pnas.1319947111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gm S, S S, L S, J M, C K, He V. Two wnt genes in Caenorhabditis elegans. Oncogene. 1993;8:1857–1864. [PubMed] [Google Scholar]
- Gorrepati L, Krause MW, Chen W, Brodigan TM, Correa-Mendez M, Eisenmann DM. Identification of Wnt Pathway Target Genes Regulating the Division and Differentiation of Larval Seam Cells and Vulval Precursor Cells in Caenorhabditis elegans. G3: Genes|Genomes|Genetics. 2015;5:1551–1566. doi: 10.1534/g3.115.017715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin J, King RS. The long and the short of Wnt signaling in C. elegans. Curr Opin Genet Dev. 2008;18:362–367. doi: 10.1016/j.gde.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP Mediates Activation of a Mitochondrial Unfolded Protein Response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Herman MA, Vassilieva LL, Horvitz HR, Shaw JE, Herman RK. The C. elegans gene lin-44, which controls the polarity of certain asymmetric cell divisions, encodes a Wnt protein and acts cell nonautonomously. Cell. 1995;83:101–110. doi: 10.1016/0092-8674(95)90238-4. [DOI] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Frankino PA, Paul JW, Tronnes SU, Dillin A. A Futile Battle? Protein Quality Control and the Stress of Aging. Dev Cell. 2018;44:139–163. doi: 10.1016/j.devcel.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- Jackson BM, Abete-Luzi P, Krause MW, Eisenmann DM. Use of an Activated Beta-Catenin to Identify Wnt Pathway Target Genes in Caenorhabditis elegans, Including a Subset of Collagen Genes Expressed in Late Larval Development. G3: Genes|Genomes|Genetics. 2014;4:733–747. doi: 10.1534/g3.113.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen HC, Coudreuse DYM, Betist MC, van de Water S, Zivkovic D, Clevers HC. The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 2002;16:1291–1302. doi: 10.1101/gad.981802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. The Biology of Proteostasis in Aging and Disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, Van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt Signaling Pathway in Development and Disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science (80−) 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle Mitohormesis Promotes Longevity via Systemic Repression of Insulin Signaling. Cell. 2013;155 doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CL, Baum PD, Gu M, Jorgensen EM, Clark SG, Garriga G. C. elegans AP-2 and Retromer Control Wnt Signaling by Regulating MIG-14/Wntless. Dev Cell. 2008;14:132–139. doi: 10.1016/j.devcel.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt Signaling and an APC-Related Gene Specify Endoderm in Early C. elegans Embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The Taxonomy of Developmental Control in Caenorhabditis elegans. Science (80−) 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Sawa H, Lobel L, Horvitz HR. The Caenorhabditis elegans gene lin-17, which is required for certain asymmetric cell divisions, encodes a putative seven-transmembrane protein similar to the Drosophila frizzled protein. Genes Dev. 1996;10:2189–2197. doi: 10.1101/gad.10.17.2189. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Elo JM, Pietiläinen KH, Hakonen AH, Sevastianova K, Korpela M, Isohanni P, Marjavaara SK, Tyni T, Kiuru-Enari S, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XXBP-1 Is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Clayton Carter J, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPRmt. Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bellen HJ. The retromer complex in development and disease. Development. 2015;142:2392–2396. doi: 10.1242/dev.123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang X, Fujioka H, Hoppel C, Whone AL, Caldwell MA, Cullen PJ, Liu J, Zhu X. Parkinson’s disease-associated mutant VPS35 causes mitochondrial dysfunction by recycling DLP1 complexes. Nat Med. 2016;22:54–63. doi: 10.1038/nm.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo J, Kenyon C. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol Cell. 1999;4:851–858. doi: 10.1016/s1097-2765(00)80394-9. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Nusse R. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- Yang PT, Lorenowicz MJ, Silhankova M, Coudreuse DYM, Betist MC, Korswagen HC. Wnt Signaling Requires Retromer-Dependent Recycling of MIG-14/Wntless in Wnt-Producing Cells. Dev Cell. 2008;14:140–147. doi: 10.1016/j.devcel.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. 2010:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Isack NR, Glodowski DR, Liu J, Chen CCH, Xu XZS, Grant BD, Rongo C. RAB-6.2 and the retromer regulate glutamate receptor recycling through a retrograde pathway. J Cell Biol. 2012;196:85–101. doi: 10.1083/jcb.201104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. vps-35(uth13) is required for the induction of cell-non-autonomous UPRmt in animals expressing Q40 in neurons. Related to Figure 1.
(A) Representative photomicrographs of vps-35(uth13) suppressed hsp-6p∷gfp induction in the animals expressing Q40∷YFP in neurons.
(B) Immunoblots of GFP expression in neuronal Q40∷YFP; hsp-6p∷gfp animals in the presence or absence of vps-35(uth13). Anti-tubulin serves as a loading control.
(C) A schematic of the two mutant alleles uth13 and ok1880 of vps-35 in C. elegans.
(D) vps-35(ok1880) was not required for the induction of UPRER reporter hsp-4p∷gfp when animals were treated with tunicamycin. Synchronized L4 animals were treated with 25ng/μl tunicamycin (Tm) or DMSO and rotated in an incubator at 20°C for 4h prior to imaging.
(E) Representative photomicrographs of vps-35(ok1880) which had no influence on UPRER reporter hsp-4p∷gfp induction in animals expressing spliced XBP-1 in neurons.
(F) vps-35(ok1880) is not required for the induction of the cytosolic heat shock stress response reporter hsp-16.2p∷gfp. Synchronized Day 1 adult animals were placed at 34°C for 20min and allowed to recover for 6h prior to imaging.
(G) Representative photomicrographs of vps-35(ok1880) which had no influence on UPRcyt reporter sod-3p∷gfp in animals expressing HSF-1 in neurons.
Figure S2. Other cargo proteins retrieved by the retromer complex are not required for the induction of cell-non-autonomous UPRmt. Related to Figure 2.
(A) Representative photomicrographs of Day 2 adult animals overexpressing DRP-1 in neuronal Q40∷YFP; hsp-6p∷gfp background.
(B) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp grown on empty vector, eat-3, or fzo-1 RNAi background from hatching.
(C) Representative photomicrographs of Day 2 adult animals expressing neuronal Q40∷YFP; hsp-6p∷GFP in WT, glr-1(n2461), ced-1(e1735), sma-6(wk7), and drp-1(tm1108) animals.
(D) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, mig-14(ga62), dpy-23(e840), or mom-1 RNAi background grown on cco-1 RNAi from hatching.
(E) Representative photomicrographs of overexpressing MIG-14 in vps-35(ok1880); hsp-6p∷gfp; rgef-1p∷Q40∷yfp background animals. The posterior region of the intestine where hsp-6p∷gfp is suppressed or induced is shown.
Figure S3. Other Wnt ligands are not required for the induction of cell-non-autonomous UPRmt in animals expressing Q40 in neurons. Related to Figure 3
(A) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷gfp in WT, lin-44(n1792), cwn-1(ok546), cwn-2(ok895), or mom-2 RNAi background.
(B) Quantification of hsp-6p∷gfp expression in neuronal Q40∷YFP animals in the WT, lin-44(n1792), cwn-1(ok546), or cwn-2(ok895) background as shown in (A). (*** denotes p<0.0001 via t-test. Error bars indicate SEM, n ≥ 20 worms,).
(C) Representative photomicrographs of animals expressing neuronal Q40∷YFP; hsp-6p∷GFP in WT, lin-44(n1792), cwn-1(ok546), cwn-1(ok546), or mom-2 RNAi background grown on cco-1 RNAi from hatching.
(D) Quantitative PCR of hsp-6 mRNA level. Synchronized Day 1 adult animals of WT, rgef-1p∷Q40∷yfp, egl-20(n585), or rgef-1p∷Q40∷yfp;egl-20(n585) were collected for qPCR.
*p < 0.05, **p < 0.01 via t-test. Error bars indicate the SEM from four biological replicates.
Figure S4. Expression of Wnt ligand/EGL-20 had a specific effect on UPRmt. Related to Figure 4.
(A) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT and two additional egl-20 transgenic animals. Strains used in this figure are ythIs4[rgef-1p∷egl-20; myo-2p∷tdTomato] and ythIs2[gly-19p∷egl-20; myo-2p∷tdTomato].
(B) Representative photomicrographs of DVE-1∷GFP reporter animals in WT, ythIs4[rgef-1p∷egl-20; myo-2p∷tdTomato], or ythIs2[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(C) Representative photomicrographs of rgef-1p∷egl-20∷mCherry animals and gly-19p∷egl-20∷mCherry animals in a WT background. Images were taken at Day 1 of adulthood. EGL-20 was specifically expressed in the neurons with rgef-1 promoter and expressed in the intestine with gly-19 promoter.
(D) Representative photomicrographs of hsp-6p∷gfp reporter animals in WT, ythEx43[myo-2p∷egl-20∷mCherry], ythEx39[myo-3p∷egl-20∷mCherry] or ythEx44[lin-26p∷egl-20∷mCherry] background. EGL-20 expression in the pharynx (myo-2 promoter), in the muscle cells (myo-3 promoter), or in the hypodermal cells (lin-26 promoter) are not able to induce the expression of the hsp-6p∷gfp reporter.
(E) Transcript levels of egl-20 in animals expressing egl-20 in neurons (ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato]), in the intestine (ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato]), or in the Wnt-producing cells (ythIs8[egl-20p∷egl-20∷mCherry; pRF6(rol-6)]) at Day 1 of adulthood were measured by QPCR. Results are shown relative to transcript levels in WT animals with error bars indicate SEM from three biological replicates. (** denotes p < 0.01, * denotes p <0.05 via-t test)
(F) Representative photomicrographs of hsp-4p∷gfp UPRER reporter animals in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(G) Representative photomicrographs of hsp-16.2p∷gfp UPRcyt reporter animals in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background.
(H) Representative photomicrographs of the mitochondrial morphology of muscle cells in WT, ythIs3[rgef-1p∷egl-20; myo-2p∷tdTomato], and ythIs1[gly-19p∷egl-20; myo-2p∷tdTomato] background, which was marked by myo-3p∷GFP(mit). Scale bar represents 10 m.
(I) Representative photomicrographs of ythIs3[gly-19p∷egl-20; myo-2p∷tdTomato]; hsp-6p∷gfp reporter animals grown on EV, atfs-1 RNAi, dve-1 RNAi, lin-65 RNAi, or jmjd-1.2 RNAi from hatching. Images were taken of Day 1 adult animals.
(J) Immunoblot of hsp-6p∷GFP expression in animals grown on EV, atfs-1 RNAi, dve-1 RNAi, lin-65 RNAi, or jmjd-1.2 RNAi from hatching.
Figure S5. Other Wnt receptors are not required for the induction of cell-non-autonomous UPRmt in animals expressing Q40∷YFP in neurons. Related to Figure 5.
(A) Representative photomicrographs of neuronal Q40∷YFP; hsp-6p∷gfp animals in WT, lin-17(sy277), or mom-5 RNAi background.
(B) Representative photomicrographs of hsp-6p∷gfp animals in WT, mig-1(e1787), bar-1 RNAi, or pop-1 RNAi background grown on cco-1 RNAi from hatching as indicated. hsp-6p∷gfp expression was not affected in mig-1(e1787), bar-1 RNAi, and pop-1 RNAi animals.
(C) Representative photomicrographs of animals expressing DVE-1∷GFP reporter grown on EV or pry-1 RNAi from hatching as indicated. DVE-1∷GFP is accumulated in the pry-1 RNAi animals.
(D) Quantification of the number of intestinal nuclei with DVE-1 expression. The genotypes are as in (C). (*** denotes p< 0.0001 via t-test. Error bars indicate SEM, n ≥ 30 worms).
(E) Representative photomicrographs of animals expressing hsp-6p∷GFP grown on EV or lin-23 RNAi from hatching. hsp-6p∷GFP expression was induced in lin-23 RNAi animals.
(F) Immunoblot of GFP expression in hsp-6p∷gfp animals grown on EV or lin-23 RNAi from hatching. The genotypes are as in (E). Anti-tubulin serves as a loading control.
Figure S6. Expression of vps-35 in Wnt/EGL-20 producing cells strongly rescued the suppression of the hsp-6p∷gfp reporter in the vps-35 mutants with neuronal Q40 expression. Related to Figure 5.
(A) Representative photomicrographs of vps-35(ok1880); neuronal Q40∷YFP; hsp-6p∷gfp animals with or without egl-20p∷vps-35, rgef-1p∷vps-35, gly-19p∷vps-35 rescue as indicated.
(B) Quantification of hsp-6p∷gfp expression in vps-35(ok1880); neuronal Q40∷YFP; hsp-6p∷gfp animals with or without egl-20p∷vps-35, rgef-1p∷vps-35, gly-19p∷vps-35 rescue. The genotypes are as in (A). (*** denotes p< 0.0001 via t-test, ns denotes p > 0.05 via t-test. Error bars indicate SEM, n ≥ 30 worms)
Figure S7. EGL-20 is expressed in neurons in C. elegans. Related to Figure 7.
(A) Representative photomicrographs of animals expressing neuronal EGL-20; hsp-6p∷gfp in WT, ser-4(ok152), ser-7(tm1325), ser-1(ok345), or ser-1(ok345); ser-7(tm1325) double mutants background.
(B) Representative photomicrographs of animals expressing neuronal EGL-20; dve-1p∷dve-1∷gfp in WT or mod-1(ok103) background.
(C) Representative photomicrographs of egl-20p∷egl-20∷mcherry in WT, vps-35(ok1880), or mig-14(ga62) background.
(D) Quantitative PCR of egl-20 mRNA level. Synchronized Day 1 adult animals of WT, rgef-1p∷Q40∷yfp were collected for qPCR. ns denotes p > 0.05 via t-test. Error bar indicates the SEM from six biological replicates.
(E) Immunoblots of mCherry and YFP expression in egl-20p∷egl-20∷mcherry animals in the presence or absence of neuronal Q40. Anti-tubulin serves as a loading control.
(F) Quantitative PCR of Wnt target genes mRNA level. Synchronized Day 1 adult animals of WT and rgef-1p∷Q40∷yfp were collected for qPCR. *p < 0.05, **p < 0.01, ns denotes p > 0.05 via t-test. Error bars indicates the SEM from six biological replicates.
(G) Representative of three center images of confocal photomicrographs of animals expressing egl-20p∷mcherry or egl-20p∷egl-20p∷mCherry in combination with rgef-1p∷Q40∷yfp. Scale bar represents 50 m.
(H) Representative photomicrographs of animals expressing egl-20p∷egl-20p∷mCherry in combination with rgef-1p∷yfp or rgef-1p∷Q40∷yfp. Scale bar represents 50 m.


