SUMMARY
Overnutrition disrupts circadian metabolic rhythms by mechanisms that are not well understood. Here we show that diet-induced obesity (DIO) causes massive remodeling of circadian enhancer activity in mouse liver, triggering synchronous high amplitude circadian rhythms of both fatty acid (FA) synthesis and oxidation. SREBP expression was rhythmically induced by DIO, leading to circadian FA synthesis and, surprisingly, FA oxidation (FAO). DIO similarly caused a high amplitude circadian rhythm of PPARα, which was also required for FAO. Provision of a pharmacological activator of PPARα abrogated the requirement of SREBP for FAO (but not FA synthesis), suggesting that SREBP indirectly controls FAO via production of endogenous PPARα ligands. The high amplitude rhythm of PPARα imparted time-of-day-dependent responsiveness to lipid-lowering drugs. Thus, acquisition of rhythmicity for non-core clock components PPARα and SREBP1 remodels metabolic gene transcription in response to overnutrition and enables a chronopharmacological approach to metabolic disorders.
Graphical abstract
INTRODUCTION
Circadian rhythmicity enables organisms to predict and adapt to daily environmental changes by impacting various physiological and behavioral processes (Bass and Lazar, 2016; Takahashi, 2017). This has a genetic basis in a molecular clock, which was first discovered in Drosophila melanogaster (Konopka and Benzer, 1971) and has proven to be comprised of transcription factors (TFs) whose interlocking positive and negative feedback loops maintain rhythmic transcription (Sehgal, 2017). In mammals, the core clockwork is complicated by additional negative feedback loops, and this clock mechanism exists in nearly all cells of the body (Balsalobre et al., 1998; Yoo et al., 2004) .
The physiological clockwork is influenced by environmental factors that define modern societies. Circadian misalignment among night shift workers and individuals with sleep disorders is increasingly recognized as a risk factor for metabolic disorders including type 2 diabetes, cardiovascular disease, hypertension and cancer (Takahashi et al., 2008). This is in part due to a dissonance between the body’s endogenous timing and the artificial bombardment with light and food at different times of day (Asher and Sassone-Corsi, 2015). The nutritive environment also impacts global transcriptional and epigenomic circadian rhythms, and recent studies have described massive changes in circadian gene expression in diet-induced obesity (DIO) (Eckel-Mahan et al., 2013; Kohsaka et al., 2007). However, the underlying mechanisms by which the nutritive environment impacts circadian rhythmicity are not well understood.
In recent years, it has become clear that core clock TFs regulate the expression not only of each other but also of hundreds of output genes (Koike et al., 2012; Menet et al., 2014; Papazyan et al., 2016b). Although the clock mechanism is ubiquitous, the outputs are tissue-specific, as determined by tissue-specific cistromes of the core TFs (Perelis et al., 2015; Zhang et al., 2015). The cistromes include functional enhancer sites, where TF binding leads to regulation of gene transcription over large genomic distances (Levine et al., 2014). Circadian enhancer activity has been reported on a genome-wide scale (Fang et al., 2014; Koike et al., 2012; Lam et al., 2013; Perelis et al., 2015), and has been exploited to identify the TFs controlling rhythmic gene expression in different phases in a specific tissue (Fang et al., 2014). In liver, for example, the core clock repressive TF Rev-erbα controls circadian gene transcription directly in its own phase, and indirectly by regulating the expression E4BP4, a non-core TF that is expressed with a circadian rhythm in a different phase (Fang et al., 2014).
Here we report that DIO leads to extensive remodeling of circadian enhancers, with new or massively enhanced circadian rhythms at enhancers linked to genes involved in both FA synthesis and oxidation. We demonstrate that DIO-selective circadian rhythmicity of the lipogenic transcription factor Sterol Regulatory Element-Binding Protein (SREBP) (Horton et al., 2002) was required not only for the induction of circadian FA synthesis but, surprisingly, for FA oxidation. DIO also led to oscillation of Peroxisome Proliferated Activated Receptor α (PPARα), a major regulator of FA oxidation (Kersten, 2014), and pharmacological activation of PPARα was able to bypass the requirement of SREBP for diet-induced FA oxidation. Moreover, lipid lowering therapy with a PPARα agonist was more efficacious when administered at the daily peak of PPARα expression. Thus, the acquisition of rhythmic expression and function of non-core clock TFs PPARα and SREBP remodels metabolic gene transcription in response to a challenging nutritive environment and enables a chronopharmacological approach to metabolic disorders.
RESULTS
DIO reprograms circadian hepatic enhancers and gene transcription
To determine the transcriptional effects of a DIO (60% fat, high sucrose diet) on mouse liver, we performed Global Run-On sequencing (GRO-seq) around the clock and compared the results with those on normal chow (NC). Of 1,722 gene bodies whose transcription oscillated significantly in NC, only 278 maintained circadian rhythm in DIO livers, with over half of these remaining the same phase (Figure 1A). By contrast, 1,621 gene body transcripts were circadian in DIO livers, with 1343 being induced by the diet (Figures 1A, 1B, and Table S1A). A major advantage of GRO-seq is the simultaneous determination of enhancer activities based on eRNA transcription (Li et al., 2016), which correlates with enhancer activity (Kim et al., 2010). When all circadian eRNAs were mapped to the nearest circadian gene body transcription, the phases of each enhancer-gene pair were highly correlated (r = 0.85) (Figure S1A). A positive correlation was observed between eRNA expression and the enhancer mark H3K27ac, although the dynamic range of GRO-seq was much greater (Figure S1B). Circadian eRNAs were identified in NC and DIO livers, and filtered for H3K27Ac enrichment relative to genomic background to more stringently identify enhancers (Figure S1C). Remarkably, DIO led to remodeling of more than two-thirds of NC circadian enhancers, with 3,461 enhancers gaining circadian activity in DIO (Figures 1C, 1D, and Table S1B) while 5642 enhancers lost circadian rhythm in the overnutritive condition. Thus, DIO globally remodels the circadian enhancer landscape.
Figure 1. DIO reprograms circadian hepatic enhancers and gene transcription.
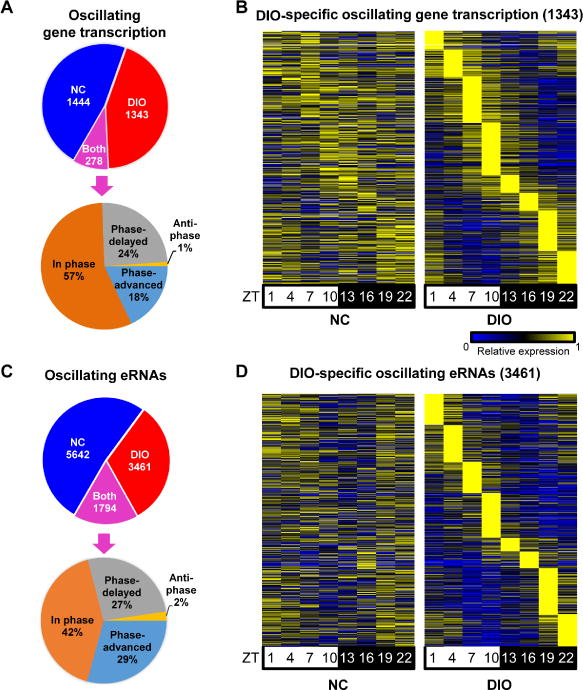
(A and C) Identification of circadian gene transcription (A) and eRNAs (C) in NC, DIO and both conditions (upper panel) and phase analysis of “both” gene transcription (A) and eRNAs (C) (lower panel). JTK_CYCLE (Hughes et al., 2010), p ≤ 0.01 for genes and p ≤ 0.05 for eRNAs, 21 ≤ period (t) ≤ 24 hr, peak to trough ratio > 1.5. (B and D) Heat map of the relative transcription of DIO-specific circadian gene transcription (B) and eRNAs (D) throughout the day in NC (left) or DIO (right). The color bar indicates the scale used to show the expression of gene across eight time points, with the highest expression normalized to 1. See also Figure S1 and Table S1.
We first mined eRNAs to uncover TFs responsible for transcripts whose circadian rhythm was lost in DIO. These eRNAs were most enriched at the phase surrounding ZT22 and enriched for the Rev-erb/RORE motif (Figure S1D). Interestingly, circadian expression of Rev-erbα is modestly attenuated in livers of DIO mice (Figure S1E and S1F), which could explain some though probably not all of the lost circadian enhancer activity. We then turned our focus to circadian gene transcription that was gained in DIO mice.
DIO induces circadian de novo lipogenesis
Compared with NC, DIO-specific circadian gene transcription was strikingly increased at ZT10 (5 PM) (Figure 2A). The DIO-specific circadian transcripts were enriched for metabolic pathways, especially those related to nonalcoholic fatty liver disease (NAFLD), amino acid degradation, glycerophospholipid metabolism, insulin resistance, and FA metabolism (Figure 2B and Figure S2A). By contrast, analysis of the common oscillating transcripts revealed a strong enrichment for circadian rhythm pathways (Figure S2B). Core clock gene expression was quite similar between NC and DIO (Figure S2C) although DIO mice eat a higher percentage of their total caloric intake in the light (Figure S2D) as previously reported (Kohsaka et al., 2007). Consistent with its effect on nascent gene transcription as well as with prior studies (Kohsaka et al., 2007), DIO induced the levels and the rhythmicity of expression of genes mediating hepatic de novo lipogenesis (DNL), including Fasn, Acaca, Acly and Elovl5 (Figure 2C). Tracing deuterated water into newly synthesized FA to quantify the rate of hepatic FA synthesis in vivo demonstrated that the enhanced rhythmicity of DNL gene expression reflected physiological flux, as DIO induced the magnitude in addition to the absolute rate of DNL in the liver (Figure 2D).
Figure 2. DIO induces circadian de novo lipogenesis.
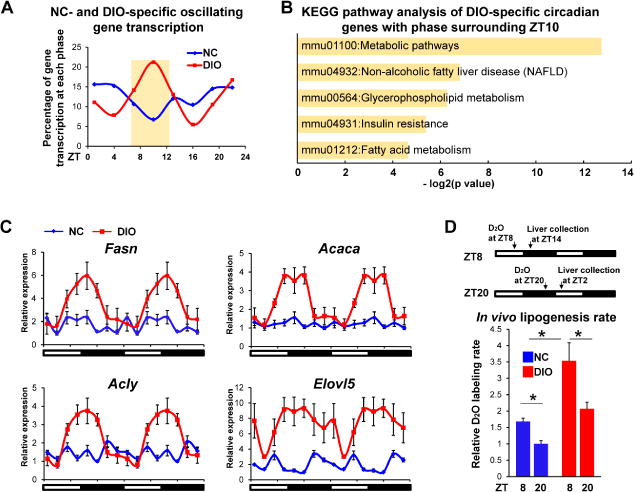
(A) Percentage of gene transcription at each phase that oscillated only in livers of NC or DIO mice. (B) KEGG pathways enriched in DIO-specific circadian genes with phase surrounding ZT10. (C) Relative expression of genes involved in DNL pathway in livers of NC or DIO mice was determined by RT-qPCR. Data are expressed as the mean ± SEM (n = 4-6 per time point) and normalized to the minimal expression in NC mice of the day. Data are double-plotted for better visualization. (D) In vivo fatty acid synthesis rates were measured in livers of mice after oral gavage of D2O either at ZT8 or at ZT20. Data are presented as mean ± SEM. *p < 0.05 in Student’s t-test (n = 5 mice per group). See also Figure S2.
DIO-induced circadian expression of SREBP mediates changes in DNL
Consistent with the gene transcription, DIO-induced circadian eRNAs were also enriched between ZT 7-13 as compared with NC controls (Figure 3A). Unbiased analysis revealed enrichment of several TF motifs at the sites of ZT 7-13 eRNAs, with PPAR and SREBP motifs emerging from two different algorithms (Figure S3A and S3B). The motifs for other E-box binding proteins were not enriched at these eRNA loci (Figure S3C). Consistent with this, Integrated Analysis of Motif Activity and Gene Expression (IMAGE) analysis (Madsen et al., 2017) predicted that SREBP factors contributed to the expression of eRNAs whose peak around ZT10 was synchronous with potential target gene transcription (Figure 3B). IMAGE analysis also predicted that PPARα was the likely TF at the PPAR motifs. However, we first tested the adipocyte-predominant, lipogenic transcription factor PPARγ (Rosen et al., 2000) which also binds to the PPARE and had been previously suggested to mediated the DIO-induced rhythmic expression of DNL genes (Eckel-Mahan et al., 2013; Murakami et al., 2016).
Figure 3. DIO-induced circadian expression of SREBP mediates changes in DNL.
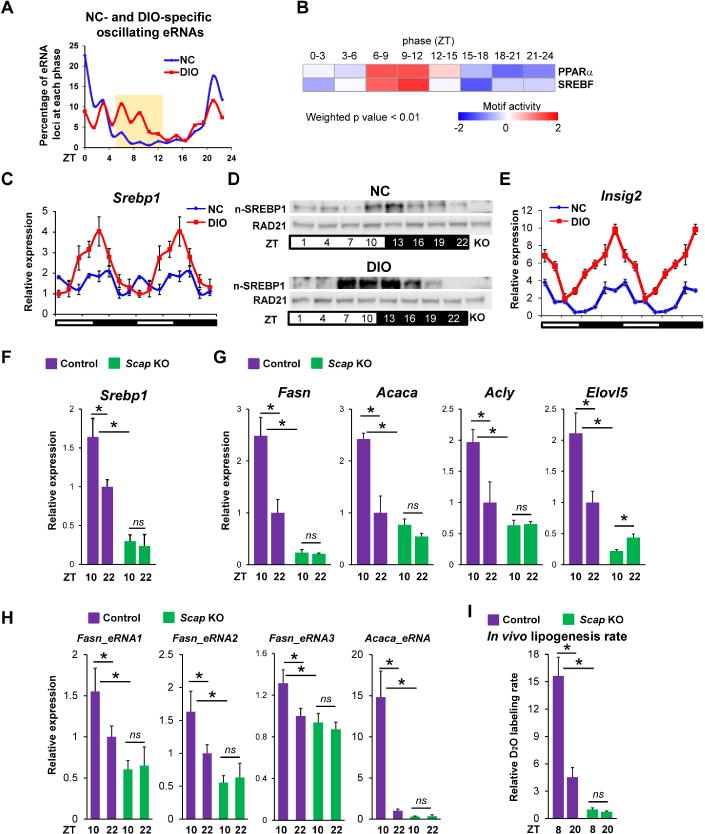
(A) Percentage of eRNAs that oscillated only in livers of NC or DIO mice. (B) Heat map of motif activities generated by IMAGE (Madsen et al., 2017) based on enriched eRNAs and gene transcription with phase between ZT7-13 in eight eRNA phases (weighted p value < 0.01). Out- of-phase eRNAs and genes were used as background. (C) Relative mRNA expression of Srebp1 in livers of NC and DIO mice. RT-qPCR data are double-plotted for better visualization (n = 4-6 per time point). (D) Nuclear form SREBP1 (n-SREBP1) protein level in livers of NC and DIO mice (n = 4-6 per time point). Antibody specificity is confirmed by liver of Scap knockout mice (KO). (E) Relative mRNA expression of Insig2 in livers of NC and DIO mice. RT-qPCR data are double-plotted for better visualization (n = 4-6 per time point). (F-H) Relative mRNA expression of Srebp1 (F), genes involved in DNL pathway (G) and eRNAs nearby indicated genes (H) in livers of control and Scap knockout mice under DIO. RT-qPCR data are expressed as the mean ± SEM (n = 4-6 per group). (I) In vivo fatty acid synthesis rates were measured in livers of mice after oral gavage of D2O either at ZT8 or at ZT20 in livers of control and Scap knockout DIO mice. Data are presented as mean ± SEM. ns: p > 0.05 and *p < 0.05 in Student’s t-test (n = 5 mice per group). See also Figure S3.
Consistent with prior reports (Eckel-Mahan et al., 2013), DIO induced rhythmicity of Pparγ mRNA expression (Figure S3D), although even in DIO liver the absolute expression of Pparγ was extremely low relative to white adipose tissue (Figure S3E). To determine whether this low level of hepatic Pparγ expression was required for DIO-induced DNL rhythmicity, we delivered Cre recombinase driven by the hepatocyte-specific human thyroxine binding globulin (AAV8-TBG-Cre) to livers of mice with floxed Pparγ alleles to abrogate Pparγ gene expression (Figure S3F). Deletion of Pparγ caused no significant changes in the amplitude or rhythmicity of DNL genes (Figure S3G), indicating that Pparγ is not required for DIO-mediated circadian changes in hepatic lipogenic gene expression.
We next turned our attention to lipogenic factors other than PPARγ that bind motifs enriched in the DIO-induced enhancers. The highly enriched E-box motif is bound by lipogenic factor SREBP (Kim et al., 1995; Osborne, 2000) and, indeed, DIO induced a high amplitude rhythmicity of Srebp1 (Figure 3C). By contrast, although the expression of other DNL regulators, such as Chrebp and Srebp2, which also binds to the SREBP motif, were mildly increased in DIO, their circadian rhythmicity was not enhanced (Figure S3H). Importantly, the rhythmicity of active, nuclear SREBP1 was also induced by DIO (Figure 3D). Nuclear localization of SREBP1 is controlled by INSIG2, which prevents the proteolytic processing and nuclear localization of SREBP (Engelking et al., 2005; Yabe et al., 2002). Insig2 gained high amplitude, circadian rhythmic expression in the livers of DIO mice in a phase opposite to the circadian increase in nuclear SREBP1 (Figure 3E). The circadian trough in INSIG2 may contribute to the peak amount and timing of nuclear SREBP1, whose feed-forward activation of SREBP1 gene expression (Amemiya-Kudo et al., 2000) likely magnifies the circadian rhythm of Srebp1 mRNA.
To evaluate the role of SREBP1 in DIO-mediated rhythmic expression of DNL pathway, AAV8-TBG-Cre was used for hepatocyte-specific deletion of SCAP, an adaptor protein that is required for SREBP activation and stability (Hua et al., 1996). As expected, deletion of SCAP led to a dramatic reduction of Srebp1 expression in DIO mice (Figure 3F), and this was accompanied by a marked attenuation of the rhythmic expression of DNL genes (Figure 3G) and corresponding eRNAs (Figure 3H). Importantly, the magnitude and rhythmicity of de novo lipogenesis were also SCAP-dependent (Figure 3I). SREBP2 and its target genes Hmgcr and Mvk were also down-regulated in SCAP deficient livers, but the expression of these genes was not circadian (Figure S3I).
DIO induces circadian fatty acid oxidation
In addition to DNL, the GRO-seq analysis also identified newly circadian nascent transcription of genes involved in FA oxidation (FAO). Indeed, both the absolute levels and circadian rhythmicity of Acox1, Aldh3a2, Acaa1 and Hadh were dramatically induced by DIO (Figure 4A). In addition to these genes that directly contribute to FAO, DIO also induced circadian rhythmicity of the gene encoding Adipose Triglyceride Lipase (Atgl), which contributes to the production of ligand for the master transcriptional regulator of FAO, PPARα (Figure 4B) (Ong et al., 2011; Zechner et al., 2012). Indeed, the rate of FAO in mouse DIO-liver homogenates was up-regulated, as previously reported in human (Zhu et al., 2011) and, importantly, increased at ZT10 relative to ZT22 (Figure 4C), suggesting that the processes of DNL and FAO were synchronized, as were their gene expression patterns. Interestingly, in white adipose tissue the same DNL and FAO genes did not acquire circadian rhythmicity in DIO, indicating that these dietary effects were liver-specific (Figure S4).
Figure 4. DIO induces circadian fatty acid oxidation.
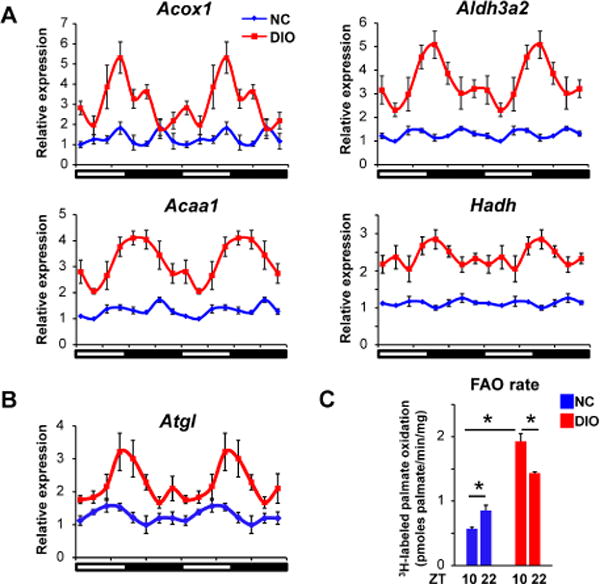
(A-B) Relative mRNA expression of genes involved in FAO pathway (A) and Atgl (B) in livers of NC and DIO mice (n = 4-6 per time point). Data are expressed as the mean ± SEM and double-plotted for better visualization. (C) Rate of 3H-labeled palmiate oxidation in liver homogenates. The livers were collected at ZT10 and ZT22 in NC and DIO mice. Data are presented as mean ± SEM. *p < 0.05 in Student’s t-test. See also Figure S4.
DIO-induced circadian expression of PPARα mediates the acquired rhythm of FAO
Given the enrichment of PPAR binding motifs in DIO-induced circadian eRNAs, we considered whether the gain of expression and rhythmicity of FAO gene expression could be due to PPARα. Indeed, DIO induced Pparα and magnified its circadian rhythm with peak expression at ZT10 (Figure 5A). By contrast, the circadian rhythm of Pparδ was not enhanced and its peak expression was at ZT22 (Figure S5A). To explore the potential function of PPARα in DIO-induced circadian transcription, we examined the expression of oscillating eRNAs with peak expression at ZT10 surrounding PPARα binding sites, and noted that these eRNAs have higher expression at ZT10 than at ZT22 in DIO, but not in NC-fed mice (Figure 5B). Furthermore, in DIO, the transcription of genes predicted by IMAGE to be PPARα targets was also circadian with peak expression surrounding ZT10 (Figure 5C). The binding of PPARα at sites near FAO genes was circadian in DIO but not in NC mice (Figure 5D), and was specific since it was not observed in livers depleted of PPARα (Figure S5B). Moreover, DIO-induced circadian rhythms of FAO genes (Figure 5E) and corresponding eRNA expression (Figure S5C) were attenuated in livers lacking PPARα. Physiologically, PPARα was also necessary for the circadian rhythm and magnitude of FAO (Figure 5F).
Figure 5. DIO-induced circadian expression of PPARα mediates the acquired rhythm of FAO.
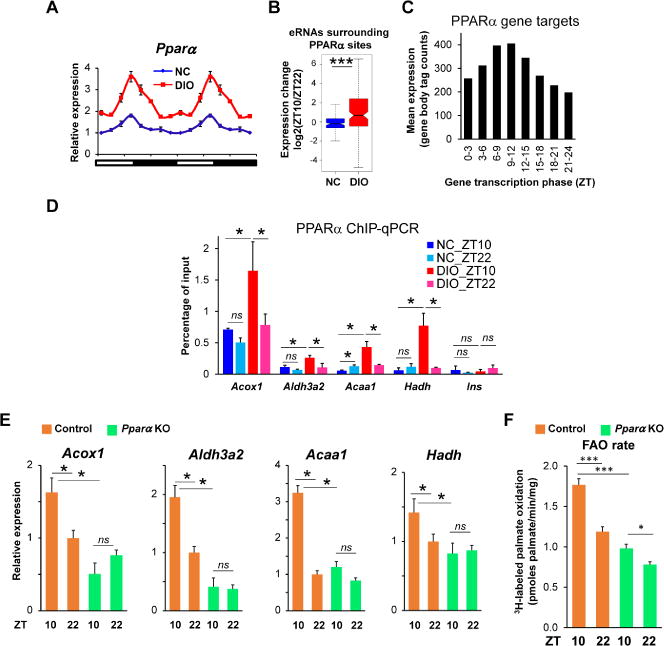
(A) Relative mRNA expression of Pparα in livers of NC and DIO mice (n = 4-6 per time point). Data are double-plotted for better visualization. (B) Expression changes (ZT10 vs ZT22) in livers of NC mice (blue) and DIO mice (red) for eRNAs surrounding PPARα binding sites. ***p < 0.001 in Student’s t-test. (C) Average gene body transcription of genes predicted by IMAGE to be regulated by PPARα. Bar plot shows average GRO-seq tag counts for targeted genes. (D) PPARα ChIP-qPCR for Acox1, Aldh3a2, Acaa1, Hadh and Ins from livers in NC or DIO mice, which were collected at ZT10 or ZT22 (n = 4-5 per group). (E) Relative mRNA expression of Acox1, Aldh3a2, Acaa1 and Hadh in livers of control or Pparα knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). RT-qPCR data are presented as the mean ± SEM. ns: p > 0.05 and *p < 0.05 in Student’s t-test. (F) Rate of 3H-labeled palmate oxidation liver homogenates of control and Pparα knockout DIO mice, which were collected at ZT10 or ZT22. Data are presented as the mean ± SEM. *p < 0.05 and ***p < 0.001 in Student’s t-test. See also Figure S5.
SREBP1 activates FAO genes by promoting PPARα transcriptional activity
As the DIO-induced circadian rhythmicity of FAO genes was similar in phase to that of DNL genes, which were dependent upon SCAP/SREBP, we wondered how the loss of SCAP/SREBP would affect FAO. Remarkably, although SREBP1 is best known as a lipogenic TF, its depletion via Scap deletion led to a marked downregulation and attenuation of rhythmicity of the DIO-induced FAO genes (Figure 6A) and corresponding eRNA that were directly controlled by PPARα (Figure S6A). Moreover, loss of SCAP dramatically decreased basal FAO rate at both ZT10 and ZT22, and abrogated the rhythmicity of FAO rate (Figure 6B).
Figure 6. SREBP1 activates FAO genes by promoting PPARα transcriptional activity.
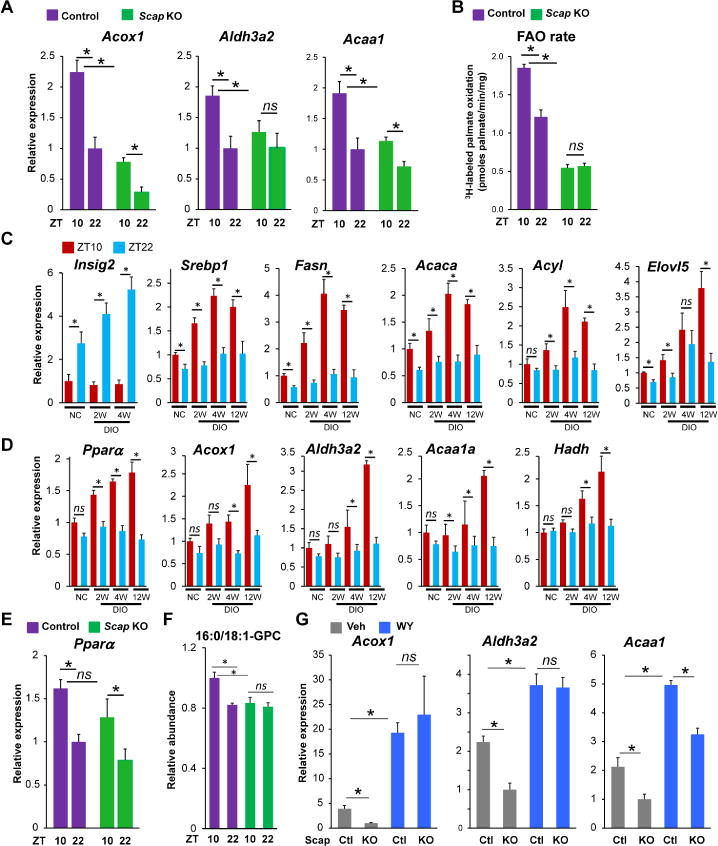
(A) Relative mRNA expression of FAO genes, including Acox1, Aldh3a2 and Acaa1 in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). (B) Rate of 3H-labeled palmate oxidation liver homogenates of control and Scap knockout DIO mice, which were collected at ZT10 or ZT22. (C) Relative mRNA expression of Insig2, Srebp1 and Srebp1 target genes at ZT10 or ZT22 in the livers of normal chow mice followed by 2, 4 and 12 weeks on the hypernutritive diet (n = 3-5 per group). (D) Relative mRNA expression of PPARα and its target genes at ZT10 or ZT22 in the livers of normal chow mice followed by 2, 4 and 12 weeks on the hypernutritive diet (n = 3-5 per group). (E) Relative mRNA expression of Pparα in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). (F) Relative abundance of 16:0/18:1-GPC in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 4 per group). (G) Relative expression of FAO genes in livers of control and Scap knockout DIO mice with or without receiving WY (n = 4 mice per group). RT-qPCR data are presented as mean ± SEM. * p < 0.05 in Student’s t-test. See also Figure S6.
Since these observations were made after 12 weeks of the obesogenic diet, we next explored the chronology of the acquisition of circadian DNL and FAO gene expression. Interestingly, the induction of rhythmicity of Insig2, Srebp1 and DNL genes was apparent after 2 weeks (Figure 6C). These effects were independent of changes serum insulin (Figure S6B), and were earlier than observed for genes in the FAO pathway, whose enhanced expression and rhythmicity tended to be later (Figure 6D). Although these genes were controlled by PPARα, the level and circadian rhythm of Pparα expression were not significantly altered in the absence of SREBPs (Figure 6E), leading us to hypothesize that SREBP was controlling the generation of an endogenous activating ligand of PPARα. In support of this, both Atgl and Fasn, whose circadian rhythm and expression were induced in DIO, have been suggested to contribute to endogenous PPARα ligand formation (Chakravarthy et al., 2005; Haemmerle et al., 2011). Although there are likely to be multiple endogenous ligands for PPARα, there is evidence that 16:0/18:1-GPC fulfills the criteria (Chakravarthy et al., 2009), and interestingly 16:0/18:1-GPC levels were rhythmic, with higher concentration at ZT10 than ZT22, in an SREBP-dependent manner (Figure 6F). If SREBP was controlling an endogenous PPARα ligand, then treatment with an exogenous PPARα agonist should rescue the effect of Scap deletion on FAO. Indeed, WY-14,643 (WY) was able to overcome the inhibition of FAO gene expression in the mice lacking hepatic SCAP (Figure 6G). By contrast, WY treatment did not induce Fasn in the absence of SCAP (Figure S6C), consistent with direct effects of SREBP1 on DNL. These data support the hypothesis that SREBP promotes FAO by contributing to the generation of endogenous ligands for PPARα.
Pharmacological lipid lowering is more effective at the peak of PPARα expression
PPARα agonists are used in the clinic as a therapy for hyperlipidemia (Frick et al., 1987; Rubins et al., 1999). Most patients with this condition are obese (Schauer et al., 2017), yet current dosing strategies do not consider the circadian expression of PPARα in DIO. We therefore tested whether PPARα agonist therapy might be more efficacious at certain times of the day by using WY, whose elimination half-life is ~1 hour after gavage dosing (NTP, 2007). Indeed, a single oral gavage dose of WY caused more robust expression of FAO genes and pre-mRNAs (reflecting nascent transcription) at ZT8 (Pparα peak) than at ZT20 (Pparα trough) (Figure 7A). We then performed a 3-week study, providing WY to DIO mice by daily gavage either at ZT8 or ZT20 with a dose (3mg/kg) shown to not alter food intake (Karimian Azari et al., 2013), in 6-day cycles with a gap day after each cycle. Mice were euthanized at ZT10 after the third cycle, such that the time after last dose was 50 hours for mice dosed at ZT8 and 38 hours for ZT20-treated mice. Remarkably, livers of mice treated with WY at ZT8 exhibited reduced lipid accumulation compared with those dosed at ZT20 (Figure 7B). WY also reduced hepatic triglycerides more when dosed at ZT8 than at ZT20 (Figure 7C). Moreover, serum triglyceride levels, which are the main biomarker of PPARα agonist therapy, were lower when the drug was administered at ZT8 (Figure 7D). By contrast, expression of core clock genes and Srebp1 was not statistically different between treatment and control groups (Figure S7). Thus, in DIO mice, provision of pharmacological ligand at the time of peak PPARα expression causes maximal reduction of liver and serum triglycerides.
Figure 7. Pharmacological lipid lowering is more effective at the peak of PPARα expression.
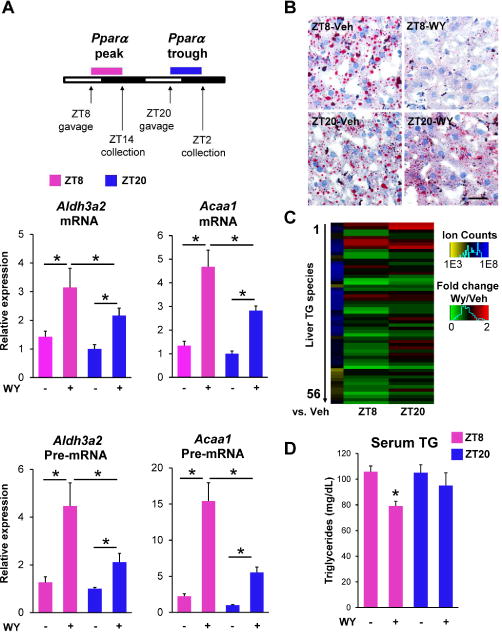
(A) RT-qPCR showing the effect of WY (3mg/kg) on the mature mRNA and pre-mRNA of Acaa1 and Aldh3a2 at ZT8 and at ZT20 (n = 4-5 mice per group). RT-qPCR data are presented as mean ± SEM, and *p < 0.05 in Student’s t-test. (B) Representative Oil Red O staining of livers from DIO mice with oral gavage of vehicle or WY (3 mg/kg) at ZT8 or ZT20 for three weeks. Scale bar, 50 um. (C) Heatmap displays down-regulated hepatic triglycerides (TG) species upon WY treatment at ZT8 or ZT22 as detected via mass spectrometry. Scale bar (green vs. red) represents fold change for WY vs. vehicle and scale bar (yellow vs. blue) represents average ion counts of each triglycerides species in liver with vehicle at ZT8. (D) Serum triglyceride measurements in mice described in (B). Data are presented as mean ± SEM, and *p < 0.05 between vehicle and WY treatments at ZT8 in Student’s t-test. See also Figure S7 and Table S2.
DISCUSSION
We have demonstrated that an obesogenic diet induces widescale remodeling of circadian enhancers and gene transcription. Remarkably, rather than affecting the core clock TFs, DIO led to a marked and synchronous circadian oscillation of the anabolic TF SREBP1 and the catabolic TF PPARα, and corresponding rhythms of DNL and FAO, respectively. The two processes are linked by SREBP, which directly controls DNL and promotes the activity of PPARα.
For over 40 years, it has been appreciated that, in mice, hepatic DNL is circadian and is exacerbated by obesity (Hems et al., 1975). Yet the underlying mechanism for the metabolic perturbation remained unknown. The circadian control of DNL on normal chow is determined in part by the nuclear receptor Rev-erbα, which is a core clock component (Bugge et al., 2012; Cho et al., 2012; Preitner et al., 2002) that can also directly repress lipogenic genes in a circadian manner (Feng et al., 2011). Here we demonstrate that the increased circadian DNL in obesity is due to an acquired high amplitude circadian rhythm of lipogenic TF SREBP1. More broadly, our results demonstrate that the non-core clock TFs can be recruited as circadian regulators of metabolism.
SREBP and PPARα are also required for the circadian expression of eRNAs, which reflect the active enhancers, near some DNL and FAO genes. We also observed SREBP regulates the expression of several eRNAs, whose loci surround Fasn gene. This suggests functional redundancy amongst enhancers for regulation of target gene (Hong et al., 2008; Osterwalder et al., 2018; Soccio et al., 2011) can be modulated by single TF SREBP. Comparing DIO-selective eRNAs and DIO-lost eRNAs in light phase did not reveal the motifs for TFs previously suggested to entrain feeding/fasting cycles and insulin response, such as FOXO1 (Vollmers et al., 2009). We recognize that phase-specific enrichment could be masked if the majority of binding was at promoters rather than enhancers, since GRO-seq cannot identify enhancer elements at promoter regions because promoter region transcription is typically divergent. It will be interesting to profile eRNA expression surrounding binding sites of these TFs in livers of NC and DIO mice. In sum, unbiased motif analysis in DIO-selective eRNAs led us to identify the motif sequences of SREBP and PPARα as selectively circadian in DIO and positively correlated with gene expression of SREBP1 and PPARα. Loss-of-function studies indicates these TFs are required for the circadian expression of eRNAs near genes involved in DNL and FAO pathways.
DNL and FAO are important for the orchestration of hepatic lipid metabolism under physiological conditions. In NAFLD, DNL is enhanced and thought to contribute to hepatic lipid accumulation, while increasing FAO via activation of PPARs is used in the clinic (Staels et al., 2013; Ye et al., 2001). However, emerging data suggest a beneficial effect of DNL in preventing lipotoxicity. For example, ectopic expression of nuclear SREBP1c prevents lipotoxic liver damage in HDAC3 and SCAP (SREBP cleavage-activating protein) hepatic double knockout mice (Papazyan et al., 2016a). The present study suggests that DNL has a beneficial effect by dynamically providing endogenous activators of PPARα in the lipid-overloaded liver of DIO mice. FASN has previously been shown to be required for endogenous generation of the putative PPARα ligand 16:0/18:1-GPC (Chakravarthy et al., 2009) in a diet containing no fat. Here we show that an obesogenic diet high in fat and sucrose leads to circadian production of this putative ligand in a Scap-dependent manner. We speculate that the DIO-induced synchronization of opposing hepatic lipid metabolic processes is an adaptive response to the overnutritive environment, although clearly they are not sufficiently homeostatic since fatty liver and insulin resistance develop under the same conditions. Whether this is partially protective, or ultimately maladaptive, for example by generating ROS or other toxic products, remains to be determined.
Chronotherapy refers to the principle of administering drugs at times when they are most effective and/or best tolerated to enhance effectiveness and reduce toxicity (Ohdo, 2010). There are some precedents in the field of lipid metabolism, where statins have been reported to be more effective in the evening due to the maximized cholesterol synthesis rate (Saito et al., 1991). However, several studies have reported that dosing time does not affect statins efficiency (Izquierdo-Palomares et al., 2016; Kim et al., 2013; Martin et al., 2002; Plakogiannis et al., 2005). PPARα agonists have clinical application in the treatment of lipid disorders, but they also have side-effects including toxicity in muscle (Hodel, 2002). In light of our observation of a markedly circadian oscillation of PPARα expression on DIO, we noted increased efficacy of a short-acting PPARα agonist at ZT8 versus ZT20. These results support the chronotherapeutic notion that, due to the rhythmicity of PPARα in DIO, pharmacological lipid lowering is more effective at specific times of day. Indeed, recent studies have shown that Pparα expression oscillates in human liver (Anafi et al., 2017; Ueda et al., 2004), further supporting the potential role of PPARα agonists as chronotherapeutic agents.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BrdU | Santa Cruz Biotechnology | Cat# sc-32323, RRID:AB_626766 |
| H3K27ac | Abcam | Cat# ab4729, RRID:AB_2118291 |
| PPARα | Santa Cruz Biotechnology | Cat# sc-398394 |
| SREBP1c | Abcam | Cat# ab3259, RRID:AB_303650 |
| RAD21 | Cell Signaling Technology | Cat# 4321S, RRID:AB_1904106 |
| Chemicals and Reagents | ||
| TRIzol | ThermoFisher Scientific | Cat# 10296010 |
| CircLigase ssDNA Ligase | Epicentre | Cat# CL4111K |
| BrUPT | Sigma | Cat# B7166 |
| cOmplete, EDT-free Protease Inhibitor Cocktail | Roche | Cat# 11873580001 |
| SUPERase-In RNAse Inhibitor | ThermoFisher Scientific | Cat# AM2696 |
| Deuterium oxide | Sigma | Cat# 435767 |
| WY 14,643 | Cayman | Cat# 70730 |
| 9,10-3H-palmitoyl-CoA | PerkinElmer | Cat# NET043001MC |
| Low Range ssRNA Ladder | New England Biolabs | Cat# N0364S |
| APE 1 | New England Biolabs | Cat# M0282S |
| TrackIt 25 bp DNA Ladder | Thermo Fisher Scientific | Cat# 10488-022 |
| Novex TBE-Urea Gels, 10%, 10 well | Thermo Fisher Scientific | Cat# EC6875BOX |
| Novex TBE Gels, 10%, 12 well | Thermo Fisher Scientific | Cat# EC62752BOX |
| Novex TBE Running Buffer (5x) | Thermo Fisher Scientific | Cat# LC6675 |
| ChIP DNA Clean & Concentrator- Capped column | Zymo Research | Cat# D5205 |
| DNA LoBind Microcentrifuge Tube | Eppendorf | Cat# 30108.051 |
| Exonuclease I (E. coli) | New England Biolabs | Cat# M0293L |
| Novex TBE-Urea Sample Buffer (2x) | Thermo Fisher Scientific | Cat# LC6876 |
| Novex Hi-Density TBE Sample Buffer (5x) | Thermo Fisher Scientific | Cat# LC6678 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080-051 |
| Deoxynucleotide (dNTP) Solution Mix | New England Biolabs | Cat# N0447S |
| SYBR Gold Nucleic Acid Gel Stain | Thermo Fisher Scientific | Cat# S11494 |
| Phusion Hot Start II DNA Polymerase | Thermo Fisher Scientific | Cat# F-549L |
| E. coli Poly (A) Polymerase | New England Biolabs | Cat# M0276S |
| UltraPure Glycogen | Thermo Fisher Scientific | Cat# 10814010 |
| TURBO DNA-free Kit | Thermo Fisher Scientific | Cat# AM1907 |
| Micro Bio-Spin P-30 Gel Columns | Bio-Rad | Cat# 732-6250 |
| RNA Fragmentation Reagents | Thermo Fisher Scientific | Cat# AM8740 |
| T4 Polynucleotide Kinase | New England Biolabs | Cat# Y904L |
| SSPE (20x) | Thermo Fisher Scientific | Cat# AM9767 |
| NuPAGE 4-12% Bis-Tris Protein Gels, 1.0 mm, 10-well | Thermo Fisher Scientific | Cat# NP0321 BOX |
| NuPAGE MOPS SDS Running Buffer (20X) | Thermo Fisher Scientific | Cat# NP0001 |
| NuPAGE LDS Sample Buffer (4X) | Thermo Fisher Scientific | Cat# NP0008 |
| HistoPrep Media | Thermo Fisher Scientific | Cat# SH75-125D |
| AG 1-X8 formate resin | Bio-Rad | Cat# 1401443 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74106 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368813 |
| Triglyceride Assay Kit | StanBio | Cat# 2100-430 |
| Deposited Data | ||
| GRO-seq in DIO | This study | GEO: GSE108878 |
| H3K27ac ChIP-seq | This study | GEO: GSE108878 |
| GRO-seq in NC | (Fang et al., 2014) | GEO: GSE59486 |
| H3K27ac ChIP-seq | (Koike et al., 2012) | GEO: GSE39860 |
| PPARα ChIP-seq | (Lee et al., 2014) | GEO: GSE61817 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson labs | 000664 |
| Mouse: C57BL/6J DIO | Jackson labs | 380050 |
| Mouse: Scap floxed | Timothy Osborne | (Matsuda et al., 2001) |
| Mouse: Pparγfloxed | Jackson labs | 004584 |
| Mouse: Pparα −/− | Jackson labs | 008154 |
| Oligonucleotides | ||
| See Table S3 for qPCR primer list | ||
| See Table S4 for ChIP-qPCR primer list | ||
| Software and Algorithms | ||
| Bowtie 1.1.1 | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| SAMtools 1.1 | (Li et al., 2009) | http://samtools.sourceforge.net/ |
| Bedtools 2.19 | (Quinlan and Hall, 2010) | http://bedtools.readthedocs.org/en/latest/ |
| edgeR | (Robinson et al., 2010) | https://bioconductor.org/ |
| pheatmap | https://cran.r-project.org/web/packages/pheatmap/ | |
| HOMER v4.6 | (Heinz et al., 2010) | http://homer.salk.edu/homer/ |
| JTK_CYCLE | (Hughes et al., 2010) | https://github.com/mfcovington/jtk-cycle |
| IMAGE | (Madsen et al., 2017) | https://github.com/JesperGrud/IMAGE |
| R 3.1.0 | www.r-project.org/ | |
| Perl v5.16 | https://www.perl.org | |
| GATHER | (Chang and Nevins, 2006) | http://changlab.uth.tmc.edu/gather/gather.py |
| Adobe Illustrator | Adobe | Adobe.com |
| Other | ||
| AAV-TBG-GFP | UPenn Vector Core | N/A |
| AAV-TBG-Cre | UPenn Vector Core | N/A |
| Rodent Diet with 60% kcal% fat | Research Diets | Cat# D12492 |
| Normal chow | LabDiet | Cat# 5010 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mitchell A. Lazar, M.D., Ph.D. (lazar@pennmedicine.upenn.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild type C57BL/6J male mice and DIO C57BL/6J male mice were obtained from Jackson Labs. DIO C57BL/6J male mice were fed with 12-weeks rodent diet with 60% kcal% fat (Table S5). Scap floxed mice were previously described (Matsuda et al., 2001) and obtained from T.F. Osborne following backcrossing to the C57BL/6J genetic background. Pparγ floxed mice and Pparα −/− mice were obtained from Jackson Labs on a C57BL/6J background (Lee et al., 2014; Wang et al., 2013). All mice were housed under a 12 hr light and 12 hr dark cycle (lights on at 7 a.m. [zeitgeber time 0, ZT0] and lights off at 7 p.m. [ZT12]). For DIO experiments, male mice were given a rodent diet with 60% kcal% fat for 12 weeks starting at 8 weeks of age. Control age-matched mice were fed normal chow. The UPenn Vector Core generated the AAV vectors (AAV-TBG-GFP for control and AAV-TBG-Cre for knockout). AAVs were intravenously injected with 1.5 x 1011 genome copies (GC) per mouse at 8 weeks of age prior to the start of DIO experiments. For single WY treatment, mice were dosed at 3mg/kg WY via oral gavage either at ZT8 or ZT20. Livers were collected 6 hr after oral gavage. For 3 weeks WY treatment, the drug application regimen was oral gavage either at ZT8 or ZT20 daily for 6 day cycles of WY with a gap day after each cycle. Mice were euthanized at ZT10 after the third cycle, such that the time after the last dose was 50 hours for mice dosed at ZT8 and 38 hr for mice dosed at ZT20. All animal care and use procedures followed the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania in accordance with the guidelines of the NIH.
GRO-seq
GRO-seq was performed using livers from DIO mice, collected every three hours throughout the day following the protocol described previously (Fang et al., 2014). Briefly, livers were first subjected to dounce homogenization in cold swelling buffer (10 mM Tris pH 7.5, 2 mM MgCl2, 3 mM CaCl2, 2 U/ml Superase-In). After centrifugation at 400 g, the nuclei were extracted using lysis buffer (swelling buffer with 10% glycerol and 1% Igepal). After washing twice with lysis buffer, the nuclei were resuspended in freezing buffer (50 mM Tris pH 8.3, 40% glycerol, 5 mM MgCl2, 0.1 mM EDTA) at a concentration of 10 × 107 nuclei/ml. 4 × 107 nuclei were used for each library. The nuclear run-on reaction was incubated in run-on buffer (10 mM Tris pH 8.0, 5 mM MgCl2, 1 mM DTT, 300 mM KCl, 200 U/ml Superase-In, 1% Sarkosyl, 500 mM ATP, GTP and Br-UTP, 2 mM CTP) for 7 min at 30°C. Nascent transcripts were enriched with anti-BrUTP antibodies. After 10 min RNA hydrolysis and reverse transcription, all products were used for GRO-seq library preparations in parallel to reduce batch effects.
GRO-seq data processing
Data processing was carried out as described previously (Fang et al., 2014). After trimming to clear low-quality base, adaptor, and poly-A tailing sequence, the GRO-seq sequencing reads were aligned to the mm9 genome using Bowtie version 1.1.1. Uniquely mapped reads were extended to 150bp in the 5′ to 3′ direction and used for downstream analysis.
To quantify gene transcription, GRO-seq signal in gene bodies was measured using GRO-seq reads mapped to the sense strand of the gene in a 10kb window (+2kb to +12kb relative to the TSS) within the Refseq annotated gene body. Smaller genes between 2kb and 12kb in length were quantified using smaller window sizes, from +2kb to the transcription end site (TES). For genes shorter than 2kb, the entire gene body was used for the quantification. The mapped reads within each gene quantification window were counted using HOMER (Heinz et al., 2010) and expressed as read per kb per ten million reads (RPKTM). Genes with transcription levels greater than 5 RPKTM were considered actively transcribed. eRNA identification and quantification in DIO were performed according to previously established protocols (Fang et al., 2014). Data quality and normalization factors are shown in Table S1C, and the Pearson correlation coefficients of whole transcriptome (gene or eRNA) suggest a high degree of consistency among all samples.
Identification of NC- and DIO-specific circadian gene transcripts and eRNAs
To determine NC- and DIO-specific gene transcripts, the time-ordered RPKTM of actively transcribed genes was duplicated and inputted to JTK_CYCLE (Hughes et al., 2010) as a 16-time point data set. This allowed thorough identification of oscillation patterns starting at each different time point. Oscillating transcripts were defined as those with JTK_CYCLE p ≤ 0.01, maximum RPKTM value > 5, oscillation amplitude (peak/trough) > 1.5, and oscillation period within the range of 21 to 24 hr. Based on the p value from the output of JTK_CYCLE, we determined circadian transcription in the NC, DIO and Both conditions. We further identified enriched pathways for DIO-specific circadian genes with phase surrounding ZT10 using GATHER (Chang and Nevins, 2006).
eRNA calling was performed as previously described (Fang et al., 2014). To determine NC- and DIO-specific eRNAs, de novo identified eRNAs in DIO were pooled together with eRNAs in NC to prepare a master eRNA peak set. Next, to call high confidence enhancers, we overlapped the identified eRNA loci with the abundance of another enhancer mark, H3K27ac. The filter used was a requirement that the H3K27Ac signal to be 3 times higher than that of matched 2 kb control regions. eRNA peaks whose loci were enriched by these criteria for H3K27ac levels were considered to be high confidence active enhancers. eRNA level was quantified by calculating GRO-seq reads mapped within ± 500bp of the eRNA locus center using HOMER, and normalized to RPKTM. For unidirectional intergenic eRNAs, tag count was performed on the DNA strand where the eRNA peak was identified. For intragenic eRNAs, tag count was performed on the antisense strand of the coding gene as previously described (Fang et al., 2014). Then, the DIO-, NC-specific and “Both” condition circadian eRNAs were determined by JTK_CYCLE as performed for gene body transcription, except that JTK_CYCLE p ≤ 0.05 was used to account for the higher signal to noise inherent among short transcripts.
Motif mining and IMAGE analysis
To identify TFs enriched in the loci of DIO-selective eRNAs with phase surrounding ZT10, eRNAs within 200 kb of DIO-selective circadian genes in phase ZT 7-13 were used for motif analysis using HOMER (Heinz et al., 2010). eRNAs within 200 kb of DIO-selective circadian genes out of phase ZT7-13 were used as background. The same setting was using for IMAGE analysis (Madsen et al., 2017). Motifs were collected from CIS-BP (Weirauch et al., 2014), HOMER (Heinz et al., 2010) and HOCOMOCO (Kulakovskiy et al., 2016). Only motifs that could be mapped to mouse transcription factors were included, and redundant motifs were removed. Motifs for each TF were clustered, merged, and trimmed using methods as previously described (Madsen et al., 2017). DIO-selective circadian eRNAs and genes with phase ZT7-13 were used to compare eRNAs and genes out of phase of ZT7-13 using IMAGE with default parameters based on enhancer activities and gene expressions. In order to show the dynamic changes in motif activity, R package “pheatmap” was used. The average GRO-seq tag counts of target genes (p value < 0.05) for a given transcription factor (PPARα and SREBF) were plotted using eight phases of GRO-seq data from livers of DIO mice.
Western Blot and Gene Expression Analysis
For western blot, nuclear fractionation was performed to determine nSREBP1 levels in mouse livers as described previously (Papazyan et al., 2016a). Lysates were resolved in 4%-12% Bis-Tris NUPAGE gradient gels with MOPS running buffer. Proteins were transferred onto polyvinylidene difluoride membranes and blotted with indicated antibodies.
For gene expression analysis, total RNA was extracted from liver tissue using Trizol reagent followed by RNeasy Mini Kit. The RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit and analyzed by qPCR. Gene expression was normalized to the mRNA levels of the housekeeping gene Arbp and the level of the gene of interest in the control samples.
Feeding Behavior Monitoring
Feeding behavior was studied using the BioDAQ Episodic Food Intake Monitor for mice (BioDAQ, Research Diets, Inc., New Brunswick, NJ), which allows continuous monitoring of meal patterns in undisturbed mice with minimal human interference. NC or DIO mice were habituated for one week to single housing and fed with normal chow or obesogenic diet through a food hopper in regular housing cages with environmental enrichment and bedding material. Water was provided ad libitum from regular water bottles. The feeding behavior were monitored continuously for three days. Data are expressed as the mean ± SEM (n = 4-6 per group). *p < 0.05 in Student’s t-test.
In vivo Lipogenesis Assay
Six hours prior to liver collection (ZT8 or ZT20), mice were provided 4.5 μl/g body weight D2O every two hours for three times. Livers were quickly dissected and snap frozen in liquid nitrogen with a pre-cooled Wollenberger clamp. Then, the frozen liver samples were ground at liquid nitrogen temperature with a Cryomill (Retsch, Newtown, PA). The resulting tissue powder was weighed (10 mg). Tissue powder was incubated with 1 mL of 0.3 M KOH in 90% methanol at 80°C for 1 hr in a 2 mL glass vial for saponificati on. Formic acid (0.1 mL) was then added for neutralization. The saponified fatty acids were extracted by adding 0.5 mL of hexane, vortexing, and transferring the top hexane layer to a new glass vial. Samples were then dried under a stream of N2 and dissolved in 1 mL of isopropanol:methanol (1:1, v/v) solution for LC-MS analysis. Separation was performed by reversed-phase ion-pairing chromatography on a C8 column coupled to negative-ion mode, full-scan LC-MS at 1-Hz scan time and 100,000 resolving power (stand-alone orbitrap; Thermo Fischer Scientific). Data analysis with MAVEN software and natural isotope correction were performed as previously described (Zhang et al., 2017).
16:0/18:1-GPC measurement
Analysis of lipids were performed as described previously (Feng et al., 2018). The resulting tissue powder was weighed in an Eppendorf tube (10 mg). The tissue powder was then dissolved with 1 mL of 0.1 M HCl/MeOH (50/50). Next, 0.5 mL of chloroform was added and vortexed for 10 seconds. After centrifugation at 14,000 g and 4°C for 10 min, the bottom layer was collected with a glass syringe and transferred to a glass MS vial. Samples were then dried under a stream of N2, dissolved in 1 mL of isopropanol:methanol (1:1, v/v) solution and injected into a 1290 Infinity UHPLC system coupled to an Agilent 6550 iFunnel Q-TOF mass spectrometer. To cover both the positive charged (ESI+) and negative charged (ESI−) species, each sample was analyzed twice using the same LC gradient but with different mass spectrometer ionization modes. The LC separation was performed on an Agilent Poroshell 120 EC-C18 column (150 × 2.1 mm, 2.7 m particle size) with a flow rate of 150 L/min. Solvent A was 1 mM ammonium acetate + 0.2% acetic acid in water/methanol (90:10). Solvent B was 1 mM ammonium acetate + 0.2% acetic acid in methanol/2-propanol (2:98). The solvent gradient in volume ratios was as follows: 0–2 min, 25% B; 2–4 min, 25 to 65% B; 4–16 min, 65 to 100% B; 16–20 min, 100% B; 20–21 min, 100 to 25% B; 21–27 min, 25% B. Data obtained by QTOF were first converted into mzXML format and then processed using MAVEN software (http://maven.princeton.edu) to obtain the signal intensity of ∼200 lipid species. The data were filtered by interquartile range and normalized to the total intensity for further analysis. The peak identity of 16:0/18:1-GPC was confirmed by MS/MS.
Quantification of β-oxidation rate
300 μg of protein suspension was added to 1 ml of Krebs-Ringer bicarbonate buffer containing fatty acid-free bovine serum albumin (2.5 mg/ml), 2.5 mM palmitic acid, 10mM carnitine, and 4 mCi of 9,10-3H-palmitoyl-CoA. The mixture was rocked for 2 hr in the dark at 37°C, followed by Folch-based separation of 9,10-3H-palmitoyl-CoA and 3H2O. The aqueous phase was collected, and proteins were precipitated by the addition of 10% tricarboxylic acid followed by centrifugation at 8000 g for 10 min at room temperature. The remaining radioactive palmitoyl-CoA was eliminated by strong anion exchange chromatography with AG 1-X8 formate resin. The effluent containing the 3H2O was collected and used for scintillation counting.
ChIP
ChIP assays were carried out as described previously (Zhang et al., 2015). Briefly, mouse livers were harvested, minced, and cross-linked in 1% formaldehyde for 20 min followed by quenching with 1/20 volume of 2.5 M glycine solution for 5 min and two washes with 1× PBS. Chromatin fragmentation was performed by sonication in lysis buffer (50 mM Tris-HCL at pH 8.0, 0.1% SDS, 10 mM EDTA) using a Bioruptor (Diagenode). Proteins were immunoprecipitated in ChIP buffer using anti-PPARα antibody or anti-H3K27ac overnight. Cross-linking was reversed overnight at 65°C in SDS buffer (50 mM Tris-HCL, 10 mM EDTA, 1% SDS at pH 8), and DNA was isolated using phenol/chloroform/isoamyl alcohol. Precipitated DNA was analyzed by qPCR or high-throughput sequencing.
ChIP-seq
H3K27ac ChIP experiments were performed independently on liver samples from NC or DIO mice harvested at ZT10 or ZT22. DNA was amplified according to the ChIP-seq sample preparation guide provided by Illumina using adaptor oligo and primers from Illumina, enzymes from New England Biolabs, and PCR purification kit and MinElute kit from Qiagen. All ChIP-seq libraries were sequenced on Hiseq 2500 (single-end 50bp) by the Functional Genomics Core of the Penn Diabetes Research Center.
ChIP-seq data processing
Sequenced reads were aligned to the mouse reference genome (mm9) using Bowtie version 1.1.1 (Langmead et al., 2009) with parameters ‘-n 1 -m 1’. Only one uniquely aligning read per genomic position was kept for further analysis. Peak calling was performed with HOMER (Heinz et al., 2010). Data quality and ChIP efficiency are shown in the Table S1D.
Metabolic Profiling and Histology
For hepatic triglyceride profiling, liver powder (5 mg) was dissolved in 1 mL of 0.1 M HCl in 50:50 methanol:H2O. Then, 0.5 mL of chloroform was added to the mixture and vortexed. Samples were then centrifuged at 16,000 g for 5 min and the chloroform phase at the bottom was transferred to a glass vial as the first extract using a Hamilton syringe. 0.5 mL chloroform was added to the remaining material and the extraction was repeated to get the second extract. The combined extract was dried under a stream of N2 and dissolved in 1 mL of isopropanol:methanol (1:1, v/v) solution for LC-MS analysis. Triglyceride species were detected with a positive ion mode on a Q Exactive Plus mass spectrometer coupled to a Vanquish UHPLC System (ThermoFisher Scientific, San Jose, CA). The LC separation was achieved on an Agilent Poroshell 120 EC-C18 column (150 x 2.1 mm, 2.7 μm particle size) at a flow rate of 150 μL/min. The gradient was 0 min, 25% B; 2 min, 25% B; 4 min, 65% B; 16 min, 100% B; 20 min, 100% B; 21 min, 25% B; 27 min, 25% B. Solvent A is 1 mM Ammonium acetate + 0.2% acetic acid in H2O:methanol (90:10). Solvent B is 1 mM Ammonium acetate + 0.2% acetic acid in Methanol:2-propanol (2:98).
For Oil Red O staining, fixed tissues were snap frozen in HistoPrep media and sectioned. The sections were stained with 0.5% Oil Red O in propylene glycerol overnight and then in hematoxylin for 5 sec. The procedures were performed by the Penn Digestive Disease Center Morphology Core.
Serum triglyceride levels were measured using a Triglyceride Assay Kit (StanBio) following the manufacturer’s protocol.
QUANTIFICATION AND STATISTICAL ANALYSIS
Error bars represent the SEM, and statistical significance was determined by unpaired two-tailed Student t-test; a p value of ≤ 0.05 was considered significant. Statistical tests were performed using R or MS Excel.
DATA AND SOFTWARE AVAILABILITY
The GEO accession number for the GRO-seq and ChIP-seq data reported in this paper is GSE108878.
Supplementary Material
Figure S1. DIO reprograms circadian hepatic enhancers and gene transcription, Related to Figure 1.
(A) Phase correlation between DIO-mediated circadian eRNAs and nearest circadian gene transcription. Each row has a unique pair of eRNA (red dot) and gene transcription (blue dot). Data are double plotted for better visualization. (B) Changes of tag counts between ZT10 and ZT22 either from GRO-seq or H3K27ac ChIP-seq in livers of DIO mice at eight eRNA phase groups. (C) Protocol for identifying NC- and DIO-specific eRNAs. Detailed description can be found in Extended Experimental Procedures. (D) Motif enrichment at sites of lost circadian eRNAs with phase ZT18 - ZT24 from HOMER (Heinz et al., 2010). (E-F) REV-ERBα protein level in livers of NC and DIO mice. Density quantifications are expressed as the mean ± SEM (n = 4 per time point).
Figure S2. DIO induces circadian de novo lipogenesis, Related to Figure 2.
(A-B) KEGG pathways that are enriched (p value < 0.05) in DIO-selective (A) or “both” (B) circadian transcripts in livers of NC and DIO mice. (C) Relative mRNA expression of circadian genes in livers of NC and DIO mice. RT-qPCR data are expressed as the mean ± SEM (n = 4-6 per time point). Data are double-plotted for better visualization. (D) Diurnal rhythm (Percentage of Food Intake in the Light Period) of feeding behavior in NC or DIO mice (n = 4-5 per group). NC and DIO mice were put into BioDAQ cage for one week’s adaptation and the data were collected on 3 consecutive days. Data are expressed as the mean ± SEM. *p < 0.05 in Student’s t-test.
Figure S3. DIO-induced circadian expression of SREBP mediates changes in DNL, Related to Figure 3.
(A) Motif enrichment at sites of eRNAs with indicated phase in NC or DIO mice from HOMER (Heinz et al., 2010). (B) Motif enrichment at sites of ZT 7-13 eRNAs from HOMER (Heinz et al., 2010) and IMAGE (Madsen et al., 2017). (C) Motif enrichment at sites of eRNAs loci containing the SREBP motif in Figure 3B from HOMER (Heinz et al., 2010. (D) Relative mRNA expression of Pparγ in livers of NC and DIO mice (n = 4-6 per time point). (E) Pparγ expression in liver, iWAT and eWAT of NC mice or in liver of DIO mice (n = 5 per group). (F-G) Relative mRNA expression of Pparγ (F) and DNL genes including Fasn, Acaca, Acly and Elovl5 (G) in livers of control or Pparγ knockout DIO mice. (n = 4-6 per time point). (H) Relative mRNA expression of Chrebp and Srebp2 in livers of NC and DIO mice. (n = 4-6 per time point). Data are expressed as the mean ± SEM. Data are double plotted for better visualization. (I) Relative mRNA expression of Srebp2 and its target genes in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group).
Figure S4. DIO induces circadian fatty acid oxidation, Related to Figure 4.
Relative mRNA expression of genes involved DNL pathway (A), FAO pathway (B) and core clock components (C) in epididymal white adipose tissue, which were collected at ZT10 and ZT22 in NC and DIO mice (n = 3-5 mice per group). RT-qPCR data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test.
Figure S5. DIO-induced circadian expression of PPARα mediates the acquired rhythm of FAO, Related to Figure 5.
(A) Relative mRNA expression of PPARδ in livers of NC and DIO mice (n = 4-6 per time point). Data are double-plotted for better visualization. (B) PPARα ChIP-qPCR for Acox1, Aldh3a2, Acaa1, Hadh and Ins in livers of control or Pparα knockout livers in DIO mice, which were collected at ZT10 (n = 3-5 per group). (C) Relative eRNA expression, whose loci near Acox1 gene, in livers of control or Pparα knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). RT-qPCR data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test.
Figure S6. SREBP1 activates FAO genes by promoting PPARα transcriptional activity, Related to Figure 6.
(A) Relative eRNA expression, whose loci near Acox1 gene, in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). (B) Serum insulin concentration of normal chow mice followed by 2 and 4 weeks on the hypernutritive diet (n = 3-5 per group). Data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test. (C) Relative expression of Fasn in livers of control and Scap knockout DIO mice with or without receiving WY. RT-qPCR data are presented as mean ± SEM (n = 4 mice per group). ns: p > 0.05, *p < 0.05 and **p < 0.01 in Student’s t-test.
Figure S7. Pharmacological lipid lowering is more effective at the peak of PPARα expression, Related to Figure 7.
Relative expression of Rev-erbα, Cry1 and Srebp1 at ZT8 and at ZT20 in livers of DIO mice with or without receiving WY. RT-qPCR data are presented as mean ± SEM (n = 4-5 mice per group). ns: p > 0.05 in Student’s t-test.
HIGHLIGHTS.
Diet-induced obesity (DIO) globally remodels circadian enhancers in liver
Opposing lipid pathways develop synchronous high amplitude circadian rhythms in DIO
DIO-enhanced circadian transcription of lipid genes requires SREBP and PPARα
Pharmacological lipid lowering is more effective at peak expression of PPARα in DIO
Overnutrition enforces circadian rhythmicity non-core clock genes SREBP and PPARα such that pharmacological lipid lowering is more effective when PPARα is highest.
Acknowledgments
We thank Daniel Cohen for his comments on the manuscript, and Yee Hoon Foong, Yong Hoon Kim, Jarrett Remsberg and other members of the Lazar lab for technical support and valuable discussions. We are grateful to Jesper Grud Skat Madsen and Susanne Mandrup for advice on using the IMAGE program. We thank the Functional Genomics Core, Radioimmunoassay/Biomarkers Core, and the Viral Vector Core of the Penn Diabetes Research Center (P30 DK19525) for next-generation sequencing and virus preparation, respectively. We also acknowledge the Penn DRC Metabolomics Core at Princeton (DK19525). This work was supported by the JPB Foundation (to M.A.L.) as well as by National Institutes of Health grants (R01-DK045586, to M.A.L.; R01-HL54926 to H.I.; R01-DK098542, to D.S. and F32DK116519 to D.G.). C.J. and W.X. was supported by American Diabetes Association Training Grant (1-17-PDF-076 to C.J. and 1-18-PDF-132 to W.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.G. and M.A.L. conceptualized the study, interpreted data and wrote the manuscript, which was revised and approved by all authors. D.G. performed GRO-seq experiments and bioinformatics analysis. Y.X. and P.C.B performed RT-qPCR experiments. P.C.B. and R.P. performed western blot experiments. D.G., Y.X. and D.S. performed H3K27ac ChIP-seq experiments. C.J. and J.D.R designed and performed FA synthesis rate studies. P.T.D. and H.I. designed and performed FAO rate studies. B.F. provide helps to GRO-seq analysis. C.J. performed motif analysis. D.G. and W.H. performed Oil red O assay. Y.X., P.C.B., D.G. and W.H. assisted with animal husbandry.
DECLARATION OF INTERESTS
M.A.L. and J.D.R. are consultants to Pfizer, Inc.
References
- Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci U S A. 2017;114:5312–5317. doi: 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, Turk J, Semenkovich CF. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, Li WP, Horton JD, Goldstein JL, Brown MS. Schoenheimer effect explained–feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J Clin Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Gan L, Yang CS, Liu AB, Lu W, Shao P, Dai Z, Sun P, Luo Z. Effects of Stigmasterol and beta-Sitosterol on Nonalcoholic Fatty Liver Disease in a Mouse Model: A Lipidomic Analysis. J Agric Food Chem. 2018;66:3417–3425. doi: 10.1021/acs.jafc.7b06146. [DOI] [PubMed] [Google Scholar]
- Frick MH, Elo O, Haapa K, Heinonen OP, Heinsalmi P, Helo P, Huttunen JK, Kaitaniemi P, Koskinen P, Manninen V, et al. Helsinki Heart Study: primary-prevention trial with gemfibrozil in middle-aged men with dyslipidemia. Safety of treatment, changes in risk factors, and incidence of coronary heart disease. N Engl J Med. 1987;317:1237–1245. doi: 10.1056/NEJM198711123172001. [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hems DA, Rath EA, Verrinder TR. Fatty acid synthesis in liver and adipose tissue of normal and genetically obese (ob/ob) mice during the 24-hour cycle. Biochem J. 1975;150:167–173. doi: 10.1042/bj1500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel C. Myopathy and rhabdomyolysis with lipid-lowering drugs. Toxicol Lett. 2002;128:159–168. doi: 10.1016/s0378-4274(02)00010-3. [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Palomares JM, Fernandez-Tabera JM, Plana MN, Anino Alba A, Gomez Alvarez P, Fernandez-Esteban I, Saiz LC, Martin-Carrillo P, Pinar Lopez O. Chronotherapy versus conventional statins therapy for the treatment of hyperlipidaemia. Cochrane Database Syst Rev. 2016;11:CD009462. doi: 10.1002/14651858.CD009462.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimian Azari E, Leitner C, Jaggi T, Langhans W, Mansouri A. Possible role of intestinal fatty acid oxidation in the eating-inhibitory effect of the PPAR-alpha agonist Wy-14643 in high-fat diet fed rats. PLoS One. 2013;8:e74869. doi: 10.1371/journal.pone.0074869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S. Integrated physiology and systems biology of PPARalpha. Mol Metab. 2014;3:354–371. doi: 10.1016/j.molmet.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Spotts GD, Halvorsen YD, Shih HM, Ellenberger T, Towle HC, Spiegelman BM. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim MK, Seo HS, Hyun MS, Han KR, Cho SW, Kim YK, Hoon Park S. Efficacy and safety of morning versus evening dose of controlled-release simvastatin tablets in patients with hyperlipidemia: a randomized, double-blind, multicenter phase III trial. Clin Ther. 2013;35:1350–1360 e1351. doi: 10.1016/j.clinthera.2013.06.020. [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, Ba-Alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2016;44:D116–125. doi: 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Madsen JGS, Rauch A, Van Hauwaert EL, Schmidt SF, Winnefeld M, Mandrup S. Integrated analysis of motif activity and gene expression changes of transcription factors. Genome Res. 2017 doi: 10.1101/gr.227231.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PD, Mitchell PD, Schneck DW. Pharmacodynamic effects and pharmacokinetics of a new HMG-CoA reductase inhibitor, rosuvastatin, after morning or evening administration in healthy volunteers. Br J Clin Pharmacol. 2002;54:472–477. doi: 10.1046/j.1365-2125.2002.01688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Korn BS, Hammer RE, Moon YA, Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 2014;28:8–13. doi: 10.1101/gad.228536.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Tognini P, Liu Y, Eckel-Mahan KL, Baldi P, Sassone-Corsi P. Gut microbiota directs PPARgamma-driven reprogramming of the liver circadian clock by nutritional challenge. EMBO Rep. 2016;17:1292–1303. doi: 10.15252/embr.201642463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NTP. National Toxicology Program (NTP) Technical Report on the Toxicity Studies of Wy-14-643. Toxicity Report Series. 2007;62 [Google Scholar]
- Ohdo S. Chronotherapeutic strategy: Rhythm monitoring, manipulation and disruption. Adv Drug Deliv Rev. 2010;62:859–875. doi: 10.1016/j.addr.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology. 2011;53:116–126. doi: 10.1002/hep.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazyan R, Sun Z, Kim YH, Titchenell PM, Hill DA, Lu W, Damle M, Wan M, Zhang Y, Briggs ER, et al. Physiological Suppression of Lipotoxic Liver Damage by Complementary Actions of HDAC3 and SCAP/SREBP. Cell Metab. 2016a;24:863–874. doi: 10.1016/j.cmet.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazyan R, Zhang Y, Lazar MA. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol. 2016b;23:1045–1052. doi: 10.1038/nsmb.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350:aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plakogiannis R, Cohen H, Taft D. Effects of morning versus evening administration of atorvastatin in patients with hyperlipidemia. Am J Health Syst Pharm. 2005;62:2491–2494. doi: 10.2146/ajhp050072. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- Rubins HB, Robins SJ, Collins D, Fye CL, Anderson JW, Elam MB, Faas FH, Linares E, Schaefer EJ, Schectman G, et al. Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial Study Group. N Engl J Med. 1999;341:410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- Saito Y, Yoshida S, Nakaya N, Hata Y, Goto Y. Comparison between morning and evening doses of simvastatin in hyperlipidemic subjects. A double-blind comparative study. Arterioscler Thromb. 1991;11:816–826. doi: 10.1161/01.atv.11.4.816. [DOI] [PubMed] [Google Scholar]
- Schauer PR, Bhatt DL, Kirwan JP, Wolski K, Aminian A, Brethauer SA, Navaneethan SD, Singh RP, Pothier CE, Nissen SE, et al. Bariatric Surgery versus Intensive Medical Therapy for Diabetes - 5-Year Outcomes. N Engl J Med. 2017;376:641–651. doi: 10.1056/NEJMoa1600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A. Physiology Flies with Time. Cell. 2017;171:1232–1235. doi: 10.1016/j.cell.2017.11.028. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Tuteja G, Everett LJ, Li Z, Lazar MA, Kaestner KH. Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol Endocrinol. 2011;25:694–706. doi: 10.1210/me.2010-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B, Rubenstrunk A, Noel B, Rigou G, Delataille P, Millatt LJ, Baron M, Lucas A, Tailleux A, Hum DW, et al. Hepatoprotective effects of the dual peroxisome proliferator-activated receptor alpha/delta agonist, GFT505, in rodent models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Hepatology. 2013;58:1941–1952. doi: 10.1002/hep.26461. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Minami Y, Honma S, Honma K, Iino M, Hashimoto S. Molecular-timetable methods for detection of body time and rhythm disorders from single-time-point genome-wide expression profiles. Proc Natl Acad Sci U S A. 2004;101:11227–11232. doi: 10.1073/pnas.0401882101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Mullican SE, DiSpirito JR, Peed LC, Lazar MA. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARgamma. Proc Natl Acad Sci U S A. 2013;110:18656–18661. doi: 10.1073/pnas.1314863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci U S A. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JM, Doyle PJ, Iglesias MA, Watson DG, Cooney GJ, Kraegen EW. Peroxisome proliferator-activated receptor (PPAR)-alpha activation lowers muscle lipids and improves insulin sensitivity in high fat-fed rats: comparison with PPAR-gamma activation. Diabetes. 2001;50:411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science. 2015;348:1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen L, Liu L, Su X, Rabinowitz JD. Chemical Basis for Deuterium Labeling of Fat and NADPH. J Am Chem Soc. 2017;139:14368–14371. doi: 10.1021/jacs.7b08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Baker SS, Liu W, Tao MH, Patel R, Nowak NJ, Baker RD. Lipid in the livers of adolescents with nonalcoholic steatohepatitis: combined effects of pathways on steatosis. Metabolism. 2011;60:1001–1011. doi: 10.1016/j.metabol.2010.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. DIO reprograms circadian hepatic enhancers and gene transcription, Related to Figure 1.
(A) Phase correlation between DIO-mediated circadian eRNAs and nearest circadian gene transcription. Each row has a unique pair of eRNA (red dot) and gene transcription (blue dot). Data are double plotted for better visualization. (B) Changes of tag counts between ZT10 and ZT22 either from GRO-seq or H3K27ac ChIP-seq in livers of DIO mice at eight eRNA phase groups. (C) Protocol for identifying NC- and DIO-specific eRNAs. Detailed description can be found in Extended Experimental Procedures. (D) Motif enrichment at sites of lost circadian eRNAs with phase ZT18 - ZT24 from HOMER (Heinz et al., 2010). (E-F) REV-ERBα protein level in livers of NC and DIO mice. Density quantifications are expressed as the mean ± SEM (n = 4 per time point).
Figure S2. DIO induces circadian de novo lipogenesis, Related to Figure 2.
(A-B) KEGG pathways that are enriched (p value < 0.05) in DIO-selective (A) or “both” (B) circadian transcripts in livers of NC and DIO mice. (C) Relative mRNA expression of circadian genes in livers of NC and DIO mice. RT-qPCR data are expressed as the mean ± SEM (n = 4-6 per time point). Data are double-plotted for better visualization. (D) Diurnal rhythm (Percentage of Food Intake in the Light Period) of feeding behavior in NC or DIO mice (n = 4-5 per group). NC and DIO mice were put into BioDAQ cage for one week’s adaptation and the data were collected on 3 consecutive days. Data are expressed as the mean ± SEM. *p < 0.05 in Student’s t-test.
Figure S3. DIO-induced circadian expression of SREBP mediates changes in DNL, Related to Figure 3.
(A) Motif enrichment at sites of eRNAs with indicated phase in NC or DIO mice from HOMER (Heinz et al., 2010). (B) Motif enrichment at sites of ZT 7-13 eRNAs from HOMER (Heinz et al., 2010) and IMAGE (Madsen et al., 2017). (C) Motif enrichment at sites of eRNAs loci containing the SREBP motif in Figure 3B from HOMER (Heinz et al., 2010. (D) Relative mRNA expression of Pparγ in livers of NC and DIO mice (n = 4-6 per time point). (E) Pparγ expression in liver, iWAT and eWAT of NC mice or in liver of DIO mice (n = 5 per group). (F-G) Relative mRNA expression of Pparγ (F) and DNL genes including Fasn, Acaca, Acly and Elovl5 (G) in livers of control or Pparγ knockout DIO mice. (n = 4-6 per time point). (H) Relative mRNA expression of Chrebp and Srebp2 in livers of NC and DIO mice. (n = 4-6 per time point). Data are expressed as the mean ± SEM. Data are double plotted for better visualization. (I) Relative mRNA expression of Srebp2 and its target genes in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group).
Figure S4. DIO induces circadian fatty acid oxidation, Related to Figure 4.
Relative mRNA expression of genes involved DNL pathway (A), FAO pathway (B) and core clock components (C) in epididymal white adipose tissue, which were collected at ZT10 and ZT22 in NC and DIO mice (n = 3-5 mice per group). RT-qPCR data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test.
Figure S5. DIO-induced circadian expression of PPARα mediates the acquired rhythm of FAO, Related to Figure 5.
(A) Relative mRNA expression of PPARδ in livers of NC and DIO mice (n = 4-6 per time point). Data are double-plotted for better visualization. (B) PPARα ChIP-qPCR for Acox1, Aldh3a2, Acaa1, Hadh and Ins in livers of control or Pparα knockout livers in DIO mice, which were collected at ZT10 (n = 3-5 per group). (C) Relative eRNA expression, whose loci near Acox1 gene, in livers of control or Pparα knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). RT-qPCR data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test.
Figure S6. SREBP1 activates FAO genes by promoting PPARα transcriptional activity, Related to Figure 6.
(A) Relative eRNA expression, whose loci near Acox1 gene, in livers of control or Scap knockout DIO mice, which were collected at ZT10 or ZT22 (n = 3-5 per group). (B) Serum insulin concentration of normal chow mice followed by 2 and 4 weeks on the hypernutritive diet (n = 3-5 per group). Data are presented as the mean ± SEM, ns: p > 0.05, and *p < 0.05 in Student’s t-test. (C) Relative expression of Fasn in livers of control and Scap knockout DIO mice with or without receiving WY. RT-qPCR data are presented as mean ± SEM (n = 4 mice per group). ns: p > 0.05, *p < 0.05 and **p < 0.01 in Student’s t-test.
Figure S7. Pharmacological lipid lowering is more effective at the peak of PPARα expression, Related to Figure 7.
Relative expression of Rev-erbα, Cry1 and Srebp1 at ZT8 and at ZT20 in livers of DIO mice with or without receiving WY. RT-qPCR data are presented as mean ± SEM (n = 4-5 mice per group). ns: p > 0.05 in Student’s t-test.


