Abstract
The tumor microenvironment can be hypoxic, acidic, and deficient in nutrients. This can cause the metabolism of tumor cells as well as the neighboring stromal cells to be remodelled to facilitate tumor survival, proliferation, and metastasis. Abnormal tumor lipid metabolism is a fairly new field, which has received attention in the past few years. Cross-talk between tumor cells and tumor associated stromal cells modulates the high metabolic needs of the tumor. Fatty acid turnover is high in tumor cells to meet the energy as well as synthetic requirements of the growing tumor. Lipolysis of lipids stored in lipid droplets was earlier considered to be solely carried out by cytosolic lipases. However recent studies demonstrate that lipophagy (autophagic degradation of lipids by acidic lipases) serves as an alternate pathway for the degradation of lipid droplets. Involvement of lipophagy in lipid turnover makes it a crucial player in tumorigenesis and metastasis. In this review we discuss the metabolic reprogramming of tumor cells with special focus on lipid metabolism. We also address the lipid turnover machinery in the tumor cell, especially the lipophagic pathway. Finally, we integrate the current understanding of lipophagy with tumor lipid metabolism.
Keywords: Tumor, tumor microenvironment, lipid metabolism, lipid droplets, lipophagy
Introduction
It is now well established that metabolic adaptations are important for cancer cell survival and proliferation [1]. The tumor microenvironment (TME) and oncogenic events (oncogene activation and loss of tumor suppressors) are critical for metabolic adaptation of cancer cells. The TME is composed of blood vessels, fibroblasts, immune cells, and proteins that compose the extracellular matrix. The TME is characterized by heterogeneous regions of poor oxygen supply (hypoxia) and nutrient deprivation surrounding the tumor cells. In proliferating tumor cells, oxygen demand is far greater than oxygen supply. This can creates regions of hypoxia which is one characteristic of solid tumors [2]. Insufficient diffusion of oxygen in these regions caused by increased distance between the tumor cells and vasculature can markedly influence cancer growth. Hypoxia triggers molecular changes resulting in metabolic adaptations essential for survival of cancer cells. The first adaptive metabolic reprogramming in tumor metabolism was identified to be alterations in glucose metabolism [3; 4]. Tumor cells exhibit the Warburg effect characterized by increased glucose uptake and increased rate of glycolysis leading to lactate production [5]. Malignant cancer cells tend to increase the rate of glycolysis followed by metabolism to pyruvate, which is converted to lactate by lactate dehydrogenase even in the presence of oxygen [5; 6; 7]. This is one of the reasons underlying the acidosis which is another distinctive attribute of the TME. Metabolic reprogramming is also triggered in stromal cells, so that they are able to synthesize glutamine from unconventional sources in the nutrient scarce TME [8]. Glutamine is significant in cancer because of its ability to donate its carbon and amino-nitrogen to metabolites involved in amino-acid and nucleotide metabolism and other growth-promoting pathways [9]. Though altered glycolysis and glutamine metabolism are hallmarks of cancer, lipid metabolic abnormalities in tumor cells have become increasingly recognized in the past few years [10; 11; 12].
Lipids, including sterols, di-/tri-acylglycerols and phospholipids are integral part of biological membranes and are also used for energy storage, production, and cellular signalling. Fatty acids (FA) are indispensable for lipid biosynthesis. They are a diverse class of molecules consisting of hydrocarbon chains of varying lengths and different degrees of unsaturation which comprise the hydrophobic tails of phospholipids and glycolipids. Phospholipids (phosphatidylcholine and phosphatidylethanolamine) and glycolipids along with cholesterol are major components of biological membranes and markedly influence membrane fluidity. FAs also comprise of the hydrophobic tails of diacylglycerol (DAG) and phosphatidylinositol-3,4,5-trisphosphate (PIP3), which are important signalling molecules. During sufficient nutrient availability, FAs are stored in the adipose tissue as triglycerides (TG), and when energy is depleted, TGs are degraded to release FAs. FAs can then undergo oxidation to release energy in the form of high energy phosphate; at a much greater equivalent as compared to that produced by carbohydrates.
Lipids are stored in lipid droplets (LDs) by cells when energy supplies are high (e.g., after eating). Degradation of lipids stored in LDs occurs in response to starvation to meet the energy needs of the cell or when there is an abundance of dietary lipids. Neutral lipolysis and autophagy are two pathways the cell uses to metabolize fat stored in LDs [13]. Autophagic degradation of lipids or lipophagy is a fairly recent discovery, first reported in 2009 [14]. Since then, it has been reported in a wide variety of cells [15; 16; 17; 18; 19], and in different species [20; 21; 22]. The ubiquitous nature of lipophagy across species suggests a conserved role of autophagy in cellular lipid metabolism. In this review, we describe the remodulation of the lipid metabolic profile that occurs in cancer cells. We note the role of TME in this process, and how this modulation of lipid metabolism is one of the underlying factors for tumor metastasis. Finally, we present the role of autophagy in mobilization of stored lipids in the form of LDs and its effect on cancer progression.
Lipid metabolism in the tumor microenvironment
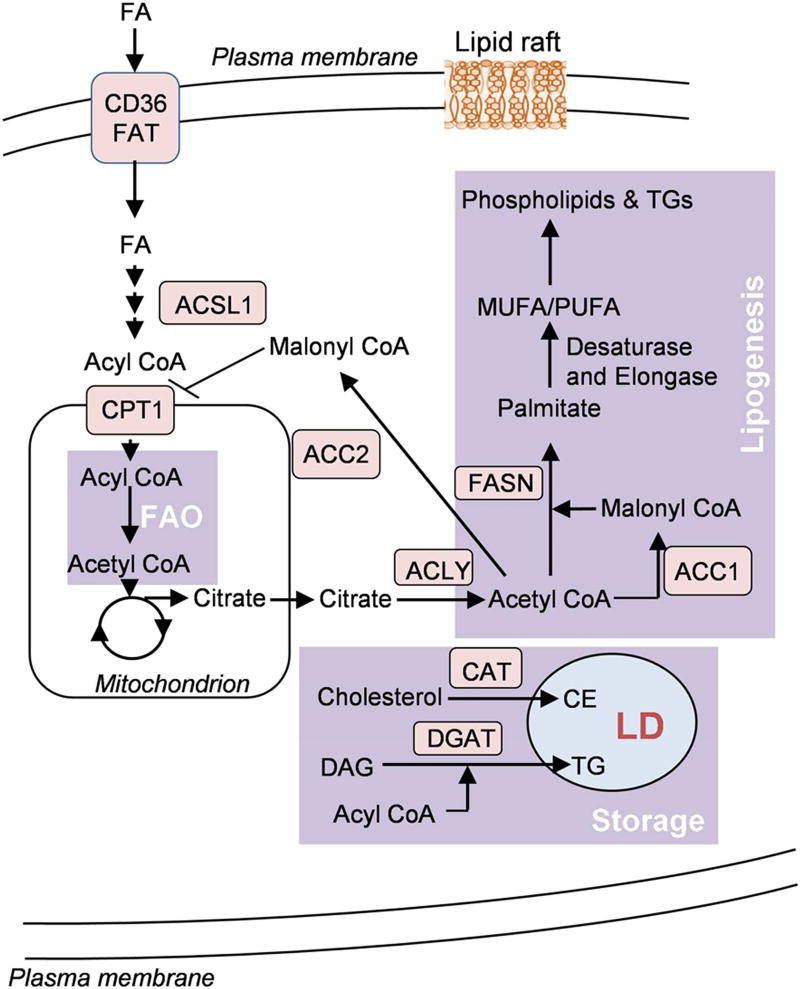
Alteration in lipid metabolism, especially FA synthesis (FAS) and fatty acid oxidation (FAO), has increasingly become recognized as an important metabolic rewiring phenomenon in tumor cells [23] (Figure 1). Cytosolic acetyl CoA serves as a substrate for FA (FA) synthesis. Acetyl-CoA is generated when ATP citrate lyase (ACLY) catalyzes the conversion of citrate to oxaloacetate in the cytoplasm. High ACLY expression has been observed in gastric adenocarcinoma patients [24]. Moreover, genetic suppression of ACLY can reduce cell proliferation and invasion in prostate, lung, osteosarcoma and cervical cancers cells [25]. Acetyl CoA can be carboxylated to malonyl CoA and this reaction is catalysed by the enzyme acetyl-CoA carboxylase (ACC). ACC catalyses the rate limiting step of FAS pathway. There are two isoforms of ACC. The cytosolic ACC1 is present in lipogenic tissues, while the mitochondrial outer membrane bound ACC2 is predominantly present in lipid oxidizing tissues. Because of their differential subcellular location, the two isoforms perform different metabolic functions. ACC1 is involved in lipid synthesis and ACC2 in inhibition of lipid degradation. Enhanced expression of ACC1 has been linked to vascular invasion and disease recurrence in hepatocellular carcinoma patients [26]. Several preclinical studies have also shown the effectiveness of anticancer therapies targeting ACC1 [27; 28].
Figure 1.
A simplified schematic representation of lipid metabolism in tumor cells. Tumor cells show altered lipid metabolic networks involving catabolism (fatty acid oxidation (FAO)), biosynthesis pathways (de novo lipogenesis) and storage as lipid droplets (LDs). Cancer cells show increased fatty acid (FA) uptake by fatty acid translocase (FAT), CD36, which then undergo β-oxidation in the mitochondrial matrix. Acetyl CoA, the end product of FAO pathway, can then either enter TCA cycle or may be transported to the cytosol in the form of citrate for fatty acid synthesis. Excess acyl CoA can undergo esterification with cholesterol or diacylglycerol (DAG) to form cholesterol ester (CE) or triacylglycerol (TG) and is stored in the form of lipid droplets.
Fatty-acid synthase (FASN) is a multi-enzyme protein complex that catalyzes de novo biosynthesis of saturated fatty acids (SFA). The complex utilizes one acetyl-CoA molecule and a sequential addition of seven molecules of malonyl-CoA to form palmitic acid (16:0). Several studies have reported overexpression of FASN in a number of cancers including breast, prostate, ovarian, and colorectal [29; 30; 31]. Further, inhibition of FASN can inhibit proliferation, migration and invasion of hepatocellular carcinoma cells [32].
The end product of the FASN catalysed de novo lipogenesis pathway, palmitate, can be elongated by ELOVL6 (elongation of very long chain fatty acids protein 6) or be desaturated by SCDs (stearoyl-CoA desaturases). SCDs catalyze the conversion of SFA to Δ-9 monounsaturated FAs (MUFAs); e.g., conversion of palmitic acid (16:0) to palmitoleic acid (16:1) and conversion of stearic acid (18:0) to oleic acid (18:1). Humans are reported to have two isoforms of SCD, SCD1 and SCD5 [33]. SCD1 has been shown to be highly expressed in lung adenocarcinoma and was found to be important for cell proliferation, migration and invasion. Moreover, the study suggested SCD1 as a potential biomarker of lung adenocarcinoma [34]. A separate study showed delayed tumor growth in colorectal cancer mouse model on SCD1 inhibitor administration [35]. However, it is important to note that clinically, SCD1 inhibitors have not always proven to be effective therapeutics in humans [36].
In addition to enhanced de novo lipogenesis, tumor cells can acquire FAs through lipolysis to support growth. LPL is a key enzyme for extracellular lipolysis. The enzyme is responsible for hydrolysis of the TGs in circulating chylomicrons and very-low-density lipoprotein (VLDL). The FAs released by hydrolysis of circulating TGs can be taken up by the cells via CD36, a transmembrane channel protein for exogenous free FA uptake. High LPL activity was reported in non-small cell lung cancer tissue [37]. LPL has also been identified as a biomarker for poor prognosis in chronic lymphocytic leukemia [38]. In a separate study, LPL was found to be upregulated in triple negative breast cancer cell lines [39]. Further, the study reported overexpression of CD36, in cancer cell lines. These findings suggest that certain cancer cells use LPL and CD36 to acquire diet-derived FA from the bloodstream by lipolysis which can fuel their growth.
FAs obtained through lipolysis are broken down through mitochondrial fatty acid β-oxidation pathway to meet the energy requirement of the rapidly proliferating cells [40]. FAO involves a cyclical series of reactions that result in the oxidation of β-carbon of the fatty acid. Each cycle of FAO leads to shortening of fatty acids by two carbons and generation of NADH, FADH2 and acetyl CoA. NADH and FADH2, generated by FAO, enter the electron transport chain (ETC) to produce ATP. FAO plays an important role in satisfying the energy needs of cancer cells and ensuring their survival and proliferation in acidic and hypoxic tumor environment. Evidence suggests that cancer cells chronically exposed to acidosis show increased FAO [41]. Due to acidosis, cancer cells show dramatic reduction of glucose derived acetyl-CoA. In such conditions, FAO contributes to the generation of acetyl-CoA which drives the tricarboxylic acid (TCA) cycle for energy production [41].
In addition to the role of FAO in driving the TCA cycle, acetyl CoA produced during the process of FAO is also used for FAS in cancer cells. This is supported by the findings that ACC1 (involved in FAS) is frequently found to be upregulated [30; 42] and ACC2 (involved in inhibition of FAO) inhibited in various cancers [43]. FAs are activated to form acyl CoA in the cytoplasm before they undergo FAO. Acyl CoA is transported to the mitochondrial matrix with the help of carnitine palmitoyltransferase 1 (CPT1) where FAO occurs. CPT1 is considered as the rate limiting enzyme for FAO. CPT1C, an isoform of CPT1, is known to be primarily expressed in the brain [44]. Zaugg and colleagues reported that CPT1C has oncogenic potential [45]. The study reported that CPT1C expression in cancer cells promoted FAO and ATP production, tumor growth, and rescued the cells from metabolic stress [45]. More recently, CPT1C was identified as a biomarker and key regulator of cancer cell senescence through mitochondria-associated metabolic reprogramming. The study went on to suggest that inhibition of CPT1C may be used as a therapeutic strategy for cancer treatment through induction of tumor senescence [46]. Unlike CPT1C, CPT1A, another isoform of CPT1, is expressed in many tissues and most abundantly in the liver. CPT1A is reported to promote metastasis in alveolar rhabdomyosarcoma cells by promoting cell motility [47]. Further, inhibition of CPTIA can result in impaired cancer cell proliferation in acute myeloid leukemia [48; 49].
Metabolic reprogramming by cells in tumor microenvironment
The TME is a highly complex combination of tumor and the neighboring cellular and molecular components. Cellular components of TME include stromal cells (blood and lymphatic endothelial cells, cancer associated fibroblasts (CAFs)), tumor-infiltrating lymphocytes (B cells, T cells and NK cells) and myeloid populations (dendritic cells, myeloid-derived suppressor cells and macrophages) [50]. Molecular components of the TME include chemokines, cytokines, extracellular matrix and the soluble immunosuppressive molecules. The cellular and molecular components of TME co-ordinately act to maintain a condition of chronic inflammation and immune suppression which promotes tumorigenesis, progression, invasion and immune-evasion of tumor [51]. There is a cross-talk between neighboring cells in stromal and cancer cells, and this is essential for cancer cell survival and proliferation.
It has been reported in colon and ovarian cancers that endothelial cells trigger metabolic reprogramming by inducing an over-expression of poly-unsaturated fatty acids (PUFA) and glycerophospholipids in cancer cells [52]. These changes in fatty acid metabolism are essential for tumor progression and aggression [52]. Recently, CAFs have also been shown to enhance lipid synthesis due to over-expression of FASN. Cancer cells over-express fatty acid transporter protein 1, which mediates uptake of FAs from CAFs [53]. Tumor associated dendritic cells are responsible for initiation and maintenance of immune responses. Studies suggest that tumor associated dendritic cells show abnormal lipid accumulation induced by ER stress response factor XBP1. Lipid accumulated dendritic cells were found to be ineffective in presenting tumor-associated antigens thereby promoting kidney, thyroid, ovarian and head and neck cancer [54; 55].
The lipid-activated transcription factors, peroxisome proliferator-activated receptors (PPARs) are a group of nuclear receptors that play a role in regulating lipid homeostasis. The PPAR family comprises of three members: PPARα, PPARβ/δ, and PPARγ. Numerous studies have established the role of PPARα in lipid and lipoprotein metabolism [56; 57]. PPARα has a central role for increasing FAO in hepatocytes [58]. PPARβ/δ promotes FA catabolism in skeletal muscle and may suppress macrophage derived inflammation [59]. In cancers, PPARβ/δ plays an important role in apoptotic cell clearance. In mice, dead cancer cells polarize tumor activated macrophages into an IL-10 expressing, pro-tumor phenotype by inducing FASN and PPARβ/δ activation [60]. However, this has not been demonstrated in a human model to date. This is important to note because the role of PPARβ/δ in cancer remains controversial, with some studies showing that activating this PPAR promotes cancer while others indicate that activating this PPAR isoform inhibits cancer [61; 62; 63]. PPARγ induces differentiation of preadipocytes into adipocytes and stimulates triglyceride storage [64]. Several studies indicate that activation of PPARγ suppresses tumor development and progression by making the TME less hospitable for tumor growth and metastasis [65; 66]. PPARγ and FASN expression have recently been reported to be positively correlated with human prostate cancer [67]. However, similar to PPARβ/δ, the role of PPARγ in some cancers remains controversial. For example, there are ongoing clinical trials examining whether PPARγ agonists may be effective for preventing or treating cancer [63]. Another lipid activated transcription factor, the liver X receptor (LXR), also plays an important role in modulating the TME. Apoptotic tumor cells containing oxysterols activate LXR in macrophages causing suppression of dendritic cell migration and recruitment of neutrophils in tumors resulting in tolerance and immunosuppression [68].
The role of lipid metabolism in metastasis
The metastatic potential of cancer cells positively correlates with the expression of genes involved in fatty acid synthesis, oxidation and intracellular lipid storage. Metabolic alterations are important in metastasis of melanoma, breast and prostate cancers. Enzymes involved in lipid metabolism have been shown to play a role in metastasis. For example, SCD and long chain fatty acyl synthetase (ACSL) 1 and 4 cooperate to induce epithelial to mesenchymal transition resulting in an increased invasion potential of colon cancer cells [69]. This is further supported by findings that inhibition of these enzymes restores epithelial features and restricts cancer cell growth. More recently, a study reported over-expression of genes involved in fatty acid uptake (CAV1, CD36, PPARα) and de novo lipogenesis (MLXIPL) in metastatic tumors [70]. A separate study reported an association between metastatic potential and levels of several phospholipids including phosphatidylcholine (PC), phosphatidylinositol (PI), phosphatidylethanolamine (PE) and phosphatidylserine (PS) in metastatic breast cancer cell line. The study reported that higher metastatic potential strongly correlated with higher levels of PS 18:0/20:4, PI 18:0/20:4, and PC 18:0/20:4. On the other hand, lower metastatic potential correlated with lower levels of PE 18:1/18:1 and PI 18:0/18:1 [71].
Dietary lipid dependence of metastasis-initiating cells has recently been reported in melanoma- and breast cancer. Inhibition of fatty acid translocase CD36 was shown to impair metastasis [72]. Conversely, over-expression of CD36 significantly increased lymph node metastasis of oral squamous cell carcinoma cell lines with low metastatic potential [72]. These findings establish fatty acid receptor CD36 as a marker and functional driver of metastasis in a lipid metabolism-dependent manner. CD36 has been associated with progression and poor prognosis in several other tumor types, such as glioblastoma and hepatocellular carcinoma. Metastatic-initiating cells are also characterized by a distinct lipid metabolic signature related to fatty acid degradation, de novo lipogenesis and lipid storage. In invasive ductal carcinoma, acetyl-CoA synthetase 2 (ACSS2), an enzyme that catalyzes the conversion of acetate to acetyl-CoA, was found to be overexpressed under hypoxic and lipid depleted conditions. ACSS2 increases acetate consumption and thereby fatty acid biosynthesis in harsh TME and scarcity of alternate carbon sources for lipogenesis [73].
Lipid rafts are highly dynamic lipid microdomains in the plasma membrane. They are known to be rich in cholesterol and sphingolipids. The highly dynamic nature of lipid rafts are known to be important for cancer metastasis [74; 75; 76; 77; 78]. Edelfosine, a synthetic analog of ether phospholipid, has been shown to accumulate in lipid rafts and induce apoptosis in several hematopoietic cancer cells [79; 80]. Palmitoylation of the CD44 antigen, a cell-surface glycoprotein cell adhesion and migration, facilitates its sequestration in lipid rafts and was shown to influence migration of invasive breast cancer cells [75]. A separate study showed that cholesterol depletion disrupts lipid rafts, and enhances microdomain-dependent CD44 shedding, thereby suppressing tumor cell metastasis in human glioma and pancreatic cancer cells [77].
Lipophagy in cancer
As discussed above, both lipid uptake and de novo lipogenesis can be enhanced in cancer cells. Complementary lipid degradation mechanisms are required to serve the purpose of energy generation, biosynthesis of membranes, and synthesis of other biomolecules.
Lipid droplets
The surplus lipids in a cell do not exist in the form of non-esterified free fatty acids (FFAs) because FFAs have a potential for cytotoxicity at high concentration [81]. Therefore, cells store excess fatty acids and cholesterol in the form of neutral, inert biomolecules such as sterol esters and TGs in cellular structure called lipid droplets or LDs [82; 83]. LDs are intracellular deposits of lipids surrounded by a layer of phospholipids and finally separated from the hydrophilic cytosol by structural proteins known as perilipins (PLINs) [84; 85; 86]. Stored lipids in the form of LDs with sizes ranging from 0.1–10 µm are present in all cells, however in the adipose tissue they coalesce to form a single large droplet with sizes up to 100 µm. Apart from size, the content of LDs in adipose tissue also differs from other cells. While lipids are stored in the core of LDs as cholesterol and triglycerides, only triglycerides are predominant in the core of LDs in adipose tissue [87; 88]. Cancer cells are characterized with upregulation of FAS and FA uptake, leading to increased accumulation of LDs. This is supported by studies which show that a phenotype of abundant LDs in cells is a marker for colorectal cancer [89] and is also associated with lung cancer [90]. Recent studies have also linked LD abundance with increased aggressiveness [91], as well as to chemotherapy resistance [92] of tumors.
Lipophagy
The mobilization of lipids from LDs is mediated by lipolysis (Figure 2). The dynamic interactions between cytosolic lipases with inhibitory proteins (also present in cytosol) and perilipins regulates the rate of lipolysis [93]. Cells initiate lipolysis to meet their energy requirement as well as on overabundance of lipids to prevent stores from being compromisingly enlarged. Previously, mobilization of lipids in LDs by lipolysis was perceived to be entirely carried out by LD associated lipases. However, recent findings indicate that autophagy serves as an alternate pathway [14]. Autophagy is a catabolic process which involves self degradation of cellular components [94]. Degradation of cellular organelles and proteins is the most well studied function of autophagy. It has been well established that under nutrient stress, autophagy degrades cellular components to provide the cells with essential building blocks as well as to satisfy its energy needs. Most of these studies were focused on protein degradation by autophagy to release amino acids which could be later used as building blocks or to provide substrates for energy. However, recent studies suggest that autophagy has a role in degradation of lipids to release free fatty acids which are a highly efficient source of energy as compared to amino acids or carbohydrates. Lipophagy was initially demonstrated in hepatocytes [14; 95]. Since this original observation a variety of cell types like adipocytes [19], enterocytes[18], glial cells [17], T cells [16], neurons [15] have also exhibited lipophagy under conditions of lipotoxicity or nutrient stress. Interestingly, lipophagy has also been reported in rice [96], yeast [22], C.elegans [21] as well as some fungi [20]. Therefore, lipophagy seems to be a well-conserved mechanism of lipid degradation.
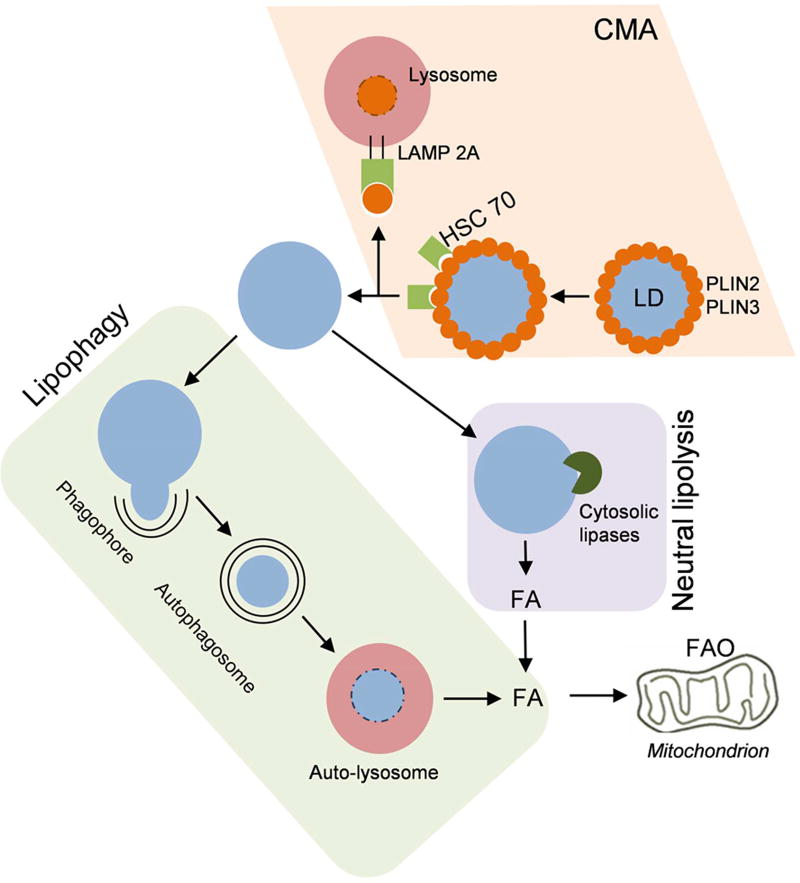
Figure 2.
Schematic representation of Lipolysis: Lipid droplet surface proteins PLIN2 and PLIN3 are degraded via chaperone mediated autophagy (CMA). Consequently the LD surface is exposed for the action of neutral cytosolic lipases and lipophagic machinery. A double membrane engulfs a part of or whole LD, thus forming autophagosome which fuses with lysosome to form autolysosome. Lysosomal acid lipases act on the lipids to form free fatty acids. Cytosolic lipases directly act on LD surface to degrade lipids to fatty acids. Subsequently, β-oxidation of fatty acids takes place in mitochondria to generate energy and metabolic intermediates.
Singh and colleagues showed that lysosomes do not directly fuse with LDs [14]. LDs as a whole or a part thereof is encapsulated in a double membrane autophagosome which then provides its components to lysosomes for degradation [14]. It has been proposed that LD surface proteins (PLINs) act as gatekeepers of LDs, with their degradation being a pre-requisite for lipolysis to occur. Degradation of PLINs has been shown to correlate with lipolysis [97; 98] with both being induced under starvation. The former group showed that polyubiquitination of PLINs tags them for proteasomal degradation. Kaushik and Cuervo show that PLINs are targeted for chaperone mediated autophagic (CMA) degradation [97]. HSC70 binds with PLIN2 and PLIN3 by recognising a canonical pentapeptide motif present in both proteins. The HSC70 and PLIN complex binds with lysosome associated membrane protein 2A (LAMP2A), leading to their uptake and simultaneous degradation by lysosome. Kaushik and Cuervo show that blocking of CMA results in decreased association of cytosolic lipases and autophagic machinery with LDs and thereby reduced LD degradation [97]. Thus, cross-talk between proteolytic and lypolytic machinery of cell regulates LD turnover. The mechanism underlying the recognition of LDs by autophagic machinery and involvement of any receptors still remain unanswered. Poly-ubiquitination with specific lysine linkages are a well established tag for lysosomal degradation of proteins. However, the role of poly-ubiquitination in lipophagy remains unclear. Interestingly, an Ube2g2 (E2 ubiquitin conjugase G2) interacting protein, ancient ubiquitous protein (AUP1) has been shown to associate with LDs. This may be involved in tagging LDs for lysosomal degradation [99]. However, as LDs are known to sequester several proteins [100], the specificity of this interaction to LD degradation needs to be further studied. Apart from polyubiquitination as a signal for specific autophagic degradation of LDs, autophagy selective receptors may also be involved in conferring selectivity to lipophagy. One such receptor of interest is Huntingtin, which acts as a scaffold for component recognition by autophagy [101]. Importantly, cells lacking functional Huntingtin exhibit lipid accumulation [102]. Therefore, the possibility of Huntingtin as a lipophagic receptor for LD recognition should be explored. A separate study reports that LC3, an autophagic protein involved in autophagosome biogenesis is capable of binding the phospholipid, cardiolipin [103]. It may therefore be presumed that LC3 may directly recognize LD lipids. Further studies are therefore warranted to establish the molecular mechanisms behind specific recognition of LDs by lipophagic machinery.
The relationship between autophagy and cancer is complex and unclear. Autophagy is reported to show both positive and negative effects on tumor progression [104]. Autophagy catabolizes damaged proteins and organelles like mitochondria and peroxisomes, which are potential sources of ROS. Autophagy thus protects the cell from oxidative stress damage and chronic inflammation [105; 106]. In the presence of intracellular stress, autophagy eliminates the damaged and toxic cellular components to ensure cellular homeostasis. Mathew and colleagues showed that the absence of autophagy led to an accumulation of protein aggregates and ER chaperones which led to an activation of the DNA damage response [107]. In addition to preventing genetic mutations, autophagy may also hinder the growth of mutation bearing cancer cells [88]. Therefore, as a cytoprotective mechanism which plays the role of cell’s garbage disposal system, autophagy may prevent tumor initiation and progression. However, autophagy is responsible for the metabolic plasticity which is characteristic of cancer cells. Therefore, autophagy may play an essential role in survival of cancer cells in oxygen, pH, and nutrient stress [108; 109; 110].
Lipophagy also plays a dual pro- and anti-cancer role. A tumor suppressor function has been ascribed to lysosomal acid lipase (LAL), with their activity associated with tumorigenesis and metastasis [111]. LAL deficiency results in an abnormal haematopoiesis leading to an abundance of immature myeloid-derived suppressor cells (MSDCs). MSDCs mediate suppression of immune surveillance and thus evasion of host immunity by the tumor [112]. In addition, it is reported that LAL deficiency induced MSDCs can directly stimulate tumorigenesis and metastasis [113]. By contrast, a tumor suppressor role for LAL is suggested by studies showing that expression of LAL improves lipid metabolism, as well as reduces metastasis in lung and liver cancer [114; 115]. Another recent study reported that lipophagy mediates ER stress induced apoptosis [116]. Moreover, lipophagy impairment correlates with poor patient prognosis and survival [117; 118; 119]. Apart from its tumor suppressor role, lipophagy-dependent degradation of lipids, may provide the rapidly proliferating cancer cells with energy substrates and intermediates for synthesis of biomolecules, thus helping them survive [120]. These studies though still preliminary, provide an insight into the significance of hitherto underappreciated role of lipophagy in cancer metabolism. Thus, it is clear that further studies are needed to determine the role of lipophagy in cancer, which could vary depending on tumor type, and the stage of tumorigenesis.
It is now well established that metabolic adaptations are important for cancer cell survival and proliferation. The TME has an essential role in the metabolic adaptation of cancer cells. To meet the nutrient requirements of the rapid cell proliferation of cancer cells, multiple substrates other than glucose are likely needed. It has been suggested that lipids may be a major alternate fuel supporting cancer cell proliferation. Redirecting lipid metabolism in tumor cells and its role in tumor progression and metastasis has received widespread attention in recent years. The role of autophagy in degradation of lipids stored in the form of lipid droplets in now increasingly recognized in the regulation of lipid homeostasis. Despite major advancement in our understanding of lipophagy, many questions still remain unanswered. (i) Whether cross-talk occurs between neutral lipolysis and lipophagy, and if so, under what circumstances? (ii) Is selectivity and specificity involved in degradation of LDs by lipophagy? (iii) Is a subset of lipids preferentially degraded by lipophagy over others? (iv) What are the structural and functional characteristics of the lipophagic machinery? Finally, characterization of the mechanistic details of lipophagy perturbations in tumor progression is required to fully take advantage of its potential as a target for novel cancer chemoprevention and chemotherapy.
Supplementary Material
Highlights.
Alterations in lipid metabolism modulate tumor development and progression
Lipophagy plays a central role in regulating lipid homeostasis in tumors
The role of lipophagy in cancer remains underappreciated
Acknowledgments
This work was supported in part by the following grants: ES0221896 (A.D.P), ES028288 (A.D.P.), CA124533 (J.M.P.), CA140369 (J.M.P.), and the USDA National Institute of Food and Agriculture Federal Appropriations under Project 4607.
ABBREVIATIONS
- ACC
Acetyl-CoA carboxylase
- ACLY
ATP citrate lyase
- ACSS
Acetyl-CoA synthetase
- AUP1
Ancient ubiquitous protein
- CAF
Cancer associated fibroblast
- CMA
Chaperone mediated autophagy
- CPT
Carnitine palmitoyltransferase 1
- DAG
diacylglycerol
- ER
Endoplasmic reticulum
- FA
Fatty acid
- FAO
Fatty acid oxidation
- FAS
Fatty acid synthesis
- FASN
Fatty-acid synthase
- FAT
Fatty acid translocase
- FFA
Free fatty acid
- HSC70
Heat shock cognate 71 kDa protein
- LAL
Lysosomal acid lipase
- LAMP
Lysosomal-associated membrane protein
- LD
Lipid droplets
- LPL
Lipoprotein lipase
- LXR
Liver X receptor
- MSDC
Myeloid-derived suppressor cells
- MUFA
Monounsaturated fatty acid
- PIP3
Phosphatidylinositol-3,4,5-trisphosphate
- PLIN
Perilipin
- PPAR
Peroxisome proliferator-activated receptors
- PUFA
Poly-unsaturated fatty acid
- ROS
Reactive oxygen species
- SCD
Stearoyl-CoA desaturase
- SFA
Saturated FA
- SFA
Saturated fatty acid
- TG
Triglycerides
- TME
Tumor microenvironment
- Ube2g2
E2 ubiquitin conjugase G2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Alberghina L, Stephanopoulos G, Chiaradonna F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–30. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 7.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–70. [PubMed] [Google Scholar]
- 8.Yang L, Achreja A, Yeung TL, Mangala LS, Jiang D, Han C, Baddour J, Marini JC, Ni J, Nakahara R, Wahlig S, Chiba L, Kim SH, Morse J, Pradeep S, Nagaraja AS, Haemmerle M, Kyunghee N, Derichsweiler M, Plackemeier T, Mercado-Uribe I, Lopez-Berestein G, Moss T, Ram PT, Liu J, Lu X, Mok SC, Sood AK, Nagrath D. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016;24:685–700. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina MA, Sanchez-Jimenez F, Marquez J, Rodriguez Quesada A, Nunez de Castro I. Relevance of glutamine metabolism to tumor cell growth. Mol Cell Biochem. 1992;113:1–15. doi: 10.1007/BF00230880. [DOI] [PubMed] [Google Scholar]
- 10.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–91. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–61. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. 2010;1801:381–91. doi: 10.1016/j.bbalip.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–684. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–83. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol. 2010;185:7349–57. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khaldoun SA, Emond-Boisjoly MA, Chateau D, Carriere V, Lacasa M, Rousset M, Demignot S, Morel E. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol Biol Cell. 2014;25:118–32. doi: 10.1091/mbc.E13-06-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW., Jr Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–30. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen LN, Bormann J, Le GT, Starkel C, Olsson S, Nosanchuk JD, Giese H, Schafer W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet Biol. 2011;48:217–24. doi: 10.1016/j.fgb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Lapierre LR, Gelino S, Melendez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011;21:1507–14. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beloribi-Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian X, Hu J, Zhao J, Chen H. ATP citrate lyase expression is associated with advanced stage and prognosis in gastric adenocarcinoma. Int J Clin Exp Med. 2015;8:7855–60. [PMC free article] [PubMed] [Google Scholar]
- 25.Xin M, Qiao Z, Li J, Liu J, Song S, Zhao X, Miao P, Tang T, Wang L, Liu W, Yang X, Dai K, Huang G. miR-22 inhibits tumor growth and metastasis by targeting ATP citrate lyase: evidence in osteosarcoma, prostate cancer, cervical cancer and lung cancer. Oncotarget. 2016;7:44252–44265. doi: 10.18632/oncotarget.10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang MD, Wu H, Fu GB, Zhang HL, Zhou X, Tang L, Dong LW, Qin CJ, Huang S, Zhao LH, Zeng M, Wu MC, Yan HX, Wang HY. Acetyl-coenzyme A carboxylase alpha promotion of glucose-mediated fatty acid synthesis enhances survival of hepatocellular carcinoma in mice and patients. Hepatology. 2016;63:1272–86. doi: 10.1002/hep.28415. [DOI] [PubMed] [Google Scholar]
- 27.Rios Garcia M, Steinbauer B, Srivastava K, Singhal M, Mattijssen F, Maida A, Christian S, Hess-Stumpp H, Augustin HG, Muller-Decker K, Nawroth PP, Herzig S, Berriel Diaz M. Acetyl-CoA Carboxylase 1-Dependent Protein Acetylation Controls Breast Cancer Metastasis and Recurrence. Cell Metab. 2017;26:842–855. e5. doi: 10.1016/j.cmet.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Jones JE, Esler WP, Patel R, Lanba A, Vera NB, Pfefferkorn JA, Vernochet C. Inhibition of Acetyl-CoA Carboxylase 1 (ACC1) and 2 (ACC2) Reduces Proliferation and De Novo Lipogenesis of EGFRvIII Human Glioblastoma Cells. PLoS One. 2017;12:e0169566. doi: 10.1371/journal.pone.0169566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 30.Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin Cancer Res. 1997;3:2115–20. [PubMed] [Google Scholar]
- 31.Chavarro JE, Kenfield SA, Stampfer MJ, Loda M, Campos H, Sesso HD, Ma J. Blood levels of saturated and monounsaturated fatty acids as markers of de novo lipogenesis and risk of prostate cancer. Am J Epidemiol. 2013;178:1246–55. doi: 10.1093/aje/kwt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hao Q, Li T, Zhang X, Gao P, Qiao P, Li S, Geng Z. Expression and roles of fatty acid synthase in hepatocellular carcinoma. Oncol Rep. 2014;32:2471–6. doi: 10.3892/or.2014.3484. [DOI] [PubMed] [Google Scholar]
- 33.Ntambi JM, editor. Stearoyl-CoA Desaturase Genes in Lipid Metabolism. Springer-Verlag; New York: 2013. [Google Scholar]
- 34.Huang L. Evaluating the Performance of a New Model for Predicting the Growth of Clostridium perfringens in Cooked, Uncured Meat and Poultry Products under Isothermal, Heating, and Dynamically Cooling Conditions. J Food Sci. 2016;81:M1754–65. doi: 10.1111/1750-3841.13356. [DOI] [PubMed] [Google Scholar]
- 35.Chen L, Ren J, Yang L, Li Y, Fu J, Tian Y, Qiu F, Liu Z, Qiu Y. Stearoyl-CoA desaturase-1 mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci Rep. 2016;6:19665. doi: 10.1038/srep19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Dales NA, Winther MD. Opportunities and challenges in developing stearoyl-coenzyme A desaturase-1 inhibitors as novel therapeutics for human disease. J Med Chem. 2014;57:5039–56. doi: 10.1021/jm401516c. [DOI] [PubMed] [Google Scholar]
- 37.Podgornik H, Sok M, Kern I, Marc J, Cerne D. Lipoprotein lipase in non-small cell lung cancer tissue is highly expressed in a subpopulation of tumor-associated macrophages. Pathol Res Pract. 2013;209:516–20. doi: 10.1016/j.prp.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Kaderi MA, Kanduri M, Buhl AM, Sevov M, Cahill N, Gunnarsson R, Jansson M, Smedby KE, Hjalgrim H, Jurlander J, Juliusson G, Mansouri L, Rosenquist R. LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia. Haematologica. 2011;96:1153–60. doi: 10.3324/haematol.2010.039396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuemmerle NB, Rysman E, Lombardo PS, Flanagan AJ, Lipe BC, Wells WA, Pettus JR, Froehlich HM, Memoli VA, Morganelli PM, Swinnen JV, Timmerman LA, Chaychi L, Fricano CJ, Eisenberg BL, Coleman WB, Kinlaw WB. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol Cancer Ther. 2011;10:427–36. doi: 10.1158/1535-7163.MCT-10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–32. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corbet C, Pinto A, Martherus R, Santiago de Jesus JP, Polet F, Feron O. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016;24:311–23. doi: 10.1016/j.cmet.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Yahagi N, Shimano H, Hasegawa K, Ohashi K, Matsuzaka T, Najima Y, Sekiya M, Tomita S, Okazaki H, Tamura Y, Iizuka Y, Nagai R, Ishibashi S, Kadowaki T, Makuuchi M, Ohnishi S, Osuga J, Yamada N. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur J Cancer. 2005;41:1316–22. doi: 10.1016/j.ejca.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 43.Camarda R, Zhou AY, Kohnz RA, Balakrishnan S, Mahieu C, Anderton B, Eyob H, Kajimura S, Tward A, Krings G, Nomura DK, Goga A. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–32. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrasco P, Jacas J, Sahun I, Muley H, Ramirez S, Puisac B, Mezquita P, Pie J, Dierssen M, Casals N. Carnitine palmitoyltransferase 1C deficiency causes motor impairment and hypoactivity. Behav Brain Res. 2013;256:291–7. doi: 10.1016/j.bbr.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, Kalliomaki T, Elia A, Luo X, Nadeem V, Bungard D, Yalavarthi S, Growney JD, Wakeham A, Moolani Y, Silvester J, Ten AY, Bakker W, Tsuchihara K, Berger SL, Hill RP, Jones RG, Tsao M, Robinson MO, Thompson CB, Pan G, Mak TW. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25:1041–51. doi: 10.1101/gad.1987211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Chen Y, Guan L, Zhang H, Huang Y, Johnson CH, Wu Z, Gonzalez FJ, Yu A, Huang P, Yang S, Chen P, Fan X, Huang M, Bi H. Carnitine palmitoyltransferase 1C regulates cancer cell senescence through mitochondria-associated metabolic reprograming. Cell Death Differ. 2018 doi: 10.1038/s41418-017-0013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L, Wang YD, Wu J, Cui J, Chen T. Carnitine palmitoyltransferase 1A (CPT1A): a transcriptional target of PAX3-FKHR and mediates PAX3-FKHR-dependent motility in alveolar rhabdomyosarcoma cells. BMC Cancer. 2012;12:154. doi: 10.1186/1471-2407-12-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricciardi MR, Mirabilii S, Allegretti M, Licchetta R, Calarco A, Torrisi MR, Foa R, Nicolai R, Peluso G, Tafuri A. Targeting the leukemia cell metabolism by the CPT1a inhibition: functional preclinical effects in leukemias. Blood. 2015;126:1925–9. doi: 10.1182/blood-2014-12-617498. [DOI] [PubMed] [Google Scholar]
- 49.Lee EA, Angka L, Rota SG, Hanlon T, Mitchell A, Hurren R, Wang XM, Gronda M, Boyaci E, Bojko B, Minden M, Sriskanthadevan S, Datti A, Wrana JL, Edginton A, Pawliszyn J, Joseph JW, Quadrilatero J, Schimmer AD, Spagnuolo PA. Targeting Mitochondria with Avocatin B Induces Selective Leukemia Cell Death. Cancer Res. 2015;75:2478–88. doi: 10.1158/0008-5472.CAN-14-2676. [DOI] [PubMed] [Google Scholar]
- 50.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–82. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 51.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 52.Halama A, Guerrouahen BS, Pasquier J, Satheesh NJ, Suhre K, Rafii A. Nesting of colon and ovarian cancer cells in the endothelial niche is associated with alterations in glycan and lipid metabolism. Sci Rep. 2017;7:39999. doi: 10.1038/srep39999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopes-Coelho F, Andre S, Felix A, Serpa J. Breast cancer metabolic cross-talk: Fibroblasts are hubs and breast cancer cells are gatherers of lipids. Mol Cell Endocrinol. 2017 doi: 10.1016/j.mce.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 54.Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE, Gupta D, Holcomb K, Ellenson LH, Caputo T, Lee AH, Conejo-Garcia JR, Glimcher LH. ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell. 2015;161:1527–38. doi: 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, Corzo A, Cho HI, Celis E, Lennox B, Knight SC, Padhya T, McCaffrey TV, McCaffrey JC, Antonia S, Fishman M, Ferris RL, Kagan VE, Gabrilovich DI. Lipid accumulation and dendritic cell dysfunction in cancer. Nat Med. 2010;16:880–6. doi: 10.1038/nm.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol. 2015;62:720–33. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 57.Gervois P, Torra IP, Fruchart JC, Staels B. Regulation of lipid and lipoprotein metabolism by PPAR activators. Clin Chem Lab Med. 2000;38:3–11. doi: 10.1515/CCLM.2000.002. [DOI] [PubMed] [Google Scholar]
- 58.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–80. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–7. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park J, Lee SE, Hur J, Hong EB, Choi JI, Yang JM, Kim JY, Kim YC, Cho HJ, Peters JM, Ryoo SB, Kim YT, Kim HS. M-CSF from Cancer Cells Induces Fatty Acid Synthase and PPARbeta/delta Activation in Tumor Myeloid Cells, Leading to Tumor Progression. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 61.Peters JM, Yao PL, Gonzalez FJ. Targeting Peroxisome Proliferator-Activated Receptor-beta/delta (PPARbeta/delta) for Cancer Chemoprevention. Curr Pharmacol Rep. 2015;1:121–128. doi: 10.1007/s40495-015-0026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters JM, Gonzalez FJ, Muller R. Establishing the Role of PPARbeta/delta in Carcinogenesis. Trends Endocrinol Metab. 2015;26:595–607. doi: 10.1016/j.tem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–95. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ferre P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes. 2004;53(Suppl 1):S43–50. doi: 10.2337/diabetes.53.2007.s43. [DOI] [PubMed] [Google Scholar]
- 65.Chang TH, Szabo E. Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor gamma in non-small cell lung cancer. Cancer Res. 2000;60:1129–38. [PubMed] [Google Scholar]
- 66.Keshamouni VG, Han S, Roman J. Peroxisome proliferator-activated receptors in lung cancer. PPAR Res. 2007;2007:90289. doi: 10.1155/2007/90289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad I, Mui E, Galbraith L, Patel R, Tan EH, Salji M, Rust AG, Repiscak P, Hedley A, Markert E, Loveridge C, van der Weyden L, Edwards J, Sansom OJ, Adams DJ, Leung HY. Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci U S A. 2016;113:8290–5. doi: 10.1073/pnas.1601571113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Traversari C, Sozzani S, Steffensen KR, Russo V. LXR-dependent and -independent effects of oxysterols on immunity and tumor growth. Eur J Immunol. 2014;44:1896–903. doi: 10.1002/eji.201344292. [DOI] [PubMed] [Google Scholar]
- 69.Sánchez-Martínez R, Cruz-Gil S, de Cedrón MG, Álvarez-Fernández M, Vargas T, Molina S, García B, Herranz J, Moreno-Rubio J, Reglero G, Pérez-Moreno M, Feliu J, Malumbres M, de Molina AR. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget. 2015;6:38719–38736. doi: 10.18632/oncotarget.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nath A, Chan C. Genetic alterations in fatty acid transport and metabolism genes are associated with metastatic progression and poor prognosis of human cancers. Sci Rep. 2016;6:18669. doi: 10.1038/srep18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim HY, Lee KM, Kim SH, Kwon YJ, Chun YJ, Choi HK. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget. 2016;7:67111–67128. doi: 10.18632/oncotarget.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, Berenguer A, Prats N, Toll A, Hueto JA, Bescos C, Di Croce L, Benitah SA. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017;541:41–45. doi: 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 73.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, McGarry L, James D, Shanks E, Kalna G, Saunders RE, Jiang M, Howell M, Lassailly F, Thin MZ, Spencer-Dene B, Stamp G, van den Broek NJ, Mackay G, Bulusu V, Kamphorst JJ, Tardito S, Strachan D, Harris AL, Aboagye EO, Critchlow SE, Wakelam MJ, Schulze A, Gottlieb E. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murai T. Lipid Raft-Mediated Regulation of Hyaluronan-CD44 Interactions in Inflammation and Cancer. Front Immunol. 2015;6:420. doi: 10.3389/fimmu.2015.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Babina IS, McSherry EA, Donatello S, Hill AD, Hopkins AM. A novel mechanism of regulating breast cancer cell migration via palmitoylation-dependent alterations in the lipid raft affiliation of CD44. Breast Cancer Res. 2014;16:R19. doi: 10.1186/bcr3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murai T. The role of lipid rafts in cancer cell adhesion and migration. Int J Cell Biol. 2012;2012:763283. doi: 10.1155/2012/763283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murai T, Maruyama Y, Mio K, Nishiyama H, Suga M, Sato C. Low cholesterol triggers membrane microdomain-dependent CD44 shedding and suppresses tumor cell migration. J Biol Chem. 2011;286:1999–2007. doi: 10.1074/jbc.M110.184010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim KB, Yi JS, Nguyen N, Lee JH, Kwon YC, Ahn BY, Cho H, Kim YK, Yoo HJ, Lee JS, Ko YG. Cell-surface receptor for complement component C1q (gC1qR) is a key regulator for lamellipodia formation and cancer metastasis. J Biol Chem. 2011;286:23093–101. doi: 10.1074/jbc.M111.233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gajate C, Mollinedo F. Lipid rafts, endoplasmic reticulum and mitochondria in the antitumor action of the alkylphospholipid analog edelfosine. Anticancer Agents Med Chem. 2014;14:509–27. doi: 10.2174/1871520614666140309222259. [DOI] [PubMed] [Google Scholar]
- 80.Castro BM, Fedorov A, Hornillos V, Delgado J, Acuna AU, Mollinedo F, Prieto M. Edelfosine and miltefosine effects on lipid raft properties: membrane biophysics in cell death by antitumor lipids. J Phys Chem B. 2013;117:7929–40. doi: 10.1021/jp401407d. [DOI] [PubMed] [Google Scholar]
- 81.Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:319–36. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- 82.Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–85. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 83.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–8. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 84.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–59. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 86.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204:635–46. doi: 10.1083/jcb.201311051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–10. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli R, Rajamanickam VP, Malerba M, De Angelis F, Falqui A, Carbone E, Todaro M, Medema JP, Stassi G, Di Fabrizio E. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2015;33:35–44. doi: 10.1002/stem.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Menard JA, Christianson HC, Kucharzewska P, Bourseau-Guilmain E, Svensson KJ, Lindqvist E, Indira Chandran V, Kjellen L, Welinder C, Bengzon J, Johansson MC, Belting M. Metastasis Stimulation by Hypoxia and Acidosis-Induced Extracellular Lipid Uptake Is Mediated by Proteoglycan-Dependent Endocytosis. Cancer Res. 2016;76:4828–40. doi: 10.1158/0008-5472.CAN-15-2831. [DOI] [PubMed] [Google Scholar]
- 91.Nieva C, Marro M, Santana-Codina N, Rao S, Petrov D, Sierra A. The lipid phenotype of breast cancer cells characterized by Raman microspectroscopy: towards a stratification of malignancy. PLoS One. 2012;7:e46456. doi: 10.1371/journal.pone.0046456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rak S, De Zan T, Stefulj J, Kosovic M, Gamulin O, Osmak M. FTIR spectroscopy reveals lipid droplets in drug resistant laryngeal carcinoma cells through detection of increased ester vibrational bands intensity. Analyst. 2014;139:3407–15. doi: 10.1039/c4an00412d. [DOI] [PubMed] [Google Scholar]
- 93.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 95.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–78. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, Kitahata N, Ando D, Ishikawa M, Wada S, Miyao A, Hirochika H, Shimada H, Makino A, Saito K, Ishida H, Kinoshita T, Kurata N, Kuchitsu K. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014;10:878–88. doi: 10.4161/auto.28279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–70. doi: 10.1038/ncb3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu G, Sztalryd C, Londos C. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta. 2006;1761:83–90. doi: 10.1016/j.bbalip.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 99.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Johnson MR, Ke Z, Chen L, Welte MA. Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr Biol. 2014;24:1485–91. doi: 10.1016/j.cub.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rui YN, Xu Z, Patel B, Chen Z, Chen D, Tito A, David G, Sun Y, Stimming EF, Bellen HJ, Cuervo AM, Zhang S. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–75. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–61. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 103.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Wang KZQ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376–91. doi: 10.1007/s10495-008-0307-5. [DOI] [PubMed] [Google Scholar]
- 105.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 106.Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–73. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhasz G, Bilder D, Brech A, Stenmark H, Rusten TE. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gammoh N, Fraser J, Puente C, Syred HM, Kang H, Ozawa T, Lam D, Acosta JC, Finch AJ, Holland E, Jiang X. Suppression of autophagy impedes glioblastoma development and induces senescence. Autophagy. 2016;12:1431–9. doi: 10.1080/15548627.2016.1190053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yan C, Zhao T, Du H. Lysosomal acid lipase in cancer. Oncoscience. 2015;2:727–8. doi: 10.18632/oncoscience.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qu P, Du H, Wilkes DS, Yan C. Critical roles of lysosomal acid lipase in T cell development and function. Am J Pathol. 2009;174:944–56. doi: 10.2353/ajpath.2009.080562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao T, Du H, Ding X, Walls K, Yan C. Activation of mTOR pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal(−/−) mice. Oncogene. 2015;34:1938–48. doi: 10.1038/onc.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao T, Ding X, Du H, Yan C. Lung Epithelial Cell-Specific Expression of Human Lysosomal Acid Lipase Ameliorates Lung Inflammation and Tumor Metastasis in Lipa(−/−) Mice. Am J Pathol. 2016;186:2183–2192. doi: 10.1016/j.ajpath.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Du H, Zhao T, Ding X, Yan C. Hepatocyte-Specific Expression of Human Lysosome Acid Lipase Corrects Liver Inflammation and Tumor Metastasis in lal(−/−) Mice. Am J Pathol. 2015;185:2379–89. doi: 10.1016/j.ajpath.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mukhopadhyay S, Schlaepfer IR, Bergman BC, Panda PK, Praharaj PP, Naik PP, Agarwal R, Bhutia SK. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radic Biol Med. 2017;104:199–213. doi: 10.1016/j.freeradbiomed.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 117.Xu G, Jiang Y, Xiao Y, Liu XD, Yue F, Li W, Li X, He Y, Jiang X, Huang H, Chen Q, Jonasch E, Liu L. Fast clearance of lipid droplets through MAP1S-activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival. Oncotarget. 2016;7:6255–65. doi: 10.18632/oncotarget.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gomez de Cedron M, Ramirez de Molina A. Microtargeting cancer metabolism: opening new therapeutic windows based on lipid metabolism. J Lipid Res. 2016;57:193–206. doi: 10.1194/jlr.R061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.