Abstract
Purpose
To develop a comprehensive genetic test for female and male infertility in support of medical decisions during assisted reproductive technology (ART) protocols.
Methods
We developed a next-generation sequencing (NGS) gene panel consisting of 87 genes including promoters, 5′ and 3′ untranslated regions, exons, and selected introns. In addition, sex chromosome aneuploidies and Y chromosome microdeletions were analyzed concomitantly using the same panel.
Results
The NGS panel was analytically validated by retrospective analysis of 118 genomic DNA samples with known variants in loci representative of female and male infertility. Our results showed analytical accuracy of > 99%, with > 98% sensitivity for single-nucleotide variants (SNVs) and > 91% sensitivity for insertions/deletions (indels). Clinical sensitivity was assessed with samples containing variants representative of male and female infertility, and it was 100% for SNVs/indels, CFTR IVS8-5T variants, sex chromosome aneuploidies, and copy number variants (CNVs) and > 93% for Y chromosome microdeletions. Cost analysis shows potential savings when comparing this single NGS assay with the standard approach, which includes multiple assays.
Conclusions
A single, comprehensive, NGS panel can simplify the ordering process for healthcare providers, reduce turnaround time, and lower the overall cost of testing for genetic assessment of infertility in females and males, while maintaining accuracy.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1204-7) contains supplementary material, which is available to authorized users.
Keywords: Infertility, Next-generation sequencing, Clinical genetic testing, Diagnostic
Introduction
It is estimated that 48 million couples were affected by infertility in 2010, and there has not been any significant improvement in infertility levels between 1990 and 2010 [1–3]. In the USA, 12% of women aged 15–44 have impaired fecundity. Increasingly, more couples rely on assisted reproductive technologies (ART) to get pregnant and have children, and in the USA, 231,936 ART cycles were performed in 2015 [4].
A significant proportion of infertility cases are due to genetic defects. Male infertility accounts for 50% of infertility cases [5], with known genetic factors accounting for 15–30% of male infertility [6]. Chromosomal alterations [7], inversions [8], translocations [9], Y chromosome microdeletions [10], and gene mutations (for example single-nucleotide variants (SNVs) in CFTR [11]) are the main genetic variants causing male infertility. In females, infertility is a more heterogeneous condition. While genetics clearly play a role, these effects are mostly polygenic, making it difficult to define a single genetic cause. The two most common female factor conditions, ovulatory dysfunction (25%) and endometriosis (15%), have familial predisposition, suggesting a genetic basis [12]. In addition, sex chromosome alterations [13] and several single gene mutations have been described impacting female fertility [14, 15], causing conditions like hypogonadotropic hypogonadism, premature ovarian insufficiency, endometriosis, and polycystic ovarian syndrome (reviewed in [12]).
Traditionally, several assays are needed to make a definitive genetic diagnosis of infertility, which makes the process expensive and slow. For example, in males, a variety of techniques are necessary for analysis of genetics: sex chromosome aneuploidies are detected by cytogenetic tests like karyotyping; Y chromosome microdeletions are detected by polymerase chain reaction (PCR)-based methods; and CFTR mutations are detected by Sanger DNA sequencing. However, a shotgun approach in which all genetic tests are ordered for all patients is not recommended because cost is prohibitive [16]. In females, success rates vary depending on many factors with age being the most important. An infertility evaluation includes very diverse tests including blood and urine hormone levels, imaging, and for women with unexplained infertility issues, genetic tests like karyotyping or DNA sequencing of selected genes.
Next-generation sequencing (NGS) permits the simultaneous interrogation of multiple disease-causing variants in many genes, allowing expanded genetic diagnostics to be routinely used in medical practice. This is already a reality in other medical fields, like oncology [17] and heart disease [18], where panel testing allows for the most comprehensive assessment of genetic etiologies. NGS is also very cost-effective as it allows for the detection of very different types of variants (for example, SNVs, small indels, large Y chromosome deletions, and sex chromosome aneuploidies) by using a single test in combination with multiple bioinformatics algorithms to process these diverse data. Here, we present the development of an NGS panel and bioinformatics pipeline for the detection of genetic variants with direct impact on female and male infertility. We also present a cost comparative analysis of current approaches in comparison with the NGS test.
Materials and methods
Subjects and samples
Samples of genomic DNA with previously identified variants in the genes interrogated by the sequencing panel were obtained as purified DNA from the Human Genetic Cell Repository (National Institute of General Medical Sciences) at the Coriell Institute for Medical Research (Camden, NJ) or the Indiana Biobank (Indianapolis, IN) or extracted from saliva samples collected by DNAsimple (Philadelphia, PA). All samples submitted to our diagnostics laboratory as saliva were manually extracted using the Qiagen QIAamp Mini Kits following the manufacturer’s instructions. DNA was quantified using Quant-iT Picogreen dsDNA Assay Kit (Life Sciences) and a Varioskan LUX (Thermo Scientific). Supplementary Table S1 describes all samples used in this study, with biorepository numbers, as well as their previous characterization results and rationale for inclusion. All samples presented here are de-identified (HHS 45 CFR part 46.101(b)(4)). IRB approval to handle de-identified samples was obtained through ASPIRE (Santee, CA), protocol IRB-R-003.
Fertility panel description
A targeted next-generation sequencing panel consisting of 87 genes related to infertility disorders was created (Supplementary Table S2). Genes relevant to disease phenotype were included based on relationships described in Online Mendelian Inheritance in Man (OMIM), Human Phenotype Ontology (HPO), GeneReviews, and primary literature. The panel included all coding exons, splice sites, promoter regions, 5′ untranslated regions (UTRs), and 3′ UTRs for each of the genes. Clinically relevant noncoding (intronic) regions that contained previously described pathogenic variants, as reported in ClinVar NIH database, were also included. The total size of the gene panel is 1,444,982 bp. In addition, selected regions from the Y chromosome (928,649 bp) were included to allow for the detection of Y chromosome microdeletions. Finally, the panel also includes 188 genes that we considered “research” genes, with links to infertility and which we wanted to follow for research purposes (Supplementary Table S3).
Next-generation DNA sequencing
All laboratory procedures were performed in a Clinical Laboratory Improvement Amendments (CLIA) laboratory. DNA samples were prepared for sequencing using HyperPlus Library Preparation Kit (Roche, Indianapolis, IN) and sequenced on a NextSeq500 (Illumina, San Diego, CA), following the manufacturer’s instructions. A detailed sequencing protocol is provided in Supplementary Methods section.
Variant identification and classification
All bioinformatics algorithms were implemented within the Elements™ platform (Phosphorus, New York, NY). FASTQ files were produced from each sequencing run and processed using the germline calling pipeline (version 2.03.01.30066) in DRAGEN (Edico Genome, San Diego, CA). Variants identified by NGS were confirmed by an orthogonal method (microarrays or Sanger sequencing). After confirmation, each variant was classified as pathogenic, likely pathogenic, variant of unknown significance (VUS), likely benign, or benign, following the American College of Medical Genetics (ACMG) guidelines [19].
Orthogonal confirmation
A custom Affymetrix Axiom array was used to confirm all the reportable variants including SNVs, indels, CNVs, Y chromosome microdeletions, and sex chromosome aneuploidies. Microarrays were processed following the manufacturer’s instructions. To confirm variants not included in the microarray, Sanger sequencing was used. Orthogonal analysis was also performed for cases when there was discrepancy between expected and received NGS results. If both NGS and confirmatory results agreed, the results were counted as concordant. Detailed protocols are described in the Supplementary Methods section.
FMR1 testing
FMR1 testing was performed with 120 ng genomic DNA per sample and the AmplideX PCR CE FMR1 kit (Asuragen, Austin, TX), following the manufacturer’s instructions. PCR products were separated in a 3500XL capillary electrophoresis system (Applied Biosystems, Foster City, CA) using conditions described in the FMR1 kit manual.
Results
Gene panel design
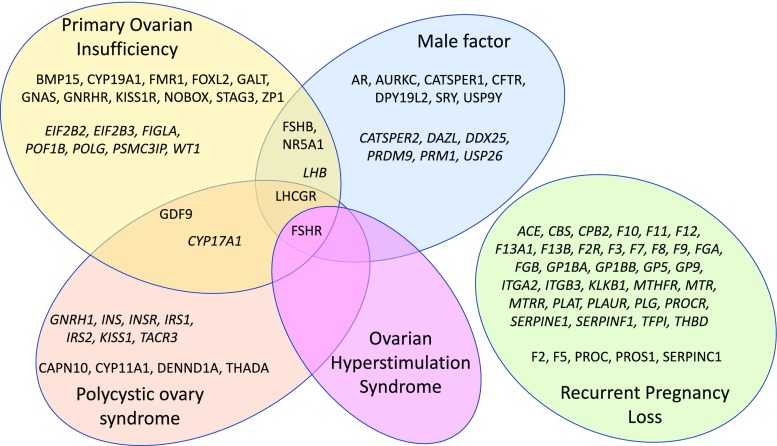
To maximize the clinical impact of our comprehensive infertility genetic test, we focused only on genes with demonstrated impact on infertility phenotype. Genes were classified as “diagnostic” when variants in them were reported to cause infertility across multiple populations, as supported by multiple publications from different laboratories, demonstrating a direct relationship with infertility. Genes were classified as “informative” when variants in them were reported to be associated with infertility, but the causality link has not been unequivocally established. For male infertility, the panel includes Y chromosome microdeletions, CFTR mutations, and sex chromosome aneuploidies [6]. For female infertility, the panel includes sex chromosome aneuploidies and genes in which variants have been associated with recurrent pregnancy loss caused by thrombophilia, primary ovarian insufficiency, polycystic ovary syndrome, and ovarian hyperstimulation syndrome. The gene list, as well as the rationale used for their selection, is shown in Fig. 1 and Supplementary Table S2. Figure 2 describes the laboratory and analysis workflow.
Fig. 1.
Description of the NGS panel by gene content, organized by the main infertility indications. Genes classified as “diagnostic” are shown in standard font, and genes classified as “informative” are shown in italic font
Fig. 2.
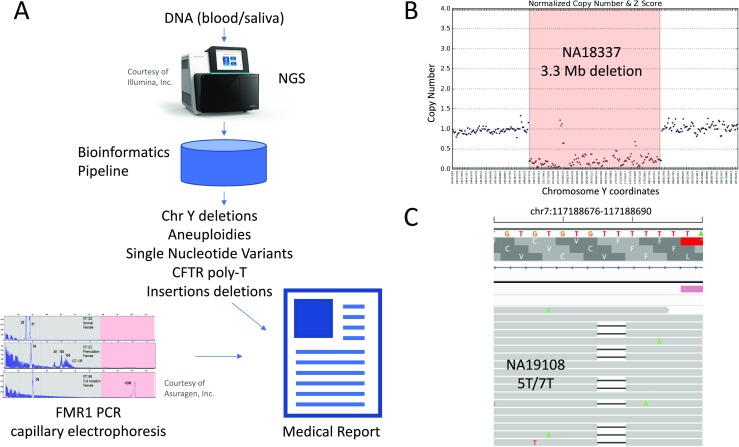
a Outline of our NGS test. A DNA sample (saliva or blood) is sequenced by NGS and processed by our custom bioinformatics pipeline. Y chromosome microdeletions, sex chromosome aneuploidies, CFTR IVS8-5T polymorphism, indels, and SNVs are called. Variants are interpreted by expert curators and a medical report is generated. In parallel, FMR1 testing is performed using PCR and capillary electrophoresis, and results are incorporated into the medical report. b Example of Y chromosome microdeletion called in sample NA18337. c Example of IVS8-5T tract detection in sample NA19108, heterozygous for 5T/7T
Analytical and clinical validation
To validate panel performance and determine analytical sensitivity, specificity, and accuracy, we sequenced 24 samples from the 1000 Genomes (1000G) project [20] for which the location of SNVs and indels are known (Supplementary Table S1). Representative sequencing quality control statistics are shown in Supplementary Table S4. NGS results from these validation samples were compared to known 1000G variants (Table 1). Microarray analysis and/or Sanger sequencing was performed to assess the discrepancies of SNVs/indels between results produced by the NGS panel and previously known 1000G data. Analytical sensitivity of the test was > 99% for SNVs and > 91% for indels, and specificity was > 99% for both SNVs and indels. Final accuracy for SNVs and indels was 99.98 and 99.42%, respectively.
Table 1.
Performance characteristics of the NGS test. PPV, positive predictive value; NPV, negative predictive value; CNV, copy number variant
| Variant type | True positives | True negatives | False positives | False negatives | |
| snp | 4578 | 257,918 | 2 | 40 | |
| Indel | 137 | 2106 | 0 | 13 | |
| Variant type | Accuracy | Sensitivity | Specificity | PPV | NPV |
| snp | 99.98% | 99.13% | 99.98% | 99.96% | 99.98% |
| Indel | 99.42% | 91.33% | 99.39% | 100.00% | 99.39% |
| Batch type | Concordant | Discordant | Concordance | ||
| Intra-batch | 588 | 17 | 97.19% | ||
| Inter-batch | 427 | 28 | 93.85% |
To determine clinical sensitivity in the detection of CNVs, sex chromosome aneuploidies and Y chromosome microdeletions, we used 34 samples with 38 known variants (Supplementary Table S1). The set of variants previously known for these samples is limited, so specificity could not be assessed. The analysis correctly detected 3/3 CNVs, 19/19 sex chromosome aneuploidies, and 15/16 Y chromosome microdeletions (Supplementary Tables S5 and S6, Table 1 and Fig. 2). In three cases (NA20435, NA18333, and NA22031), Y chromosome microdeletions of smaller size were found by NGS when compared to previously known data (Supplementary Table S6); however, microarray analysis confirmed the previously reported microdeletion size. In one case (NA20434), NGS data located the microdeletion 1.17 Mbp downstream of the previously known location, and microarray analysis confirmed NGS result. Sample NA12662 has a sex chromosome aneuploidy (duplication of X chromosome) and a Y chromosome microdeletion ((22769319-27097245)x0). The sex chromosome aneuploidy was identified, but the Y chromosome microdeletion was missed. Thus, clinical sensitivity for sex chromosome aneuploidies and CNVs is 100% (22/22), and for Y chromosome microdeletions, it is 93.75% (15/16).
Next, 11 DNA samples with 17 known SNVs/indels were processed (Table 2). Known variants were confirmed in 16/17 cases. Sample NA02795 was reported with a pathogenic variant GALT c.130G>A; p.Val44Met. However, this variant was not identified by NGS and Sanger sequencing did not detect it either; therefore, based on NGS and Sanger sequencing concordance, we did not count this case against the sensitivity. Thus, clinical sensitivity for SNVs/indels by our assay was 100%.
Table 2.
Clinical sensitivity was determined by sequencing SNVs/indels of known variants
| Sample | Previously reported | NGS test call |
|---|---|---|
| NA11763 | GALT c.591A>G; p.Gln188Arg | GALT NM_000155.3:c.563A>G; p.Gln188Arg |
| NA11763 | GALT c.1025C>T; p.Arg333Trp | GALT NM_000155.3:c.997C>T; p.Arg333Trp |
| NA14899 | F5 c.1691G>A; p.Arg506Gln (homozygous) | F5 NM_000130.4:c.1691G>A; p.Arg506Gln |
| IndianaBiobank02 | SERPINC1 c.1148_1149delTC; p.Leu383Profs*10 | SERPINC1 NM_000488.3:c.1148_1149delTC; p.Leu383Profs*10 |
| NA00422 | GALT c.563A>G; p.Gln188Arg | GALT NM_000155.3:c.563A>G; p.Gln188Arg |
| NA00422 | GALT c.1030C>A; p.Gln344Lys | GALT NM_000155.3:c.1030C>A; p.Gln344Lys |
| NA02795 | GALT c.425T>A; p.Met142Lys | GALT NM_000155.3:c.425T>A; p.Met142Lys |
| NA02795 | GALT c.130G>A; p.Val44Met | None |
| DNAsimple01 | CFTR c.1584G>A; p.Glu528= | CFTR NM_000492.3:c.1584G>A; p.Glu528= |
| NA02796 | GALT c.512T>C; p.Phe171Ser | GALT NM_000155.3:c.512T>C; p.Phe171Ser |
| NA02796 | GALT c.404C>T; p.Ser135Leu | GALT NM_000155.3:c.404C>T; p.Ser135Leu |
| NA16643* | F5 c.1691G>A; p.Arg506Gln | F5 NM_000130.4:c.1601G>A; p.Arg534Gln |
| NA17431 | GALT c.591A>G; p.Gln188Arg | GALT NM_000155.3:c.563A>G; p.Gln188Arg |
| NA17431 | GALT c.612T>C; p.Leu195Pro | GALT NM_000155.3:c.584T>C; p.Leu195Pro |
| NA16000 | MTHFR c.677C>T; p.Ala222Val | MTHFR NM_005957.1:c.677C>T; p.Ala222Val |
| NA16000 | F2 c.20210G>A (homozygous) | F2 NM_000506.4:c.*97G>A |
| DNAsimple02 | F2 c.20210G>A | F2 NM_000506.4:c.*97G>A |
*The official designation of F5 c.1691G>A is NM_000130.4:c.1601G>A; NP_000121.2:p.Arg534Gln
CFTR intron 8 poly dT (IVS8-5T)
Varied lengths of a thymidine (T)-tract (5, 7, or 9T) are found in front of the splice-acceptor site of intron 8 of the CFTR gene in males with congenital bilateral absence of the vas deferens [21]. The length correlates with the efficiency of exon 9 splicing. This polymorphism is detected clinically by allele-specific multiplex PCR [22]. In order to determine sensitivity of the NGS panel, we analyzed 72 samples from the 1000G project. When 1000G data was used as reference, the NGS panel correctly detected the IVS8 alleles in 67/72 cases, and for the 5 discrepant cases, Sanger sequencing demonstrated that the NGS panel call was the correct one (Supplementary Table S7). Therefore, our assay had 100% sensitivity to detect the CFTR IVS8-5T allele.
FMR1 testing
CGG triplet expansions are known to cause fragile X syndrome, and alleles in the pre-mutation range have been associated with increased risk for premature ovarian failure [23]. Current NGS protocols based on hybridization enrichment for targeted panels do not accurately quantify the number of repeats. Therefore, to complement the NGS panel with FMR1 analysis, we used an already available protocol based on PCR amplification and capillary electrophoresis detection of CGG repeats [24]. Test sensitivity was determined by analysis of 26 samples harboring different CGG repeat expansions (Supplementary Table S8). We correctly identified all alleles previously classified as “mutation” (> 200 CGG repeats), “pre-mutation” (55 to 199 CGG repeats), and “normal” (< 45 CGG repeats). However, the assay misclassified one allele in the “intermediate” range (between 45 and 54 CGG repeats) as “pre-mutation” (NA20236, 53 vs 55 CGG repeats, Supplementary Table S8). Given that the rate of expansions of intermediate alleles is not well understood, testing at-risk relatives of individuals with an intermediate allele may determine the stability of the allele in the family [25].
Cost analysis
Currently, after semen analysis, in cases where severe abnormal semen analysis is observed, genetic testing for variants associated with male infertility is performed with multiple assays. For example, CFTR IVS8-5T is detected by allele-specific multiplex PCR [22], Y chromosome microdeletions are detected by PCR of sequence-tagged sites [26], and sex chromosome aneuploidies are detected by traditional karyotyping or microarrays [27]. NGS can detect all these variants using a single test. In order to determine the economic impact, we performed a cost analysis for the male fertility panel, using average pricing available from reference laboratories performing these assays (Table 3 and Supplementary Table S9). Overall, the savings to providers could be as much as $2723 per case, $3322 if performed by multiple assays vs $599 if performed by NGS, which translates in savings of > 550%. Furthermore, NGS testing has a shorter turnaround time because there is no need to do reflex testing. The only exception may be cases with complex chromosomal rearrangements and mosaics or translocations/inversions that do not result in gain/loss of DNA and are, therefore, not captured by the NGS panel.
Table 3.
Cost comparison between traditional diagnostic methods for male infertility and NGS diagnostic test
| Variant | Methodology | CPT code | Cost ($US) |
|---|---|---|---|
| CFTR IVS8 poly T tract | Multiplex polymerase chain reaction, Sanger sequencing or NGS | 81,224 | 396 |
| CFTR mutations | Sanger sequencing of CFTR gene | 81,223 | 1430 |
| Sex chromosome aneuploidies | Karyotype | 88,262 | 946 |
| Y chromosome microdeletion | PCR-based analysis of 14 different regions along the length of the Y chromosome | 81,403 | 660 |
| Total cost: | 3322 | ||
| NGS infertility test | NGS sequencing, includes confirmation with orthogonal methods (microarray or Sanger) | 81,224, 81,223, 88,262, 81,403 | 599 |
Discussion
We present a comprehensive genetic test based on NGS that covers the main infertility indications [6, 12]: the female panel analyzes genes associated with increased risks for female infertility, including primary ovarian insufficiency, polycystic ovary syndrome, sex chromosome aneuploidy, ovarian hyperstimulation syndrome, and thrombophilia-related pregnancy loss (Fig. 1). The male panel includes Y chromosome microdeletions, congenital absence of the vas deferens, sex chromosome aneuploidy, and other causes of male factor infertility (Fig. 1).
From a clinical care perspective, this NGS test has the ability to influence key decisions in patient management following infertility diagnosis. In the case of female infertility patients, a complement or earlier diagnosis of predisposition to severe diminished ovarian response or premature ovarian failure may help providers to better guide patients to gamete cryopreservation in anticipation of future pregnancies. For patients found to have a definitive diagnosis of polycystic ovary syndrome, it has been shown that patients who receive a gonadotropin-releasing hormone antagonist protocol undergoing controlled ovarian stimulation experience reduced risk of ovarian hyperstimulation syndrome [28].
For male infertility patients, it is a routine for IVF laboratories or reproductive urologists to perform semen analysis for sperm count, motility, and morphology. For those men with results indicative of severe oligospermia or azoospermia, it is a routine to reflex to genetic tests to help explain etiology and guide treatment. Depending on the nature of the mutations, providers are able to better determine whether male patients are candidates for surgical retrieval of sperm [29]. Reduced cost and faster turnaround time in genetic analysis will help patients reach the proper treatment more efficiently.
Genetic analysis is currently performed using many different platforms: SNVs and indels are detected by Sanger sequencing or NGS, sex chromosome aneuploidies by karyotyping or microarrays, Y chromosome microdeletions by multiplex PCR of sequence tags, and CFTR intron 8 polymorphism by allele-specific multiplex PCR. NGS allows for the detection of these different variants using a single platform with excellent sensitivity and specificity. Out of 127 different events, the NGS panel complemented with orthogonal confirmation methods accurately detected almost all of them: 17/17 SNVs/indels, 3/3 CNVs, 19/19 sex chromosome aneuploidies, 15/16 Y chromosome microdeletions, and 72/72 CFTR IVS8 polymorphic sites (5 of them IVS8-5T). In three Y chromosome microdeletion cases, the NGS panel identified microdeletions of smaller size; however, an analysis using microarrays confirmed the previously known size in all three cases. The NGS panel, thus, identified accurately most Y chromosome microdeletions (one Y chromosome microdeletion was missed), but orthogonal methods are required to confirm the microdeletion size. In summary, overall clinical sensitivity for all variants is 99.21% (126/127).
Our study has several limitations. Sample size is limited, and even though we processed samples representing all types of variants, the final number of samples in each group is small. For example, we only processed three clinical samples containing CNVs. Therefore, given limited sample size, performance characteristics must be interpreted with caution. In addition, this is a retrospective study, so it serves as a proof of concept, but a prospective study (currently ongoing at our center) will be needed to confirm our findings.
In its current form, the NGS panel cannot detect balanced translocations; therefore, it would miss reciprocal translocations and Robertsonian translocations, which are known to cause infertility in 0.9% of men [9]. Furthermore, the NGS panel cannot detect complex chromosomal rearrangements like derivative chromosomes or mosaicism, even though these often involve gain or loss of DNA. We tested several samples with derivative chromosomes and mosaicism and the current NGS panel could not accurately assess these variants. For example, samples IndianaBiobank01 and NA21681 in Supplementary Table S5 contain a derivative X,Y chromosome and a 6,Y translocation, respectively, and both were missed. In these cases, a karyotype is needed, which could be used as reflex after the NGS test. We are currently investigating the possibility to incorporate detection of these abnormalities as part of this NGS test [30].
Another limitation is the detection of FMR1 variant caused by triplet CGG expansion, varying from 20 to > 900 repeats. It is not possible to accurately identify triplet expansions by using PCR-enriched NGS. New methods involving PCR-free libraries allow diagnosis [31], but it would require a parallel library preparation and sequencing reaction, significantly increasing cost. Therefore, currently, a separate PCR assay to determine FMR1 mutation is needed to assess fragile-X mutation.
The direct consequence of using a single NGS assay instead of multiple diagnostic assays, each one detecting a class of DNA variants, is a potential reduction in cost and turnaround time required to make a definitive diagnosis, with savings that could be up to $2723 per case ($3322 for traditional methods vs $599 for NGS testing). Integrating testing into a single assay simplifies test ordering and result tracking for the clinician and decreases cost to the patient by reducing the number of assays that need to be performed, analyzed, and reported.
Conclusions
We have developed a comprehensive genetic test for female and male infertility that achieves excellent clinical sensitivity, but at a fraction of the cost of traditional methods of testing, and a shorter turnaround time than current methods.
Electronic supplementary material
(XLSX 208 kb)
(DOCX 130 kb)
Acknowledgements
We thank Sara Bristow for her initial scientific input, Santiago Munne for his scientific support, and Ed O’Neill for editorial comments on the manuscript.
Authors’ contributions
SP, AB, and OP participated in study design and data analysis.
SP, BP, MA, GK, MJ, CC, YC, RF, MN, CH, AP, RW, AB, and OP participated in data collection, interpretation of results, and drafting the manuscript.
Funding
This study was fully funded by Phosphorus, Inc.
Compliance with ethical standards
All authors in this manuscript are full time employees of Phosphorus, or own Phosphorus stock.
References
- 1.Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, National SGA. Regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9:1–12. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mansour R, Ishihara O, Adamson GD, Dyer S, de Mouzon J, Nygren KG, et al. International Committee for Monitoring Assisted Reproductive Technologies world report: assisted reproductive technology 2006. Hum Reprod. [Internet] 2014;29:1536–1551. doi: 10.1093/humrep/deu084. [DOI] [PubMed] [Google Scholar]
- 3.Chandra A, Copen CE, Stephen EH. Infertility and impaired fecundity in the United States, 1982–2010: data from the National Survey of Family Growth. Natl Health Stat Report [Internet] 2013;67:1–19. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. http://nccd.cdc.gov/drh_art. Assist. Reprod. Technol. Rep. 2015.
- 5.Thonneau P, Marchand S, Tallec A, Ferial M, Ducot B, Lansac J, et al. Incidence and main causes of infertility in a resident population (1 850 000) of three French regions (1988–1989)*. Hum Reprod. 1991;6(6):811–816. doi: 10.1093/oxfordjournals.humrep.a137433. [DOI] [PubMed] [Google Scholar]
- 6.Neto FTL, Bach PV, Najari BB, Li PS, Goldstein M. Genetics of male infertility. Curr Urol Rep [Internet]. Current Urology Reports. 2016;17:70–82. doi: 10.1007/s11934-016-0627-x. [DOI] [PubMed] [Google Scholar]
- 7.Groth KA, Skakkebæk A, Høst C, Gravholt CH, Bojesen A. Klinefelter syndrome—a clinical update. J Clin Endocrinol Metab. 2013;98:20–30. doi: 10.1210/jc.2012-2382. [DOI] [PubMed] [Google Scholar]
- 8.Sasagawa I, Ishigooka M, Kubota Y, Tomaru M, Hashimoto T, Nakada T. Pericentric inversion of chromosome 9 in infertile men. Int Urol Nephrol [Internet] 1998;30:203–207. doi: 10.1007/BF02550578. [DOI] [PubMed] [Google Scholar]
- 9.Mau-Holzmann UA. Somatic chromosomal abnormalities in infertile men and women. Cytogenet Genome Res. 2005;111:317–336. doi: 10.1159/000086906. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Chen Z, Ni Y, Li Z. CFTR mutations in men with congenital bilateral absence of the vas deferens (CBAVD): a systemic review and meta-analysis. Hum Reprod. 2012;27:25–35. doi: 10.1093/humrep/der377. [DOI] [PubMed] [Google Scholar]
- 12.Mallepaly R, Butler PR, Herati AS, Lamb DJ. Genetic basis of male and female infertility. Monogr Hum Genet. 2017;21:1–16. doi: 10.1159/000477275. [DOI] [Google Scholar]
- 13.Grynberg M, Bidet M, Benard J, Poulain M, Sonigo C, Cédrin-Durnerin I, Polak M. Fertility preservation in Turner syndrome. Fertil Steril. 2016;105:13–19. doi: 10.1016/j.fertnstert.2015.11.042. [DOI] [PubMed] [Google Scholar]
- 14.Fu YH, Kuhl DPA, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkert AJMH, Holden JJA, Fenwick RG, Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 15.Huang H-L, Lv C, Zhao Y-C, Li W, He X-M, Li P, et al. Mutant ZP1 in familial infertility. N Engl J Med [Internet] 2014;370:1220–1226. doi: 10.1056/NEJMoa1308851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stahl PJ, Schlegel PN. Genetic evaluation of the azoospermic or severely oligozoospermic male. Curr Opin Obstet Gynecol [Internet] 2012;24:221–228. doi: 10.1097/GCO.0b013e3283558560. [DOI] [PubMed] [Google Scholar]
- 17.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol [Internet]. Nat Publ Group. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celestino-Soper PBS, Gao H, Lynnes TC, Lin H, Liu Y, Spoonamore KG, et al. Validation and utilization of a clinical next-generation sequencing panel for selected cardiovascular disorders. Front Cardiovasc Med [Internet] 2017;4:1–11. doi: 10.3389/fcvm.2017.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med [Internet] 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature [Internet]. 2015;526:68–74. Available from: http://www.nature.com/nature/journal/v526/n7571/fig_tab/nature15393_SF1.html%5Cn10.1038/nature15393%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/26432245 [DOI] [PMC free article] [PubMed]
- 21.Zielenski J, Patrizio P, Corey M, Handelin B, Markiewicz D, Asch RTLCFTR. Gene variant for patients with congenital absence of vas deferens. Am J Hum Genet. 1995;51:958–960. [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman KJ, Heim RA, Knowles MR, Silverman LM. Rapid characterization of the variable length polythymidine tract in the cystic fibrosis (CFTR) gene: association of the 5T allele with selected CFTR mutations and its incidence in atypical sinopulmonary disease. Hum Mutat. 1997;10:108–115. doi: 10.1002/(SICI)1098-1004(1997)10:2<108::AID-HUMU3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 23.Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen GJB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Thomas Caskey C, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- 24.Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, Hagerman PJ, Stenzel TT, Hadd AG, Latham GJ, Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Carvajal I, Posadas BL, Pan R, Raske C, Hagerman PJ, Tassone F. Expansion of an FMR1 grey-zone allele to a full mutation in two generations. J. Mol. Diagnostics [Internet]. American Society for Investigative Pathology and Association for Molecular Pathology. 2009;11:306–310. doi: 10.2353/jmoldx.2009.080174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler JM, Schoske R, Vallone PM, Kline MC, Redd AJ, Hammer MF. A novel multiplex for simultaneous amplification of 20 Y chromosome STR markers. Forensic Sci Int. 2002;129:10–24. doi: 10.1016/S0379-0738(02)00195-0. [DOI] [PubMed] [Google Scholar]
- 27.Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–745. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambalk CB, Banga FR, Huirne JA, Toftager M, Pinborg A, Homburg R, van der Veen F, van Wely M. GnRH antagonist versus long agonist protocols in IVF: a systematic review and meta-analysis accounting for patient type. Hum Reprod Update. 2017;23:560–579. doi: 10.1093/humupd/dmx017. [DOI] [PubMed] [Google Scholar]
- 29.Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb, and AZFc regions. Hum Reprod. 2003;18:1660–1665. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
- 30.Liang D, Wang Y, Ji X, Hu H, Zhang J, Meng L, et al. Clinical application of whole-genome low-coverage next-generation sequencing to detect and characterize balanced chromosomal translocations. Clin Genet [Internet] 2017;91:605–610. doi: 10.1111/cge.12844. [DOI] [PubMed] [Google Scholar]
- 31.Dolzhenko E, van Vugt JJFA, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. [Internet] 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 208 kb)
(DOCX 130 kb)