Abstract
Purpose
Cell-free DNA (cfDNA) which is present in the blastocoel cavity of embryos is believed to result from physiological apoptosis during development. This study assessed cfDNA content and caspase-3 protease activity in day-5 IVF blastocysts to determine if there was a correlation with embryo morphology.
Methods
Day-5 IVF blastocysts were scored according to the Gardner and Schoolcraft system (modified to generate a numerical value) and cfDNA was collected following laser-induced blastocoel collapsing prior to cryopreservation in 25 μL of media. cfDNA was quantified via fluorospectrometry and apoptotic activity was assessed via a caspase-3 protease assay using a fluorescent peptide substrate. Data were compared by linear regression.
Results
A total of 32 embryos were evaluated. There was a significant (p < 0.01) and positive correlation (cfDNA = 104.753 + (11.281 × score); R2 = 0.200) between embryo score and cfDNA content. A significant (p < 0.05) and positive correlation (cfDNA = 115.9 + (0.05 × caspase-3); R2= 0.128) was observed between caspase-3 activity and cfDNA levels. There was no significant relationship between caspase-3 activity and embryo morphology score.
Conclusions
This study provides further evidence that cfDNA is present in blastocoel fluid, can be quantified, and positively correlates with embryonic morphology. There is also evidence that at least a portion of the cfDNA present is from intracellular contents of embryonic cells that underwent apoptosis. Additional studies are warranted to determine other physiological sources of the cfDNA in blastocyst fluid and to determine the relationship with cfDNA content, embryo morphology, and chromosomal ploidy status plus implantation potential.
Keywords: Cell-free DNA, Blastocyst, Apoptosis, Caspase-3, Morphology
Introduction
In vitro fertilization (IVF) accounts for 99% of assisted reproductive technology (ART) procedures. The main challenge of IVF ultimately is to select the single best embryo that has the greatest implantation potential and subsequently, the greatest chance of developing full-term. To optimize embryo implantation potential, embryo grading via subjective and objective embryo morphology is currently used; however, it has limitations in determining overall embryonic viability [1]. Some of these limitations include comparing, grouping, and stratifying embryos statistically in order to determine the most viable embryo for implantation [2].
To increase the success of selecting a viable embryo, preimplantation genetic testing for aneuploidy (PGT-A) assesses the ploidy status of the embryo(s). However, PGT-A is invasive due to its biopsy of trophectoderm (TE) cells from the embryo itself and researchers are looking for alternative ways to determine embryo viability, including cell-free DNA analysis that has recently emerged as a promising approach.
The presence of cell-free DNA (cfDNA) in the circulation was first documented in adult serum in 1948 [3]. The source of the cfDNA was thought to be derived from circulatory lysed cells as well as apoptotic events [4]. Fetal cfDNA was then discovered in the maternal circulation, which prompted researchers to look further at cfDNA from the embryo. Cell-free DNA has since been found to be present within the amniotic fluid as well as within the yolk sac of the embryo [5]. This correlated with the idea that DNA would be present wherever there is apoptotic activity and, or tissue remodeling, such as the developing preimplantation stage embryo [6]. Blastocoel fluid has been assessed for the presence of cfDNA and one study recently discovered that cell-free genomic DNA was found in about 90% of blastocoel fluid samples harvested during the vitrification process of IVF [7]. Thus, it was determined that cfDNA could potentially be quantified or even used for PGT-A by sampling the day-5 blastocoel fluid of an embryo, termed blastocentesis [8].
The presence of blastocoel cfDNA has led to the question of what is its purpose in embryonic development. Cell-free DNA has been previously studied in the context of cancer and has been correlated with apoptotic and necrotic cell events, which can be measured via caspase activity [9]. Abnormal embryo development has also been associated with altered caspase-3 levels [10]. Studies also suggest that the amount of cfDNA may represent genetic corrections. Tobler et al. [8] reported that euploid blastocysts can marginalize aneuploid cells to the blastocoel fluid during early embryogenesis. Thus, it has been postulated that competent embryos may be identified via quantifying the cfDNA content released by the embryo [11]. Therefore, this study’s objectives were to (1) determine if cfDNA is present in blastocoel fluid, (2) determine if it correlates with embryonic morphology, and (3) determine if it correlates with caspase-3 protease activity.
Materials and methods
Blastocoel fluid was obtained following standardized IVF procedures including fertilization of the eggs via intracytoplasmic sperm injection and embryo culture (Institutional Review Board approved) from two collaborating institutions (University of Texas Health Center, San Antonio, TX; Premier Fertility, High Point, NC) prior to being shipped to USCSOM Greenville for cfDNA and caspase-3 analysis. The blastocyst-stage embryos were graded (single individual at each institution providing the samples) via the Gardner and Schoolcraft System [2]. Briefly, day-5 blastocysts are observed and are assigned a numerical score 1 to 6 based on the degree of expansion and hatching, from early blastocyst having a blastocoel cavity with less than half of the total volume (grade 1) to being totally hatched from the zona pellucida (grade 6). Inner cell mass (ICM) and trophectoderm (TE) are assessed and assigned with alphabetical score A (best; ICM, a tightly packed ICM with many cells; TE, many cells forming a cohesive epithelium), B (moderate quality; ICM, loosely grouped ICM with several cells; TE, few cells forming a loose epithelium), and C (relatively poor; ICM, very few cells and disorganized; TE, very few large cells). For example, a fully expanded blastocyst with a tightly packed ICM and multicellular epithelium would be graded “4AA.”
An algorithm was created to convert the Gardner and Schoolcraft systems alphabetical (ICM and TE grades) and numerical (expansion grade) values to a single numerical score. Rehman et al. [12] devised a numerical score for embryos by assigning a number to each category of the Gardner scale and multiplying them (i.e., a “4AB” graded embryo would become 4 × 4 × 3 = 48). Our new algorithm included a weighted significance to determine the most viable embryo as follows: weighted embryo morphology score = (expansion grade × 3) + (ICM grade × 2) + (TE grade × 1).
Blastocoel fluid from 32 embryos following laser-induced blastocoel collapsing was collected prior to cryopreservation. Individual blastocysts were placed in separate medium (Global Total; LifeGlobal Group, Guilford, CT) drops (25 μL each) under oil and a laser pulse was directed at the cellular junctions between trophectoderm cells. As cell junctions open, pressure is released and the blastocyst collapsed in and upon itself, inducing the blastocoel fluid (~ 0.001 μL) [13] to be extruded into the 25-μL drop of medium. The collapsed blastocyst was removed and cryopreserved. Each individual medium drop (25 μL) containing the blastocoel fluid (~ 0.001 μL) was collected, mixed (via pipetting), and stored at − 70 °C for further analysis. Non embryo-conditioned media (HTF + 10% human synthetic serum) served as a control.
To quantify the cfDNA concentration in the blastocoel fluid, an AccuBlue NextGen dsDNA Quantification Kit (Biotium, Fremont, CA) was used with emissions detected by a NanoDrop 3300 fluorospectrometer (Thermo Fisher Scientific, Waltham, MA) per manufacturer instructions. Briefly, the quantitation kit uses a fluorescent dye that binds specifically to dsDNA; therefore, the quantity of dsDNA can be determined by measuring fluorescence intensity combined with a standard curve of known DNA concentrations. After a standard curve was generated, 2 μL of conditioned media for each embryo was quantified independently. Assay sensitivity is 2 pg with a linear range of 1–500 ng/mL for the AccuBlue NextGen dsDNA Quantification Kit and the NanoDrop 3300 fluorospectrometer. The specificity of the AccuBlue NextGen dsDNA is 10-fold more sensitive for dsDNA than that for ssDNA or RNA [14].
Once the cfDNA had been quantified, the blastocoel fluid was then analyzed for caspase-3 activity using an in vitro assay. Detection of caspase-3 protease activity in the blastocoel fluid may indicate that apoptosis occurred within the embryo, whereby the intracellular contents of cells that underwent apoptosis were discarded into the blastocoel cavity. A caspase-3 cellular assay kit (Enzo Life Sciences, Farmingdale, NY) was used to assess protease activity that utilizes a fluorogenic peptide that active caspase-3 cleaves due to its high specificity for the peptide’s amino acid sequence. If the fluorogenic peptide substrate is cleaved by active caspase-3, then emission of the substrate can be detected in the 420–460-nm wavelength range. Ten microliters of each blastocoel fluid sample was added to individual wells containing assay buffer in a black-bottom 96-well plate (both provided in the kit). Human, recombinant caspase-3 (provided in the kit) was used as a positive control and was added to control wells containing buffer in the same 96-well plate. Wells containing buffer alone (no enzyme or blastocoel fluid) served as a blank/background reading. As per manufacturer’s instructions, 30 μM of Ac-DEVD-AMC (caspase-3-specific fluorogenic substrate) was added to individual wells in the 96-well plate. The 96-well plate was then run on a Tecan Infinite M1000 fluorescence intensity program to analyze the emission of fluorogenic peptide cleavage (Ex 360 nm/Em 460 nm). The emissions generated (AFU) from each well were measured every minute over a total of 45 min. The value that was recorded was from when the emissions stabilized. The emissions were subtracted from the background buffer emission and then analyzed in conjunction with the cfDNA concentration data.
Statistical analyses were performed using SigmaPlot 14.0 (SysStat Software, Inc., San Jose, CA). Linear regression was used to determine the statistical significance between embryo morphology scores, cfDNA concentrations, and caspase-3 protease activities. Differences were considered significant if P < 0.05.
Results
Blastocoel fluid from a total of 32 day-5 embryos were analyzed as described with the mean values presented in Table 1. No differences were observed between the two IVF centers; therefore, the data was combined into a single data set. The table reports the total number of embryos used (32) and the average embryo score based on the algorithm described earlier (21.125). The percentage of blastocoel fluid samples that were positive for cfDNA was also recorded (100%) along with the mean cfDNA concentration (133.55 ng/mL) and the percentage of blastocoel fluid samples that were positive for caspase-3 protease activity (78.1%).
Table 1.
Means for weighted embryo morphology score, blastocoel cfDNA content, and capase-3 protease activity for 32 day-5 embryos
| Morphology score (mean) | Percent positive for cfDNA | cfDNA content (ng/mL) | Percent positive for caspase-3 | Caspase activity (AFU) |
|---|---|---|---|---|
| 21.125 | 100 | 133.55 | 78.1 | 383.8 |
Embryo morphology scores ranged from 15 to 27, with a mean of 21.125. cfDNA content (ng/mL) ranged from a low of 32.3 ng/mL to a high of 315.3, with a mean of 133.55. Caspase-3 protease activity (AFU) ranged from undetectable levels to a high of 2226.6 AFU, with a mean of 383.8 AFU.
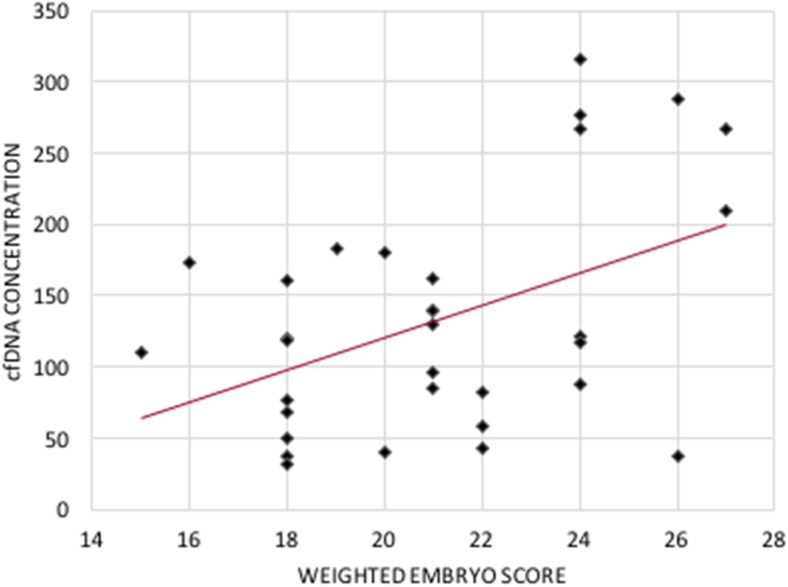
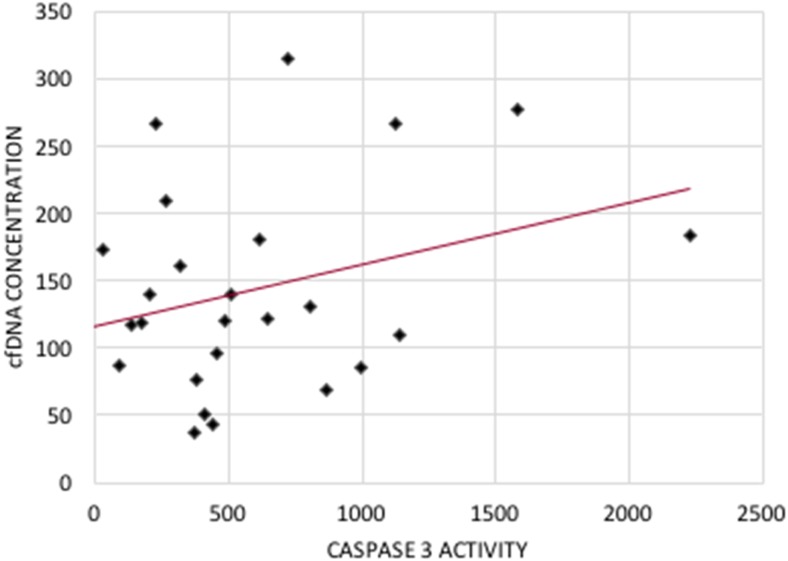
The maximum embryo morphology score of 27 had a cfDNA concentration of 266.5 ng/mL and the minimum embryo score of 15 had a cfDNA concentration of 109.9 ng/mL (Fig. 1). There was a significant (p < 0.01) and positive correlation [cfDNA = 104.753 + (11.281 × score); R2 = 0.200] between embryo morphology score and cfDNA. There was a significant (p < 0.05) and positive correlation [cfDNA = 115.9 + (0.05 × caspase − 3); R2 = 0.128] between caspase-3 protease activity and cfDNA (Fig. 2). The weighed embryo score was also compared to the caspase-3 protease activity; however, there was no significant relationship between the two.
Fig. 1.
Scatter plot of the weighted embryo score against cfDNA concentration for each of the 32 embryos. cfDNA = 104.753 + (11.281 × score); R2 = 0.200; P < 0.01
Fig. 2.
Scatter plot of the caspase-3 activity (AFU) against cfDNA concentration for all 32 embryos. cfDNA = 115.9 + (0.05 × caspase − 3); R2 = 0.128; P < 0.05
Discussion
To develop the algorithm used in this paper, several studies were evaluated. Rehman et al. [12] devised a numerical score for embryos by assigning a number to each category of the Gardner scale and multiplying them (for example, 4AB would become 4 × 4 × 3 = 48) [12]. While this numerical score did correlate with implantation potential, live birth did not [2]. The algorithm developed in this project modified this calculation to reflect literature studies that found that the blastocyst expansion and inner cell mass grades predicted live birth rates while there was no association of trophectoderm score with live birth rates [15]. However, the importance of the trophectoderm has been debated in other published papers [16].
Palini et al. [7] discovered that cell-free DNA was found in about 90% of blastocoel fluid samples produced from IVF. Our study provides further evidence that cfDNA is present in blastocoel fluid and is quantifiable supporting this previous research. Our study also indicates that cfDNA positively correlates with a high embryonic morphology score, which suggests that the better the embryo morphology, the higher the cfDNA concentration.
One theory as to why this occurs comes from Tobler et al. [8] that reported that euploid blastocysts can marginalize aneuploid cells to the blastocoel fluid during early embryogenesis. Therefore, the cfDNA would be representative of aneuploid cells that a viable embryo had marked for apoptosis. Another theory is that cfDNA could represent normal development from the breaking and repairing of DNA as the embryo develops, and thus higher cfDNA concentrations could represent remodeling of the viable embryo. These avenues of potential causes will be further researched in future studies.
Cell-free DNA has also been previously correlated with apoptotic events, which can be measured via caspase activity [9]. In our study, we discovered that there is detectable caspase-3 protease activity in the blastocoel fluid and that it positively correlates with cfDNA. The active caspase-3 in this fluid likely originated in embryonic cells undergoing apoptosis as this active enzyme is required for this programmed cell death process. The intracellular contents of the cells that underwent apoptosis were discarded into the blastocoel fluid which included the active caspase-3 enzyme. Our results confirm that cfDNA in the blastocoel fluid may be present, in part, due to apoptosis. However, due to the undetectable amounts in seven samples, this suggests that apoptosis might not be the complete explanation of cfDNA presence. Other mechanisms may include another caspase pathway (e.g., caspase-8), cellular remodeling, or autophagy. In relation to the caspase-3 activity, Spanos et al. [17] were able to identify BAX and BCL2 in blastocysts using immunohistochemistry and further detected caspase activity after compaction at the morula and blastocyst stages. This finding correlates with our detection of caspase-3 activity in the blastocoel fluid. More studies are needed to further elucidate the role of cfDNA in blastocoel fluid.
Overall, our results confirm that cfDNA resides in blastocoel fluid and its’ presence is possibly due in part to apoptosis during preimplantation embryo development. Further defining the role of cfDNA in embryogenesis will allow for more avenues to determine the best viable embryo in the future. This study’s results have the possibility of effecting the way in which the best embryos are selected for implantation, with further confirmation of viable embryos with higher cfDNA concentrations.
Compliance with ethical standards
The authors declare that they have no conflict of interest.
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Heitmann R, Hill M, Richter K, DeCherney A, Wildra E. The simplified SART embryo scoring system is highly correlated to implantation and live birth in single blastocyst transfers. J Assist Reprod Genetics. 2013;30:563–567. doi: 10.1007/s10815-013-9932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban B, Gardner DK. Morphological assessment of blastocyst stage embryos: types of grading systems and their reported outcomes. In: Gardner D, Sakkas D, Seli E, Wells D, editors. Human Gametes and Preimplantation Embryos: assessment and diagnosis. New York: Springer Science and Media; 2013. pp. 31–43. [Google Scholar]
- 3.Mandel P, Metais P. Les acides nucleiques du plasma sanguine chez l’homme. C.R. Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- 4.Norwitz ER, Levy B. Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol. 2013;6:48–62. [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, Craig JA, Chudova DI, Devers PL, Jones KW, Oliver K, Rava RP, Sehnert AJ, CARE Study Group DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 6.Jurisicova A, Acton BM. Deadly decisions: the role of genes regulating programmed cell death in human preimplantation embryo development. Reproduction. 2004;128(3):281–291. doi: 10.1530/rep.1.00241. [DOI] [PubMed] [Google Scholar]
- 7.Palini S, Galluzzi L, De Stefani S, Bianchi M, Wells D, Magnani M, Bulletti C. Genomic DNA in human blastocoel fluid. Reprod Biomed Online. 2013;26:603–610. doi: 10.1016/j.rbmo.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Tobler KJ, Zhao Y, Ross R, Benner AT, Xu X, Du L, Broman K, Thrift K, Brezina PR, Kearns WG. Blastocoel fluid from differentiated blastocysts harbors embryonic genomic material capable of a whole-genome deoxyribonucleic acid amplification and comprehensive chromosome microarray analysis. Fertil Steril. 2015;104(2):418–425. doi: 10.1016/j.fertnstert.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer F, Hesche R, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 10.Fabian D, Bukovská A, Juhás S, Koppel J. Apoptotic processes and DNA cytosine methylation in mouse embryos arrested at the 2-cell stage. Zygote. 2009;17(3):269–279. doi: 10.1017/S0967199409005413. [DOI] [PubMed] [Google Scholar]
- 11.Traver S, Assou S, Scalici E, Haouzi D, Al-Edani T, Belloc S, Hamamah S. Cell-free nucleic acids as non-invasive biomarkers of gynecological cancers, ovarian, endometrial and obstetric disorders and fetal aneuploidy. Hum Reprod Update. 2014;20(6):905–923. doi: 10.1093/humupd/dmu031. [DOI] [PubMed] [Google Scholar]
- 12.Rehman KS, Bukulmez O, Langley M, Carr BR, Nackley AC, Doody KM. Late stages of embryo progression are a much better predictor of clinical pregnancy than early cleavage in intracytoplasmic sperm injection and in vitro fertilization cycles with blastocyst-stage transfer. Fertil Steril. 2007;87:1041–1052. doi: 10.1016/j.fertnstert.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro BS, Daneshmand ST, Garner FC, Aguirre M, Thomas S. Large blastocyst diameter, early blastulation, and low preovulatory serum progesterone are dominant predictors of clinical pregnancy in fresh autologous cycles. Fertil Steril. 2008;90:302–309. doi: 10.1016/j.fertnstert.2007.06.062. [DOI] [PubMed] [Google Scholar]
- 14.Bruijns BB, Tiggelaar RM, Gardeniers JG. Fluorescent cyanine dyes for the quantification of low amounts of dsDNA. Anal Biochem. 2016;511:74–79. doi: 10.1016/j.ab.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Subira J, Craig J, Turner K, Bevan A, Ohuma E, McVeight E, Child T, Fatum M. Grade of the inner cell mass, but not trophectoderm, predicts live birth in fresh blastocyst single transfers. Hum Fertil. 2016;19(4):254–261. doi: 10.1080/14647273.2016.1223357. [DOI] [PubMed] [Google Scholar]
- 16.Ahlstrom A, Westin C, Reismer E, Wikland M, Hardarson T. Trophectoderm morphology: an important parameter for predicting live birth after single blastocyst transfer. Hum Reprod. 2011;26(12):3289–3296. doi: 10.1093/humrep/der325. [DOI] [PubMed] [Google Scholar]
- 17.Spanos S, Rice S, Karagiannis P, Taylor D, Becker DL, Winston RM, Hardy K. Caspase activity and expression of cell death genes during development of human preimplantation embryos. Reproduction. 2002;124:353–363. doi: 10.1530/rep.0.1240353. [DOI] [PubMed] [Google Scholar]