Abstract
Objective
To screen differentially expressed lncRNAs involved in OHSS. OHSS is defined as ovarian hyperstimulation syndrome. It is characterized as enlarged ovary and increased vascular permeability.
Design
Case-control study.
Setting
University-affiliated hospital.
Patient(s)
Patients with OHSS high risk (n = 30) and low risk (n = 30) were included in this study.
Intervention(s)
None.
Main outcome measure(s)
LncRNAs from women with OHSS high risk and low risk were used for high-throughput sequencing profiling. The eight most differentially expressed lncRNAs in granulosa cells were validated by semi-quantitative reverse transcription-polymerase chain reaction analysis.
Result(s)
A total of 23,815 lncRNAs were detected and 482 were differentially expressed (fold-change ≥2; p < 0.05, FDR value < 0.001), of which 205 were upregulated and 277 were downregulated. Lnc-SEC16B.1-6, lnc-SNURF-13, lnc-LGR6-6, and lnc-H2AFY2-2 were up-regulated, while lnc-BRD2-2, lnc-HSPA6-2, and lnc-CLIC6-5 were downregulated significantly in granulosa cells. These results were confirmed by qRT-PCR. KEGG pathways and Gene Ontology enrichment analysis revealed that several biological processes were significantly associated. Meanwhile, the lncRNA/miRNA interaction network was established according to ceRNA network model.
Conclusion(s)
Comprehensive expression screening identified eight novel lncRNAs associated with risk factors of OHSS process. Although it is unclear how these altered lncRNAs regulate the process of OHSS, our findings suggest these lncRNAs may be novel players in OHSS development.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1199-0) contains supplementary material, which is available to authorized users.
Keywords: OHSS, lncRNA, Development
Introduction
Ovarian hyperstimulation syndrome (OHSS) is an iatrogenic and serious complication of controlled ovarian stimulation among reproductive age women in assisted reproductive technology (ART) [1, 2]. The symptoms of OHSS contains series of clinical features including ovarian enlargement, ascites, high vascular permeability, high level of 17 β-estradiol (E2), and thrombotic complications [3, 4].The severity of OHSS can be classified as mild, moderate, and severe according to the assessment of clinical manifestations, and mild OHSS can occur in up to 32% of IVF cycles [5]. Especially, in in vitro fertilization cycles, the incidence of moderate to severe OHSS can be up to 10% under the effect of exogenous gonadotropin stimulation [6]. Vascular permeability has been considered as the main cause to OHSS and vascular endothelium growth factor (VEGF) should take the responsibility to vascular permeability [7, 8]. As an inflammatory factor, VEGF can induce local capillaries leaky via binding to its receptor VEGF receptor 2 (VEGFR2) in endothelial cell membrane [9, 10]. Despite abundant achievement has been obtained of OHSS, the pathogenesis and regulatory mechanisms of OHSS are still dismal.
The rapid advancement of high-throughput sequencing technology endowed a powerful tool to broaden the understanding of gene expression mechanisms. Over the past decade, non-coding RNAs have been believed to play pivotal roles during gene expression regulation, both in and post transcriptional level [11, 12]. According to the length, non-coding RNAs can be divided into microRNAs (17–23 nt) and long non-coding RNAs (≥ 200 nt). MicroRNAs have been well studied in granulosa cells of endocrine diseases, such as polycystic ovarian syndrome (PCOS) [13, 14]. Increasing evidence has shown that lncRNAs play critical roles in a wide range of biological processes, especially in governing gene expression during cellular development and homeostasis in inflammatory disorders [15, 16]. Recently, some lncRNAs have been identified to be involved in endocrine related diseases. For instance, the abnormal expression of HI-LNC25 is closely associated with type 2 diabetes, by which regulated the expression of the Kruppel-like zinc finger transcription factor (GLIS3) mRNA [17]. Estradiol (E2) can modulate lncRNA expression in E2 receptor (ER) α-positive epithelial ovarian cancer (EOC) cells, and certain lncRNAs have been correlated with advanced cancer progression [18]. Although there are numerous studies to report endocrine-related lncRNAs, the differential lncRNA expression profiles in OHSS and how the dysfunction of lncRNAs regulate the development of OHSS remain unclear.
In this study, to identify the aberrant expression profile of lncRNAs in granulosa cells (GC) and further investigate the roles of lncRNAs in development of OHSS, genome-wide profiling of lncRNAs was applied by using high-throughput sequencing in granulosa cells between OHSS high- and low-risk patients. Moreover, to better understand the potential roles of differentially expressed lncRNAs which may be implicated in OHSS development, we further analyzed and predicted the functions of these dysregulated lncRNAs following the lncRNA-miRNA-mRNA network.
Materials and methods
Patients and sample collection
The samples and clinical data were collected from the Reproductive Center, Department of Obstetrics and Gynecology, Sun Yat-Sen Memorial Hospital (Guangzhou, China) from Sep. 2016 to May. 2017. All the patients recruited into this research suffered from infertility and received ART/IVF. All the patients satisfied the following criteria: 20–35 years old; GnRH agonist long protocol or GnRH antagonist protocol; and no coexisting inflammatory disease. And for the OHSS high-risk patients, additionally the inclusion criteria were as follows: serum AMH > 5 ng/mL; serum E2 > 3500 pg/ml on HCG day and No. of oocytes >20. The controls (OHSS low-risk patients) were confirmed to be fallopian tubal diseases through laparoscopy and hysteroscopy, or male factor infertility; and non-PCOS; serum E2<3500 pg/ml and No. of oocytes <20. Women suffering from malignancy, benign ovarian cyst including endometrioma, allergic diseases; pelvic inflammation, known chronic, systemic, metabolic, or endocrine disease excluding polycystic ovarian syndrome, were excluded from this study. A total of 30 GC samples from OHSS high-risk patients and 30 control samples from patients with OHSS low risk were recruited in this study. The first step, three patients who had actually developed moderate OHSS from OHSS high-risk patients and three patients who did not develop OHSS from OHSS low-risk patients were prepared for sequencing. All patients provided informed consent, and the study was approved by the ethics committee of Sun Yat-Sen University. The sample collection and treatment were carried out in accordance with the approved guidelines.
Granulosa cells were obtained from women undergoing oocyte retrieval for in vitro fertilization treatment. Briefly, follicular fluid was collected from all of the follicles and then was centrifuged at 350 g for 5 min. The cells were resuspended and then added to 10 mL Ficoll (Sigma-Aldrich, St. Louis, MO). After centrifugation at 450 g for 15 min, the interphase cells were collected.
RNA isolation and quality measurement
Total RNA was isolated from tissue samples with TRizol (Invitrogen) according to the manufacturer’s instructions. The quality of RNA was measured with a NanoDrop (Thermo Fisher, USA), and the concentration of RNA was evaluated with Qubit 2.0 Fluorometer (Agilent Technologies). Samples with RNA Integrity Number greater than 4 as assessed by a BioAnalyzer (Agilent) with no visible sign of genomic DNA contamination from the HS Nanochip tracings were used for total RNA library generation. The total RNA from tissue samples was used only if the ratio of the absorbance at 260 and 280 nm (A260/A280) was between 1.8 and 2.2. All RNA samples were stored at 80 °C until further use.
Preparation of strand-specific RNA-Seq libraries and RNA-Seq
Three patients who had actually developed moderate OHSS from OHSS high risk and three patients who did not develop OHSS from OHSS low-risk patients were recruited for sequencing. After extracting the total RNA from three samples respectively, equation RNA of every three samples were pooled for library construction. mRNA and non-coding RNAs are enriched by removing rRNA from the total RNA with kit. By using the fragmentation buffer, the mRNAs and non-coding RNAs are fragmented into short fragments (about 200~500 nt), then the first-strand cDNA is synthesized by random hexamer-primer using the fragments as templates, and dTTP is substituted by dUTP during the synthesis of the second strand. Short fragments are purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. After that, the short fragments are connected with adapters, then the second strand is degraded using UNG (Uracil-N-Glycosylase) finally. After agarose gel electrophoresis, the suitable fragments are selected for the PCR amplification as templates. During the QC steps, Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System are used in quantification and qualification of the sample library. At last, the library sequenced using Illumina HiSeq™ X 10 sequencer.
Mapping reads to reference genome
rRNA removed reads are mapped to reference genome using an improved version of TopHat2 [19], which can align reads across splice junction without relying on gene annotation. TopHat uses Bowtie as an alignment ‘engine’ and breaks up reads that Bowtie cannot align on its own into smaller pieces called segments [20]. TopHat infers that the read spans a splice junction and estimates where junction’s splice sites are. By processing each “initially unmappable” read, TopHat can build up an index of splice sites in the transcriptome on the fly without a priori gene or splice site annotation. RNA-Seq read alignments can not only reveal new alternative splicing junction and isoforms, but also can be used to accurately quantify gene and transcript expression, because the number of reads produced by a transcript is proportional to its abundance.
Transcripts assembling and bioinformatic analysis
Reads mapped to genome will be assembled by Cufflinks [21]; we perform Reference Annotation Based Transcripts (RABT) assembly with the reference gene annotation to compensate incompletely assembled transcripts caused be read coverage gaps in the regions of reference gene. Faux-reads were generated from reference transcripts in order to capture features in the reference that could be missing in the sequencing data due to low coverage; these reads were merged with the (aligned) sequenced reads for assembly. The set of transfrags generated in the last step was then compared with the reference transcripts to remove transfrags that were approximately equivalent to the whole or a portion of a reference transcript.
Cuffdiff is a program that uses the Cufflinks transcript quantification engine to calculate gene and transcript expression levels in more than one condition and test them for significant differences on the basis of FPKM value. It looks for differentially expressed genes by estimating how many fragments came from each isoform and then converting the counts into isoform expression levels. To find differentially expressed genes, Cuffdiff calculates expression levels in each condition for each gene by adding up the expression levels for each gene’s splice isoforms. Cuffdiff tests for differences in each gene’s expression level across conditions. It is able to estimate p values for differential expression of individual transcripts. Like other tools, it uses a negative binomial model estimated from data to obtain variance estimates from which p values are computed.
Semi-quantitative RT-PCR
To confirm the RNA-seq results by an independent technique, qPCR was used to measure expression of selected genes. Quantitative real-time PCR was performed by using an ABI PRISM 7700 DNA Sequence Detection System (Applied Biosystems) and a SYBR Green PCR kit (Applied Biosystems). Total RNA was extracted and reverse transcribed to cDNA with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). The comparative threshold cycle method was used to calculate the relative gene expression, and GAPDH was used as a control.
Statistical analysis
LncRNA with an adjusted p value < 0.05 were assigned as differentially expressed. Pheatmap R package was then used for unsupervised cluster of differentially expressed genes and experimental conditions. KOBAS [22] software was used to test the statistical enrichment of genes which differentially expressed in KEGG pathways and Gene Ontology (GO) database (http://www.geneontology.org) which include molecular function, cellular component, and biological process information. p values were adjusted using the Benjamini and Hochberg’s approach, statistically significant were assigned as adjusted p value < 0.05. LncRNAs and mRNA UTR′3 sequences were used to find the sRNA binding site by miRanda (http://www.microrna.org/) [23]. Cytoscape (http://www.cytoscape.org/) [24] was applied to build the lncRNA-sRNA-mRNA interaction network.
The Wilcoxon signed-rank test was performed to analyze significant differences regarding the expression level of lncRNAs and miRNAs between samples. Pearson’s coefficient was applied to compare the microarray data and qPCR results. Student’s t test (two-tailed) was performed for other data analysis. p < 0.05 was considered to be significant.
Results
Identification of differentially expressed lncRNAs in granulosa cells between OHSS high-risk and low-risk patients
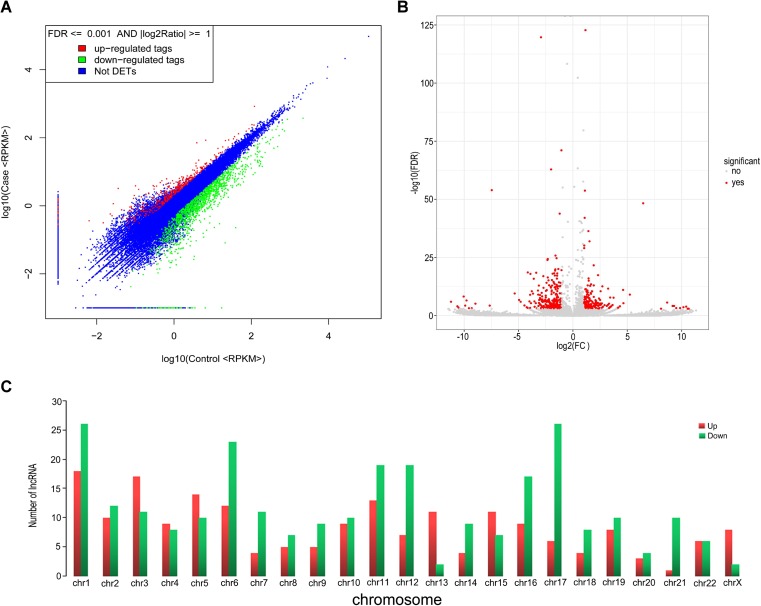
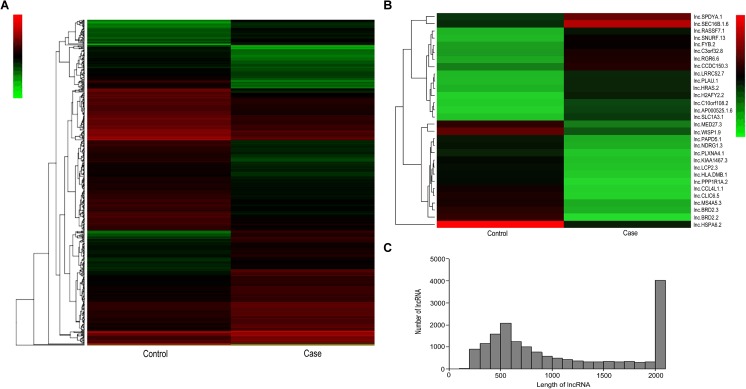
To find and identify lncRNAs which were differentially expressed in granulosa cells between OHSS high-risk and low-risk of patients, the lncRNA sequencing was performed and the RNA-Seq data was annotated using GENCODE (v19) as a reference. In our study, through RNA-sequencing in granulosa cells from OHSS high-risk patient, a total of 23,815 lncRNA genes were found (12,547 upregulated and 11,268 downregulated) and 482 lncRNA genes were found significantly differentially expressed, including 205 lncRNA genes were upregulated, whereas 277 were downregulated, compared to the granulosa cells of OHSS low-risk patient (fold-change ≥ 2, p value < 0.05, FDR value < 0.001). Differentially expressed lncRNAs were displayed through fold-change filtering (Fig. 1a). And the volcano plot filtering identified differentially changed lncRNAs with statistical significance between two groups (Fig. 1b).The frequency distribution of lncRNAs number and expression profiles between two groups were shown in Fig. 1c. Hierarchical clustering showed that lncRNAs expression pattern were distinguishable between OHSS high risk and low risk of patients samples (Fig. 2a). Of those, the top 30 up- and downregulated lncRNAs are listed in Table 1 by fold change (Fig. 2b, Table 1). The distribution of lncRNAs number and length profiles between two groups were shown in Fig. 2c.
Fig. 1.
Differences and characterizations in lncRNA expression profiles in granulosa cells between OHSS high-risk and low-risk patients. a Scatter plots are used to evaluate the difference in the expression of lncRNAs between experiment group and control group. The values plotted on X and Y axes are the averaged normalized signal values of each group (log10 scaled). The middle blue dots refer to no difference between the two groups. The lncRNAs above (red dots) and below (green dots) indicate more than 2.0-fold changes between two groups. b Volcano plots are using for visualizing differential expression between two different conditions. The vertical lines correspond to 2.0-fold (log2 scaled) up and down, respectively, and the horizontal line represents a p value of 0.05 (−log10 scaled). The red points in plot represent the differentially expressed lncRNAs with statistical significance. c The distribution of differentially expressed lncRNAs in human chromosomes
Fig. 2.
Heat map and hierarchical clustering showing expression values of all lncRNAs and the most up- and downregulated lncRNAs. Each column represents a sample, and each row represents a lncRNA. Red strip represents high relative expression, and green strip represents low relative expression. Case represents OHSS high-risk group, and control represents OHSS low-risk group. a Hierarchical cluster analysis of all lncRNAs using methods from Gene Spring software. b Hierarchical cluster analysis of the most up- and downregulated lncRNAs. c Frequency distribution of lncRNAs length. X axis: the range of lncRNAs length in granulosa cells between OHSS high-risk and low-risk patients. Y-axis: the frequency of lncRNAs with specific length
Table 1.
lncRNAs ranked by fold changes in sequencing data
| geneID | Genomic coordinates | Gene category | p value | Fold change |
|---|---|---|---|---|
| lnc-SEC16B.1-6 | chr1:178065367-178071882 | Antisense | 1.24661E-019 | 5.0695498748 |
| lnc-SNURF-13 | chr15:25658544-25658901 | Antisense | 2.37832E-006 | 4.6521528827 |
| lnc-LGR6-6 | chr1:202530389-202530835 | Intronic | 3.87352E-008 | 4.4775880466 |
| lnc-H2AFY2-2 | chr10:71960924-71961551 | lincRNA | 6.42424E-008 | 4.3990338703 |
| lnc-C3orf32-8 | chr3:8618880-8619316 | lincRNA | 4.4563E-007 | 3.938930838 |
| lnc-RASSF7-1 | chr11:557595-560106 | Antisense | 6.37496E-024 | 3.8383195777 |
| lnc-FYB-2 | chr5:39374847-39376144 | Sense overlapping | 9.19494E-006 | 3.7444157349 |
| lnc-PLAU-1 | chr10:75764895-75787673 | Intronic | 0.0001058842 | 3.434959889 |
| lnc-CCDC150-3 | chr2:197852654-197855043 | Antisense | 1.548186E-012 | 3.4323810903 |
| lnc-LRRC52-7 | chr1:164649322-164650036 | Intronic | 1.96248E-006 | 3.2905471609 |
| lnc-HRAS-2 | chr11:555660-558420 | Sense overlapping | 1.325978E-015 | 3.2403597706 |
| lnc-SPDYA-1 | chr2:29003252-29003526 | Intronic | 9.85858E-007 | 3.2167935177 |
| lnc-SLC1A3-1 | chr5:36907214-36912362 | Intronic | 6.82E-15 | 3.063612874 |
| lnc-C10orf108-2 | chr10:674578-677195 | Antisense | 2.42E-09 | 2.9501457305 |
| lnc-AP000525.1–6 | chr22:16117749-16127623 | lincRNA | 8.84488E-012 | 3.1657333031 |
| lnc-BRD2-2 | chr6:33045252-33045521 | Intronic | 1.143574E-008 | 0.118744685 |
| lnc-HSPA6-2 | chr1:161561940-161575452 | lincRNA | 1.067834E-122 | 0.1332596812 |
| lnc-CLIC6-5 | chr21:36377027-36384582 | Antisense | 6.0305E-008 | 0.1588540009 |
| lnc-CCL4L1-1 | chr17:34515328-34523870 | Antisense | 4.17992E-008 | 0.1702007152 |
| lnc-PPP1R1A-2 | chr12:54936895-54937889 | Antisense | 9.18332E-010 | 0.1811814065 |
| lnc-BRD2-3 | chr6:33045969-33046235 | Intronic | 2.56644E-006 | 0.18811658 |
| lnc-HLA-DMB-1 | chr6:32902406-32904678 | Sense overlapping | 1.138762E-026 | 0.2052420389 |
| lnc-LCP2-3 | chr5:169673241-169673861 | Sense overlapping | 5.41726E-009 | 0.2199516935 |
| lnc-KIAA1467-3 | chr12:13349720-13362504 | Sense overlapping | 2.30888E-008 | 0.2212609298 |
| lnc-MS4A5-3 | chr11:60146005-60157458 | Sense overlapping | 3.3154E-015 | 0.2231202653 |
| lnc-WISP1-9 | chr8:134067204-134070012 | Intronic | 1.612984E-065 | 0.2536645275 |
| lnc-PLXNA4-1 | chr7:131193828-131195657 | Sense overlapping | 0.000001104 | 0.2553010728 |
| lnc-MED27-3 | chr9:134600556-134600854 | Intronic | 4.6589E-006 | 0.2607330106 |
| lnc-NDRG1-3 | chr8:134500325-134501876 | Sense overlapping | 7.42412E-006 | 0.2664011195 |
| lnc-PAPD5-1 | chr16:50139752-50140295 | Sense overlapping | 3.94792E-006 | 0.2890200825 |
Validation of differentially expressed lncRNAs in granulosa cells between OHSS high-risk and low-risk patients
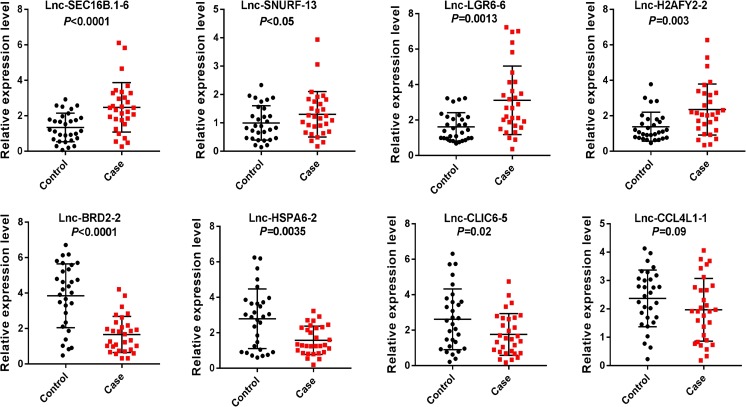
To validate the differentially expression profiles, semi-quantitative RT-PCR were performed to confirm the expression changes of eight lncRNAs, including four upregulated and four downregulated in granulosa cells from OHSS patients with high risk (n = 30) and low risk (n = 30). The results showed that these lncRNAs were significantly differentially expressed. The quantitative RT-PCR results confirmed the RNA-Seq results that lnc-SEC16B.1-6, lnc-SNURF-13, lnc-LGR6-6, and lnc-H2AFY2-2 were observed to be upregulated, while lnc-BRD2-2, lnc-HSPA6-2, and lnc-CLIC6-5 were found to be downregulated significantly in granulosa cells from OHSS high-risk patients in comparison with that in granulosa cells from OHSS low-risk patients, no statistical significance was found in lnc-CCL4L1-1 expression between two groups (Fig. 3).
Fig. 3.
Quantitative real-time PCR validation for the expression of eight lncRNAs. The expression levels of eight lncRNAs were validated by qPCR in 30 patients. Case represents OHSS high-risk group, and control represents OHSS low-risk group. Data are shown with triplicate as mean ± SEM
Construction of the lncRNA-miRNA-mRNA association network
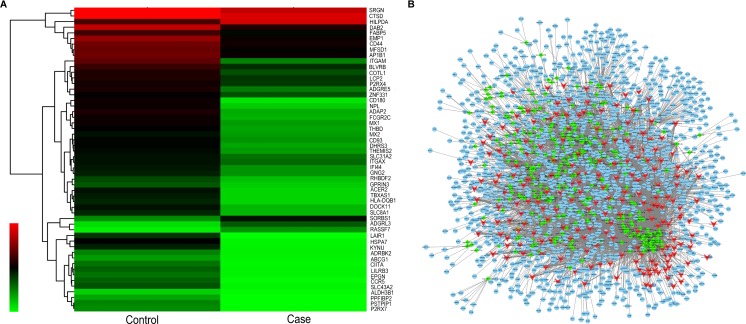
It has been reported that lncRNAs can regulate many biological processes by involved in multiple signal pathway, sponging of specific miRNAs as competing endogenous RNAs (ceRNA) and controlling downstream signaling molecular expression and activation is one of the pivotal functions [25, 26]. To determine the function and molecular mechanism of lncRNAs, the lncRNA-miRNA-mRNA interaction network was predicted based on the binding relationship between lncRNAs and their target miRNAs, which were predicted by conserved seed-matching sequence using the software for miRNA target prediction according to miRanda database. Based on the validation result above, the 8 lncRNAs were predicted according to the complementary miRNA matching sequence and a total of 1,361 miRNAs was predicted. The number of miRNA which has binding sites to lncRNAs was varied largely among the 8 lncRNAs, such as 393 miRNAs were successfully predicted to bind to lnc-HSPA6-2:9 and 45 miRNAs have at least 2 binding sites to the lnc-HSPA6-2:9; meanwhile, only 87 miRNAs can bind to lnc-SEC16B.1-6:1 and almost all of the miRNA has only one binding site to the lncRNA (Supplement Table 1).We next constructed an lncRNA-miRNA-mRNA network using 482 lncRNAs and 351 miRNAs together with their 1459 target genes. The data displayed the interaction network of each lncRNA, its potential complementary binding miRNAs, and their target genes, as shown in Fig. 4a, b.
Fig. 4.
The lncRNA/miRNA/mRNA network analysis. a Cluster analysis of the expression alternation of target genes and the target genes are displayed. b The lncRNA/miRNA/mRNA interaction network consists of 205 upregulated lncRNAs (red), 277 downregulated lncRNAs (red), and 351 predicted miRNAs (green) together with their 1459 target genes (blue)
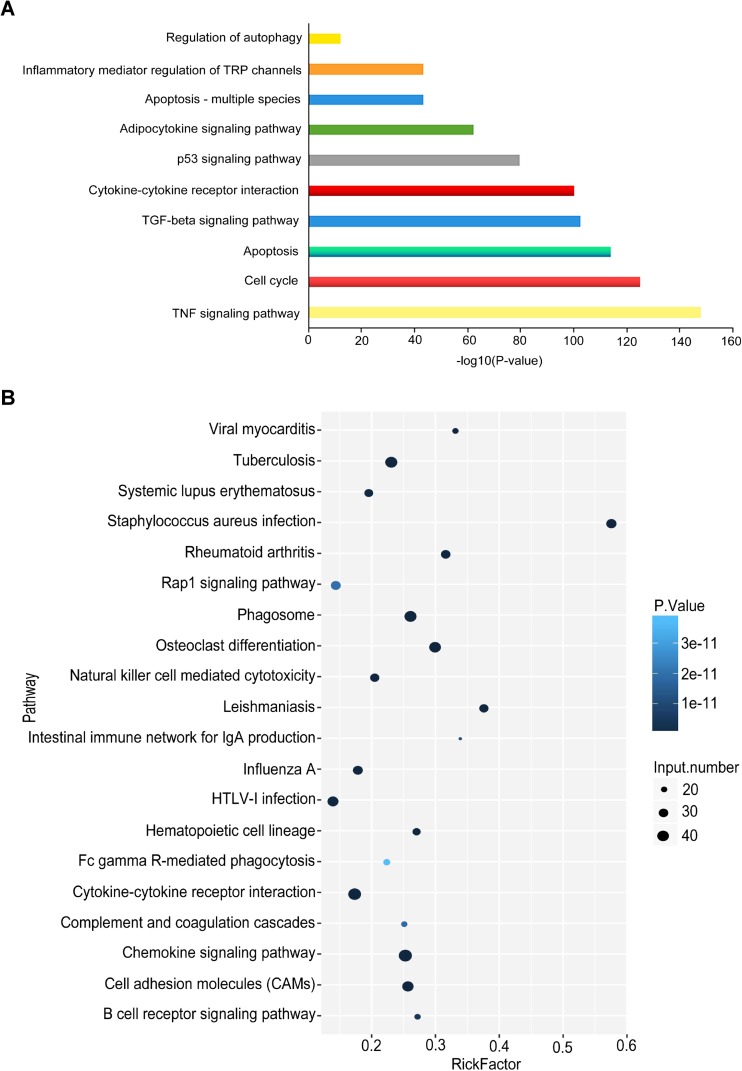
We finally explored the biological process (including cellular component, molecular function, and biological process) of deregulated lncRNAs using GO analysis and gene-enriched KEGG pathways analysis. Using GO analysis, several important pathological processes may be involved in the dysregulation of lncRNAs, including the TNF-α signaling, cell cycle, apoptosis, TGF-β signaling, cytokine-cytokine receptor interaction, and inflammatory pathway, which are closely related to the development of OHSS (Fig. 5a) [27–29].The similar result was also obtained by KEGG pathways analysis. As Fig. 5b shown, the differential expression of lncRNAs was closely associated with cell adhesion, chemokine signaling, cytokine-cytokine receptor, and phagosome pathway, which were suggested to be involved in ovarian dysfunction. These results showed that the dysregulation of lncRNAs might contribute to the development of OHSS. Further studies are necessary to confirm the binding relation, function, and molecular mechanisms of lncRNAs-miRNAs axis in OHSS pathogenesis.
Fig. 5.
Function annotations for differentially expressed lncRNAs. a Gene ontology and b KEGG pathway analysis were applied for the annotation of the cellular component, biological process, and molecular function of target genes. p < 0.05 was considered as statistically significant. In GO analysis, the vertical axis shows the annotated functions of the target genes. The horizontal axes show −log2 transformed p value and the gene number of each cluster, respectively
Discussion
OHSS has become the most common and potentially most dangerous complication of assisted reproductive technology which is caused by the development of multiple ovarian follicles [30].Thus, enlarged ovary volume, increased capillary permeability, fluid, and protein extravasation into the body of the third space are significant. It is well known that abnormal ovarian secretion of VEGF is the key of OHSS and accurate prediction of OHSS is strongly needed.
Benefit from the advances in sequencing technologies, the genome-wide profiling has extensively revealed that almost 98% of the transcriptional outputs are ncRNAs, which may play important roles in a variety of biological processes. The physiological functions of many ncRNAs are still being explored. Meanwhile, miRNAs are well studied in regulating of transcription, epigenetics modulation, and RNA-protein interaction [31].To our knowledge, several pathophysiologic mechanisms may be involved in ovarian dysfunction disease, including inflammation, oxidative stress, apoptosis, and autophagy, which were modulated by abnormal expression of ncRNAs [32]. Indeed, some miRNAs have been regarded as the potential diagnosis and therapeutic markers for ovarian diseases [33]. However, studies on the identification and functional characterization of lncRNAs in ovarian diseases, especially in OHSS, are still limited.
In this study, we first carried out genome-wide lncRNA expression profiles in the GCs of women with OHSS high and low risk by high-throughput sequencing. We found that 482 lncRNAs were significantly differentially expressed in GCs between two groups. The eight lncRNAs with the most differentially expressed genes were further validated by semi-quantitative PCR in 30 paired GC samples, and the analysis confirmed the sequencing findings. GO analysis and KEGG pathway analysis also implied that the differential expression of lncRNAs were correlated with serial of signal pathways, including cell cycle, apoptosis, autophagy, and inflammatory signaling pathway (TNF-α, TGF-β, cytokine-cytokine receptor interaction, adipocytokine signaling pathway), which were suggested to contribute to the pathogenesis of OHSS. Recently, lncRNAs are recognized to modulate downstream signaling pathway by functioning as the competing endogenous RNAs (ceRNA) to which can bind to the special cluster of miRNAs as sponges. Moreover, this regulatory model has been validated in tumorigenesis and other disease. In this study, we successfully predicted more than 100 lncRNA/miRNA interactions based on conserved seed sequence matches. And the lncRNA/miRNA interaction network was constructed using the bioinformatics tools, which may shed a light to unveil the information for ceRNA research of lncRNAs. Therefore, the interaction of lncRNAs/miRNAs needs to be further verified by more basic work.
As far as we know, this is the first study to report the expression profiles of lncRNAs in OHSS development. However, the deficiency of this study is obvious. We only have sequencing evidence to predict and analyze the roles and functions of altered lncRNAs in OHSS pathophysiology. More experiments not only in cell line, but also in animal model, are still needed to deeply reveal the underlying regulatory mechanisms of lncRNAs, which may help us to understand its functional significance in modulating OHSS.
In conclusion, our study for the first time revealed the lncRNAs expression profiles in OHSS development and the dysregulation of lncRNAs might associate with OHSS pathophysiology. With the construction of a ceRNA crosstalk network via bioinformatic analysis, we constructed a perspective to screen lncRNAs that could be involved in OHSS development. Our study also presented several possible candidate lncRNAs in GC for future diagnostic, therapeutic and functional research associated with OHSS. These candidate lncRNAs were correlated with several biological processes, suggesting that they may play important roles in the development and progression of OHSS. Our findings highlighted the possibility that lncRNAs could serve as a predictive marker for OHSS diagnosis and therapy. The limitation of this study is that the sample size of sequenced patients and validation cohort should be enlarged. More samples from patients should be recruited to verify the result in the further study.
Electronic supplementary material
(XLS 169 kb)
Acknowledgements
This study was funded by National Natural Science Funding (no. 81701519).
References
- 1.Luke B, Brown MB, Morbeck DE, Hudson SB, Coddington CC, 3rd, Stern JE. Factors associated with ovarian hyperstimulation syndrome (OHSS) and its effect on assisted reproductive technology (ART) treatment and outcome. Fertil Steril. 2010;94:1399–1404. doi: 10.1016/j.fertnstert.2009.05.092. [DOI] [PubMed] [Google Scholar]
- 2.Mourad S, Brown J, Farquhar C. Interventions for the prevention of OHSS in ART cycles: an overview of Cochrane reviews. Cochrane Database Syst Rev. 2017;1:CD012103. doi: 10.1002/14651858.CD012103.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A.a.o. Practice Committee of the American Society for Reproductive Medicine. Electronic address, M. Practice Committee of the American Society for Reproductive Prevention and treatment of moderate and severe ovarian hyperstimulation syndrome: a guideline. Fertil Steril. 2016;106:1634–1647. doi: 10.1016/j.fertnstert.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 4.Humaidan P, Nelson SM, Devroey P, Coddington CC, Schwartz LB, Gordon K, Frattarelli JL, Tarlatzis BC, Fatemi HM, Lutjen P, Stegmann BJ. Ovarian hyperstimulation syndrome: review and new classification criteria for reporting in clinical trials. Hum Reprod. 2016;31:1997–2004. doi: 10.1093/humrep/dew149. [DOI] [PubMed] [Google Scholar]
- 5.Papanikolaou EG, Pozzobon C, Kolibianakis EM, Camus M, Tournaye H, Fatemi HM, Van Steirteghem A, Devroey P. Incidence and prediction of ovarian hyperstimulation syndrome in women undergoing gonadotropin-releasing hormone antagonist in vitro fertilization cycles. Fertil Steril. 2006;85:112–120. doi: 10.1016/j.fertnstert.2005.07.1292. [DOI] [PubMed] [Google Scholar]
- 6.Delvigne A, Rozenberg S. Epidemiology and prevention of ovarian hyperstimulation syndrome (OHSS): a review. Hum Reprod Update. 2002;8:559–577. doi: 10.1093/humupd/8.6.559. [DOI] [PubMed] [Google Scholar]
- 7.Cerrillo M, Pacheco A, Rodriguez S, Gomez R, Delgado F, Pellicer A, Garcia-Velasco JA. Effect of GnRH agonist and hCG treatment on VEGF, angiopoietin-2, and VE-cadherin: trying to explain the link to ovarian hyperstimulation syndrome. Fertil Steril. 2011;95:2517–2519. doi: 10.1016/j.fertnstert.2010.12.054. [DOI] [PubMed] [Google Scholar]
- 8.Naredi N, Talwar P, Sandeep K. VEGF antagonist for the prevention of ovarian hyperstimulation syndrome: current status. Med J Armed Forces India. 2014;70:58–63. doi: 10.1016/j.mjafi.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrowski D, Szabo L, Sator M, Just A, Egarter C. Ovarian hyperstimulation syndrome is correlated with a reduction of soluble VEGF receptor protein level and a higher amount of VEGF-A. Hum Reprod. 2012;27:196–199. doi: 10.1093/humrep/der349. [DOI] [PubMed] [Google Scholar]
- 10.Scotti L, Abramovich D, Pascuali N, Irusta G, Meresman G, Tesone M, Parborell F. Local VEGF inhibition prevents ovarian alterations associated with ovarian hyperstimulation syndrome. J Steroid Biochem Mol Biol. 2014;144(Pt B):392–401. doi: 10.1016/j.jsbmb.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B, Zhang YW, Tong XH, Liu YS. Characterization of microRNA profile in human cumulus granulosa cells: identification of microRNAs that regulate notch signaling and are associated with PCOS. Mol Cell Endocrinol. 2015;404:26–36. doi: 10.1016/j.mce.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Jiang L, Huang J, Li L, Chen Y, Chen X, Zhao X, Yang D. MicroRNA-93 promotes ovarian granulosa cells proliferation through targeting CDKN1A in polycystic ovarian syndrome. J Clin Endocrinol Metab. 2015;100:E729–E738. doi: 10.1210/jc.2014-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Cao Q, Ge J, Liu C, Ma Y, Meng Y, Wang Y, Zhao X, Liu R, Li C, Wang Y, Zhong J, Ju W, Jenkins EC, Brown WT, Zhong N. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: genome wide differential expression of lncRNAs in early spontaneous abortion. Am J Reprod Immunol. 2014;72:359–375. doi: 10.1111/aji.12275. [DOI] [PubMed] [Google Scholar]
- 16.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakic N, Garcia-Hurtado J, Rodriguez-Segui S, Pasquali L, Sauty-Colace C, Beucher A, Scharfmann R, van Arensbergen J, Johnson PR, Berry A, Lee C, Harkins T, Gmyr V, Pattou F, Kerr-Conte J, Piemonti L, Berney T, Hanley N, Gloyn AL, Sussel L, Langman L, Brayman KL, Sander M, McCarthy MI, Ravassard P, Ferrer J. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu JJ, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor alpha-positive ovarian cancer progression. Oncol Rep. 2014;31:1613–1622. doi: 10.3892/or.2014.3000. [DOI] [PubMed] [Google Scholar]
- 19.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 26.Shen L, Wang Q, Liu R, Chen Z, Zhang X, Zhou P, Wang Z. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 2018;46(2):717–729. doi: 10.1093/nar/gkx1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scotti L, Irusta G, Abramovich D, Tesone M, Parborell F. Administration of a gonadotropin-releasing hormone agonist affects corpus luteum vascular stability and development and induces luteal apoptosis in a rat model of ovarian hyperstimulation syndrome. Mol Cell Endocrinol. 2011;335:116–125. doi: 10.1016/j.mce.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Chen CD, Chen HF, Lu HF, Chen SU, Ho HN, Yang YS. Value of serum and follicular fluid cytokine profile in the prediction of moderate to severe ovarian hyperstimulation syndrome. Hum Reprod. 2000;15:1037–1042. doi: 10.1093/humrep/15.5.1037. [DOI] [PubMed] [Google Scholar]
- 29.Bedarida GV, Hoffmann U, Tato F. Jugular vein thrombosis with severe local and systemic inflammation in a woman with ovarian hyperstimulation syndrome. Thromb Haemost. 2006;95:1035–1037. doi: 10.1160/TH05-12-0794. [DOI] [PubMed] [Google Scholar]
- 30.Gera PS, Tatpati LL, Allemand MC, Wentworth MA, Coddington CC. Ovarian hyperstimulation syndrome: steps to maximize success and minimize effect for assisted reproductive outcome. Fertil Steril. 2010;94:173–178. doi: 10.1016/j.fertnstert.2009.02.049. [DOI] [PubMed] [Google Scholar]
- 31.Yan Z, Shah PK, Amin SB, Samur MK, Huang N, Wang X, Misra V, Ji H, Gabuzda D, Li C. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic Acids Res. 2012;40:e135. doi: 10.1093/nar/gks395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murri M, Insenser M, Fernandez-Duran E, San-Millan JL, Escobar-Morreale HF. Effects of polycystic ovary syndrome (PCOS), sex hormones, and obesity on circulating miRNA-21, miRNA-27b, miRNA-103, and miRNA-155 expression. J Clin Endocrinol Metab. 2013;98:E1835–E1844. doi: 10.1210/jc.2013-2218. [DOI] [PubMed] [Google Scholar]
- 33.Huang X, Liu C, Hao C, Tang Q, Liu R, Lin S, Zhang L, Yan W. Identification of altered microRNAs and mRNAs in the cumulus cells of PCOS patients: miRNA-509-3p promotes oestradiol secretion by targeting MAP3K8. Reproduction. 2016;151:643–655. doi: 10.1530/REP-16-0071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS 169 kb)