Abstract
Background and Purpose
Thiazolidinediones, acting as PPAR‐γ ligands, reduce hepatic steatosis in humans and animals. However, the underlying mechanism of this action remains unclear. The purpose of this study was to investigate changes in hepatic metabolites and lipids in response to treatment with the thiazolidinedione pioglitazone in an animal model of obese Type 2 diabetes.
Experimental Approach
Male Otsuka Long‐Evans Tokushima Fatty (OLETF) rats were orally administered either vehicle (control) or pioglitazone (30 mg·kg−1) and fed a high‐fat diet (60% kcal fat) for 12 weeks. Hepatic metabolites were analysed via metabolomic and lipidomic analyses. Gene expression and PLA2 activity were analysed in livers from pioglitazone‐treated and control rats.
Key Results
OLETF rats that received pioglitazone showed decreased fat accumulation and improvement of lipid profiles in the liver compared to control rats. Pioglitazone treatment significantly altered levels of hepatic metabolites, including free fatty acids, lysophosphatidylcholines and phosphatidylcholines, in the liver. In addition, pioglitazone significantly reduced the expression of genes involved in hepatic de novo lipogenesis and fatty acid uptake and transport, whereas genes related to fatty acid oxidation were up‐regulated. Gene expression and enzyme activity of PLA2, which hydrolyzes phosphatidylcholines to release lysophosphatidylcholines and free fatty acids, were significantly decreased in the livers of pioglitazone‐treated rats compared to control rats.
Conclusions and Implications
Our results present evidence for the ameliorative effect of pioglitazone on hepatic steatosis, largely due to the regulation of lipid metabolism, including fatty acids, lysophosphatidylcholines, phosphatidylcholines and related gene‐expression patterns.
Abbreviations
- GC‐TOF
gas chromatography‐time of flight
- LPCAT
lysophosphatidylcholine acyltransferase
- LysoPC
lysophosphatidylcholine
- LysoPE
lysophosphatidylethanolamine
- MUFA
monounsaturated fatty acid
- NAFLD
non‐alcoholic fatty liver disease
- NASH
non‐alcoholic steatohepatitis
- OLETF
Otsuka Long‐Evans Tokushima Fatty
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- TG
triglyceride
- TZD
thiazolidinedione
- UPLC‐Q
ultra‐performance LC‐quadrupole
Introduction
Non‐alcoholic fatty liver disease (NAFLD) is characterized by massive triglyceride (TG) accumulation in the liver and can range from simple fatty liver (steatosis) to non‐alcoholic steatohepatitis (NASH), which is defined as hepatic steatosis with hepatocyte damage and inflammation (Ahmed et al., 2015). NASH may further progress to cirrhosis, liver failure and hepatocellular carcinoma (Ahmed et al., 2015). Despite the growing prevalence and incidence of NAFLD, definitive therapy for NAFLD and, more specifically, for NASH, has not yet been established (Younossi et al., 2016).
Thiazolidinediones (TZDs), agonists of http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=595&familyId=86&familyType=NHR, are an attractive candidate therapy for NAFLD/NASH. TZDs including http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1056 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2694 reduce hyperlipidaemia and hyperglycaemia and improve insulin resistance, which are strongly associated with NAFLD (Spiegelman, 1998). TZDs have provided histological benefit by improving steatosis and inflammation in many clinical trials (Promrat et al., 2004; Belfort et al., 2006; Sanyal et al., 2010; Cusi et al., 2016). TZDs also increase fat storage in adipose tissue, leading to the sensitization of liver and peripheral tissues to insulin (Lehrke and Lazar, 2005). In addition, TZDs promote production of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=3726, which is a well‐recognized anti‐diabetic adipokine and is associated with improved insulin sensitivity and hepatic fat accumulation through enhancement of http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=593&familyId=86&familyType=NHR and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1540 activity, fatty acid oxidation and insulin signalling (Yamauchi et al., 2002; Kadowaki et al., 2006; Awazawa et al., 2011; Blüher, 2012). More recently, the sirtuins, a family of NAD ‐dependent deacetylases and ADP‐ribosyltransferases, have been suggested to be a crucial regulator of beneficial TZD‐mediated metabolic effects in the liver (Shen et al., 2010; Yang et al., 2011; Yang et al., 2014).
However, information regarding metabolite changes related to TZD treatment is limited (Watkins et al., 2002; Rull et al., 2014; Meierhofer et al., 2014). To understand the metabolic effects and mechanisms of TZD treatment in NAFLD/NASH disease models, comprehensive MS‐based metabolomics studies on tissues and biological fluids are needed. Recently, a metabolomics approach has been used to identify metabolic changes and mechanisms in the organs and biofluids of animal models after the administration of food, medicine or a single compound (Jung et al., 2015; Park et al., 2015; Suh et al., 2016; Zhang et al., 2016). Moreover, metabolomics studies integrated other omics data, such as genomics and transcriptomics, allowing further insight into the mechanisms of obesity‐related disease treatment (Rull et al., 2014; Meierhofer et al., 2014; Takahashi et al., 2014; Aw and Fukuda, 2015).
In this study, we performed metabolomic and lipidomic profiling of liver tissue to determine the mechanisms of action of the TZD pioglitazone in the treatment of NAFLD in a rat model of obesity and Type 2 diabetes. To clarify the effects of TZDs, we further interpreted metabolome data and lipid metabolism‐related gene expression.
Methods
Animals
All animal experiments were performed in accordance with the criteria of the ‘Guide for the Care and Use of Laboratory Animals’ published by the US National Institutes of Health (National Research Council, 1996) and approved by the Ethics Committee for Animal Experiments of Kangbuk Samsung Hospital, Sungkyunkwan University. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015).
Male Otsuka Long‐Evans Tokushima Fatty (OLETF) rats (150–200 g, 4 weeks old) were purchased from Otsuka Pharmaceutical (Tokushima, Japan). These rats, which develop spontaneous obesity and diabetes mellitus, were used as a model of NAFLD (Kawano et al., 1992). Because of a http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=76 defect, the rats exhibit hyperphagia, hyperglycaemia and obesity and ultimately develop histological changes comparable to NAFLD in the liver (Song et al., 2013). Rats were housed in individual cages using heated wood chip litter as bedding material in a pathogen‐free environment and maintained in a temperature‐ and humidity‐controlled room (24 ± 2°C and 60% humidity) with a 12‐h light/dark cycle and fed standard irradiated rodent chow (11% kcal fat; LabDiet, St. Louis, MO, USA) and water ad libitum. Body weight and food intake were measured every week. At the age of 7 weeks, rats weighing 250–300 g were randomly divided into two groups: the control group (n = 10) and the treatment group (n = 8). To investigate the metabolic effects of pioglitazone on hepatic steatosis, we fed the animals a high‐fat diet (60% kcal fat, Research Diets, New Brunswick, NJ, USA) and administered either vehicle as a control or pioglitazone (30 mg·kg−1·day−1) as a treatment via stomach gavage daily for 12 weeks. The order of treatment was randomized. Pioglitazone was dissolved in 0.5% sodium carboxymethyl cellulose as a vehicle. There were no experimental losses related to toxic events following pioglitazone treatment. At the end of the experiment, rats were fasted overnight and deeply anaesthetized with an overdose of isoflurane. Blood was collected from the abdominal aorta, and then tissues were immediately dissected, weighed and stored at −80°C until further analysis.
Measurement of glucose tolerance and insulin sensitivity
The glucose tolerance test (GTT) was carried out after an 18‐h fast. For GTT, rats were orally administered glucose solution (2 g·kg−1 body weight), and blood glucose levels were measured at baseline and 15, 30, 60, 90 and 120 min after injection using the automated glucocard X‐Meter (Arkray, Kyoto, Japan). The insulin tolerance test (ITT) was also carried out after an 18‐h fast. For the ITT, rats were given an i.p. injection of human insulin (1 U·kg−1 body weight), and blood glucose levels were measured at baseline and 15, 30, 60, 90 and 120 min after injection. The AUC was calculated.
Metabolic parameters
Plasma glucose and insulin were analysed by enzymic assay (Sigma‐Aldrich, St. Louis, MO, USA and Crystal Chem, Downers Grove, IL, USA). Blood and liver TG levels were also measured by enzymic assay (Sigma‐Aldrich). Commercial kits were employed for the measurement of free fatty acids (Wako Pure Chemical Industries, Osaka, Japan) and total cholesterol (Cayman Chemical Com., Ann Arbor, MI, USA). Liver metabolic parameters were normalized to respective liver weights.
Histological analysis and NAFLD activity score
Dissected liver tissues were fixed in 10% formalin buffer overnight. The tissues were then embedded in paraffin, sliced into 4‐μm‐thick sections and stained with haematoxylin and eosin (H&E). Digital images were captured with an Olympus BX51 light microscope (100× magnification; Tokyo, Japan). A pathologist blinded to the experimental conditions evaluated the NAFLD activity score (NAS). Three features of NAFLD (steatosis, lobular inflammation and ballooning) were scored as described previously (Chang et al., 2015). NAS was calculated as the sum of scores for steatosis (0–3), ballooning (0–2) and inflammation (0–3).
Gene expression analysis
Total RNA was extracted from tissues using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). RNA was subsequently reverse‐transcribed to cDNA using a High‐Capacity RNA‐to‐cDNA Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. Gene expression levels were analysed by real‐time PCR using the LightCycler 480 System (Roche Diagnostics, Indianapolis, IN, USA) based on SYBR Green fluorescence signals, as described previously. The primer sequences used are listed in Supporting Information Table S1. The PCR parameters were as follows: pre‐denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 10 s and extension at 72°C for 20 s. mRNA expression of each target was normalized to glyceraldehyde‐3‐phosphate dehydrogenase as an internal control and expressed as fold change relative to the control group.
PLA2 activity assay
Rat tissues were homogenized in cold PBS at pH 7.4 and centrifuged at 10 000× g for 15 min at 4°C. The supernatants were collected, and the intracellular activity of http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=275 was measured with the EnzChek® Phospholipase A2 Assay Kit (Invitrogen).
Lipid peroxidation measurement
Malondialdehyde (MDA), a product of lipid peroxidation, was measured with the OxiSelect™ TBARS Assay Kit (Cell Biolabs, San Diego, CA, USA) according to the manufacturer's instructions. The extent of lipid peroxidation was expressed in μmol·mg−1 protein.
Metabolomic analysis
Each liver sample (100 mg) was extracted with 1 mL of 50% cold methanol (methanol : water, v/v) and 10 μL of internal standard (2‐chlorophenylalanine, 0.5 mg·mL−1) using an MM400 mixer mill (Retsch®, Haan, Germany) at a frequency of 30 s−1 for 10 min with a zirconium bead. After cold centrifugation (12 578× g for 10 min), the supernatant was filtered through a 0.2‐μm polytetrafluoroethylene filter and evaporated using a speed vacuum concentrator (Modulspin 31, Biotron, Korea). The final concentration of each sample was 5 mg·mL−1 using methanol for MS analysis. Gas chromatography‐time of flight (GC‐TOF)‐MS analysis was performed according to Jung et al. (2015), and 1 μL of derivatized samples was injected in splitless mode. Ultra‐performance LC‐quadrupole (UPLC‐Q)‐TOF‐MS analysis was processed as previously described by Suh et al. (2016), and 5 μL of sample was injected into UPLC‐Q‐TOF‐MS. Identification of metabolites with MS/MS fragmentation were provided in the Supporting Information Data S1.
Lipidomic analysis
For lipidomic analysis, each liver sample (100 mg) was extracted with 1 mL of solvent mixture (chloroform/methanol, 1:2, v/v) using a mixer mill (MM400, Retsch) at a frequency of 30 s−1 for 2 min. After a 1‐h incubation at room temperature, 300 μL of chloroform and 450 μL of water were added, vortexed and centrifuged (1000× g, 10 min, 4°C). The lower (organic) phase was transferred to new tube, and the upper phase was re‐extracted using 600 μL of chloroform. After vortexing and centrifugation, the lower phase was transferred to a new tube. Pooled chloroform extracts were dried to complete dryness for lipid profiling. Each dried extract was reconstituted in 100 μL of solvent mixture (methanol/chloroform, 9:1, v/v) and diluted 50‐fold with methanol/chloroform (9/1, v/v) containing 7.5 mM ammonium acetate. Lipid profiling was performed using Thermo LTQ XL ion trap MS (Thermo Fisher Scientific, West Palm Beach, FL, USA) as previously described by Shon et al. (2015). A detailed information of lipid metabolites with MS/MS fragmentation were already constructed and provided in LipidBlast libraries (Kind et al., 2013).
Measurement of total phospholipids
Separation of phospholipids via one‐dimensional TLC was performed according to Leray et al. (1987). After spraying with primuline solution on a TLC plate, we checked fluorescent spots under UV light using gel documentation.
Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2015). Metabolomics and lipidomics data were analysed by an analyst blinded to treatment status. Data were expressed as mean ± SD. Significant differences between groups (P value) were evaluated using PASW statistics 18 software (SPSS Inc., Chicago, IL, USA) via non‐parametric tests with the Mann–Whitney test. For metabolomics statistical analysis, GC‐TOF‐MS and UPLC‐Q‐TOF‐MS raw data were converted to NetCDF format (*.cdf) using LECO Chroma TOF™ software (version 4.44, LECO Corp., St. Joseph, MI, USA) and MassLynx DataBridge (version 4.1, Waters Corp., Milford, MA, USA) respectively. After raw file conversion, peak alignment was processed using MetAlign software (version 041012, http://www.metalign.nl). Alignment of spectral data obtained from ion trap mass spectrometer was performed using Genedata Expressionist MSX (Genedata AG, Basel, Switzerland). Metabolomics and lipidomics data were normalized using an internal standard. Multivariate statistical analysis including PCA and OPLS‐DA was performed using SIMCA P+ (version 12.0, Umetrics, Umea, Sweden). Significant differences between groups (P value) were evaluated using PASW statistics 18 software (SPSS Inc.) via non‐parametric tests with the Mann–Whitney Test. A heat map was constructed using MeV software (version 4.8.1, http://www.tm4.org). Fold change was calculated as the ratio of the difference between the experimental and the control groups to identify any trends in the effects of pioglitazone.
Materials
Pioglitazone was obtained as a gift from Takeda Pharmaceutical (Tokyo, Japan). All chemicals and solvents used in this study were of analytical reagent grade. Freshly distilled water was used throughout the experiments.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a, 2017b, 2017c).
Results
Effects of pioglitazone on metabolic parameters
After 3 months on a high‐fat diet, rats fed with pioglitazone gained significantly more body weight compared with rats fed with vehicle (control), in agreement with earlier studies (Tang et al., 1999; Kawaguchi et al., 2004; Collino et al., 2010). Total fat weight to body weight ratios for pioglitazone‐treated rats was significantly higher than that of control rats (Table 1), consistent with the previous finding that TZDs including pioglitazone lead to increased fat storage into adipose tissue. There was a significant increase in subcutaneous fat and epididymal fat in rats that were treated with pioglitazone (Table 1), while mesenteric and retroperitoneal fat were not significantly different from those of control rats. Rats treated with pioglitazone showed a 25% reduction in the AUC for the glucose tolerance test and a 45% reduction in the AUC for the insulin tolerance test compared with control rats (Figure 1). Pioglitazone administration also reduced blood glucose levels by 20% (Table 1). Levels of plasma TG (25%) and free fatty acids (31%) significantly decreased in pioglitazone‐treated rats compared with control rats, and a less dramatic decrease in plasma total cholesterol level was observed (14%) (Table 1).
Table 1.
The effects of pioglitazone on metabolic parameters
| Control (n = 10) | Pioglitazone (n = 8) | |
|---|---|---|
| Body weight (g) | – | – |
| Baseline | 345.6 ± 15.6 | 350.3 ± 4.5 |
| Final | 715.9 ± 27.7 | 828.6 ± 44.5a |
| Weight change | 370.3 ± 25.4 | 478.3 ± 44.3a |
| Food intake (g) | 20.4 ± 2.0 | 18.4 ± 2.2 |
| Fat pad weight (%) | 17.6 ± 1.2 | 18.9 ± 0.7a |
| Subcutaneous fat (%) | 4.7 ± 0.6 | 5.9 ± 0.5a |
| Epididymal fat (%) | 3.3 ± 0.2 | 3.6 ± 0.3a |
| Mesenteric fat (%) | 1.9 ± 0.3 | 1.9 ± 0.3 |
| Retroperitoneal fat (%) | 7.7 ± 0.9 | 7.2 ± 0.8 |
| Liver weight (%) | 2.8 ± 0.1 | 2.2 ± 0.1a |
| Plasma glucose (mM) | 6.2 ± 0.4 | 5.1 ± 0.3a |
| Plasma insulin (pM) | 288.0 ± 104.7 | 344.5 ± 41.9 |
| Plasma adiponectin (ng·mL−1) | 3.6 ± 0.5 | 5.8 ± 0.5a |
| Plasma triglycerides (mM) | 1.0 ± 0.2 | 0.8 ± 0.1a |
| Plasma free fatty acids (μM) | 442.6 ± 119.3 | 279.4 ± 74.5a |
| Plasma total cholesterol (mM) | 2.9 ± 0.4 | 2.5 ± 0.2a |
| Liver triglycerides (μmol·g−1 liver) | 127.8 ± 19.1 | 95.4 ± 35.9a |
| Liver free fatty acids (μmol·g−1 liver) | 8.5 ± 2.6 | 5.8 ± 1.3a |
| Liver total cholesterol (μmol·g−1 liver) | 6.9 ± 0.6 | 6.8 ± 0.9 |
| Liver malondealdehyde (μmol·mg−1 protein) | 41.9 ± 10.1 | 35.8 ± 3.7a |
Data are expressed as the mean ± SD. Control: HFD (n = 10); pioglitazone: 30 mg·kg−1 pioglitazone + HFD (n = 8). Fat pad weight and liver weights are expressed as a percentage of fasted body weight. Data are expressed as the mean ± SD (n = 10 for the control group; n = 8 for the PIO‐treated group).
P < 0.05, significantly different from the control group. HFD, high‐fat diet.
Figure 1.
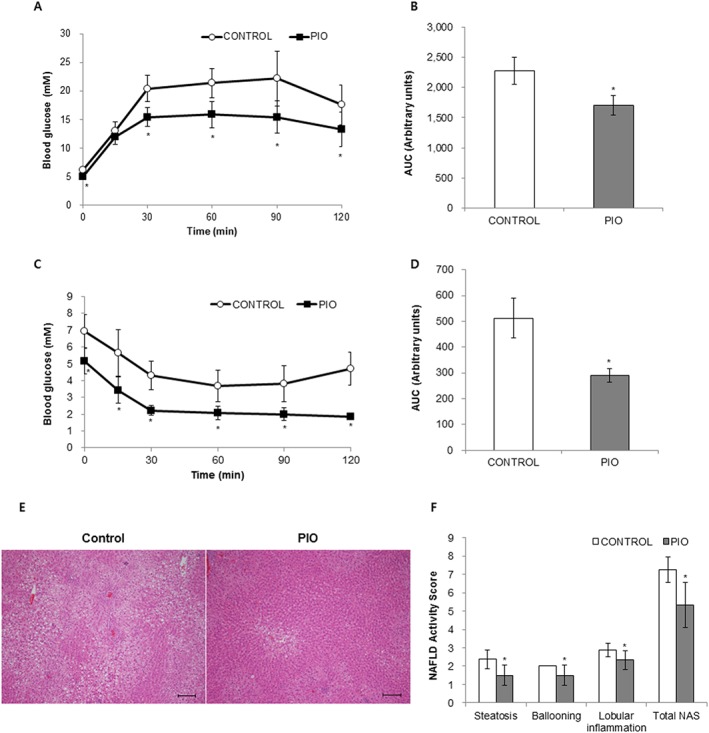
Effects of pioglitazone on oral glucose tolerance (OGTT), intraperitoneal insulin tolerance (ipITT) and hepatic steatosis. Effects of pioglitazone (PIO) on OGTT (A and B) and ipITT (C and D). Representative H&E liver sections (E, scale bar, 200 μm). The NAFLD activity score (NAS) is shown in F. Data are expressed as the mean ± SD; n = 10 for control group, n = 8 for PIO‐treated group. * P < 0.05, significantly different from control.
Effects of pioglitazone on hepatic steatosis
Despite the observed increase in body weight, the liver weight to body weight ratio was significantly decreased in pioglitazone‐treated rats compared with control rats (Table 1). As shown in Figure 1E, F, on histological analysis using H&E staining, control livers exhibited an accumulation of lipid droplets in most hepatocytes with features of both macrovesicular and microvesicular steatosis, diffuse hepatocyte ballooning and foci of inflammatory cell infiltrates throughout the lobules, indicating that a high‐fat diet resulted in severe steatosis. Pioglitazone administration significantly decreased the degree of steatosis, ballooning, lobular inflammation and total NAFLD activity score compared with vehicle treatment. Levels of hepatic TGs and free fatty acids were significantly attenuated in pioglitazone‐treated rats compared with control rats (Table 1). However, there was no significant difference in the level of total cholesterol in the livers of pioglitazone‐treated rats. The degree of lipid peroxidation based on the level of MDA, a product of lipid peroxidation, was lower in pioglitazone‐treated rat livers compared with control rat livers (Table 1).
Hepatic metabolite analysis
To identify the hepatic metabolites influenced by pioglitazone administration, we performed comprehensive metabolite profiling of rat liver using GC‐TOF‐MS, UPLC‐Q‐TOF‐MS and nanomate‐LTQ‐MS analyses with multivariate statistical analysis. The robustness and reproducibility of the MS analysis were ascertained by analysing the sample extracts in blocks of 10 runs followed by an injection of quality control sample made with pooled samples. The analytical samples were subjected to randomized runs in each block. Further, the potential analytical biases generated while sample preparation and chromatographic runs were minimized through employing the internal standards for data normalization, generating the adjusted quantitative parameters for identified metabolites.
The PCA score plot from the UPLC‐Q‐TOF‐MS dataset exhibited distinct clustering for each group, while that from GC‐TOF‐MS and nanomate‐LTQ‐MS datasets showed overlap between the two groups (Supporting Information Figure S1). As shown in Figure 2, the orthogonal partial least squares discriminant analysis (OPLS‐DA) score plots for liver samples from GC‐TOF‐MS (Figure 2A), UPLC‐Q‐TOF‐MS (Figure 2B) and nanomate‐LTQ‐MS (Figure 2C) datasets showed significant separation between control and pioglitazone groups. The fitness and prediction values of the OPLS‐DA model were evaluated using R2X, R2Y, Q2 and P values. In this study, a total of 131 metabolites were identified, and the relative level of each metabolite was converted into fold‐change in Tables 2 and 3. The levels of most amino acids, fatty acids and some carbohydrates (arabinose, adonitol, glucuronic acid, lactose and maltose) were relatively decreased by pioglitazone treatment compared with the control group. The levels of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2796 (PE) and lysophospholipids, with the exception of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5652 (lysoPE) with C18:0 and two forms of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2508 (lysoPC) with C16:0 and C18:0, were also decreased by pioglitazone treatment, while most TG levels were increased in the control group than in the pioglitazone treatment group.
Figure 2.
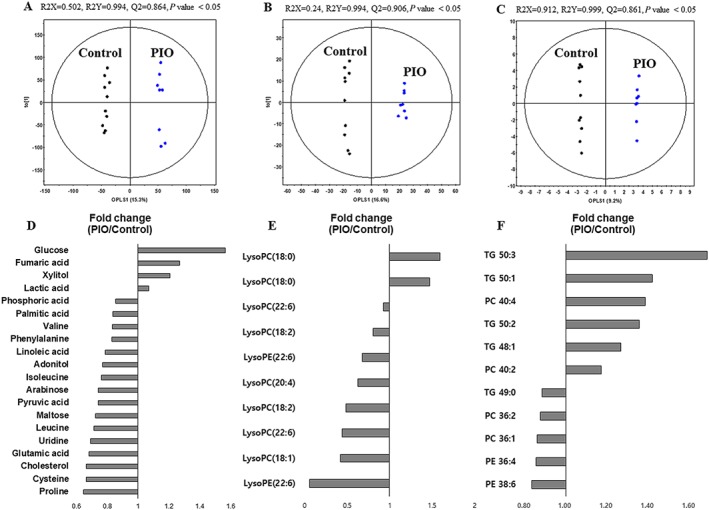
Orthogonal partial least squares discriminant analysis (OPLS‐DA) score plots from GC‐TOF‐MS, UPLC‐Q‐TOF‐MS and nanomate‐LTQ‐MS data and significantly altered hepatic metabolites between control and pioglitazone‐treated groups. OPLS‐DA score plots from GC‐TOF‐MS (A), UPLC‐Q‐TOF‐MS (B) and nanomate‐LTQ‐MS (C) data and significantly altered hepatic metabolites between control and pioglitazone(PIO)‐treated groups (D–F); n = 10 for control group, n = 8 for PIO‐treated group. Metabolites selected based on VIP > 0.7 and P < 0.05, from the OPLS‐DA model. VIP, variable importance in projection.
Table 2.
Identification of liver metabolites from GC‐TOF‐MS and UPLC‐Q‐TOF‐MS combined with multivariate analysis
| GC‐TOF‐MS | |||||||
|---|---|---|---|---|---|---|---|
| RT | Metabolites | TMS | VIP | Fold change (PIO/Control) | Identified ion (m/z) | Fragment ion | ID |
| Amino acids | |||||||
| 5.44 | Alanine | 2TMS | 0.41 | 0.93 | 116 | 73, 116, 147, 190 | STD/MS |
| 6.63 | Valine | 2TMS | 1.10 | 0.83 | 144 | 73, 100, 144, 218 | STD/MS |
| 7.18 | Leucinea | 2TMS | 1.69 | 0.72 | 158 | 73, 15, 148, 158, 205, 299 | STD/MS |
| 7.40 | Isoleucinea | 2TMS | 1.03 | 0.76 | 100 | 59, 73, 86, 100, 114, 158, 178, 218, 232 | STD/MS |
| 7.45 | Prolinea | 2TMS | 1.50 | 0.65 | 216 | 73, 100, 142, 216 | STD/MS |
| 7.53 | Glycine | 3TMS | 0.59 | 0.93 | 174 | 59, 73, 86, 100, 117, 133, 147, 158, 174, 248, 276 | STD/MS |
| 8.02 | Serine | 3TMS | 0.43 | 0.92 | 218 | 59, 73, 100, 116, 133, 147, 174, 188, 204, 218, 278 | STD/MS |
| 8.27 | Threonine | 3TMS | 0.65 | 0.87 | 117 | 57, 73, 86, 101, 117, 147, 203, 219, 291 | STD/MS |
| 9.41 | Aspartic acid | 3TMS | 0.52 | 0.67 | 232 | 73, 100, 147, 188, 202, 218, 232 | STD/MS |
| 9.46 | 5‐Oxoproline | 2TMS | 0.36 | 0.97 | 156 | 59, 73, 84, 100, 133, 147, 156, 230, 258 | STD/MS |
| 9.70 | Cysteine | 3TMS | 1.15 | 0.66 | 220 | 59, 73, 100, 116, 132, 147, 163, 204, 220, 294 | STD/MS |
| 10.19 | Glutamic acida | 3TMS | 1.74 | 0.68 | 128 | 56, 73, 84, 100, 114, 128, 147, 204, 230, 246, 258, 348 | STD/MS |
| 10.29 | Phenylalanine | 2TMS | 0.94 | 0.83 | 218 | 59, 73, 100, 117, 130, 147, 160, 177, 192, 204, 218, 266 | STD/MS |
| 11.67 | Ornithine | 4TMS | 0.28 | 0.94 | 130 | 59, 73, 86, 100, 130, 142, 174 | STD/MS |
| Organic compounds | |||||||
| 4.89 | Pyruvic acida | 1TMS | 1.43 | 0.74 | 174 | 59, 73, 89, 99, 115, 158, 174, 189 | STD/MS |
| 5.00 | Lactic acid | 2TMS | 0.71 | 1.07 | 117 | 73, 117, 147, 191 | STD/MS |
| 6.96 | Urea | 2TMS | 0.30 | 1.06 | 189 | 66, 73, 87, 100, 115, 130, 147, 157, 171, 189 | STD/MS |
| 7.84 | Fumaric acid | 2TMS | 0.99 | 1.27 | 245 | 73, 147, 217, 245 | STD/MS |
| Fatty acids and lipids | |||||||
| 13.08 | Palmitic acid | 1TMS | 1.01 | 0.84 | 117 | 73, 117, 129, 185, 313 | STD/MS |
| 14.11 | Linoleic acid | 1TMS | 1.20 | 0.79 | 103 | 67, 103, 117, 129, 147, 262, 337 | STD/MS |
| 14.27 | Stearic acid | 1TMS | 0.33 | 0.92 | 117 | 55, 73, 117, 129, 145, 185, 201, 341 | STD/MS |
| 19.68 | Cholesterol | 1TMS | 0.94 | 0.67 | 129 | 73, 129, 213, 255, 329, 368, 458 | STD/MS |
| Carbohydrates | |||||||
| 10.71 | Arabinosea | 4TMS | 1.53 | 0.74 | 103 | 59, 73, 89, 103, 117, 133, 147, 189, 217 | STD/MS |
| 10.93 | Xylitola | 5TMS | 1.09 | 1.21 | 103 | 59, 73, 103, 147, 217 | STD/MS |
| 11.06 | Adonitola | 5TMS | 1.35 | 0.77 | 217 | 59, 73, 89, 103, 129, 147, 189, 217, 319 | STD/MS |
| 12.35 | Glucosea | 5TMS | 1.67 | 1.57 | 205 | 59, 73, 89, 103, 117, 147, 189, 205, 229, 319 | STD/MS |
| 12.78 | Glucuronic acid | 5TMS | 0.41 | 0.88 | 333 | 59, 73, 89, 103, 129, 147, 292, 333 | STD/MS |
| 16.82 | Lactose | 8TMS | 0.52 | 0.86 | 204 | 73, 103, 147, 169, 204, 243, 271, 305, 361 | STD/MS |
| 17.17 | Maltosea | 8TMS | 1.42 | 0.72 | 204 | 59, 73, 103, 129, 147, 204, 243, 271, 291, 319, 361 | STD/MS |
| Nucleosides | |||||||
| 15.60 | Uridinea | 3TMS | 1.58 | 0.69 | 217 | 73, 103, 147, 169, 191, 217, 259, 299, 445 | STD/MS |
| Others | |||||||
| 6.21 | Phosphoric acid | 2TMS | 0.87 | 0.86 | 241 | 73, 89, 119, 133, 147, 163, 181, 195, 211, 225, 241, 256 | STD/MS |
| UPLC‐Q‐TOF‐MS | ||||||||
|---|---|---|---|---|---|---|---|---|
| RT | Metabolites | Identified ion (m/z) | VIP | Fold change (PIO/Control) | MW | HMDB formula | Error (mDa) | ID |
| 5.50 | Taurine‐conjugated cholic acid | 514.2845 | – | 1.09 | 515 | C26H45NO7S | 2 | Lib |
| 6.16 | Taurine‐conjugated deoxycholic acid | 498.2913 | – | 1.21 | 499 | C26H45NO6S | 2.4 | Lib |
| 6.29 | Taurine‐conjugated cholic acid | 514.2871 | – | 1.24 | 515 | C26H45NO7S | 5 | Lib |
| 7.74 | Taurine‐conjugated deoxycholic acid | 498.2892 | – | 0.91 | 499 | C26H45NO6S | 0 | Lib |
| 8.15 | LysoPE (22:6)a | 524.2798 | 1.38 | 0.06 | 525 | C27H44NO7P | 4 | Lib |
| 8.18 | LysoPC (22:6)a | 552.3109 | 1.63 | 0.44 | 567 | C30H50NO7P | −3.4 | Lib |
| 8.21 | LysoPC (18:2)a | 504.3125 | 1.90 | 0.48 | 519 | C26H50NO7P | −0.8 | Lib |
| 8.23 | LysoPC (20:4)a | 528.3077 | 1.58 | 0.63 | 543 | C28H50NO7P | 3.9 | Lib |
| 8.28 | LysoPE (22:6) | 524.2784 | 1.05 | 0.68 | 525 | C27H44NO7P | −1.2 | Lib |
| 8.31 | LysoPC (22:6) | 552.3093 | 0.33 | 0.93 | 567 | C30H50NO7P | −3.9 | Lib |
| 8.35 | LysoPE (20:4) | 500.2786 | 0.08 | 0.98 | 501 | C25H44NO7P | −3.6 | Lib |
| 8.37 | LysoPC (20:4) | 528.3110 | 0.81 | 0.93 | 543 | C28H50NO7P | 1.4 | Lib |
| 8.39 | LysoPC (18:2)a | 504.3115 | 1.18 | 0.80 | 519 | C26H50NO7P | −2.9 | Lib |
| 8.62 | LysoPC (16:0) | 480.3119 | 0.61 | 1.22 | 495 | C24H50NO7P | 1.7 | Lib |
| 8.79 | LysoPE (16:0) | 452.2778 | 0.45 | 1.20 | 453 | C21H44NO7P | −3.5 | Lib |
| 8.83 | LysoPC (16:0) | 480.3086 | 0.47 | 1.15 | 495 | C24H50NO7P | 3.3 | Lib |
| 8.87 | LysoPC (18:1)a | 506.3289 | 1.68 | 0.42 | 521 | C26H52NO7P | 3.6 | Lib |
| 9.06 | LysoPC (18:1) | 506.3273 | 0.01 | 1.00 | 521 | C26H52NO7P | −0.1 | Lib |
| 9.69 | LysoPC (18:0) | 508.3431 | 1.14 | 1.60 | 523 | C26H54NO7P | 0.3 | Lib |
| 9.89 | LysoPC (18:0) | 508.3390 | 1.08 | 1.47 | 523 | C26H55NO7P | −2.8 | Lib |
ID, identification; HMDB, The Human Metabolome Database (http://www.hmdb.ca); Lib, in house library; MS, mass fragments pattern; RT, retention time; STD, standard compounds; TMS, trimethylsilyl; VIP, variable importance in projection.
Metabolites showing significant differences (VIP > 0.7 and P value <0.05) between experimental groups.
Table 3.
Identification of liver metabolites via nanomate‐LTQ‐MS combined with multivariate analysis
| No. | Metabolites | Identified ion (m/z) | VIP | Adduct | Fold‐changeb (PIO/Control) |
|---|---|---|---|---|---|
| 1 | CE 18:3 | 664.6 | 1.20 | NH4+ | 0.77 |
| 2 | CE 20:2 | 694.5 | 0.39 | NH4+ | 1.09 |
| 3 | CE 20:3 | 692.6 | 0.44 | NH4+ | 1.07 |
| 4 | CE 20:4 | 690.5 | 0.48 | NH4+ | 1.10 |
| 5 | CE 20:5 | 688.5 | 0.07 | NH4+ | 0.99 |
| 6 | DG 34:1 | 612.4 | 0.02 | NH4+ | 1.00 |
| 7 | DG 34:2 | 610.3 | 0.02 | NH4+ | 1.00 |
| 8 | DG 36:1 | 640.6 | 1.28 | NH4+ | 1.25 |
| 9 | PC 32:1 | 754.6 | 0.47 | Na+ | 1.03 |
| 10 | PC 32:2 | 752.6 | 0.29 | Na+ | 0.97 |
| 11 | PC 34:0 | 784.8 | 1.07 | Na+ | 0.92 |
| 12 | PC 34:1 | 782.7 | 0.62 | Na+ | 0.95 |
| 13 | PC 34:2 | 780.7 | 0.19 | Na+ | 1.02 |
| 14 | PC 34:3 | 778.6 | 0.06 | Na+ | 1.00 |
| 15 | PC 35:0 | 798.7 | 0.73 | Na+ | 1.11 |
| 16 | PC 36:0 | 790.7 | 1.49 | H+ | 0.89 |
| 17 | PC 36:1a | 788.8 | 1.82 | H+ | 0.86 |
| 18 | PC 36:2a | 786.7 | 1.87 | H+ | 0.88 |
| 19 | PC 36:3a | 806.7 | 1.14 | Na+ | 0.91 |
| 20 | PC 36:4 | 804.7 | 0.30 | Na+ | 0.95 |
| 21 | PC 36:5 | 802.7 | 0.68 | Na+ | 1.05 |
| 22 | PC 38:5 | 808.7 | 0.72 | H+ | 0.95 |
| 23 | PC 38:2 | 814.8 | 0.96 | H+ | 0.94 |
| 24 | PC 38:3 | 812.8 | 0.88 | H+ | 0.94 |
| 25 | PC 38:4 | 810.7 | 1.02 | H+ | 0.93 |
| 26 | PC 38:6 | 828.7 | 0.58 | Na+ | 0.90 |
| 27 | PC 38:7 | 826.7 | 0.58 | Na+ | 1.06 |
| 28 | PC 40:2a | 842.7 | 1.76 | H+ | 1.17 |
| 29 | PC 40:4a | 860.7 | 2.30 | Na+ | 1.39 |
| 30 | PE 36:4a | 762.8 | 2.36 | Na+ | 0.86 |
| 31 | PE 38:3 | 770.6 | 0.44 | H+ | 0.96 |
| 32 | PE 38:4 | 768.6 | 0.45 | H+ | 0.95 |
| 33 | PE 38:5 | 766.6 | 1.21 | H+ | 0.91 |
| 34 | PE 38:6a | 764.5 | 2.04 | H+ | 0.83 |
| 35 | TG 42:0 | 740.5 | 0.08 | NH4+ | 1.01 |
| 36 | TG 46:0 | 796.7 | 0.09 | NH4+ | 1.01 |
| 37 | TG 46:1 | 794.7 | 1.41 | NH4+ | 0.89 |
| 38 | TG 46:2 | 792.6 | 1.30 | NH4+ | 0.85 |
| 39 | TG 48:0 | 824.7 | 0.06 | NH4+ | 1.00 |
| 40 | TG 48:1a | 822.7 | 1.58 | NH4+ | 1.27 |
| 41 | TG 48:2 | 820.6 | 0.98 | NH4+ | 1.27 |
| 42 | TG 48:3 | 818.7 | 0.48 | NH4+ | 1.03 |
| 43 | TG 49:0a | 838.8 | 1.63 | NH4+ | 0.88 |
| 44 | TG 49:1 | 836.7 | 0.84 | NH4+ | 0.94 |
| 45 | TG 49:2 | 834.7 | 0.57 | NH4+ | 0.96 |
| 46 | TG 50:0 | 852.8 | 1.41 | NH4+ | 1.32 |
| 47 | TG 50:1a | 850.7 | 1.55 | NH4+ | 1.42 |
| 48 | TG 50:2a | 848.7 | 2.17 | NH4+ | 1.36 |
| 49 | TG 50:3a | 846.7 | 2.22 | NH4+ | 1.69 |
| 50 | TG 50:4a | 844.6 | 1.53 | NH4+ | 1.37 |
| 51 | TG 51:1 | 864.7 | 1.32 | NH4+ | 1.19 |
| 52 | TG 52:0 | 880.8 | 1.00 | NH4+ | 1.20 |
| 53 | TG 52:1 | 878.8 | 0.98 | NH4+ | 1.50 |
| 54 | TG 52:2 | 876.7 | 1.07 | NH4+ | 1.57 |
| 55 | TG 52:3 | 874.7 | 1.14 | NH4+ | 1.57 |
| 56 | TG 52:4 | 872.7 | 1.12 | NH4+ | 1.49 |
| 57 | TG 52:5 | 870.7 | 1.44 | NH4+ | 1.65 |
| 58 | TG 53:1 | 892.7 | 1.40 | NH4+ | 1.29 |
| 59 | TG 53:2 | 890.7 | 1.50 | NH4+ | 1.41 |
| 60 | TG 53:3 | 888.7 | 1.34 | NH4+ | 1.37 |
| 61 | TG 53:4 | 886.6 | 1.45 | NH4+ | 1.29 |
| 62 | TG 54:3 | 902.7 | 0.81 | NH4+ | 1.39 |
| 63 | TG 54:4 | 900.7 | 0.80 | NH4+ | 1.37 |
| 64 | TG 54:5 | 898.7 | 0.78 | NH4+ | 1.34 |
| 65 | TG 54:6 | 896.7 | 0.88 | NH4+ | 1.39 |
| 66 | TG 55:0 | 922.7 | 0.50 | NH4+ | 1.21 |
| 67 | TG 55:1 | 920.7 | 0.61 | NH4+ | 1.26 |
| 68 | TG 55:2 | 918.7 | 0.86 | NH4+ | 1.25 |
| 69 | TG 55:3 | 916.7 | 0.71 | NH4+ | 1.09 |
| 70 | TG 55:4 | 914.7 | 0.90 | NH4+ | 1.12 |
| 71 | TG 55:5 | 912.7 | 1.21 | NH4+ | 1.21 |
| 72 | TG 56:1 | 934.7 | 0.10 | NH4+ | 1.01 |
| 73 | TG 56:3 | 930.7 | 0.19 | NH4+ | 0.97 |
| 74 | TG 56:4 | 928.8 | 0.64 | NH4+ | 1.23 |
| 75 | TG 56:6 | 924.7 | 0.48 | NH4+ | 1.21 |
| 76 | TG 57:2 | 946.7 | 0.11 | NH4+ | 1.04 |
| 77 | TG 57:3 | 944.6 | 0.16 | NH4+ | 1.06 |
| 78 | TG 57:4 | 942.7 | 0.10 | NH4+ | 0.98 |
| 79 | TG 58:5 | 954.8 | 0.03 | NH4+ | 0.99 |
| 80 | TG 58:6 | 952.7 | 0.02 | NH4+ | 1.01 |
VIP, variable importance in the projection.
Significantly different from OPLS‐DA models, based on VIP value (>0.7) and P <0.05.
Relative levels of metabolites were converted into fold‐change.
Hepatic metabolites that were discriminant between groups were selected based on the variable importance in projection value (>0.7) from each OPLS‐DA model and the P value (<0.05) from non‐parametric testing. Twenty‐eight metabolites, including four amino acids, one organic acid, four carbohydrates, six lysophospholipids, seven phospholipids, five TGs and uridine, were significantly different between groups (Figure 2). The levels of most amino acids and carbohydrates, except glucose and xylitol, were significantly down‐regulated in pioglitazone‐treated rats (Figure 2D). However, two carbohydrates (glucose and xylitol), two phosphatidylcholines (PCs) (40:2 and 40:4), and most TGs, except TG (49:0), were significantly up‐regulated in control rats compared to pioglitazone‐treated rats. In particular, we observed a marked alteration in lipid metabolism‐related metabolites (fatty acids, cholesterol, lysoPCs, lysoPEs, PCs, PEs and TGs) in the liver. According to Figure 2D–F, most fatty acids, lysoPCs with a relatively long acyl chain (≥18) and a double bond and phospholipids with a relatively short acyl chain (<39) were significantly decreased in the pioglitazone‐treated group compared with the control group.
Effect of pioglitazone on phospholipase activity and phospholipid biosynthesis
Lysophospholipids are mainly generated from PLA2‐catalysed hydrolysis of phospholipids (Balsinde et al., 1999). Therefore, to further explain the observed decreases in lysoPC levels in pioglitazone‐treated livers, we determined PLA2 gene expression and its activity in the liver. Among the various subtypes of PLA2, we measured the activity of cytosolic PLA2α (cPLA2α), the enzyme that catalyses the hydrolysis of membrane phospholipids to liberate lysophospholipids and arachidonic acid (C20:4), which is subsequently metabolized into eicosanoids (Balsinde et al., 1999). As shown in Figure 3A, B, pioglitazone treatment significantly decreased cPLA2α activity and tended to reduce mRNA expression of PLA2G4A, the gene encoding cPLA2α. Furthermore, we determined whether phospholipid biosynthesis was affected by pioglitazone treatment. First, we measured total PC and PE content in the liver using TLC. As shown in Figure 3C, TLC analysis revealed that hepatic total PC content was decreased in pioglitazone‐treated rats compared with control rats, whereas total PE levels in the liver were not altered among control and pioglitazone‐treated groups. We also measured the mRNA levels of all lysophosphatidylcholine acyltransferase (LPCAT) family members, which catalyse the synthesis of PC from lysoPC (Zhao et al., 2008), in the livers of control and pioglitazone‐treated rats. Figure 3D showed that LPCAT3 is the most abundant isoform of this family in the liver, and there was a lower trend in the mRNA expression of LPCAT3 in the pioglitazone‐treated group, i.e., this difference was not statistically significant. Therefore, livers of pioglitazone‐treated rats had decreased pool of phospholipid, especially PC, while LPCAT enzymes, which re‐synthesize PC from lysoPC, were not affected.
Figure 3.
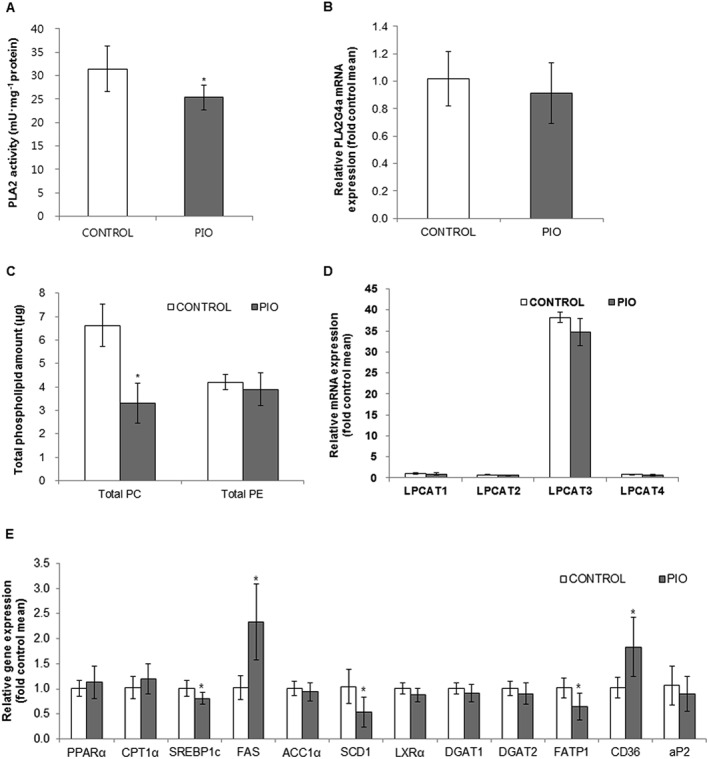
Effects of pioglitazone on hepatic PLA2 activity phospholipid biosynthesis and gene expression related to hepatic fatty acid metabolism. Effects of pioglitazone (PIO) on hepatic PLA2 activity (A), PLA2G4a gene expression (B), total phospholipid amounts (C), LPCAT gene expression (D) and gene expression related to hepatic fatty acid metabolism (E). Data are expressed as mean ± SD; n = 10 for control group, n = 8 for PIO‐treated group. * P < 0.05, significantly different from control. ACC1α, acetyl‐CoA carboxylase 1α; CPT1α, carnitine palmitoyltransferase 1α; DGAT, diglyceride acyltransferase; FAS, fatty acid synthase; CD36/FAT, fatty acid translocase; FATP1, fatty acid transport protein 1; aP2/FABP4, fatty acid binding protein 4; LXRα, liver X receptor α; SREBP1c, sterol regulatory element‐binding protein 1c; SCD1, stearoyl‐CoA desaturase‐1.
Hepatic expression of genes related to fatty acid metabolism
We further investigated whether pioglitazone affects gene expression involved in fatty acid metabolism (Figure 3E). First, we determined the expression of genes involved in de novo lipogenesis. Pioglitazone down‐regulated expression of sterol regulatory element‐binding protein 1c (SREBP1c), the master regulator of lipogenesis, and stearoyl‐CoA desaturase‐1 (SCD1), key enzymes for the synthesis of monounsaturated fatty acids (MUFAs) from saturated fatty acids (Liu et al., 2015). Gene expression of http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=602 and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2821&familyId=844&familyType=ENZYME 1 and 2, all of which are related to hepatic de novo lipogenesis, was decreased or trended lower in pioglitazone‐treated groups compared with control groups respectively. However, mRNA levels of two other SREBP1c‐target geneshttp://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2608 and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=255#1263, were up‐regulated or similar to those of untreated rats respectively. Next, we investigated whether pioglitazone increased fatty acid oxidation, thereby reducing fat accumulation in rat livers. The mRNA levels of PPAR‐α and its target gene, carnitine palmitoyltransferase 1α, showed a trend towards higher values in pioglitazone‐treated rats, compared with control rats.. We also observed that expression of genes involved in uptake and transport of fatty acids, especially long‐chain fatty acids (carbons 12–20), including http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1108 and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2534 (also called adipocyte protein 2, aP2), was significantly down‐regulated in the livers of the pioglitazone group. Thus, these observations could account for a decrease in free fatty acids, which can be taken up by hepatocytes and activate PPAR‐γ signalling in the liver. On the other hand, fatty acid translocase/CD36 mRNA expression was up‐regulated in the liver by pioglitazone treatment. Therefore, these results indicated that pioglitazone treatment may result in decreased availability of fatty acids for hepatic de novo lipogenesis and increased fatty acid oxidation, decreasing the accumulation of lipid in the liver.
Discussion
In this study, we found that pioglitazone treatment improved the plasma lipid profile including circulating free fatty acids and TGs, glucose tolerance and liver histology in OLETF rats fed a high‐fat diet, consistent with the results from previous studies (Tang et al., 1999; Kawaguchi et al., 2004; Collino et al., 2010). We also observed that pioglitazone treatment resulted in alteration of hepatic metabolite and lipid profiles related to metabolic parameters. Furthermore, we found that pioglitazone regulates the metabolic pathways involved in hepatic de novo lipogenesis, fatty acid transport and oxidation and lysophospholipid biosynthesis, which are involved in metabolite and lipid profile shifts, thus improving hepatic steatosis.
NAFLD is highly prevalent and strongly associated with metabolic disorders such as obesity, diabetes and cardiovascular disease. Recently, metabolomic studies, defined as the comprehensive quantitative and qualitative analysis of all metabolites in objects including cells, tissues or biofluids, have been performed to investigate metabolomic components related to the formation of hepatic steatosis in humans and animals (Nicholson et al., 1999; Beyoğlu and Idle, 2013). Compared with non‐steatotic livers, lipid species including cholesterol esters, TGs, diacylglycerols and sphingomyelins were elevated in steatotic liver (Beyoğlu and Idle, 2013). In addition, phosphocholine, choline, betaine and trimethylamine N‐oxide were up‐regulated in both the liver and the plasma of rodents with diet‐induced fatty liver, indicating an increased turnover of PC and PE species in the liver, thus releasing free fatty acids through the action of PLAs (Toye et al., 2007; Beyoğlu and Idle, 2013). Furthermore, increased hepatic levels of lysoPC, lysoPE and PC species have been reported for steatotic livers compared with non‐steatotic livers in humans and rodents (García‐Cañaveras et al., 2011; Han et al., 2017).
Our metabolomic and lipidomic analysis revealed that pioglitazone treatment significantly altered several hepatic metabolites and lipids including free fatty acids, lysophospholipids and phospholipids. Some of our results from metabolomic/lipidomic analysis were coincident with the earlier findings, whereas others were not. In the previous studies, levels of numerous long‐chain fatty acids such as saturated fatty acids (e.g. http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1055 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=3377) and MUFA (e.g. http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5547 and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=1054) were positively affected by a high‐fat diet (Kim et al., 2011; Liu et al., 2015). In our study, we observed that pioglitazone decreased palmitic acid and stearic acid, which suggests its role as a mediator of lipotoxicity. We could not detect hepatic MUFA in this study due to the small samples used for metabolomic analysis. However, it is conceivable that MUFA production may be decreased in the pioglitazone group via reduction of saturated fatty acids and SCD1 expression. In addition, expression of genes involved in long‐chain fatty acid uptake and transport and hepatic de novo lipogenesis was decreased. These findings suggest the attenuation of hepatic de novo lipogenesis following pioglitazone treatment.
A recent study has demonstrated elevation in lysoPCs, especially the fatty acyl group of C16:0 and C18:3, in human steatotic liver tissues (García‐Cañaveras et al., 2011). In another study on human livers, a significant increase in the level of lysoPC with C20:4 was observed in human samples during NAFLD progression (Han et al., 2017). Increased hepatic lysophospholipids have been reported in db/db mice and may cause insulin resistance (Han et al., 2011). Also, a high‐fat diet increased numerous hepatic lysophospholipids in mice (Kim et al., 2011; Kim et al., 2014; Liu et al., 2015). In the present study, pioglitazone treatment significantly decreased most hepatic lysoPCs, except those lysoPCs with fatty acyl groups of C16:0 and C18:0. Lower levels of cytosolic PLA2α activity and PLA2G4A mRNA expression in the pioglitazone‐treated group indicated a reduced potential for biosynthesis of lysophospholipids and release of free fatty acids. Meanwhile, adiponectin treatment reversed high‐fat diet‐induced increases in lysophospholipid levels in mouse liver (Liu et al., 2015). Given the observation of a significant increase in circulating adiponectin in the pioglitazone group, we cannot rule out the possibility that the effects of TZDs on lysophospholipid levels were mediated by adiponectin.
Although many studies have indicated that levels of lysoPCs are closely associated with oxidative stress and inflammation (Nishi et al., 1998; Stock et al., 2006; Schilling and Eder, 2010), this effect depends on the length and unsaturation of the fatty acyl group (Riederer et al., 2010; Hung et al., 2011). LysoPCs containing an unsaturated fatty acyl group such as C20:4 and C22:6 showed potent anti‐inflammatory activity in in vivo and in vitro models (Huang et al., 2010; Hung et al., 2011; Hung et al., 2012). Interestingly, lysoPC containing C16:0, which has pro‐inflammatory properties, enhanced glucose uptake in an insulin‐independent and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1485&familyId=533&familyType=ENZYME‐dependent manner in adipocytes and to have a glucose‐lowering effect in Type 1 and Type 2 diabetes mouse models (Yea et al., 2009). Activation of PPAR‐α also induced hepatic lysoPC (16:0) production and secretion (Takahashi et al., 2015). In addition, lysoPC (16:0) activated PPAR‐α and induced the expression of PPAR‐α target genes (Takahashi et al., 2015). Therefore, further study is needed to investigate the functions of lysoPCs in metabolic regulation.
Hepatic levels of PC species and turnover of PC and PE species are increased in steatotic livers (Toye et al., 2007; García‐Cañaveras et al., 2011; Han et al., 2011; Beyoğlu and Idle, 2013). In the present study, we observed that pioglitazone treatment significantly altered the amount and fatty acyl composition of hepatic phospholipids, in particular, PCs, as well as the total PC amount in the liver. Unlike rosiglitazone, which was previously reported not to alter the total concentration of PC in the liver (Watkins et al., 2002), pioglitazone treatment significantly reduced total PC in the rat liver. In addition, we found a relative decrease in palmitic acid (C16:0) in PC composition. However, given the lack of significant changes in gene expression of LPCAT, which resynthesizes PC from lysoPC, we cannot conclude that pioglitazone affects the turnover of PCs in the liver, and further investigation of this potential interaction is needed.
In summary, we have demonstrated here that pioglitazone changed hepatic metabolite and lipid profiles and regulated relevant metabolic pathways, which may explain the beneficial effect of pioglitazone on hepatic steatosis, induced by a high‐fat diet in OLETF rats (Figure 4). Pioglitazone reduced hepatic fatty acid levels not only by down‐regulating hepatic de novo synthesis of fatty acids but also by disrupting uptake and transport of exogenous fatty acids, which may contribute to a reduction in the availability of fatty acids for TG and phospholipid biosynthesis. In addition, pioglitazone treatment lowered the function of PLA2, but not LPCAT, leading to decreased production of lysophospholipids. Pioglitazone also increased fatty acid oxidation. These changes may help to decrease lipid accumulation in the liver.
Figure 4.
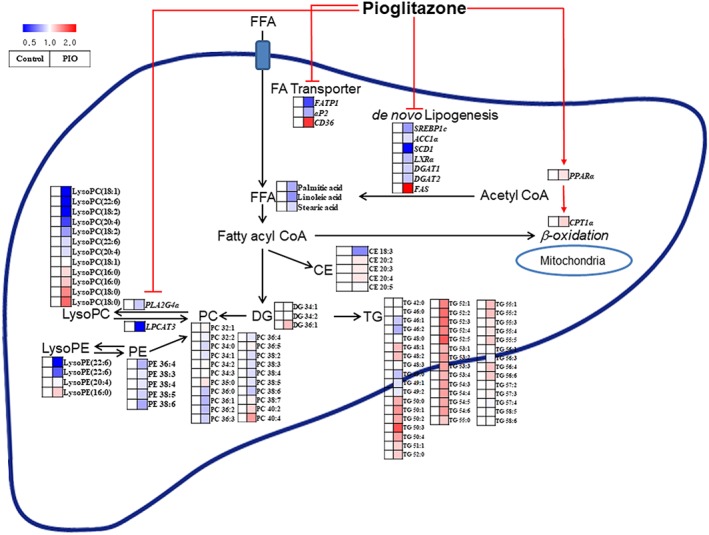
Scheme of the mechanism by which pioglitazone (PIO) regulates hepatic metabolites, lipids and related gene‐expression patterns (italicized). All metabolites, lipids and related gene‐expression patterns in the PIO‐treated groups compared to the control groups are illustrated in the heat map. Pioglitazone down‐regulates hepatic de novo synthesis of fatty acids and disrupts uptake and transport of exogenous fatty acids, which may contribute to reduced availability of fatty acids for TG and phospholipid biosynthesis in the liver. In addition, pioglitazone treatment lowers the function of PLA2, but not LPCAT, leading to decreased production of lysophospholipids. Pioglitazone also increases fatty acid oxidation. These changes may contribute to the decreased accumulation of lipid in the liver. The pathway was modified from KEGG (http://www.genome.jp/kegg). FFA, free fatty acid; FA transporter, fatty acid transporter.
One limitation of this study is the absence of a normal diet‐fed control group for comparison of hepatic metabolites. However, we validated the high‐fat diet‐induced development and improvement in hepatic steatosis by pioglitazone treatment in OLETF rats using physiological and histological assessments. Our data indicated that pioglitazone treatment changed hepatic metabolites and lipids associated with lipid metabolism in a NAFLD model, which may act as targets for diagnosis and clinical intervention in patients with NAFLD.
Author contributions
H.Y., D.H.S., C.H.L. and C.Y.P. conceived and designed the study. H.Y., D.H.S., D.H.K., E.S.J. and K.H.L. performed the experiments and procedures. H.Y., D.H.S., C.H.L. and C.Y.P. wrote the manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This http://onlinelibrary.wiley.com/doi/10.1111/bph.13405/abstract acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Table S1 Primers used for real‐time quantitative‐polymerase chain reaction
Figure S1 PCA score plots from GC‐TOF‐MS, UPLC‐Q‐TOF‐MS, and Nanomate‐LTQ‐MS. PCA score plots from GC‐TOF‐MS (A), UPLC‐Q‐TOF‐MS (B), and Nanomate‐LTQ‐MS (C) data between control and PIO groups (D) (n = 10 for control group, n = 8 for PIO‐treated group)
Data S1 Identification of metabolites with MS_MS fragmentation
Acknowledgements
We would like to thank Dr Ingu Do for assistance with histological activity and interpretation of the data. This research was supported by research funds from the Yuhan Corporation to C.Y.P. Moreover, research in the lab of C.H.L. was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (No. 2016M3A9A5923160).
Yang, H. , Suh, D. H. , Kim, D. H. , Jung, E. S. , Liu, K.‐H. , Lee, C. H. , and Park, C.‐Y. (2018) Metabolomic and lipidomic analysis of the effect of pioglitazone on hepatic steatosis in a rat model of obese Type 2 diabetes. British Journal of Pharmacology, 175: 3610–3625. 10.1111/bph.14434.
Contributor Information
Choong Hwan Lee, Email: chlee123@konkuk.ac.kr.
Cheol‐Young Park, Email: cydoctor@chol.com.
References
- Alexander SPH, Cidlowski JA, Kelly E, Marrion NV, Peter JA, Faccenda E et al (2017a). The concise guide to pharmacology 2017/2018: nuclear hormone receptor. Br J Pharmacol 174: S208–S224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017c). The Concise Guide to PHARMACOLOGY 2017/18: Transporters. Br J Pharmacol 174: S360–S446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Wong RJ, Harrison SA (2015). Nonalcoholic fatty liver disease review: diagnosis, treatment, and outcomes. Clin Gastroenterol Hepatol 13: 2062–2070. [DOI] [PubMed] [Google Scholar]
- Awazawa M, Ueki K, Inabe K, Yamauchi T, Kubota N, Kaneko K et al (2011). Adiponectin enhances insulin sensitivity by increasing hepatic IRS‐2 expression via a macrophage‐derived IL‐6‐dependent pathway. Cell Metab 13: 401–412. [DOI] [PubMed] [Google Scholar]
- Balsinde J, Balboa MA, Insel PA, Dennis EA (1999). Regulation and inhibition of phospholipase A2 . Annu Rev Pharmacol Toxicol 39: 175–189. [DOI] [PubMed] [Google Scholar]
- Belfort R, Harrison SA, Brown K, Darland C, Finch J, Hardies J et al (2006). A placebo‐controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N Engl J Med 355: 2297–2307. [DOI] [PubMed] [Google Scholar]
- Beyoğlu D, Idle JR (2013). The metabolomic window into hepatobiliary disease. J Hepatol 59: 842–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M (2012). Clinical relevance of adipokines. Diabetes Metab J 36: 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E, Kim L, Park SE, Rhee EJ, Lee WY, Oh KW et al (2015). Ezetimibe improves hepatic steatosis in relation to autophagy in obese and diabetic rats. World J Gastroenterol 21: 7754–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collino M, Aragno M, Castiglia S, Miglio G, Tomasinelli C, Boccuzzi G et al (2010). Pioglitazone improves lipid and insulin levels in overweight rats on a high cholesterol and fructose diet by decreasing hepatic inflammation. Br J Pharmacol 160: 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Curtis MJ, Bond RA, Spina D, Ahluwalia A, Alexander SP, Giembycz MA et al (2015). Experimental design and analysis and their reporting: new guidance for publication in BJP. Br J Pharmacol 172: 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusi K, Orsak B, Bril F, Lomonaco R, Hecht J, Ortiz‐Lopez C et al (2016). Long‐term pioglitazone treatment for patients with nonalcoholic steatohepatitis and prediabetes or type 2 diabetes mellitus: a randomized trial. Ann Intern Med 165: 305–315. [DOI] [PubMed] [Google Scholar]
- García‐Cañaveras JC, Donato MT, Castell JV, Lahoz A (2011). A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J Proteome Res 10: 4825–4834. [DOI] [PubMed] [Google Scholar]
- Han J, Dzierlenga AL, Lu Z, Billheimer DD, Torabzadeh E, Lake AD et al (2017). Metabolomic profiling distinction of human nonalcoholic fatty liver disease progression from a common rat model. Obesity (Silver Spring) 25: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MS, Lim YM, Quan W, Kim JR, Chung KW, Kang M et al (2011). Lysophosphatidylcholine as an effector of fatty acid‐induced insulin resistance. J Lipid Res 52: 1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS guide to pharmacology in 2018: updates and expansion to encompass the new guide to immunopharmacology. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Hung ND, Sok DE, Kim MR (2010). Lysophosphatidylcholine containing docosahexaenoic acid at the sn‐1 position in anti‐inflammatory. Lipids 45: 225–236. [DOI] [PubMed] [Google Scholar]
- Hung ND, Kim MR, Sok DE (2011). Mechanisms for anti‐inflammatory effects of 1‐[15(S)‐hydroxyeicosapentaenoyl] lysophosphatidylcholine, administered intraperitoneally, in zymosan A‐induced peritonitis. Br J Pharmacol 162: 1119–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung ND, Sok DE, Kim MR (2012). Prevention of 1‐palmitoyl lysophosphatidylcholine‐induced inflammation by polyunsaturated acyl lysophosphatidylcholine. Inflamm Res 61: 473–483. [DOI] [PubMed] [Google Scholar]
- Jung ES, Park HM, Lee KE, Shin JH, Mun S, Kim JK et al (2015). A metabolomics approach shows that catechin‐enriched green tea attenuates ultraviolet B‐induced skin metabolite alterations in mice. Metabolomics 11: 861–871. [Google Scholar]
- Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K (2006). Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest 116: 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Sakaida I, Tsuchiya M, Omori K, Takami T, Okita K (2004). Pioglitazone prevents hepatic steatosis, fibrosis, and enzyme‐altered lesions in rat liver cirrhosis induced by a choline‐deficient L‐amino acid‐defined diet. Biochem Biophys Res Commun 315: 187–195. [DOI] [PubMed] [Google Scholar]
- Kawano K, Hirashima T, Mori S, Saitoh Y, Kurosumi M, Natori T (1992). Spontaneous long‐term hyperglycemic rat with diabetic complications. Otsuka Long‐Evans Tokushima Fatty (OLETF) strain. Diabetes 41: 1422–1428. [DOI] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG, Group NCRRGW (2010). Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT et al (2011). Metabolomic analysis of livers and serum from high‐fat diet induced obese mice. J Proteome Res 10: 722–731. [DOI] [PubMed] [Google Scholar]
- Kim HY, Kim M, Park HM, Kim J, Kim EJ, Lee CH et al (2014). Lysophospholipid profile in serum and liver by high‐fat diet and tumor induction in obesity‐resistant BALB/c mice. Nutrition 30: 1433–1441. [DOI] [PubMed] [Google Scholar]
- Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O (2013). LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods 10 (8): 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrke M, Lazar MA (2005). The many faces of PPARγ. Cell 123: 993–999. [DOI] [PubMed] [Google Scholar]
- Leray C, Pelletier X, Hemmendinger S, Cazenave JP (1987). Thin‐layer chromatography of human platelet phospholipids with fatty acid analysis. J Chromatogr 420: 411–416. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sen S, Wannaiampikul S, Palanivel R, Hoo RL, Isserlin R et al (2015). Metabolomic profiling in liver of adiponectin‐knockout mice uncovers lysophospholipid metabolism as an important target of adiponectin action. Biochem J 469: 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhofer D, Weidner C, Sauer S (2014). Integrative analysis of transcriptomics, proteomics, and metabolomics data of white adipose and liver tissue of high‐fat diet and rosiglitazone‐treated insulin resistant mice identified pathway alterations and molecular hubs. J Proteome Res 13: 5592–5602. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC, Holmes E (1999). ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29: 1181–1189. [DOI] [PubMed] [Google Scholar]
- National Research Council (1996). Guide for the Care and Use of Laboratory Animals. The National Academies Press: Washington, DC. [Google Scholar]
- Nishi E, Kume N, Ueno Y, Ochi H, Moriwaki H, Kita T (1998). Lysophosphatidylcholine enhances cytokine‐induced interferon γ expression in human T lymphocytes. Circ Res 83: 508–515. [DOI] [PubMed] [Google Scholar]
- Park HM, Moon E, Lee S, Kim SY, Do SG, Kim J et al (2015). Topical application of baby‐ and adult‐aloe on ultraviolet B irradiated mouse skin with metabolite profiling. Metabolomics 11: 1219–1230. [Google Scholar]
- Promrat K, Lutchman G, Uwaifo GI, Freedman RJ, Soza A, Heller T et al (2004). A pilot study of pioglitazone treatment for nonalcoholic steatohepatitis. Hepatology 39: 188–196. [DOI] [PubMed] [Google Scholar]
- Riederer M, Ojala PJ, Hrzenjak A, Graier WF, Malli R, Tritscher M et al (2010). Acyl chain‐dependent effect of lysophosphatidylcholine on endothelial prostacyclin production. J Lipid Res 51: 2957–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rull A, Geeraert B, Aragonès G, Beltrán‐Debón R, Rodríguez‐Gallego E, García‐Heredia A et al (2014). Rosiglitazone and fanofibrate exacerbate liver steatosis in a mouse model of obesity and hyperlipidemia. A transcriptomic and metabolomics study. J Proteome Res 13: 1731–1743. [DOI] [PubMed] [Google Scholar]
- Sanyal AJ, Chalasani N, Kowdley KV, McCullough A, Diehl AM, Bass NM et al (2010). Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med 362: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling T, Eder C (2010). Importance of lipid rafts for lysophosphatidylcholine‐induced caspase‐1 activation and reactive oxygen species generation. Cell Immunol 265: 87–90. [DOI] [PubMed] [Google Scholar]
- Shen Z, Liang X, Rogers CQ, Rideout D, You M (2010). Involvement of adiponectin‐SIRT1‐AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol 298: G364–G374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon JC, Shin HS, Seo YK, Yoon YR, Shin H, Liu KH (2015). Direct infusion MS‐based lipid profiling reveals the pharmacological effects of compound K‐reinforced ginsenosides in high‐fat diet induced obese mice. J Agric Food Chem 63: 2919–2929. [DOI] [PubMed] [Google Scholar]
- Song YS, Fang CH, So BI, Park JY, Lee Y, Shin JH et al (2013). Time course of the development of nonalcoholic fatty liver disease in the Otsuka Long‐Evans Tokushima Fatty rat. Gastroenterol Res Pract 2013: 342648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM (1998). PPAR‐γ: adipogenic regulator and thiazolidinedione receptor. Diabetes 47: 507–514. [DOI] [PubMed] [Google Scholar]
- Stock C, Schilling T, Schwab A, Eder C (2006). Lysophosphatidylcholine stimulates IL‐1β release from microglia via a P2X7 receptor‐independent mechanism. J Immunol 177: 8560–8568. [DOI] [PubMed] [Google Scholar]
- Suh DH, Jung ES, Park HM, Kim SH, Lee S, Jo YH et al (2016). Comparison of metabolites variation and antiobesity effects of fermented versus nonfermented mixtures of Cudrania tricuspidata, Lonicera caerulea, and soybean according to fermentation in vitro and in vivo . PLoS One 11: e0149022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Goto T, Yamazaki Y, Kamakari K, Hirata M, Suzuki H et al (2015). Metabolomics reveal 1‐palmitoyl lysophosphatidylcholine production by peroxisome proliferator‐activated receptor α. J Lipid Res 56: 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Saito K, Jia H, Kato H (2014). An integrated multi‐omics study revealed metabolic alterations underlying the effects of coffee consumption. PLoS One 9: e91134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw W, Fukuda S (2015). Toward the comprehensive understanding of the gut ecosystem via metabolomics‐based integrated omics approach. Semin Immunol 37: 5–16. [DOI] [PubMed] [Google Scholar]
- Tang Y, Osawa H, Onuma H, Nishimiya T, Ochi M, Makino H (1999). Improvement in insulin resistance and the restoration of reduced phosphodiesterase 3B gene expression by pioglitazone in adipose tissue of obese diabetic KKAy mice. Diabetes 48: 1830–1835. [DOI] [PubMed] [Google Scholar]
- Toye AA, Dumas ME, Blancher C, Rothwell AR, Fearnside JF, Wilder SP et al (2007). Subtle metabolic and liver gene transcriptional changes underlie diet‐induced fatty liver susceptibility in insulin‐resistant mice. Diabetologia 50: 1867–1879. [DOI] [PubMed] [Google Scholar]
- Watkins SM, Reifsnyder PR, Pan HJ, German JB, Leiter EH (2002). Lipid metabolome‐wide effects of the PPARγ agonist rosiglitazone. J Lipid Res 43: 1809–1817. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S et al (2002). Adiponectin stimulates glucose utilization and fatty‐acid oxidation by activating AMP‐activated protein kinase. Nat Med 8: 1288–1295. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Choi JM, Chae SW, Kim WJ, Park SE, Rhee EJ et al (2011). Activation of peroxisome proliferator‐activated receptor γ by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS One 6: e17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SJ, Choi JM, Chang E, Park SW, Park CY (2014). Sirt1 and sirt6 mediate beneficial effects of rosiglitazone on hepatic lipid accumulation. PLoS One 9: e105456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yea K, Kim J, Yoon JH, Kwon T, Kim JH, Lee BD et al (2009). Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. J Biol Chem 284: 33833–33840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M (2016). Global epidemiology of nonalcoholic fatty liver disease‐meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology 64: 73–84. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Vaziri ND, Wei F, Cheng XL, Bai X, Zhao YY (2016). An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci Rep 6: 22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen YQ, Bonacci TM, Bredt DS, Li S, Bensch WR et al (2008). Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J Biol Chem 283: 8258–8265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used for real‐time quantitative‐polymerase chain reaction
Figure S1 PCA score plots from GC‐TOF‐MS, UPLC‐Q‐TOF‐MS, and Nanomate‐LTQ‐MS. PCA score plots from GC‐TOF‐MS (A), UPLC‐Q‐TOF‐MS (B), and Nanomate‐LTQ‐MS (C) data between control and PIO groups (D) (n = 10 for control group, n = 8 for PIO‐treated group)
Data S1 Identification of metabolites with MS_MS fragmentation


