Abstract
Background and Purpose
Activation of the human pregnane X receptor (PXR; NR1I2) has potential therapeutic uses for inflammatory bowel disease (IBD). Imperatorin (IMP), a naturally occurring coumarin, is the main bioactive ingredient of Angelica dahurica Radix, which is regularly used to treat the common cold and intestinal disorders. However, there are no data on the protective effects of IMP against IBD.
Experimental Approach
The effects of IMP on PXR‐modulated cytochrome P450 3A4 (CYP3A4) expression were assessed using a PXR transactivation assay, a mammalian two‐hybrid assay, a competitive ligand‐binding assay, analysis of CYP3A4 mRNA and protein expression levels and measurement of CYP3A4 activity using a cell‐based reporter gene assay and in vitro model. The inhibitory effects of IMP on NF‐κB activity were evaluated by a reporter assay and NF‐κB p65 nuclear translocation. The anti‐IBD effects of IMP were investigated in a dextran sulphate sodium (DSS)‐induced colitis mouse model. Colon inflammatory cytokines were assessed by elisa.
Key Results
IMP activated CYP3A4 promoter activity, recruited steroid receptor coactivator 1 to the ligand‐binding domain of PXR and increased the expression and activity of CYP3A4. PXR knockdown substantially reduced IMP‐induced increase in CYP3A4 expression. Furthermore, IMP‐mediated PXR activation suppressed the nuclear translocation of NF‐κB and down‐regulated LPS‐induced expression of pro‐inflammatory genes. Nevertheless, PXR knockdown partially reduced the IMP‐mediated inhibition of NF‐κB. IMP ameliorated DSS‐induced colitis by PXR/NF‐κB signalling.
Conclusions and Implications
IMP acts as a PXR agonist to attenuate DSS‐induced colitis by suppression of the NF‐κB‐mediated pro‐inflammatory response in a PXR/NF‐κB‐dependent manner.
Abbreviations
- AF‐2
activation function 2
- CD
Crohn's disease
- CYP3A4
cytochrome P450 3A4
- DSS
dextran sulphate sodium
- H&E
haematoxylin and eosin
- IBD
inflammatory bowel disease
- IMP
imperatorin
- iNOS
inducible NOS
- LBD
ligand‐binding domain
- MDR1
multidrug resistance 1
- MPO
myeloperoxidase
- PCN
pregnenolone‐16α‐carbonitrile
- PXR (NR1I2)
pregnane X receptor
- qRT‐PCR
quantitative RT‐PCR
- RID
receptor‐interacting domain
- SRC‐1
steroid receptor coactivator 1
- UC
ulcerative colitis
- UPLC
ultra‐HPLC
Introduction
Inflammatory bowel disease (IBD) is a chronic, idiopathic, inflammatory disease in the gastrointestinal tract caused by a dysregulated immune response to host intestinal microflora. IBD primarily presents as either ulcerative colitis (UC) or Crohn's disease (CD). Although the exact aetiology of IBD is still relatively unknown, the present prevailing view is that the development of IBD is associated with alterations in intestinal epithelial barrier function and dysregulation of the mucosal immune system (Ahmed, 2006; Kaser et al., 2010). The most common symptoms of both UC and CD are recurring abdominal pain, diarrhoea, rectal bleeding and malnutrition. Conventional drugs such as sulfasalazine exhibit some side effects (Rogler, 2010). While new biological therapies including http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5074 antagonists and integrin antagonists have improved the therapeutic effects against IBD, there are concerns regarding their potential side effects, toxicity, tolerability and high cost (Jobin, 2010; Gilroy and Allen, 2014). Thus, new therapeutic drugs for IBD need to be developed.
The http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=606 (PXR; SXR; NR1I2) is a ligand‐activated transcription factor that is predominately expressed in the liver and intestine (Kliewer et al., 2002). Once activated, PXR forms a heterodimer with the retinoid X receptor and binds to specific PXR response elements on the promoter of PXR target genes, such as cytochrome P450 3A4 (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1337) and the ATP‐binding cassette transporter ABCB1 [also known as multidrug resistance 1 (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=768)], to control their transcription (Chen, 2008). In addition to the well‐established role of PXR as a xenobiotic sensor, recent studies have extended the role of PXR to the regulation of IBD. For instance, PXR and its target genes were shown to participate in maintaining intestinal epithelial barrier integrity (Roediger and Babidge, 1997; Venkatesh et al., 2014). Specific polymorphisms in the PXR locus might cause a decrease in PXR activity and might be associated with increased susceptibility to IBD (Dring et al., 2006), suggesting a role for PXR in the pathogenesis of IBD. In addition, the reciprocal repression of PXR and NF‐κB revealed a potential molecular mechanism that links PXR and certain inflammation signalling pathways (Xie and Tian, 2006; Zhou et al., 2006). Interestingly, activating PXR in mice ameliorated dextran sulphate sodium (DSS)‐induced colitis by inhibiting the NF‐κB pro‐inflammatory pathway in the colon (Shah et al., 2007; Cheng et al., 2010), suggesting a potential role for PXR agonists as therapeutic drugs to treat IBD.
Angelica dahurica Radix is a traditional Chinese medicine that is commonly used as a herbal anti‐inflammatory medicine for the treatment of respiratory diseases and intestinal disorders (Lee et al., 2015; Hwang et al., 2017). In particular, in long‐term clinical observations, A. dahurica Radix has been shown to substantially ameliorate the swelling and atrophic spots of colonic mucosal membranes in patients with colitis (Hwang et al., 2017). The major biologically active components identified in A. dahurica Radix are coumarins, which attenuate allergic inflammatory responses (Li and Wu, 2017). Imperatorin (IMP), a naturally occurring coumarin, is one of the most abundant coumarins in A. dahurica Radix (up to 1% of total content). Several studies have indicated that IMP suppresses the expression of key pro‐inflammatory factors such as http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1250 (iNOS), http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1376, TNF‐α and NF‐κB (Guo et al., 2012; Huang et al., 2012). However, there is currently limited research analysing the potential of IMP to treat IBD. Therefore, in the present study, we hypothesized that IMP could activate the transcriptional activity of PXR and act as a PXR agonist, inducing the expression of CYP3A4 and MDR1 in a model cell‐line through a series of in vitro measurements. Additionally, we used in vitro and in vivo models to explore the effects of IMP on inflammation and IBD and to reveal the underlying mechanisms mediated by PXR activation.
Methods
Cell culture
The human intestinal epithelial cell lines HCT116 and LS174T, the human acute monocytic leukaemia cell line THP‐1 and the HEK293T cell line were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in DMEM with 10% FBS, 100 U·mL−1 penicillin, 100 μg·mL−1 streptomycin and 2 mM l‐glutamine at 37°C in a humidified incubator containing 5% CO2. The cell medium was replaced with fresh medium every 2 days. Cells were passaged upon reaching ~80% confluence.
PXR transactivation assay
HCT116 cells were transfected with pCMV6‐entry (100 ng per well), pCMV6‐mPXR (100 ng per well), pCMV6‐XL4‐hPXR (100 ng per well), pGL3‐CYP3A4‐luc (100 ng per well) and pGL3‐CMV Renilla luciferase plasmids (10 ng per well) using FuGENE 6. At 24 h after transfection in growth medium, approximately 1 × 104 live cells were plated in each well of a 96‐well culture plate and cultured for an additional 24 h in phenol red‐free DMEM supplemented with 1% charcoal–dextran‐treated FBS. Transfected cells were treated with 0.1 mL fresh supplemented culture medium containing DMSO (1% v.v‐1; vehicle control), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2765 (10 μM), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2762 (PCN; 10 μM), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2568 (2.5 μM) or IMP (6.25–25 μM) for 24 h. At the end of the treatment period, the cells were lysed, and their luminescence was determined by a luciferase assay using the Dual‐Glo luciferase reporter assay system and Enspire Multimode Plate Reader (PerkinElmer, Waltham, MA, USA).
Mammalian two‐hybrid assay
The mammalian two‐hybrid assay was performed as described previously by Lau et al. (2012). Briefly, 5 h after being plated, cultured HEK293T cells were co‐transfected with VP16‐SXR‐ligand‐binding domain (LBD; 40 ng per well), or VP16‐SXR‐ΔAF2 LBD (40 ng per well), or VP16 empty vector (40 ng per well), GAL4 steroid receptor coactivator 1‐receptor‐interacting domain ((SRC‐1‐RID), pGL4.74 (hRluc/TK) as an internal control plasmid (10 ng per well) and pFR‐luc (100 ng per well). At 24 h after transfection, the cells were treated with 0.1 mL fresh supplemented culture medium containing DMSO, IMP (6.25–25 μM) or rifampicin (10 μM) for 24 h. A Dual‐Glo luciferase reporter assay was used to measure luciferase activity.
Competitive ligand‐binding assay
A LanthaScreen time‐resolved (TR)‐FRET PXR competitive binding assay was conducted according to the manufacturer's protocol (#PV4839; Invitrogen, Carlsbad, CA, USA). Briefly, the assay was performed in a volume of 20 μL in 384‐well black plates containing GST‐hPXR ligand‐binding domain (5 nM), fluorescence‐labelled hPXR agonist (40 nM; Fluomore PXR Green), terbium‐labelled anti‐GST antibody (5 nM) in the presence of DMSO, IMP (6.25–25 μM), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2763 (10 μM) or PCN (10 μM). The reaction mixture was incubated at 25°C for 20 min, and then TR‐FRET was measured using an Enspire Multimode Plate Reader (PerkinElmer) with an excitation wavelength of 340 nm and emission wavelengths of 520 nm (fluorescein emission) and 495 nm (terbium emission). The TR‐FRET ratio was calculated by dividing the emission signal at 520 nm by that at 495 nm. The background TR‐FRET ratio was determined from wells containing the same reagents as the vehicle‐treated control group but without the PXR ligand‐binding domain. The net TR‐FRET ratio was calculated by subtracting the background TR‐FRET ratio from the TR‐FRET ratio.
RNA isolation and quantitative RT‐PCR
Total RNA was extracted from samples using TRIzol reagent. The quality and quantity of total RNA were determined using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed using the SuperScript™ VILO™ cDNA Synthesis Kit, and qPCR was carried out using TaqMan gene expression assays (Applied Biosystems, Carlsbad, CA, USA) in an ABI StepOnePlus system (Applied Biosystems) according to the manufacturer's protocol. The following TaqMan probes were used: hPXR (https://www.thermofisher.com/taqman-gene-expression/product/Hs01114267_m1?CID=&ICID=&subtype=, Mm01344139_m1), CYP3A4 (https://www.thermofisher.com/taqman-gene-expression/product/Hs00604506_m1?CID=&ICID=&subtype=), Cyp3a11 (Mm00731567_m1), MDR1 (https://www.thermofisher.com/taqman-gene-expression/product/Hs00184500_m1?CID=&ICID=&subtype=, Mm00440761_m1), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4974 (https://www.thermofisher.com/taqman-gene-expression/product/Hs01555410_m1?CID=&ICID=&subtype=), iNOS (https://www.thermofisher.com/taqman-gene-expression/product/Hs01075529_m1?CID=&ICID=&subtype=), COX‐2 (https://www.thermofisher.com/taqman-gene-expression/product/Hs00153133_m1?CID=&ICID=&subtype=), http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6757 (https://www.thermofisher.com/taqman-gene-expression/product/Hs00164932_m1?CID=&ICID=&subtype=) and GAPDH (https://www.thermofisher.com/taqman-gene-expression/product/Hs02786624_g1?CID=&ICID=&subtype=, Mm99999915_g1). For standardization and quantification, GAPDH mRNA was amplified simultaneously. The comparative Ct method was used to estimate the relative quantification for gene expression according to the following formulae: ΔCt = Ct (test gene) − Ct (GAPDH); ΔΔCt (test gene) = ΔCt (test gene in treatment group) − ΔCt (test gene in vehicle control group); mRNA expression fold change = 2−ΔΔCt, which indicates the relative mRNA level of the corresponding transcript compared with control samples.
Western blotting analysis
Samples from cells and mouse colon tissues (100–200 mg) were rinsed in cold PBS (pH 7.4), lysed, homogenized in RIPA lysis buffer containing a cocktail of protease inhibitors on ice and centrifuged at 12 000× g for 15 min at 4°C. The supernatant was collected, and the protein concentration was measured using a bicinchoninic acid assay. Equal concentrations of protein were separated using a 10% SDS‐PAGE (Bio‐Rad Laboratories, Hercules, CA, USA) and transferred to Immuno‐Blot PVDF membranes (0.2 μm; Bio‐Rad Laboratories). The membranes were then blocked with 5% BSA in Tris‐buffered saline containing 0.1% Tween 20 and incubated overnight at 4°C with primary antibodies targeting PXR (1:500), MDR1 (1:1000), CYP3A4 (1:3000), NF‐κB p65 (1:1000), phospho‐p65 (1:1000), http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1989 (1:1000), phospho‐IκBα (1:1000) and GADPH (1:3000). The blots were then incubated with HRP‐conjugated secondary antibodies and detected by enhanced chemiluminescence using Molecular Imager ChemiDOC™ XBS imaging systems (Bio‐Rad Laboratories).
Analysis of CYP3A4 activity
Ultra‐HPLC/tandem MS (UPLC‐MS/MS) (AB SCIEX™, Framingham, MA, USA) was used to determine the concentration of 1′‐hydroxymidazolam to analyse CYP3A4 catalytic activity. In the present study, LS174T cells (~100 000 cells per well) were plated in each well of a 48‐well culture plate and treated with DMSO, rifampicin, IMP or ketoconazole for 48 h; the culture medium was then removed and supplemented with fresh medium containing midazolam (10 μM). After 4 h incubation, the supernatant (100 μL) was collected, and 200 μL acetonitrile with 100 ng·mL−1 midazolam‐D4 maleate (internal standard) was added; the mixture was then vigorously vortexed and centrifuged, and 10 μL supernatant was analysed by UPLC‐MS/MS. Chromatographic separation of 1′‐hydroxymidazolam was carried out using a Waters ACQUITY BEH C18 column (2.1 × 50 mm, 1.7 μm) in a Waters ACQUITY UPLC System. The flow rate was 0.7 mL·min−1 with the following gradient elution: 0–0.2 min: 10–20% acetonitrile, maintained for 1.0 min, followed by 20% acetonitrile until 1.2 min, and a gradient to 90% acetonitrile for 0.3 min and an isocratic step to 90% acetonitrile for 0.5 min, at 2.05–2.5 min: 90–10% acetonitrile. MS/MS quantification was performed on multiple reaction monitoring mode with a positive electrospray ionization interface using [M + H]+ ions: m/z 342 → m/z 324 for 1′‐hydroxymidazolam and m/z 330 → m/z 295 for midazolam‐D4 maleate (internal standard), with a dwell time of 40 ms. The MS parameters were optimized as follows: ion spray voltage was set at +3.5 kV, the heater gas temperature was set at 350°C and declustering potential and collision energy were set at 70 and 30 V for 1′‐hydroxymidazolam and 120 and 35 V for midazolam‐D4 maleate.
Molecular modelling and docking study
Two‐ and three‐dimensional structural information of the compound was obtained from PubChem (https://pubchem. ncbi. nlm. http://nih.gov/). Molecular modelling was conducted with the SYBYL program package. IMP was set as the template. Its conformation was searched and identified by energy minimization using the MMFF94 force field with the Powell conjugate gradient minimization algorithm and a convergence criterion of 0.005 kcal·mol−1·Å−1. The MMFF94 charge was used to calculate partial atomic charges. All other parameters were set to default.
The four‐letter PDB code (1M13) of hPXR, with a resolution of 2.15 Å, was retrieved from PDB. The crystal structure of hPXR was subjected to geometry optimization through Swiss PDB viewer v4.1.0. The structure of the compound IMP was prepared for docking using SYBYL‐X2.1.1 software. The execution of molecular docking analysis and visualization were carried out using Surflex‐Dock. The co‐crystalized structure was prepared using SYBYL‐X2.1.1. Removal of solvent molecules, addition of hydrogen and AMBER7 FF99 charge calculations were then carried out. For docking simulations by SYBYL Vina, all default parameters were used.
RNA interference assay
LS174T cells were plated on six‐well plates. After reaching ~80% confluence, the cells were transiently transfected with PXR siRNA (M‐003415‐02; Dharmacon, Lafayette, CO, USA) targeting the human PXR mRNA with lipofectamine 2000. Control siRNA, a non‐targeting siRNA (D‐001210‐01‐20; Dharmacon), was used as a negative control. After 24 h transfection, cells were treated with DMSO, rifampicin or IMP for 48 h, followed by an additional incubation with or without TNF‐α (20 ng·mL−1) and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=5019 (2000 ng·mL−1) for 24 h. At the end of the incubation period, cells were collected for quantitative RT‐PCR (qRT‐PCR) and Western blot analysis, as described above.
PXR‐mediated NF‐κB inhibition reporter assay
HEK293T cells (1 × 106 cells) were transiently co‐transfected with pGL4.32 (luc2P/NF‐κB‐RE/Hygro) NF‐κB reporter, pCMV6‐entry, or pCMV6‐XL4‐hPXR and pRL‐TK using FuGENE 6. The pGL4.32 reporter is an NF‐κB reporter vector with NF‐κB response elements and firefly luciferase gene. After incubation overnight, cells were treated with DMSO, rifampicin or IMP for 24 h, followed by an additional incubation with or without TNF‐α (20 ng·mL−1) for another 24 h. A standard dual luciferase assay was carried out on the cell lysates, and the results are expressed as the fold change compared with control cells.
Immunofluorescence staining
LS174T cells or THP‐1 cells were seeded in cell imaging dishes and incubated overnight. Cells were treated with DMSO, rifampicin or IMP for 48 h followed by an additional incubation with or without TNF‐α (20 ng·mL−1) or LPS (1000 ng·mL−1) for 24 h. At the end of the incubation period, cells were fixed with 4% paraformaldehyde for 20 min at room temperature. After being washed with PBS, cells were permeabilized with 0.1% Triton X‐100 in PBS for 10 min at room temperature. After incubation with blocking buffer 5% BSA for 30 min, cells were incubated with rabbit anti‐NF‐κB p65 antibody (1:50) at 4°C overnight and then further incubated with Alexa Fluor 488‐conjugated anti‐rabbit IgG antibody (A‐21206; Invitrogen) for 1 h at room temperature. DAPI (100 ng·mL−1) in PBS was added to stain the nuclei. Fluorescence images were obtained using a confocal laser scanning microscope (Leica, Wetzlar, Germany) or a fluorescence microscope (Nikon, Tokyo, Japan).
Animals and treatment
All animal care and experimental studies were approved by and in accordance with the guidelines of the Animal Ethics Committee of Guangzhou University of Chinese Medicine. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015). Eight‐week‐old male C57BL/6 mice (18–20 g) were purchased from the Animal Laboratory Centre of Guangdong Province (Guangzhou, China). They were kept in a specific pathogen‐free animal facility at a constant temperature of 22°C and a relative humidity of 50–70%, with a 12 h light/dark cycle and free access to food and water. The mice were acclimatized to laboratory conditions for 3 days before experimentation. Experimental colitis was induced by the administration of DSS as previously described (Zhang et al., 2015b). Mice were randomly divided into seven groups: normal, IMP, DSS, DSS + IMP25, DSS + IMP50, DSS + IMP100 and DSS + PCN. Animals received a daily gavage of IMP (25, 50 or 100 mg·kg−1) or PCN (10 mg·kg−1) in 0.5% carboxymethyl cellulose from day 1 to day 10. From day 3, 2 h after the administration of IMP, mice were given 4% DSS (w.v‐1) solution dissolved in sterile distilled water (vehicle control) ad libitum for 7 days. IMP and PCN administration continued until the end of the DSS treatment period.
Evaluation of colitis
Daily observation for signs of colitis, such as weight loss, diarrhoea and rectal bleeding, was performed and recorded. Mice were killed under anaesthesia with ether using a desktop rodent anaesthesia ventilator (RWD Life Science Co., Ltd., Shenzhen, China). Colons were immediately removed, and the total length of the colon was measured. Portions of the colons were stored at −80°C for further experiments. Portions of the colons were fixed in 10% formalin, embedded in paraffin, processed for routine haematoxylin and eosin (H&E) staining of sections and then examined under a light microscope (Leica). The histological scores of H&E‐stained colon specimens were assessed by two pathologists in a blinded fashion. Histological sections were scored using a validated scoring system as described by Dou et al. (2014).
Measurement of colon/cell supernatant inflammatory cytokines
Mouse colon segments were homogenized in ice‐cold PBS. After centrifugation at 3000× g for 10 min at 4°C, colon inflammatory cytokines (such as TNF‐α and IL‐1β) in supernatant homogenates were quantified, according to the manufacturer's instructions and guidelines, using elisa kits (Cusabio, Houston, TX, USA) specific for the mouse inflammatory cytokines indicated. The results are expressed in pg·g−1 of tissue in each sample. Cell culture supernatants were collected and analysed in the same manner described above.
Determination of colon myeloperoxidase activity
To monitor the degree of inflammation, tissue http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2789 (MPO) activity, which is linearly related to neutrophil infiltration in inflamed tissues, was evaluated in the inflamed colon samples. MPO activity was measured in colon tissue samples as described by Hillegass et al. (1990), and the values are expressed as U·g−1 of tissue in each sample.
Immunohistochemistry
Mouse colon tissue samples were fixed with 10% formalin for 24 h, dehydrated and embedded in paraffin. Standard immunohistochemical procedures were performed. Tissue sections were incubated with the primary antibody (rabbit monoclonal NF‐κB p65 antibody, or rabbit polyclonal PXR antibody) at 4°C overnight, followed by incubation with a streptavidin–biotin–peroxidase‐conjugated secondary antibody for 30 min at room temperature. For the negative control, the primary antibody was replaced with 1% non‐immune serum in PBS. The immunohistochemical staining of NF‐κB p65 and PXR was scored by measuring the integrated optical density (OD) of at least three images of each slice using Image Pro‐Plus 6.0 software (Media Cybernetics, Rockville, VA, USA).
Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2018). All results in the figures and text are expressed as the mean ± SEM. The data were evaluated using GraphPad Prism Version 6.0 (GraphPad Software, La Jolla, CA, USA). The statistical significance of multiple comparisons was analysed using one‐way ANOVA followed by post hoc Bonferroni's test when F achieved P < 0.05 and there was no significant variance inhomogeneity. The difference between two groups was analysed using Student's unpaired t‐test. A value of P < 0.05 was considered statistically significant for all tests.
Materials
Rifampicin, DMSO, PCN, LPS, ketoconazole, TNF‐α and SR12813 were purchased from Sigma‐Aldrich (St. Louis, MO, USA). IMP (>98.3% purity) was obtained from the National Institute for the Control of Pharmaceutical and Biological Products (Guangzhou, China). DSS (MW: 36–50 KDa) was acquired from MP Biomedicals (Santa Ana, CA USA). DMEM, FBS, TRIzol, the LanthaScreen™ TR‐FRET PXR competitive binding assay system, Applied Biosystems Fast TaqMan Master Mix and charcoal–dextran‐treated FBS were purchased from Thermo Fisher Scientific. The pCMV6‐entry, pCMV6‐mPXR and pCMV6‐XL4‐hPXR plasmids were obtained from OriGene (Rockville, MD, USA). The NF‐κB reporter vector pGL4.32 (luc2P/NF‐κB‐RE/Hygro), FuGENE 6 and Dual‐Glo luciferase reporter assay system were purchased from Promega (Madison, WI, USA). The pGL3‐CYP3A4‐luc and pGL3‐CMV Renilla luciferase plasmids were kindly provided by Dr Ming Huang (College of Pharmacy, Sun Yet‐sen University, Guangzhou, China). The other plasmids, including GAL4 SRC1‐RID, VP16‐SXR‐LBD and VP16 SXR‐ΔAF2 LBD expression plasmids, were constructed as described previously by Takeshita et al. (2006) (GiMan Biotech, Shanghai, China). The PXR (ab85451 for human and ab192579 for mouse), CYP3A4 (ab124921 for human and ab197053 for mouse), MDR1 (ab170904), GAPDH (ab8245) and PCNA (ab18197) primary antibodies were purchased from Abcam (Cambridge, UK). NF‐κB p65 (#8242), phospho‐p65 (#3033), IκBα (#4812) and phospho‐IκBα (#2859) were purchased from Cell Signaling Technology (Danvers, MA, USA). The RT‐PCR TaqMan probes for human PXR, CYP3A4, MDR1, iNOS, COX‐2, ICAM1, GAPDH, mouse PXR, Cyp3a11, Mdr1 and GAPDH were purchased from Invitrogen. Mouse TNF‐α and IL‐1β elisa kits were purchased from R&D Systems (Minneapolis, MN, USA).
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a, 2017b, 2017c).
Results
IMP activates PXR transactivation of CYP3A4 promoter activity
It has been shown that PXR target gene expression (e.g. CYP3A4) in the liver and intestine is mediated by a broad variety of xenobiotics, including therapeutic and herbal components (Dussault et al., 2003). To characterize the regulation of PXR function by IMP, we examined its effect on hPXR‐regulated CYP3A4 promoter activity in HCT116 intestinal cells (Figure 1). The cells were transiently transfected with the CYP3A4‐luc and pCMV6‐entry plasmids, as well as hPXR or mouse PXR (mPXR), and treated with rifampicin or IMP. IMP, like rifampicin, significantly increased CYP3A4 promoter activity dose‐dependently in an hPXR‐dependent manner in HCT116 cells (Figure 1A). Likewise, IMP, like the mPXR agonist PCN, induced the transactivation of CYP3A4 promoter activity by mPXR (Figure 1B), indicating that IMP also activates mPXR. Therefore, these results indicate that IMP, like rifampicin and PCN, activates hPXR/mPXR‐mediated CYP3A4‐luc reporter activity in the HCT116 cell line.
Figure 1.
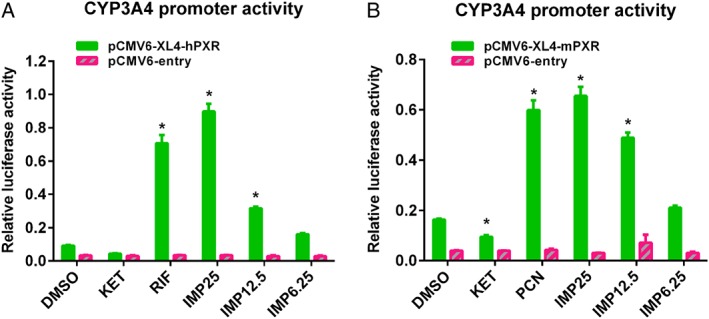
IMP activates the PXR transactivation of CYP3A4 promoter activity. HCT116 cells were transfected with pGL3‐CYP3A4‐luc (100 ng per well), as well as empty vector pCMV6‐entry (100 ng per well), (A) pCMV6‐XL4‐hPXR (100 ng per well) or (B) pCMV6‐mPXR (100 ng per well) plasmids. pGL3‐CMV Renilla luciferase control reporter vectors were used as the transfection control. After 24 h transfection, cells were treated with DMSO, rifampicin (RIF; 10 μM), ketoconazole (KET; 2.5 μM), PCN (10 μM) or IMP (6.25–25 μM) as indicated for another 24 h. CYP3A4 promoter activity is presented as relative luciferase units after being normalized against the Renilla luciferase internal control. Data are expressed as the mean ± SEM from five independent experiments. * P < 0.05 compared with the same treatment group transfected with empty vector, and the DMSO control group transfected with the receptor expression plasmid.
Interactions between IMP and hPXR
Next, we investigated the mechanism surrounding the activation of hPXR by IMP. As it has been shown that steroid receptor coactivator 1 (SRC‐1) contributes to the ligand‐induced activation of hPXR (Sui et al., 2012; Wang et al., 2013), we first examined whether IMP treatment recruited SRC‐1 to the ligand‐binding domain of hPXR in mammalian two‐hybrid assays, in which the interactions between hPXR and SRC‐1 induced by the hPXR agonist result in specific reporter activity (Sui et al., 2012; Wang et al., 2013). GAL4SRC‐1 interacted with VP16 SXR‐LBD, but not VP16, in the presence of IMP – like rifampicin (positive control) – in a dose‐dependent manner (Figure 2A). It has been reported that the C‐terminal region of the LBD [activation function 2 (AF‐2)] is conserved, and deletion or mutation in this region impairs transcriptional activity without changing either the ligand‐ or DNA‐binding affinities (Berrevoets et al., 1998; Takeshita et al., 2013). Nuclear receptor ligands induce major conformational changes in the AF‐2 region, resulting in ligand‐binding receptors interacting with coactivators (Smith and O'Malley, 2004). In the present study, we found that neither rifampicin nor IMP induced an increase in luciferase activity using the C‐terminal truncated AF2 mutant VP16‐SXR‐ΔAF2 LBD (Figure 2A), indicating that IMP‐bound PXR recruited coactivators in an AF2‐dependent manner.
Figure 2.
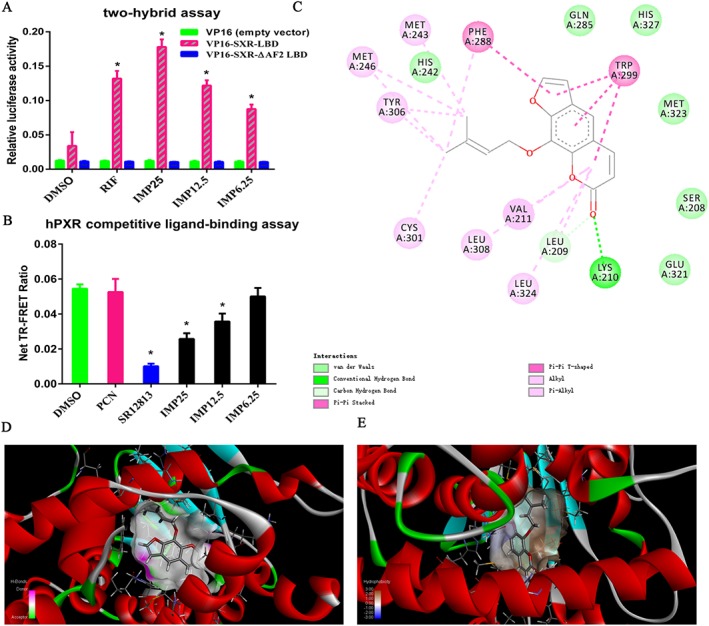
Determination of direct interactions between IMP and hPXR. (A) The effect of IMP on the recruitment of coactivator SRC‐1 to hPXR in a mammalian two‐hybrid assay. HEK293T cells were transfected with VP16‐SXR‐LBD (40 ng per well), VP16‐SXR‐ΔAF2 LBD (40 ng per well) or VP16 empty vector (40 ng per well), GAL4‐SRC‐1‐RID, pGL4.74 (hRluc/TK) (10 ng per well) and pFR‐luc (100 ng per well). Transfected cells were treated with DMSO, IMP (6.25–25 μM) or rifampicin (RIF; 10 μM). GAL4‐luc reporter activity is expressed as the relative luciferase activity after being normalized against the Renilla luciferase internal control. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the same treatment group transfected with empty vector, and the DMSO control group transfected with the receptor expression plasmid. (B) The binding of IMP to hPXR‐LBD in a TR‐FRET competitive binding assay. hPXR ligand‐binding domain (5 nM) was incubated with terbium‐labelled anti‐GST antibody (5 nM) and Fluomore PXR Green (40 nM) in the presence of DMSO, IMP (6.25–25 μM), SR12813 (10 μM) or PCN (10 μM, negative control). The net TR‐FRET ratio (520 nm/495 nm) was determined. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the DMSO control group. (C) The two‐dimensional structure of the predicted binding of IMP to hPXR (rendered in sticks). (D) Binding interactions of hPXR with surrounding residues; hydrogen bonding is represented by different colours (pink, hydrogen donors; green, hydrogen acceptors). (E) The interactions between IMP and hPXR and in terms of their hydrophobicity (more brown, more lipophilic; more blue, more hydrophilic).
The recruitment of SRC‐1 by IMP in the cell‐free system indicated that IMP, similar to rifampicin and other PXR agonists, directly binds to the hPXR‐LBD. To confirm this hypothesis, a TR‐FRET competitive hPXR‐LBD binding assay was performed.
As shown in Figure 2B, IMP significantly decreased the TR‐FRET emission ratio, dose‐dependently, compared with the vehicle control. SR12813, a potent hPXR agonist, markedly decreased the TR‐FRET emission ratio as a positive control, while PCN, the negative control, had no effect, as predicted. Furthermore, we docked small molecules to the protein and analysed the docking score. Molecular docking studies found that a compound with a high docking score (total score and Cscore) might have strong activity against the targeted receptor. The docking studies gave a total score, Cscore and unified Cscore of 8.7496, 5 and 3, respectively, indicating that IMP and hPXR may participate in many reactions. In the present work, the best‐docked conformation of IMP was analysed according to its potency. Our results show that IMP bound to the inside of the hPXR‐LBD domain, based on the hydrogen and hydrophobic interactions (Figure 2C–E). Interaction analysis revealed that IMP had hydrogen‐bonding interactions with residues Lys210 and Leu209 in different directions (Figure 2C). Furthermore, we investigated the effects of other factors, such as van der Waals forces and contacts from the surrounding residues, on the hPXR binding site. The following residues, Gln285, His327, Met323, Ser208, Glu321 and His242, potentially affect the binding of IMP to hPXR‐LBD. Evaluation of the molecular interactions showed that hydrogen bonds, hydrophobicity and van der Waals forces were the major forces contributing to the stability of the IMP‐hPXR complex. Collectively, our results show that IMP directly interacts with hPXR to facilitate the recruitment of coactivators, suggesting that IMP serves as an hPXR agonist.
IMP induces CYP3A4 and MDR1 expression and CYP3A4 activity in LS174T cells
LS174T colon adenocarcinoma cells have been shown to express endogenous hPXR, CYP3A4 and MDR1; therefore, they were used as an intestinal cell model to study the function of hPXR (Wang et al., 2013). To verify the results of the reporter gene assay, we sought to determine whether IMP could induce the expression of CYP3A4 and MDR1 in LS174T cells. Like rifampicin, IMP treatment increased the levels of CYP3A4 and MDR1 mRNA in a dose‐dependent manner (Figure 3A), with a much greater increase in CYP3A4 mRNA levels than in MDR1 mRNA levels (~7‐fold vs. ~2.3‐fold at the highest concentration of IMP). The increase in mRNA expression at the highest concentration of IMP (25 μM) was comparable with that observed with rifampicin (10 μM). However, like rifampicin, IMP had little effect on increasing the expression of hPXR mRNA. Consistent with the increase in mRNA levels, the protein levels of CYP3A4 and MDR1 also increased after treatment with IMP in a dose‐dependent manner (Figure 3B). It has been reported that increased CYP3A4 gene expression is functionally correlated to increased CYP3A4 enzymatic activity. To determine the functional relevance of the IMP‐induced increase in CYP3A4 activity, we measured the concentration of 1′‐hydroxymidazolam, a specific probe of CYP3A4‐mediated midazolam hydroxylation enzymatic activity, in LS174T cells pretreated with rifampicin, ketoconazole or IMP, followed by the addition of midazolam. Our data show that rifampicin strongly elevated CYP3A4 enzymatic activity (~2.5–2.8‐fold), whereas ketoconazole, a typical CYP3A4 enzyme inhibitor and PXR antagonist, inhibited CYP3A4 enzymatic activity as expected (Figure 3C). Consistent with the increase in CYP3A4 gene expression levels, IMP significantly increased CYP3A4 enzymatic activity in a concentration‐dependent manner (~1.35–1.85‐fold, Figure 3C). Taken together, the results show that IMP‐mediated activation of CYP3A4 is dependent on the activation of hPXR.
Figure 3.
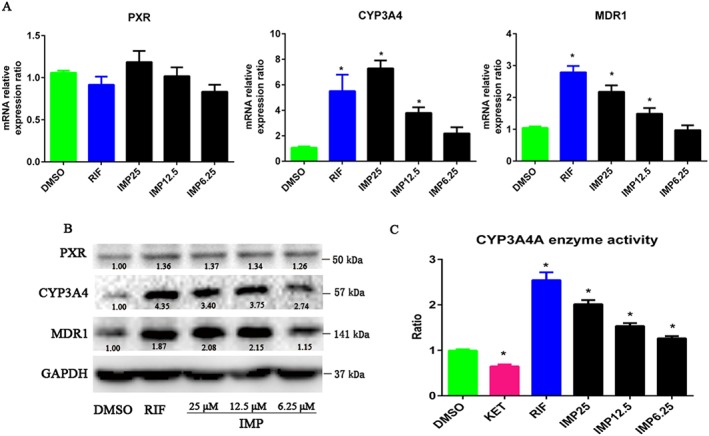
IMP induced PXR‐mediated gene expression and CYP3A4 enzymatic activity in LS174T cells. (A) Human PXR (NR1I2), CYP3A4 and MDR1 mRNA expression was analysed by qRT‐PCR in LS174T cells after treatment with DMSO, IMP (6.25–25 μM) or rifampicin (RIF; 10 μM) for 48 h. Results are expressed as fold changes compared with the DMSO control. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the DMSO control group. (B) Human PXR, CYP3A4 and MDR1 protein levels in the same cells were determined by Western blotting. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (C) IMP induced CYP3A4 enzymatic activity in LS174T cells. Cells were treated with DMSO, IMP (6.25–25 μM), ketoconazole (KET; 2.5 μM) or rifampicin (10 μM) for 48 h. CYP3A4 enzymatic activity was determined by measuring the 1′‐hydroxymidazolam concentration for specific CYP3A4‐mediated midazolam hydroxylation. The results are expressed as relative fold changes in activity compared with the vehicle control. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the DMSO control group.
Genetic knockdown of hPXR decreases IMP‐induced hPXR target gene expression
To confirm that the IMP‐mediated activation of CYP3A4 and MDR1 in LS174T cells required hPXR, we tested the effect of IMP on the expression of CYP3A4 and MDR1 when hPXR expression was knocked down in LS174T cells. After 24 h transfection with siPXR or control siRNA, cells were treated with DMSO, rifampicin or IMP for 48 h; gene expression and protein activity were analysed by qRT‐PCR and Western blotting respectively. As expected, transfection with siPXR reduced PXR expression levels by approximately 50–60% compared with control siRNA (Figure 4A, B). CYP3A4 and MDR1 mRNA levels were elevated after treatment with rifampicin and IMP in transfected control siRNA cells (Figure 4A, B). Notably, PXR knockdown largely abolished rifampicin‐ and IMP‐mediated CYP3A4 and MDR1 activation, indicating that hPXR is an essential regulator of IMP‐induced CYP3A4 and MDR1 expression (Figure 4A, B). After knocking down hPXR, the transfection of cells with siPXR resulted in a loss of IMP‐ or rifampicin‐mediated increase in CYP3A4 enzymatic activity compared with the control siRNA (Figure 4C). Therefore, hPXR is necessary for IMP to activate the expression of the hPXR target genes CYP3A4 and MDR1.
Figure 4.
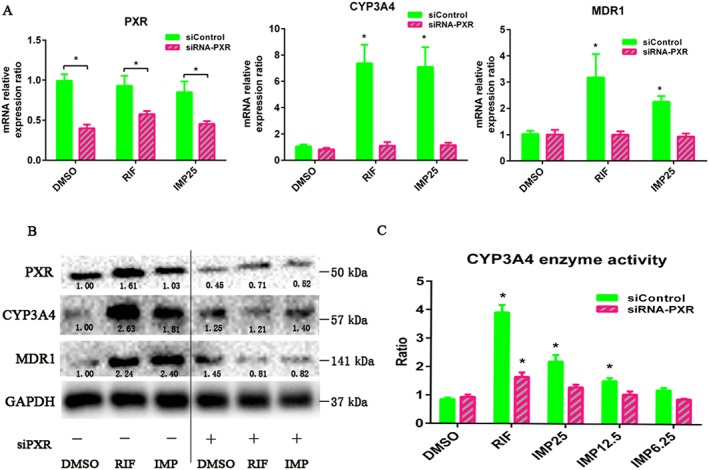
Knockdown of hPXR (NR1I2) expression decreased IMP‐induced PXR target gene expression and CYP3A4 enzymatic activity in LS174T cells. (A) LS174T cells were transiently transfected with hPXR siRNA and control non‐silencing siRNA. After transfection for 48 h, the cells were treated with DMSO, IMP (25 μM) or rifampicin (RIF; 10 μM) for another 48 h, and the mRNA expression of hPXR, CYP3A4 and MDR1 was analysed by qRT‐PCR. Results are expressed as fold changes compared with the DMSO control. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the DMSO control group. (B) Cells were transiently transfected with hPXR siRNA and control non‐silencing siRNA for 48 h and treated with DMSO, IMP (25 μM) or rifampicin (10 μM) for another 48 h. Whole‐cell lysates were collected 96 h after transfection and subjected to Western blot analysis to determine the CYP3A4, MDR1 and PXR protein levels. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (C) The knockdown of hPXR expression attenuated IMP‐induced CYP3A4 enzymatic activity in LS174T cells. After transient transfection with hPXR siRNA and control non‐silencing siRNA for 48 h, cells were treated with DMSO, IMP (6.25–25 μM) or rifampicin (10 μM) for another 48 h. CYP3A4 enzymatic activity was determined by measuring the 1′‐hydroxymidazolam concentration for specific CYP3A4‐mediated midazolam hydroxylation. The results are expressed as relative fold change in activity compared with the vehicle control. Data are expressed as mean ± SEM from five independent experiments. * P < 0.05 compared with the DMSO control group.
IMP‐mediated hPXR activation suppresses NF‐κB activity and activation and down‐regulates NF‐κB‐mediated pro‐inflammatory cytokine production
Next, we sought to determine whether IMP reduces inflammation by blocking NF‐κB activation and NF‐κB p65 nuclear translocation in vitro. Interestingly, IMP treatment dose‐dependently reversed the LPS‐induced increase in the levels of pro‐inflammatory cytokines including TNF‐α, IL‐1β and http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4998 (Figure 5A), indicating that IMP serves as a potent anti‐inflammatory agent. However, http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4975 levels were unchanged after treatment with IMP (Figure 5A). Consistent with the reduced production of pro‐inflammatory cytokines, phosphorylated NF‐κB p65 and phosphorylated IκBα protein levels were substantially suppressed by IMP treatment, although the total NF‐κB p65 protein levels remained unaffected (Figure 5B). However, IMP treatment remarkably prevented the LPS‐induced degradation of the NF‐κB inhibitor protein IκBα (Figure 5B). Furthermore, to explore whether the anti‐inflammatory role of IMP was associated with the inhibition of NF‐κB p65 nuclear translocation, we measured NF‐κB p65 protein expression levels in the nucleus by Western blotting and immunofluorescence staining respectively. Our results showed that IMP reduces NF‐κB p65 protein expression in the nucleus and blocks NF‐κB p65 translocation to the nucleus (Figure 5C, D).
Figure 5.
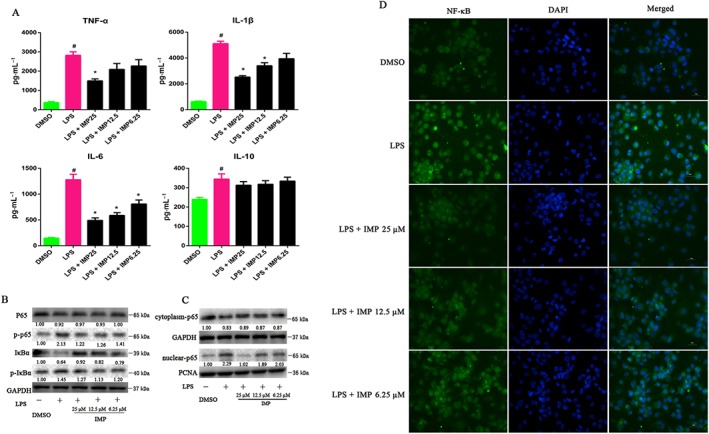
IMP decreases the production of pro‐inflammatory cytokines by supressing the NF‐κB signalling pathway in LPS‐induced THP‐1 cells. (A) THP‐1 cells were pretreated with 6.25, 12.5 or 25 μM IMP for 24 h, followed by additional exposure with or without LPS (1000 ng·mL−1) for another 24 h. Pro‐inflammatory cytokines including TNF‐α, IL‐1β, IL‐6 and IL‐10 in culture supernatants were measured by elisa. Data are expressed as the mean ± SEM from five independent experiments. # P < 0.05 compared with the DMSO control group. * P < 0.05 compared with the LPS control group. (B) The total NF‐κB p65, phospho‐p65 (p‐p65), IκBα and phospho‐IκBα (p‐IκBα) protein levels in LPS‐induced THP‐1 cells were determined by Western blotting. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (C) NF‐κB p65 protein levels in the cytoplasm and nucleus of LPS‐stimulated THP‐1 cells were determined by Western blotting. GAPDH and PCNA were used as cytoplasmic and nuclear markers respectively. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (D) NF‐κB p65 localization was performed using immunofluorescence staining and observed under a fluorescence microscope (magnification: 400×) using an anti‐NF‐κB p65 antibody (1:50) followed by an Alexa Fluor 488‐conjugated detection antibody.
Next, we investigated whether the IMP‐mediated suppression of inflammation was dependent on the activation of hPXR by IMP. Hence, a PXR‐mediated NF‐κB inhibition reporter assay was performed. Exposure to TNF‐α, an activator of the NF‐κB pathway, significantly elevated NF‐κB luciferase activity (~14‐fold) in cells transfected with the NF‐κB‐luc reporter and empty vector (Figure 6A). However, NF‐κB luciferase activity was suppressed by IMP or rifampicin treatment dose dependently after hPXR transfection (Figure 6A); no decrease in NF‐κB luciferase activity was found after treatment with IMP or TNF‐α in the empty vector (Figure 6A). More interestingly, IMP at the highest concentration of 25 μM had no effect on basal NF‐κB luciferase activity (Figure 6A). It has been shown that stimuli, including pro‐inflammatory cytokines, ROS and viral products, can induce the phosphorylation and degradation of IκBα, allowing NF‐κB to translocate to the nucleus and directly regulate the expression of target genes (Cheng et al., 2012). To further test whether the IMP‐mediated inhibition of NF‐κB luciferase activity was dependent on blocking NF‐κB p65 nuclear translocation, we performed immunofluorescence staining analysis. The nuclear levels of NF‐κB p65, due to nuclear translocation, were increased in LS174T cells treated with TNF‐α, whereas IMP or rifampicin treatment inhibited NF‐κB p65 nuclear translocation (Figure 6B).
Figure 6.
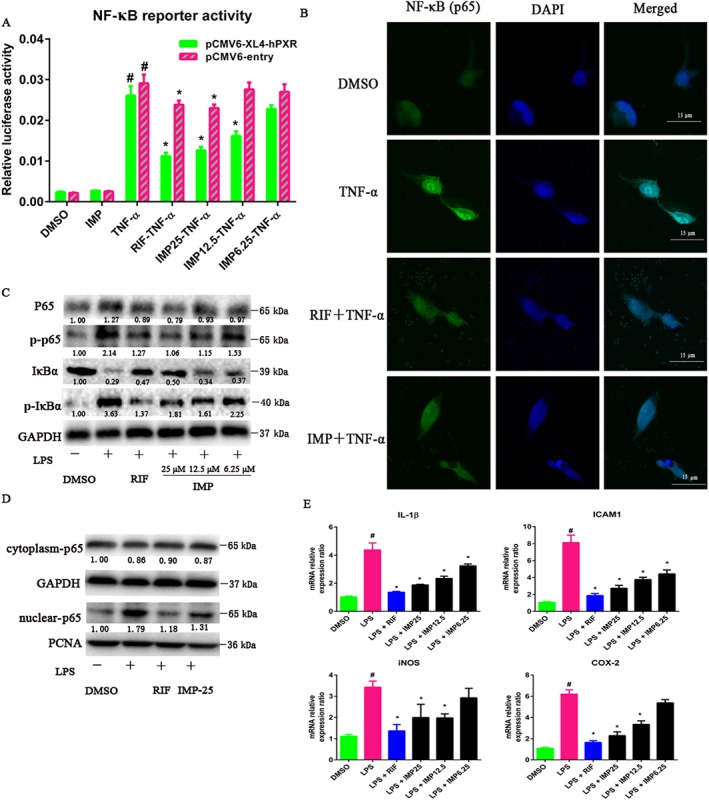
IMP suppresses LPS‐induced NF‐κB activity and down‐regulates NF‐κB‐mediated pro‐inflammatory gene expression in LS174T cells. (A) HEK293T cells were transiently transfected with pGCL4.32 (luc2P/NF‐κB‐RE/Hygro) NF‐κB reporter, pCMV6‐entry or pCMV6‐XL4‐hPXR, and pRL‐TK. After transfection overnight, cells were treated with DMSO, IMP (6.25–25 μM) or rifampicin (RIF; 10 μM) for 24 h, followed by additional incubation with or without TNF‐α (20 ng·mL−1) for another 24 h. A standard dual luciferase assay was performed on the cell lysates. Results are expressed as fold changes compared with the DMSO control. Data are expressed as mean ± SEM from five independent experiments. # P < 0.05 compared with the TNF‐α group with or without hPXR. * P < 0.05 compared with the same treatment group transfected with empty vector and the TNF‐α group transfected with the receptor expression plasmid. (B) LS174T cells were treated with DMSO, IMP (25 μM) or rifampicin (10 μM) for 48 h, followed by additional incubation with or without TNF‐α (20 ng·mL−1) for 24 h. NF‐κB p65 localization was performed using immunofluorescence staining and observed under a confocal laser scanning microscope (magnification: 630×) using an anti‐NF‐κB p65 antibody (1:50) followed by an Alexa Fluor 488‐conjugated detection antibody. (C) NF‐κB p65, phospho‐p65 (p‐p65), IκBα and phospho‐IκBα (p‐IκBα) protein levels in LS174T cells were determined by Western blotting. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (D) NF‐κB p65 protein levels in the cytoplasm and nucleus of LS174T cells were determined by Western blotting. GAPDH and PCNA were used as cytoplasm and nuclear markers respectively. The numbers at the bottom indicate the relative intensity of the protein bands, with the DMSO‐treated sample set as ‘1.00’. (E) The mRNA levels of IL‐1β, ICAM1, iNOS and COX‐2 were determined using qRT‐PCR. LS174T cells were treated with DMSO, IMP (6.25–25 μM) or rifampicin (10 μM) for 48 h, followed by additional exposure to LPS (2000 ng·mL−1) for 24 h. Results are expressed as fold changes compared with the DMSO control. Data are expressed as mean ± SEM from five independent experiments. # P < 0.05 compared with the DMSO control group. * P < 0.05 compared with the LPS control group.
Furthermore, LPS exposure resulted in a substantial decrease in IκBα protein levels, as well as a significant increase in phosphorylated NF‐κB p65 and phosphorylated IκBα protein levels (Figure 6C). In contrast, IMP or rifampicin treatment substantially increased IκBα protein levels and consequently repressed the levels of phosphorylated NF‐κB p65 and phosphorylated IκBα without affecting total NF‐κB p65 levels (Figure 6C). To further clarify the inhibitory effect of IMP on the nuclear translocation of NF‐kB p65 induced by LPS, we measured NF‐kB p65 protein levels in the cytoplasm and in the nucleus. LPS stimulation caused an increase in NF‐kB p65 protein levels in the nucleus (Figure 6D). However, IMP or rifampicin treatment significantly blocked the nuclear translocation of NF‐κB p65 (Figure 6D).
These results suggest that IMP suppresses NF‐κB activity by preventing IκBα phosphorylation/degradation and blocking the nuclear translocation of NF‐κB p65. To determine the effect of IMP on the expression of NF‐κB signalling genes, the mRNA levels of several pro‐inflammatory genes were assessed by qRT‐PCR in LS174T cells. The mRNA levels of pro‐inflammatory genes including IL‐1β, ICAM1, iNOS and COX‐2 were remarkably elevated after LPS exposure in LS174T cells (Figure 6E). Furthermore, like rifampicin, IMP dramatically reduced the LPS‐induced increase in the mRNA expression levels of these pro‐inflammatory cytokines in a dose‐dependent manner (Figure 6E). Taken together, our results suggest that IMP exerts an anti‐inflammatory effect through PXR/NF‐κB signalling.
Genetic knockdown of hPXR (NR1I2) suppresses IMP‐mediated inhibition of pro‐inflammatory gene expression
To further validate that the activation of hPXR by IMP suppresses the expression of pro‐inflammatory genes, we evaluated the effects of hPXR (NR1I2) knockdown on the IMP‐induced inhibition of pro‐inflammatory gene expression. LS174T cells were transiently transfected with siPXR or control siRNA. After transfection, the cells were treated with IMP or rifampicin, followed by additional exposure to LPS. The mRNA expression levels of pro‐inflammatory genes were determined by qRT‐PCR. In transfected control siRNA cells, the expression of pro‐inflammatory genes was significantly reduced after IMP or rifampicin treatment (Figure 7A). In contrast, the inhibitory effect on pro‐inflammatory gene expression was largely abolished in cells transfected with siPXR (Figure 7A). Consistent with the expression of pro‐inflammatory genes, hPXR knockdown almost completely abolished the inhibitory effect of IMP on the nuclear translocation of NF‐κB p65 (Figure 7B).
Figure 7.
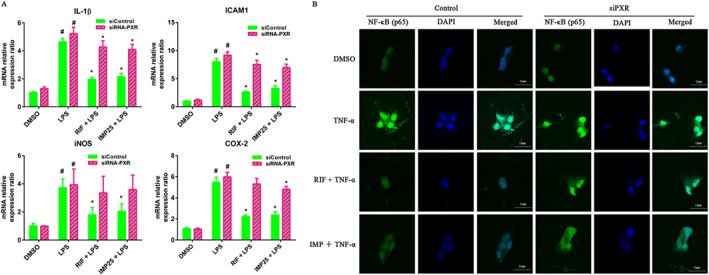
Knockdown of hPXR attenuated IMP‐mediated suppression of pro‐inflammatory gene expression in LS174T cells. (A) Cells were transiently transfected with hPXR siRNA and control non‐silencing siRNA. After transfection for 48 h, the cells were treated with DMSO, IMP (25 μM) or rifampicin (RIF; 10 μM) for another 48 h, followed by additional exposure to LPS (2000 ng·mL−1) for 24 h. The mRNA expression levels of IL‐1β, ICAM1, iNOS and COX‐2 were determined by qRT‐PCR. Results are expressed as fold changes compared with DMSO control. Data are expressed as mean ± SEM from five independent experiments. # P < 0.05 compared with DMSO group. * P < 0.05 compared with the same treatment group. (B) LS174T cells were transiently transfected with hPXR siRNA and control non‐silencing siRNA for 24 h and then treated with DMSO, IMP (25 μM) or rifampicin (10 μM) for another 48 h, followed by an additional incubation with or without TNF‐α (20 ng·mL−1) for 24 h. NF‐κB p65 localization was performed using immunofluorescence staining and observed under a confocal laser scanning microscope (magnification: 630×) using an anti‐NF‐κB p65 antibody (1:50) followed by an Alexa Fluor 488‐conjugated detection antibody.
IMP attenuates DSS‐induced colitis
Several reports have indicated that IMP attenuates LPS‐induced inflammation in vitro (Guo et al., 2012; Huang et al., 2012). It remains unclear, however, whether IMP is also effective against experimental colitis in vivo. To address this question, we extended our study to confirm the effects of IMP against DSS‐induced colitis in mice. DSS administration induced a significant loss of body weight, bloody diarrhoea, colon shortening and neutrophil infiltration in our mouse model (Figure 8). Interestingly, IMP treatment strongly ameliorated DSS‐induced body weight loss (Figure 8A), bloody diarrhoea (Figure 8B) and colon shortening (Figure 8C, D). In addition, histological studies showed that IMP treatment of mice with DSS‐induced colitis caused a significant improvement in colon crypt structures and alleviated the histological damage associated with inflammation (Figure 8E, F).
Figure 8.
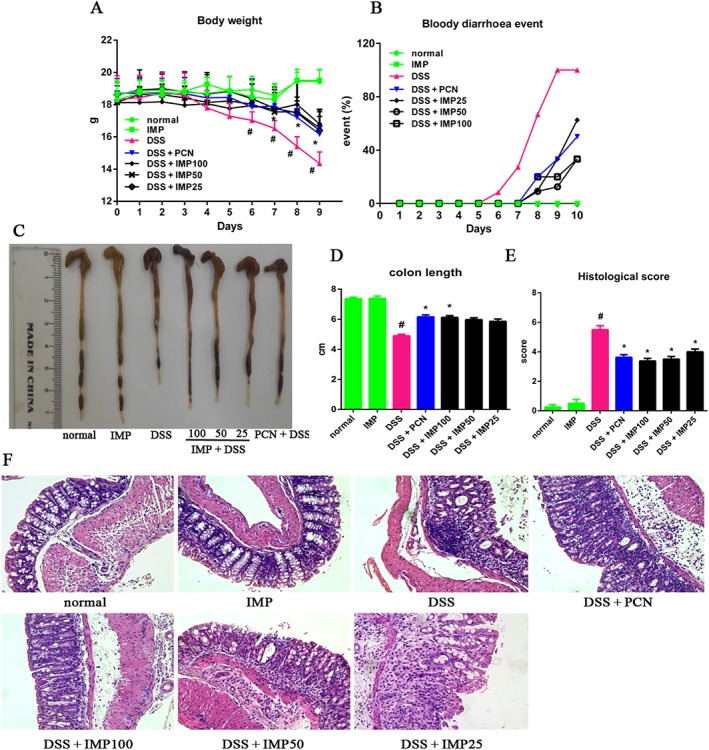
IMP ameliorated DSS‐induced colitis in mice. (A) Body weight changes after the onset of DSS‐induced colitis. (B) Mice were evaluated for the occurrence of bloody diarrhoea after DSS exposure. Data are expressed as the percentage of total mice at different time points during DSS exposure. (C) Macroscopic observation of colon shortening. (D) Colon length. (E) Histological score for H&E‐stained colon sections. (F) Representative H&E‐stained colon sections (magnification: 200×). Data are expressed as mean ± SEM for each group of mice (n = 8). # P < 0.05 compared with the normal group. * P < 0.05 compared with the DSS group.
IMP activates the PXR and inhibits NF‐κB signalling in inflamed mouse colons
We finally sought to determine whether the anti‐inflammatory effect of IMP was related to the activation of the PXR and to the repression of NF‐κB signalling in inflamed mouse colons. First, we investigated whether IMP activates mPXR signalling in healthy mice, even though substantial research has identified IMP as a PXR agonist in vitro. Therefore, we investigated the expression levels of mPXR in the colons of healthy mice. IMP treatment for 7 days at a dosage of 100 mg·kg−1 notably increased Cyp3a11 and Mdr1 expression with little effect on the gene expression of mPXR (Figure 9A), indicating that IMP activates mPXR signalling in vivo. In line with the results of previous studies (Kusunoki et al., 2014; Hu et al., 2015; Zhang et al., 2015a), treatment of mice with DSS for 7 days notably down‐regulated the expression of colonic mPXR signalling and inhibited the increased expression of its target genes, Cyp3a11 and Mdr1, in the colon (Figure 9B, C). However, treatment with IMP strongly attenuated the DSS‐induced decline in colonic mPXR expression (Figure 9B, C). Moreover, immunoblot analysis showed that the DSS‐induced phosphorylation of NF‐κB p65 and IκBα was also blocked by IMP treatment (Figure 9D), suggesting that IMP‐mediated mPXR activation could effectively suppress DSS‐induced colonic inflammation in mice. Consistent with the activation of mPXR and the suppression of NF‐κB signalling, our immunohistochemical results also showed that IMP treatment significantly enhanced mPXR (Nr1i2) expression and down‐regulated NF‐κB p65 protein expression in inflamed colons compared with mice treated with DSS alone (Figure 9E). Finally, IMP treatment significantly decreased the DSS‐induced up‐regulation of pro‐inflammatory cytokines such as TNF‐α and IL‐1β (Figure 9B) and suppressed the DSS‐triggered activation of MPO activity (Figure 9B), which is a biochemical marker of lipid peroxidation. Therefore, IMP, like the mouse PXR agonist PCN, exhibited inhibitory effects against DSS‐induced colitis in mice, as expected (Figure 9). In summary, our data indicate that IMP‐mediated mPXR activation significantly suppresses DSS‐induced colitis by affecting the PXR/NF‐κB signalling pathways.
Figure 9.
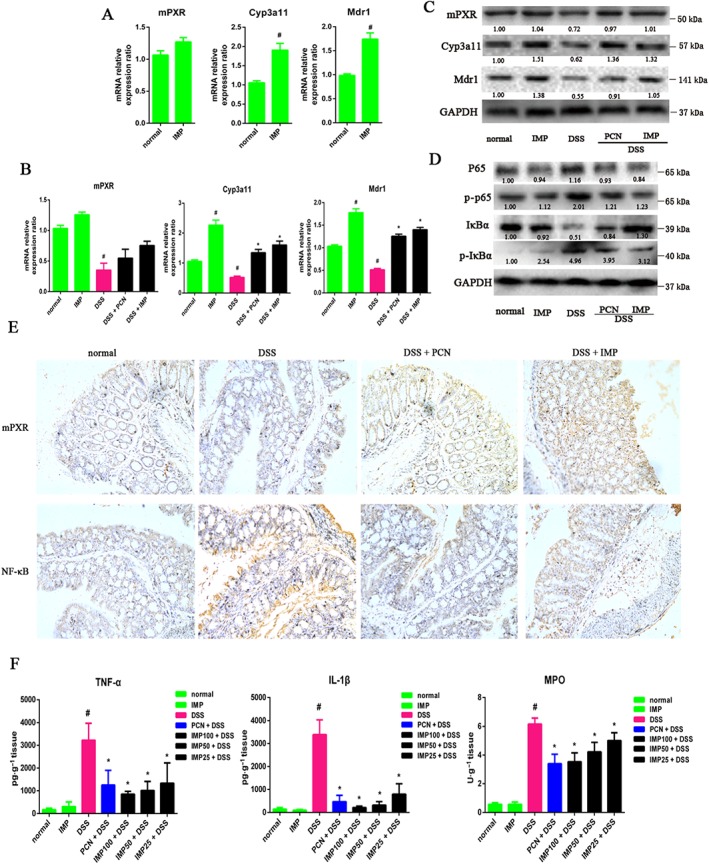
IMP inhibited NF‐κB pro‐inflammatory signalling by the activation of mPXR in DSS‐induced inflamed mouse colons. (A) IMP activated mPXR signalling in healthy mice. mPXR (Nr1i2), Cyp3a11 and Mdr1 mRNA expressions were analysed by qRT‐PCR in healthy mouse colon tissues after treatment with physiological saline (normal) or IMP at a dose of 100 mg·kg−1 for 7 days. Results are expressed as fold changes compared with the normal control. Data are expressed as mean ± SEM from five independent experiments. # P < 0.05 compared with the normal group. (B) mPXR, Cyp3a11 and Mdr1 mRNA expression levels were analysed by qRT‐PCR in colon tissues from DSS‐induced mice colitis after treatment with IMP or PCN for 9 days. Gene expression levels are expressed as fold changes compared with the normal control. # P < 0.05 compared with the normal group. * P < 0.05 compared with the DSS group. (C) Protein levels were analysed by Western blotting. The numbers at the bottom indicate the relative intensity of the protein bands, with the normal‐treated sample set as ‘1.00’. (D) Western blotting was performed in inflamed colon tissues to determine the activity of NF‐κB p65, phospho‐p65 (p‐p65), IκBα and phospho‐IκBα (p‐IκBα). The numbers at the bottom indicate the relative intensity of the protein bands, with the control‐treated sample set as ‘1.00’. (E) Representative images of immunohistochemical staining of mPXR and NF‐κB in mouse colon sections (magnification: 200×). (F) TNF‐α, IL‐1β and MPO concentrations in mouse colon sections. Data are expressed as mean ± SEM for each group of mice (n = 5). # P < 0.05 compared with the normal group. * P < 0.05 compared with the DSS group.
Discussion
To our knowledge, this is the first study showing that IMP can increase CYP3A4 expression in an hPXR‐dependent manner. IMP‐mediated PXR activation significantly inhibited NF‐κB activity and decreased the expression of several inflammatory factors. Furthermore, IMP treatment significantly increased intestinal mPXR (Nr1i2) expression and inhibited the phosphorylation of NF‐κB p65, leading to a reduction in the accumulation of pro‐inflammatory cytokines and a reduction in the hallmarks of colitis in DSS‐treated mice, such as weight loss, bloody diarrhoea, colon shortening and histological damage. Collectively, our findings suggest that IMP‐modulated PXR activation ameliorates the effects of DSS‐induced colitis by suppressing the PXR‐mediated NF‐κB pro‐inflammatory signalling pathway.
The concentrations and dosage of IMP administered in our cell culture model and mouse colitis model were estimated based on information from previous reports (Zhang et al., 2017). As the major active component of A. dahurica Radix, IMP accounts for 0.4–1.0% of its content. A. dahurica Radix is a widely used Chinese medicine for its anti‐inflammatory effects, and its average daily consumption in China is around 30 g (~120–300 mg IMP), based on the Chinese Pharmacopoeia (Commission CP, 2010). Given that the average body weight of a Chinese adult is 60 kg, the theoretical dosage for mice should be 22–55 mg·kg−1. In the present study, we treated DSS‐induced colitis mice with IMP at a dosage of 25, 50 or 100 mg·kg−1 to determine the reasonable dosage for the treatment of colitis in mice. Additionally, assuming that the plasma volume is ~2.5–3 L in the average adult (Anderson and Anderson, 2002), this theoretical range of IMP plasma concentration should be 148–370 μM. Considering the fact that the absorption efficacy and the first‐pass effect of the intestine and liver usually occur after IMP administration (Chen et al., 2015), the concentration of plasma IMP might be far less than the theoretical plasma concentration (148–370 μM). Consistent with our hypothesis, a recent preclinical study showed that the mean plasma concentration of IMP was approximately 2587 ± 365 ng·mL−1 (8.2–10.8 μM) after a single p.o. dose of 4.5 g·kg−1 A. dahurica Radix extract (equivalent to 74.1 mg·kg−1 IMP) by gastric gavage in rats (Chen et al., 2015). Similarly, the maximum mean plasma concentration of IMP was ~3174 ng·mL−1 (11.8 μM) after a single dose of i.v. injection of IMP (5 mg·kg−1) in healthy beagle dogs (Wang et al., 2012a). Therefore, the concentration of IMP at used in the present study, 6.25–25 μM, might be within a physiologically relevant range for humans. Indeed, the concentration of 12.5 μM used in the cell model in our present study, which was similar to previously reported physiological concentrations of IMP (10.8 and 11.8 μM), was high enough to stimulate CYP3A4 and MDR1 expression.
PXR is a key xenobiotic receptor regulating the metabolism and excretion of both xenobiotics and endobiotics by regulating the expression of drug‐metabolizing enzymes and drug transport (Kliewer et al., 2002). Once activated by a ligand, PXR heterodimerizes with the retinoid X receptor and binds to xenobiotic response elements located in the promoter regions to regulate the expression of PXR target genes (Wang et al., 2012c). Recently, it has been shown that the expression of PXR and its target genes is strongly associated with the integrity of the intestinal epithelial barrier in the gastrointestinal tract, which plays a key role in the pathogenesis of IBD (Venkatesh et al., 2014; Garg et al., 2016). In a previous study, MDR1 deficiency impaired mitochondrial homeostasis and promoted intestinal inflammation (Ho et al., 2018), causing colitis to spontaneously develop in mice (Panwala et al., 1998). CYP3A4 expression was decreased in inflamed small intestines of children with CD compared with the non‐inflamed duodenum (Shakhnovich et al., 2016). Several polymorphisms in the PXR locus, which alter PXR activity, have contributed to sharp increases in the risk of susceptibility to IBD (Dring et al., 2006; Glas et al., 2011). Moreover, previous studies have reported a significant decrease in PXR expression and its target genes in the inflamed terminal ileum compared with the non‐inflamed duodenum (Dring et al., 2006; Glas et al., 2011; Shakhnovich et al., 2016), suggesting a role for PXR in the pathogenesis of IBD. Emerging data have consistently revealed that the activation of PXR by naturally occurring components significantly attenuates DSS‐induced colitis (Dou et al., 2014; Sehirli et al., 2015), indicating a curative role of PXR agonists against IBD.
There is limited experimental data on the effects of IMP as a PXR agonist in human cells. A recent report showed that IMP could inhibit the expression of P‐gp (MDR1), leading to the promotion of rhodamine 123 accumulation in MDCK‐MDR1 (Liao et al., 2016), an hPXR‐deficient human renal cell line, revealing the role of PXR‐independent MDR1 regulation. It is possible that other PXR‐independent mechanisms are associated with the regulation of MDR1 by IMP, because multiple mechanisms, including tumour suppressor protein p53, hypoxia‐inducible factor‐1 and epigenetic mechanisms, have been reported to regulate the expression of MDR1 (Chen, 2010). Consistent with the inhibition of MDR1, IMP was found to directly inhibit the activity of CYP3A4, to a slightly lesser extent, in human/rat liver microsomes (Kimura et al., 2010; Cao et al., 2013). In contrast, several studies have shown that IMP treatment could increase CYP3A11 activity in mice liver microsomal samples (Kleiner et al., 2008; Wang et al., 2012b); however, it was unclear whether its expression was PXR dependent. Consistent with these studies, our results clearly demonstrated that CYP3A4 expression was activated by IMP through a PXR‐dependent mechanism. The direct binding of IMP to hPXR‐LBD in the TR‐FRET competitive assay was an unambiguous indicator that IMP is an hPXP agonist (Figure 2B). Furthermore, the molecular docking study for predicting the mode of IMP binding to hPXR‐LBD showed that IMP had hydrogen‐bonding interactions with the residues Lys210 and Leu209 in different directions. Taken together, the data show that IMP acts as an hPXR agonist to induce CYP3A4 expression in a PXR‐dependent manner.
The aetiology of IBD still remains unknown; however, it is generally accepted that an increase in the activity of intestinal pro‐inflammatory cytokines such as TNF‐α and IL‐1β plays an important role in the pathogenesis of IBD (Shah et al., 2007). It has been reported that NF‐κB is a master transcriptional regulator of pro‐inflammatory cytokines (Christian et al., 2016). Mutual crosstalk between PXR and NF‐κB might represent a target mechanism for the anti‐inflammatory properties of PXR (Zhou et al., 2006). Not only did the activation of PXR inhibit NF‐κB activity and the expression of NF‐κB target genes but also the activation of NF‐κB activity by LPS suppressed PXR activity and its expression, resulting in a decrease in the expression of PXR‐mediated downstream target genes. For instance, PCN/rifaximin activation of mouse/human PXR was found to protect against DSS‐induced IBD in wild‐type or humanized PXR mice, but not in PXR‐null mice, via PXR/NF‐κB signalling (Shah et al., 2007; Cheng et al., 2010). NF‐κB normally remains bound to the inhibitory protein IκBα in the cytoplasm. Following activation signals such as pro‐inflammatory cytokines, IκBα is rapidly phosphorylated and degraded, allowing NF‐κB to translocate to the nucleus, where it directly regulates the expression of its target genes (Christian et al., 2016). It has been reported that IMP decreases LPS‐induced inflammation in vitro by inhibiting the expression of NF‐κB‐mediated pro‐inflammatory genes such as iNOS, COX‐2 and TNF‐α (Guo et al., 2012; Huang et al., 2012). Previous studies have also found that IMP significantly inhibits the TNF‐α‐induced expression of NF‐κB target genes by suppressing IκB phosphorylation and NF‐κB p65 nuclear translocation (Wang et al., 2017), indicating that IMP is a potent inhibitor of NF‐κB. Moreover, in vivo experiments showed that IMP reduced the expression of cytokines and improved histopathological features by blocking activation of the IκBα/NF‐κB pathway (Sun et al., 2012; Yang et al., 2015). Consistent with previous reports, we found that IMP markedly decreased the release of LPS‐induced pro‐inflammatory cytokines by suppressing the activation and nuclear translocation of NF‐κB in THP‐1 cells, suggesting that IMP is a potent anti‐inflammatory reagent. Given that immune cells such as THP‐1 are unrelated to PXR signalling, to further explore how and whether IMP‐regulated NF‐κB suppression affected IMP‐mediated PXR activation, we established an LPS‐induced LS174T cell inflammation model that displayed endogenous PXR signalling. In the present study, like in THP‐1 cells, we consistently found that IMP significantly inhibited NF‐κB activity by blocking NF‐κB p65 nuclear translocation in LS174T cells. This attenuating effect of IMP was partiality lost in cells where the expression of PXR was silenced, suggesting an essential role of PXR in the effects of IMP. Consistent with the in vitro data, our in vivo data showed that IMP treatment activated mPXR (Nr1i2) signalling and inhibited the NF‐κB p65‐regulated pro‐inflammatory pathway, resulting in a reduced production of pro‐inflammatory cytokines (TNF‐α and IL‐1β). Taken together, our results show that IMP exerts a strong anti‐inflammatory effect by an effect on the PXR/NF‐κB signalling pathway.
In summary, this is the first study to demonstrate a protective effect of IMP on DSS‐induced mouse colitis. The anti‐inflammatory mechanisms of IMP are potentially associated with the activation of PXR and the inhibition of NF‐κB‐mediated pro‐inflammatory signalling.
Author contributions
C.L. and S.M. conceived and designed the study; M.L., G.Z., C.Z., F.L., X.H., S.B., X.H., C.L., C.Z., S.M. and C.L. performed the research; M.L., G.Z. and C.L. analysed the data; C.Z. and X.H. contributed the new methods and models; and M.L. and C.L. wrote the paper.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This http://onlinelibrary.wiley.com/doi/10.1111/bph.13405/abstract acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grants 81773969, 81460659, 81573566, 81673872 and 81173535), the High‐level Talents Project of Guangdong Province (grant AFD018171Z0377) and the Science Program for Overseas Scholar of Chinese Medicine (grant XH20160103).
Liu, M. , Zhang, G. , Zheng, C. , Song, M. , Liu, F. , Huang, X. , Bai, S. , Huang, X. , Lin, C. , Zhu, C. , Hu, Y. , Mi, S. , and Liu, C. (2018) Activating the pregnane X receptor by imperatorin attenuates dextran sulphate sodium‐induced colitis in mice. British Journal of Pharmacology, 175: 3563–3580. 10.1111/bph.14424.
References
- Ahmed FE (2006). Role of genes, the environment and their interactions in the etiology of inflammatory bowel diseases. Expert Rev Mol Diagn 6: 345–363. [DOI] [PubMed] [Google Scholar]
- Alexander SPH, Cidlowski JA, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017a). The Concise Guide to PHARMACOLOGY 2017/18: Nuclear hormone receptors. Br J Pharmacol 174: S208–S224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017c). The Concise Guide to PHARMACOLOGY 2017/18: Transporters. Br J Pharmacol 174: S360–S446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Anderson NG (2002). The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1: 845–867. [DOI] [PubMed] [Google Scholar]
- Berrevoets CA, Doesburg P, Steketee K, Trapman J, Brinkmann AO (1998). Functional interactions of the AF‐2 activation domain core region of the human androgen receptor with the amino‐terminal domain and with the transcriptional coactivator TIF2 (transcriptional intermediary factor 2). Mol Endocrinol 12: 1172–1183. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zhong YH, Yuan M, Li H, Zhao CJ (2013). Inhibitory effect of imperatorin and isoimperatorin on activity of cytochrome P450 enzyme in human and rat liver microsomes. Zhongguo Zhong Yao Za Zhi 38: 1237–1241. [PubMed] [Google Scholar]
- Chen L, Jian Y, Wei N, Yuan M, Zhuang X, Li H (2015). Separation and simultaneous quantification of nine furanocoumarins from Radix Angelicae dahuricae using liquid chromatography with tandem mass spectrometry for bioavailability determination in rats. J Sep Sci 38: 4216–4224. [DOI] [PubMed] [Google Scholar]
- Chen T (2008). Nuclear receptor drug discovery. Curr Opin Chem Biol 12: 418–426. [DOI] [PubMed] [Google Scholar]
- Chen T (2010). Overcoming drug resistance by regulating nuclear receptors. Adv Drug Deliv Rev 62: 1257–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Shah YM, Gonzalez FJ (2012). Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci 33: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Shah YM, Ma X, Pang X, Tanaka T, Kodama T et al (2010). Therapeutic role of rifaximin in inflammatory bowel disease: clinical implication of human pregnane X receptor activation. J Pharmacol Exp Therapeut 335: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian F, Smith EL, Carmody RJ (2016). The regulation of NF‐κB subunits by phosphorylation. Cell 5: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commission CP (2010). Pharmacopoeia of the People's Republic of China. Beijing: Chemical Industry Press. [Google Scholar]
- Curtis MJ, Alexander S, Cirino G, Docherty JR, George CH, Giembycz MA et al (2018). Experimental design and analysis and their reporting II: updated and simplified guidance for authors and peer reviewers. Br J Pharmacol 175: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou W, Zhang J, Li H, Kortagere S, Sun K, Ding L et al (2014). Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR‐dependent pathway. J Nutr Biochem 25: 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dring MM, Goulding CA, Trimble VI, Keegan D, Ryan AW, Brophy KM et al (2006). The pregnane X receptor locus is associated with susceptibility to inflammatory bowel disease. Gastroenterology 130: 341–348. [DOI] [PubMed] [Google Scholar]
- Dussault I, Yoo HD, Lin M, Wang E, Fan M, Batta AK et al (2003). Identification of an endogenous ligand that activates pregnane X receptor‐mediated sterol clearance. Proc Natl Acad Sci U S A 100: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Zhao A, Erickson SL, Mukherjee S, Lau AJ, Alston L et al (2016). Pregnane X receptor activation attenuates inflammation‐associated intestinal epithelial barrier dysfunction by inhibiting cytokine‐induced myosin light‐chain kinase expression and c‐Jun N‐terminal kinase 1/2 activation. J Pharmacol Exp Therapeut 359: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy L, Allen PB (2014). Is there a role for vedolizumab in the treatment of ulcerative colitis and Crohn's disease? Clin Exp Gastroenterol 7: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glas J, Seiderer J, Fischer D, Tengler B, Pfennig S, Wetzke M et al (2011). Pregnane X receptor (PXR/NR1I2) gene haplotypes modulate susceptibility to inflammatory bowel disease. Inflamm Bowel Dis 17: 1917–1924. [DOI] [PubMed] [Google Scholar]
- Guo W, Sun J, Jiang L, Duan L, Huo M, Chen N et al (2012). Imperatorin attenuates LPS‐induced inflammation by suppressing NF‐κB and MAPKs activation in RAW 264.7 macrophages. Inflammation 35: 1764–1772. [DOI] [PubMed] [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillegass LM, Griswold DE, Brickson B, Albrightson‐Winslow C (1990). Assessment of myeloperoxidase activity in whole rat kidney. J Pharmacol Methods 24: 285–295. [DOI] [PubMed] [Google Scholar]
- Ho GT, Aird RE, Liu B, Boyapati RK, Kennedy NA, Dorward DA et al (2018). MDR1 deficiency impairs mitochondrial homeostasis and promotes intestinal inflammation. Mucosal Immunol 11: 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Wang Y, Chen Z, Ma Z, You Q, Zhang X et al (2015). The protective effect of piperine on dextran sulfate sodium induced inflammatory bowel disease and its relation with pregnane X receptor activation. J Ethnopharmacol 169: 109–123. [DOI] [PubMed] [Google Scholar]
- Huang GJ, Deng JS, Liao JC, Hou WC, Wang SY, Sung PJ et al (2012). Inducible nitric oxide synthase and cyclooxygenase‐2 participate in anti‐inflammatory activity of imperatorin from Glehnia littoralis . J Agric Food Chem 60: 1673–1681. [DOI] [PubMed] [Google Scholar]
- Hwang YH, Yang HJ, Ma JY (2017). Simultaneous determination of three furanocoumarins by UPLC/MS/MS: application to pharmacokinetic study of Angelica dahurica Radix after oral administration to normal and experimental colitis‐induced rats. Molecules 22: E416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobin C (2010). Probiotics and ileitis: could augmentation of TNF/NFκB activity be the answer? Gut Microbes 1: 196–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Zeissig S, Blumberg RS (2010). Inflammatory bowel disease. Annu Rev Immunol 28: 583–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2010). Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Ito H, Ohnishi R, Hatano T (2010). Inhibitory effects of polyphenols on human cytochrome P450 3A4 and 2C9 activity. Food Chem Toxicol 48: 429–435. [DOI] [PubMed] [Google Scholar]
- Kleiner HE, Xia X, Sonoda J, Zhang J, Pontius E, Abey J et al (2008). Effects of naturally occurring coumarins on hepatic drug‐metabolizing enzymes in mice. Toxicol Appl Pharmacol 232: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM (2002). The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev 23: 687–702. [DOI] [PubMed] [Google Scholar]
- Kusunoki Y, Ikarashi N, Hayakawa Y, Ishii M, Kon R, Ochiai W et al (2014). Hepatic early inflammation induces downregulation of hepatic cytochrome P450 expression and metabolic activity in the dextran sulfate sodium‐induced murine colitis. Eur J Pharm Sci 54: 17–27. [DOI] [PubMed] [Google Scholar]
- Lau AJ, Yang G, Yap CW, Chang TK (2012). Selective agonism of human pregnane X receptor by individual ginkgolides. Drug Metab Dispos Biol Fate Chemicals 40: 1113–1121. [DOI] [PubMed] [Google Scholar]
- Lee K, Min SS, Ham I, Choi HY (2015). Investigation of the mechanisms of Angelica dahurica root extract‐induced vasorelaxation in isolated rat aortic rings. BMC Complement Altern Med 15: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wu L (2017). Coumarins from the roots of Angelica dahurica cause anti‐allergic inflammation. Exp Ther Med 14: 874–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao ZG, Tang T, Guan XJ, Dong W, Zhang J, Zhao GW et al (2016). Improvement of transmembrane transport mechanism study of imperatorin on P‐glycoprotein‐mediated drug transport. Molecules 21: E1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwala CM, Jones JC, Viney JL (1998). A novel model of inflammatory bowel disease: mice deficient for the multiple drug resistance gene, mdr1a, spontaneously develop colitis. J Immunol 161: 5733–5744. [PubMed] [Google Scholar]
- Roediger WEW, Babidge W (1997). Human colonocyte detoxification. Gut 41: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler G (2010). Gastrointestinal and liver adverse effects of drugs used for treating IBD. Best Pract Res Clin Gastroenterol 24: 157–165. [DOI] [PubMed] [Google Scholar]
- Sehirli AO, Cetinel S, Ozkan N, Selman S, Tetik S, Yuksel M et al (2015). St. John's wort may ameliorate 2,4,6‐trinitrobenzenesulfonic acid colitis off rats through the induction of pregnane X receptors and/or P‐glycoproteins. J Physiol Pharmacol 66: 203–214. [PubMed] [Google Scholar]
- Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ (2007). Pregnane X receptor activation ameliorates DSS‐induced inflammatory bowel disease via inhibition of NF‐κB target gene expression. Am J Physiol Gastrointest Liver Physiol 292: 1114–1122. [DOI] [PubMed] [Google Scholar]
- Shakhnovich V, Vyhlidal CA, Friesen C, Hildreth A, Daniel J, Singh V et al (2016). Decreased pregnane X receptor expression in children with active Crohn's disease. Drug Metab Dispos 44: 1066–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, O'Malley BW (2004). Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev 25: 45–71. [DOI] [PubMed] [Google Scholar]
- Sui Y, Ai N, Park SH, Riospilier J, Perkins JT, Welsh WJ et al (2012). Bisphenol A and its analogues activate human pregnane X receptor. Environ Health Perspect 120: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chi G, Soromou LW, Chen N, Guan M, Wu Q et al (2012). Preventive effect of imperatorin on acute lung injury induced by lipopolysaccharide in mice. Int Immunopharmacol 14: 369–374. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Igarashi‐Migitaka J, Koibuchi N, Takeuchi Y (2013). Mitotane induces CYP3A4 expression via activation of the steroid and xenobiotic receptor. J Endocrinol 216: 297–305. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Inagaki K, Igarashimigitaka J, Ozawa Y, Koibuchi N (2006). The endocrine disrupting chemical, diethylhexyl phthalate, activates MDR1 gene expression in human colon cancer LS174T cells. J Endocrinol 190: 897–902. [DOI] [PubMed] [Google Scholar]
- Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP et al (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll‐like receptor 4. Immunity 41: 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Lv Y, Wang Z, Ma J, Mi C, Li X et al (2017). Imperatorin efficiently blocks TNF‐α‐mediated activation of ROS/PI3K/Akt/NF‐κB pathway. Oncol Rep 37: 3397–3404. [DOI] [PubMed] [Google Scholar]
- Wang L, Lu W, Shen Q, Wang S, Zhou H, Yu L et al (2012a). Simultaneous determination of imperatorin and its 2 metabolites in dog plasma by using liquid chromatography‐tandem mass spectrometry. J Pharm Biomed Anal 70: 640–646. [DOI] [PubMed] [Google Scholar]
- Wang X, Lou YJ, Wang MX, Shi YW, Xu HX, Kong LD (2012b). Furocoumarins affect hepatic cytochrome P450 and renal organic ion transporters in mice. Toxicol Lett 209: 67–77. [DOI] [PubMed] [Google Scholar]
- Wang YM, Lin W, Chai SC, Wu J, Su SO, Schuetz EG et al (2013). Piperine activates human pregnane X receptor to induce the expression of cytochrome P450 3A4 and multidrug resistance protein 1. Toxicol Appl Pharmacol 272: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, Chen T (2012c). Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin Drug Metab Toxicol 8: 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Tian Y (2006). Xenobiotic receptor meets NF‐κB, a collision in the small bowel. Cell Metab 4: 177–178. [DOI] [PubMed] [Google Scholar]
- Yang IJ, Lee DU, Shin HM (2015). Anti‐inflammatory and antioxidant effects of coumarins isolated from Foeniculum vulgare in lipopolysaccharide‐stimulated macrophages and 12‐O‐tetradecanoylphorbol‐13‐acetate‐stimulated mice. Immunopharmacol Immunotoxicol 37: 308–317. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cao L, Wang H, Cheng X, Wang L, Zhu L et al (2015a). Ginsenosides regulate PXR/NF‐κB signaling and attenuate dextran sulfate sodium‐induced colitis. Drug Metab Dispos Biological Fate Chemicals 43: 1181–1189. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Wang B, Ren G, Sun A, Deng C et al (2015b). Notoginsenoside R1 attenuates experimental inflammatory bowel disease via pregnane X receptor activation. J Pharmacol Exp Ther 352: 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li W, Abudureheman A, Cheng T, Peng P (2017). Imperatorin possesses notable antiinflammatory activity in vitro and in vivo through inhibition of the NFκB pathway. Mol Med Rep 16: 8619–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A et al (2006). Mutual repression between steroid and xenobiotic receptor and NF‐κB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest 116: 2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]


