Abstract
Background and Purpose
Intestinal ischaemia‐reperfusion (II/R) injury is a serious clinical problem. Here we have investigated novel mechanisms and new drug targets in II/R injury by searching for microRNAs regulating such injury.
Experimental Approach
We used hypoxia/reoxygenation (H/R) of IEC‐6 cell cultures and models of II/R models in rats and mice. Microarray assays were used to identify target miRNAs from rat intestinal. Real‐time PCR, Western blot and dual luciferase reporter assays, and agomir and antagomir in vitro and in vivo were used to assess the effects of the target miRNA on II/R injury.
Key Results
The miR‐351‐5p was differentially expressed in our models and it targeted MAPK13 and sirtuin‐6. This miRNA reduced levels of sirtuin‐6 and AMP‐activated protein kinase phosphorylation, and activated forkhead box O3 (FoxO3α) phosphorylation to cause oxidative stress. Also, miR‐351‐5p markedly reduced MAPK13 level, activated polycystic kidney disease 1/NF‐κB signal and increased NF‐κB (p65). Moreover, miR‐351‐5p up‐regulated levels of Bcl2‐associated X, cytochrome c, apoptotic peptidase activating factor 1, cleaved‐caspase 3 and cleaved‐caspase 9 by reducing sirtuin‐6 levels to promote apoptosis. In addition, miR‐351‐5p mimic in IEC‐6 cells and agomir in mice aggravated these effects, and miR‐351‐5p inhibitor and antagomir in mice alleviated these actions.
Conclusions and Implications
Our data showed that miR‐351‐5p aggravated II/R injury by promoting intestinal mucosal oxidative stress, inflammation and apoptosis by targeting MAPK13 and sirtuin‐6.These data provide new insights into the mechanisms regulating II/R injury, and of miR‐351‐5p could be considered as a novel therapeutic target for such injury.
Abbreviations
- AMPK
AMP‐activated protein kinase
- FoxO3α
forkhead box O3
- GO
gene ontology
- H/R
hypoxia/reoxygenation
- HE
haematoxylin‐eosin
- ICAM‐1
intercellular cell adhesion molecule‐1
- II/R
intestinal ischaemia‐reperfusion
- MODS
multiple organ dysfunction syndrome
- MPO
myeloperoxidase
- MnSOD
manganese‐dependent SOD
- MDA
malondialdehyde
- NC
negative control
- PKD1
polycystic kidney disease 1
- PTEN
phosphatase and tensin homologue deleted on chromosome ten
- Sirt6
Sirtuin‐6
- SIRS
systemic inflammatory response syndrome
- T‐SOD
total SOD
- UTRs
untranslated regions
Introduction
Intestinal ischaemia‐reperfusion (II/R) injury, a serious clinical problem, appears as a result of trauma, shock, organ transplantation and end‐organ failure (Berlanga et al., 2002; Pope et al., 2012). II/R can cause systemic inflammatory response syndrome (SIRS), distant organ damage and multiple organ dysfunction syndrome (MODS) (Dwivedi et al., 2007; Goldsmith et al., 2012). Thus, it is critically important to investigate novel mechanisms and drug targets that could regulate II/R to facilitate the development of therapeutic strategies in this event.
Several biological processes including oxygen free radical accumulation, lipid peroxidation, apoptosis, cell injury and inflammation have been considered as the mediators of ischaemia‐reperfusion (I/R)‐induced intestinal mucosal damage (Watanabe et al., 2008; Marcus et al., 2013). Among them, oxygen free radicals caused by II/R injury can directly damage intestinal mucosa, increase the permeability of systemic microcirculation, destroy cell membrane, trigger lipid peroxidation and eventually lead to cell apoptosis or necrosis (Sukhotnik et al., 2009). At the same time, the uncontrolled inflammatory response is an important mechanism underlying SIRS (Deitch et al., 2006). Furthermore, in the early stage of II/R, large amounts of oxygen free radicals, intracellular calcium overload and mitochondrial dysfunction can induce cell apoptosis and then cause damage to the intestinal epithelium (Liu et al., 2015). However, the molecular mechanisms of II/R injury are not fully understood. Hence, it is important to investigate the underlying mechanisms for development of effective therapies against the disease.
MicroRNAs (miRNAs) have the functions of regulating gene expression through binding with the 3′ untranslated regions (UTRs) of their targeted mRNAs (Li et al., 2013). Several miRNAs including miR‐682, miR‐34a‐5p and miR‐146a are known to affect II/R injury. MiR‐682 can down‐regulate phosphatase and tensin homologue deleted on chromosome ten (PTEN) in intestinal epithelial cells to ameliorate II/R injury (Liu et al., 2016). Inhibition of miR‐34a‐5p can alleviate II/R‐induced ROS accumulation and apoptosis via activating sirtuin (Sirt)1 signalling (Wang et al., 2016). MiR‐146a can down‐regulate IL‐1 receptor‐associated kinase 1 against II/R injury (Chassin et al., 2012).
In order to identify the miRNAs relevant to a pathological condition, such II/R, it is important to find the miRNAs that are differentially expressed in biological samples taken from that condition. An effective method of such analysis is the microarrays assay, a high‐throughput screening method, has been widely used for investigating drug targets and mechanisms of diseases (Li et al., 2017; Yuan et al., 2017). Thus, the aim of the present work was to find the differentially expressed miRNAs associated with II/R injury by microarrays assay and then to investigate the molecular mechanisms and drug targets for the prevention and treatment of II/R injury.
Methods
Cell culture
The IEC‐6 cells were obtained from Shanghai Institutes for Biological Sciences (Shanghai, China) and cultured in DMEM (Gibco, California, USA) supplemented with 5% FBS, streptomycin (100 mg·mL−1) and penicillin (100 IU·mL−1) in a humidified atmosphere of 5% CO2 and 95% O2 at 37°C. DMEM was replaced every 3 days in all experiments.
Cell hypoxia/reoxygenation (H/R) model
The IEC‐6 cells incubated in Krebs–Ringer bicarbonate buffer were transferred to an incubator containing humidified atmosphere equilibrated with 1% O2, 94% N2 and 5% CO2 at 37°C for 2 h to simulate hypoxia condition. After that, the reoxygenation for different times (0.5, 1, 3, 6 and 12 h) was carried out by replacing the buffer with normal medium and normoxic conditions.
Cell viability assay
The IEC‐6 cells were seeded in 96‐well plates at a density of 5 × 103 cells per mL for 12 h. Then, the cells were subjected to the H/R procedure. At the ending of H/R, 10 μL of MTT (5 mg·mL−1) solution was added to each well and the medium was removed after incubation for 4 h at 37°C. Afterwards, a total of 150 μL of DMSO was added to each well and a POLA Rstar OPTIMA multi‐detection microplate reader (BioRad, San Diego, CA, USA) was used to determine the absorbance at 490 nm.
ROS assay in vitro
The IEC‐6 cells were seeded in 24‐well plates at a density of 5 × 103 cells per mL and the H/R experiment was performed. DCFH‐DA was diluted with a serum‐free medium to a final concentration at 10 μM. Then, the cell medium was removed and an appropriate volume of diluted DCFH‐DA was added. After that, the cells were washed three times with serum‐free medium to remove DCFH‐DA, and the images were captured by fluorescence microscopy (Olympus, Japan) with 200× magnification.
Animals
All animal care and experimental procedures were carried out in accordance with legislation regarding the Use and Care of Laboratory Animals (People's Republic of China ) and approved by the Animal Care and Use Committee of Dalian Medical University. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny et al., 2010; McGrath and Lilley, 2015). All the animal studies complied with the principles of replacement, refinement or reduction (the 3Rs).
Forty‐eight male Wistar rats weighing 170–210 g (8 weeks old) and 48 male C57BL/6J mice weighing 18–20 g (4 weeks old) were supplied by the Experimental Animal Center at Dalian Medical University (Dalian, China) (SCKK: 2013‐0006). The animals were randomly divided into six groups (n = 8): control groups and II/R groups. The animals were group‐housed with 2–3 rats per cage on a 12 h light/dark cycle in a temperature‐controlled (25 ± 2°C) room with free access to water and food, and allowed 1 week to acclimatize before experiment. The experiments were performed by investigators blinded to the group treatments.
II/R‐induced intestinal injury models in vivo
The animals were fasted for 24 h and anaesthetized with urethane (1.0 g kg‐1, i.p.), and the abdominal cavity was opened. The superior mesenteric artery and its adjacent tissues were cautiously isolated and clamped with a small vascular clamp for 45 min ischaemia, followed by reperfusion for 45, 90, 180, 360 and 720 min. After reperfusion, the animals were killed with an anaesthetic overdose and the whole intestine was rapidly removed and washed. The intestinal tissues were stored at −80°C.
Histopathological examination
The animals were killed after reperfusion, and then the intestinal samples were removed, washed with PBS at 4°C, fixed in 4% paraformaldehyde, embedded in paraffin, sliced (5 μm thickness) and stained with haematoxylin and eosin (H&E). II/R‐induced mucosal injury was evaluated, according to Chiu's score, using a light microscope (Leica DM4000B, Germany). These evaluations were performed by investigators blinded to the treatment groups
Measurement of MDA, MPO and T‐SOD levels in vivo
The levels of MDA, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2789 and total SOD (T‐SOD) in intestinal tissues of rats and mice were detected using the kits according to the manufacturer's instructions.
TUNEL assay in vivo and in vitro
TUNEL staining was performed using the assay kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. The paraffin tissue sections were dewaxed and immersed in protease K working fluid (10 μg·mL−1) for 20 min at 37°C. After washing three times with PBS, the sections were incubated with 50 μL (5 μL TdT + 45 μL fluorescein labelled dUTP solution) of TUNEL mixed solution for 1 h in the dark and wet box at 37°C. In vitro experiments, after washing with PBS for three times, the IEC‐6 cells were fixed by 4% paraformaldehyde for 30 min and incubated in 0.1% Triton X‐100 for 10 min. Then, 100 μL of TUNEL mixed solution was added into the sample for 1 h in the dark and wet box at 37°C. Finally, the images were captured by a fluorescent microscopy (Olympus, Japan) with 200× magnification.
miRNA array and analysis
The total RNA samples were isolated from the intestinal tissues of rats (n = 5) using Trizol (Invitrogen, California, USA) and purified with RNeasy mini kit (QIAGEN, Germany) according to the manufacturer's instructions. RNA quality and quantity was measured by using nanodrop spectrophotometer (ND‐1000, Nanodrop Technologies, Delaware, USA), and RNA integrity was determined by gel electrophoresis. RNA labelling and array hybridization were carried out according to Exiqon's manual as follows: After quality control, the miRCURY™ Hy3™/Hy5™ Power labelling kit (Exiqon, Vedbaek, Denmark) was used for miRNA labelling according to the manufacturer's guideline. The Hy3™‐labelled samples were hybridized on the miRCURYTM LNA Array (v.18.0) (Exiqon) according to the array manual, after stopping the labelling procedure. The slide images were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA). Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged, and the miRNAs with the intensities ≥30 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the median normalization. After normalization, those miRNAs which were significantly differentially expressed between model and control groups were identified ‐ fold change>1.5 fold ; P < 0.05 . Finally, hierarchical clustering was performed to show distinguishable miRNA expression profiling among samples.
Validation of the differentially expressed miRNAs
The levels of some differentially expressed miRNAs of interest were examined by real‐time PCR assay. Total miRNA samples were obtained from IEC‐6 cells and rats using SanPrep Column microRNA Extraction Kit according to the manufacturer's protocol. The forward (F) and reverse (R) primers of miRNAs are given in Table S1. The relative levels of the miRNAs were determined based on a two‐step qRT‐PCR using miRNA First Strand cDNA Synthesis (Tailing Reaction) and MicroRNAs qPCR Kit (SYBR Green Method) (Sangon, Shanghai) microRNA Detection Kit according to the manufacturer's protocol. U6 was used for normalization of miRNAs.
Prediction of target genes
The validated miRNAs were screened by three databases (mirbase, mirdb and miranda), and the potential target gene tree was obtained. Each target gene must be retrieved in at least two databases.
Dual luciferase reporter transfection assay
The amplified primers were designed according to the http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1502 (Rattus) 3'UTR and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=848#2712 (Rattus) 3'UTR sequences information, and the 3'UTR sequences of MAPK13 and Sirt6 genes were amplified by PCR using C6 genomic DNA as the template. Then, the sequences were cloned into the pmiR‐RB‐REPORT™ double luciferase reporter vector (Reported fluorescence for hRluc, corrected fluorescence for hluc as the internal reference), and the constituted plasmids were titled as MAPK13‐WT and Sirt6‐WT. The miR‐351‐5p binding site mutations were introduced using the Multisite‐Quick change (Stratagene, USA) according to the Ribobio's protocol and cloned into the pMIR‐RB‐REPORTTM vector (Ribobio, China), and the reconstituted plasmids were called MAPK13‐mutant (MUT) and Sirt6‐MUT. The IEC‐6 cells were seeded in 24‐well plates at a density of 5 × 103 cells per mL for 24 h and then co‐transfected with miR‐mimics and MAPK13‐MUT or Sirt6‐MUT, miR‐mimics negative control (NC) and MAPK13‐MUT or Sirt6‐MUT, miR‐ mimics and MAPK13‐WT or Sirt6‐WT, or miR‐mimics NC and MAPK13‐WT or Sirt6‐WT. After 48 h transfection, the luciferase activities were assessed according to the Dual‐Luciferase Reporter Assay protocol using a Veritas 96‐well Microplate Luminometer with substrate dispenser (Promega, Wisconsin, USA).
Immunofluorescence assay
The paraffin tissue sections, dewaxed and incubated in a humidified box overnight at 4°C with the rabbit anti‐MAPK13 and anti‐Sirt6 with 1:100 dilution after blocking nonspecific protein binding, were incubated with Alexa fluorescein‐labelled secondary antibody for 1 h at 37°C and then stained with DAPI (5 μg·mL−1) for 10 min. Formalin‐fixed cells were assessed at the same as tissue sections described above. The images were captured by a fluorescence microscopy (Olympus, Japan) with 200× magnification.
Quantitative real‐time PCR assay
The total RNA samples were obtained from IEC‐6 cells and intestine tissues of rats and mice using TransZol according to the manufacturer's protocol. Each RNA sample was reverse transcribed into cDNA using the kit of TransStart Top Green qPCR SuperMix. The forward (F) and reverse (R) primers of RNA are given in Table S2. The CT value of genes among the data from each sample was normalized to β‐actin.
Western blotting assay
The total protein samples from IEC‐6 cells and the intestinal tissues of rats and mice were extracted according to the manufacturer's instructions. Proteins were loaded on SDS‐PAGE (10–15%), transferred to PVDF membranes (Millipore, USA), blocked with 5% dried skim milk (Boster Biological Technology, China) for 2 h and incubated with primary antibodies (Table S3) overnight at 4°C. Then, the membranes were individually incubated for 2 h at room temperature with HRP‐conjugated antibody at a 1:5000 dilution. Enhanced chemiluminescence method and ChemiDoc XRS (Bio‐Rad, USA) were used to detect the expression levels of the proteins. Intensity values, expressed as the relative protein expression, were normalized to β‐actin.
Mimic and inhibitor transfection of miR‐351‐5p in vitro
The IEC‐6 cells were seeded in 24‐well plates at a density of 5 × 103 cells per mL. The miR‐351‐5p mimic or inhibitor (Ribobio, China) was tested according to the manufacturer's protocol. MiR‐351‐5p mimic (8 nM, final concentration) or inhibitor (25 nM, final concentration) in 50 μL of Opti‐MEM was mixed with 1 μL of lipofectamine in 50 μL of Opti‐MEM (Ambion, USA). The IEC‐6 cells were transfected with the complexes and maintained for 6 h, fed with original medium and cultured in normoxic conditions for another 24 h. Then, H/R (hypoxia for 2 h and reoxygenation for 1 h)‐induced injury was carried out. An equal concentration of scrambled non‐targeting control (mimic or inhibitor NC) was used for non‐sequence‐specific effect in the experiments. A range of variables, including cell viability, mRNA levels of inflammatory factors, ROS level, apoptosis and the expression levels of miR‐351‐5p and some proteins associated with the signals were assayed.
Agomir and antagomir transfection of miR‐351‐5p in mice
C57BL/6 mice were randomly divided into six groups (n = 5): Control group, Agomir NC group, Antagomir NC group, II/R group, II/R + Agomir group, and II/R + Antagomir group. Agomir (20 nmol) and antagomir (50 nmol) were injected into the tail vein and then subjected to ischaemia for 45 min and reperfusion for 90 min. Agomir‐NC (agomir NC) and antagomir‐NC (antagomir NC) were also injected into tail vein. After II/R, the mice were killed and the intestinal tissues were obtained. Samples were examined histopathologically and mRNA levels of inflammatory factors, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2789 level, apoptosis and the expression levels of miR‐351‐5p and some proteins associated with the signals were determined.
Data and statistical analysis
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2018). Data are expressed as the mean ± SD. GraphPad Prism 6.01 software (Graphpad Software, Inc, CA, USA) was used to handle these data and only when a minimum of n = 5 independent samples was acquired. Among them, some data were normalized for controlling unwanted sources of variation. Statistical analysis was performed with one‐way ANOVA followed by Tukey's post hoc test when comparing multiple independent groups, and when comparing two different groups, the unpaired t‐test was carried out. Post hoc tests were run only if F achieved P < 0.05 and there was no significant variance inhomogeneity. Statistical significance was considered to be P < 0.05.
Materials
Tissue Protein Extraction Kit was purchased from KEYGEN Biotech. Co., Ltd. (Nanjing, China). ROS assay kit, bicinchoninic acid protein assay kit and cell lysis buffer kit were obtained from Beyotime Institute of Biotechnology (Jiangsu, China). Malondialdehyde (MDA), myeloperoxidase (MPO) and total SOD (T‐SOD) assay kits were obtained from Nanjing Jiancheng Institute of Biotechnology (Nanjing, China). 3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT) was provided by Roche Diagnostics (Basel, Switzerland). DAPI was obtained from Sigma (St. Louis, MO, USA). Double‐Luciferase Reporter Assay Kit was purchased from Promega Biotech Co., Ltd. (Beijing, China). Lipofectamin2000, TransZolTM, TransScript® All‐in‐One First‐Strand cDNA Synthesis SuperMix for qPCR (One‐Step gDNA Removal), TransStart® Top Green qPCR SuperMix were purchased from Beijing TransGen Biotech Co., Ltd. (Beijing, China). SanPrep Column MicroRNA Mini‐Preps Kit, MicroRNA First Strand cDNA Synthesis Kit and MicroRNAs Quantitation PCR Kit were purchased from Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The plasmids were purchased from RiboBio. Co., Ltd. (Guangzhou, China).
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander et al., 2017a, 2017b).
Results
H/R and II/R caused damage in vivo and in vitro
The damage caused to IEC‐6 cells after exposure to the H/R procedures with different reperfusion times (30, 60, 180, 360 and 720 min) is shown in Figure 1A. Apoptosis was increased and the morphology of IEC‐6 cells was significantly changed, with the most marked changes at 60 min reoxygenation. We also assessed the acute intestinal injury in rats and mice after different reperfusion times at 45, 90, 180, 360 and 720 min (Figure 1B). After 90 min of reperfusion, a large number of inflammatory cells and necrosis were found in animals subjectd to II/R, compared with control groups. The intestinal damage in rats and mice was at its peak after reperfusion for 90 min based on Chiu's score and H&E staining.
Figure 1.
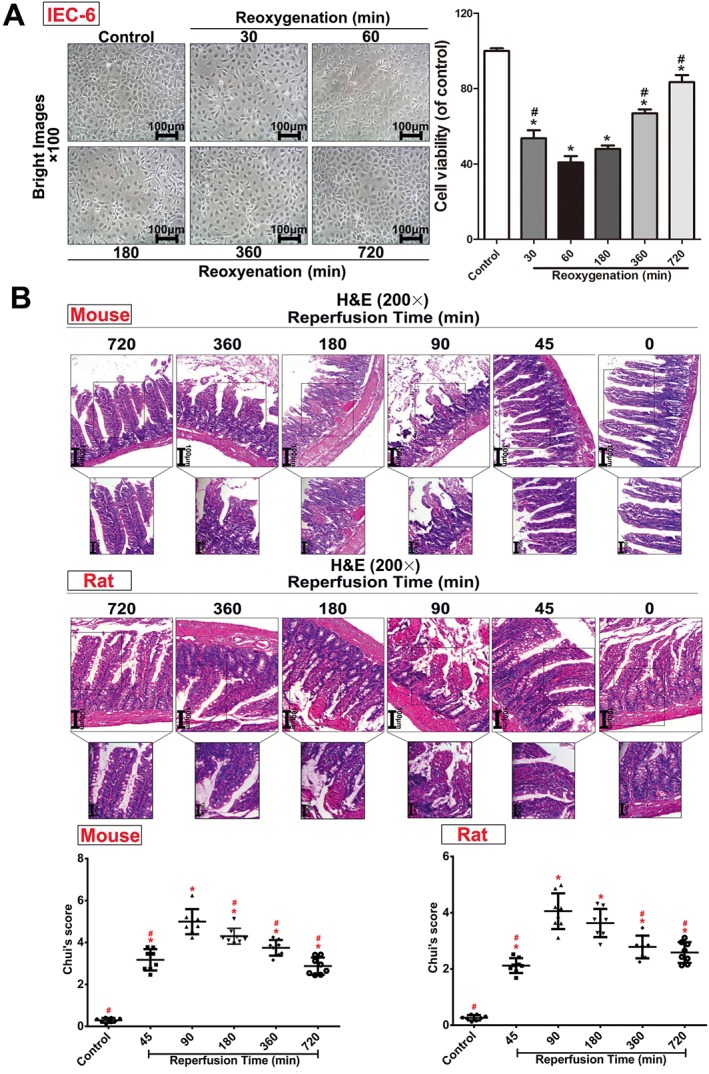
H/R injury in IEC‐6 cells and II/R injury in rats and mice. (A) Cellular morphology and cell viabilities of IEC‐6 cells after H/R injury (n = 5). (B) II/R injury in rats and mice assessed by H&E staining (200× magnification) and Chui's score (n = 8). All data are expressed as the mean ± SD. * P < 0.05, significantly different from control group; # P < 0.05, significantly different from the 60 min reoxygenation or 90 min reperfusion group.
H/R and II/R‐caused oxidative stress, inflammation and apoptosis
As shown in Figure 2A–B, levels of MDA and MPO in intestinal mucosa of rats and mice were increased from 45 to 720 min of reperfusion compared with control groups and markedly decreased from 180 to 720 min of reperfusion compared with the peak after 90 min of reperfusion. The levels of T‐SOD were decreased from 45 to 720 min of reperfusion compared with control groups and markedly increased from 180 to 720 min of reperfusion compared with 90 min of reperfusion. As shown in Figure 2C, the mRNA levels of http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=4974, TNF‐α and intercellular cell adhesion molecule‐1 (http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=6757) were significantly increased from 45 to 720 min of reperfusion compared with control groups both in vitro and in vivo and then achieved the highest levels at 60 min of reoxygenation in IEC‐6 cells and at 90 min of reperfusion in animals. The numbers of apoptotic cells reached the highest levels at 60 min of reoxygenation in vitro and at 90 min of reperfusion in vivo (Figure 2D).
Figure 2.
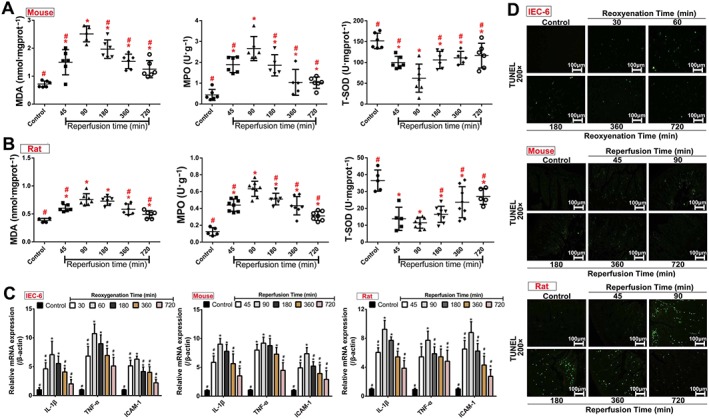
Oxidative stress, inflammation and cell apoptosis after H/R and II/R injury in vivo and in vitro. (A–B) The levels of MDA, MPO and T‐SOD in intestinal tissues of rats and mice after II/R injury (n = 8). (C) The mRNA levels of IL‐1β, TNF‐α and ICAM‐1 in vitro and in vivo (n = 5). (D) Cell apoptosis after H/R and II/R injury in vivo and in vitro based on TUNEL assay (n = 5). All data are expressed as the mean ± SD. * P < 0.05, significantly different from control group; # P < 0.05, significantly different from the 60 min reoxygenation or 90 min reperfusion group.
Identification of the differentially expressed miRNAs
The heat map (Figure 3A) showed the results of a two‐way hierarchical clustering of the samples, and a total of 30 differentially expressed miRNAs were found by the microarray assay. The details of the up‐regulated and down‐regulated miRNAs are listed in Tables S4–S5. Among them, 18 up‐regulated miRNAs (miR‐23a‐5p, miR‐300‐5p, miR‐21‐3p, miR‐344b‐1‐3p, miR‐351‐5p, miR‐874‐3p, miR‐144‐3p, miR‐3572, miR‐27a‐5p, miR‐183‐3p, miR‐294, miR‐451‐5p, miR‐483‐3p, miR‐883‐5p, miR‐125b‐2‐3p, miR‐3573‐3p, miR‐485‐3p and miR‐490‐5p) and 12 down‐regulated miRNAs (miR‐181c‐5p, miR‐187‐3p, miR‐532‐5p, miR‐194‐5p, miR‐203a‐3p, miR‐17‐1‐3p, miR‐192‐5p, miR‐28‐3p, miR‐140‐5p, miR‐142‐3p, miR‐29c‐3p and miR‐362‐3p) with clear statistical significance (P<0.05 different from control values) were identified. Furthermore, nine miRNAs ‐ miR‐144‐3p, miR‐3572, miR‐27a‐5p, miR‐142‐3p, miR‐23a‐5p, miR‐21‐3p, miR‐294, miR‐351‐5p and miR‐362‐3p ‐ associated with oxidative stress, inflammation and apoptosis (Romay et al., 2015; Wang et al., 2015; da Silva et al., 2016; Dixit et al., 2016; Brea et al., 2017; Chen et al., 2017; Li et al., 2018) were selected for validation by real‐time PCR assay in IEC‐6 cells and rats. These results, shown in Figure 3B, were consistent with the results of the microarray assays.
Figure 3.
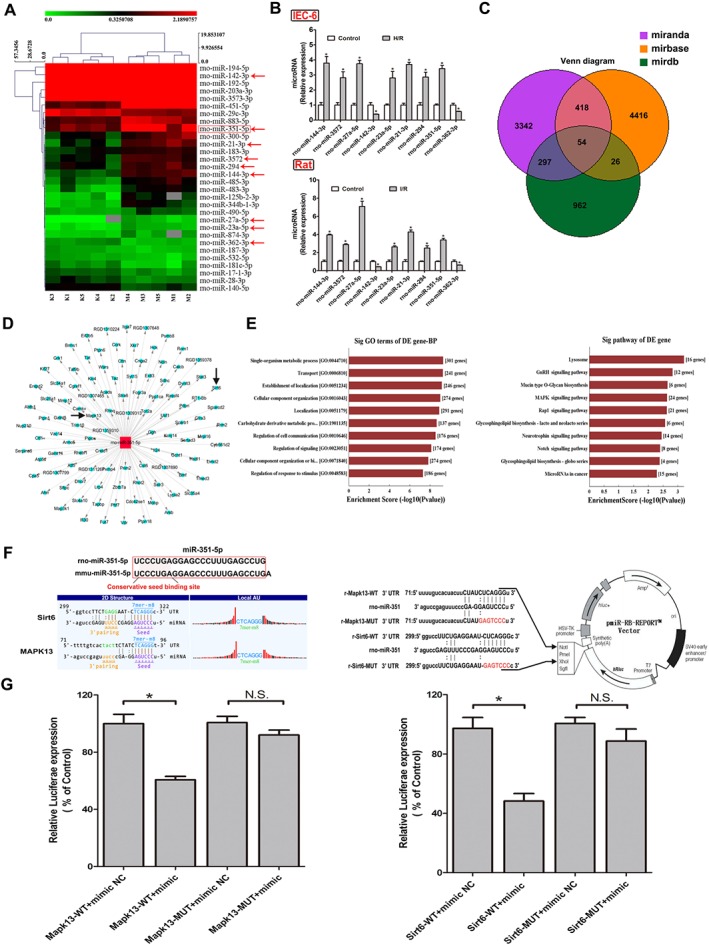
The differentially expressed miRNAs from intestinal tissue of rats after II/R injury based on microarrays assay. (A) Hierarchical clustering analysis of the differentially expressed miRNAs (changes >1.5‐fold and P < 0.05) between II/R and control groups in rats (n = 5). (B) Validation of the nine differentially expressed miRNAs in rats and IEC‐6 cells by real‐time PCR assay (n = 5). (C) Diagram of the search for the target genes of miRNAs in three databases. (D) The target genes of miR‐351‐5p screened from the database. (E) Biological processes and signal pathways of the target genes of miR‐351‐5p. (F) Diagram of miR‐351‐5p conservative seed binding sites on the 3′UTRs of the target genes (Sirt6 and MAPK13) and the construction diagram of double luciferase reporter genes (The mutant sequences used in double luciferase reporter genes are in red). (G) The relative luciferase expression with Sirt6 3′UTR or MAPK13 3′UTR after co‐transfection with miR‐351‐5p mimic or NC in IEC‐6 cells (n = 5). All data are expressed as the mean ± SD. * P < 0.05, significantly different from control group.
Identification of the miRNA to be studied
The target genes of the nine validated miRNAs were predicted, and the results are shown in Figure 3C. The three circular areas are the numbers of the target genes of the nine miRNAs, and the candidate target genes of miR‐351‐5p are shown in the tree diagram (Figure 3D). In addition, we also found that the target genes of miR‐351‐5p should be MAPK13 and Sirt6 (Figure 3D), which are known to play important roles in regulating oxidant stress, inflammation and apoptosis. The miR‐315‐5p has been confirmed as a novel element targeting PTEN to control the pro‐inflammatory environment in angiotensin II‐treated cells (da Silva et al., 2016). However, there are no publications of the effects of miR‐315‐5p in II/R injury. Therefore, miR‐351‐5p was chosen as the target miRNA in the present work. Meanwhile, the candidate target genes of miR‐351‐5p were subjected to gene ontology (GO) and pathway assays. As shown in Figure 3E, the gene‐Biology Process showed the target genes of miR‐351‐5p to be associated with many biological processes and signal pathways able to alter cell proliferation, apoptosis, oxidative stress and inflammation. Therefore, the roles of miR‐351‐5p in oxidative stress, apoptosis and inflammation induced by II/R were assessed.
MAPK13 and Sirt6 are the target genes of miR‐351‐5p
The homology of miR‐351‐5p genes of rats and mice reached to 96% (rno‐miR‐351‐5p: UCCCUGAGGAGCCCUUUGAGCC UGA; mmu‐miR‐351‐5p: UCCCUGAGGAGCC‐CUUUGAGCCUG), and the construction schematic diagram of MAPK13 and Sirt6 pmiR‐RB‐REPORT™ double luciferase reporter vector is shown in Figure 3F. Double luciferase reporter assay results shown in Figure 3G demonstrated that the relative luciferase expression of MAPK13‐WT + mimic group was significantly decreased compared with MAPK13‐WT + mimic NC group. However, compared with MAPK13‐MUT + mimic NC group, the relative luciferase expression of MAPK13‐WUT+ mimic group had no obvious change. The double luciferase reporter test of Sirt6 showed the same results as MAPK13 (Figure 3G). Therefore, MAPK13 and Sirt6 showed specific binding sites with miR‐351‐5p, confirming that they were target genes of miR‐351‐5p.
miR‐351‐5p regulates MAPK13 signal to adjust inflammation
The expression levels of miR‐351‐5p in vivo and in vitro are shown in Figure 4A. In IEC‐6 cells, the levels of miR‐351‐5p were strikingly increased at 30, 60 and 180 min of reoxygenation and reached a peak value at 60 min of reoxygenation. In mice and rats, the levels of miR‐351‐5p were significantly increased at 45–720 min of reperfusion and reached the highest levels at 90 min of reperfusion. As shown in Figure 4B, the levels of MAPK13 were clearly decreased in experimental groups, compared with control groups, and the lowest values were found at 60 min of reoxygenation in IEC‐6 cells and at 90 min of reperfusion in animals, based on immunofluorescence and Western blotting assays. In addition, using as substrate the protein polycystic kidney disease 1 (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1489), the activity of the MAPK13 signalling pathway was measured and the results showed that levels of p‐PKD1 (Ser916) or (Ser744/748) and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=996 were markedly increased in model groups compared with control groups with the peak values at 60 min of reoxygenation in IEC‐6 cells, and at 90 min of reperfusion in animals (Figure 4C). Therefore, miR‐351‐5p could regulate inflammation during II/R injury by targeting MAPK13.
Figure 4.

miR‐351‐5p aggravates inflammatory response by inhibiting MAPK13. (A) The expression of miR‐351‐5p in IEC‐6 cells, rats and mice. (B) The levels of MAPK13 in IEC‐6 cells, rats and mice based on immunofluorescence staining (200× magnification). (C) Levels of the proteins, MAPK13, p‐PKD1 (Ser916) and (Ser744/748), PKD1 and NF‐κB (P65) in vitro and in vivo. All data are expressed as the mean ± SD (n = 5).* P < 0.05, significantly different from control group, # P < 0.05, significantly different from the 60 min reoxygenation or 90 min reperfusion group.
miR‐351‐5p regulates Sirt6 signalling to alter oxidative stress
As shown in Figure 5A, levels of Sirt6 were clearly decreased in model groups compared with control groups. We also measured the activity of the Sirt6/AMP‐activated protein kinase (http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1540)/forkhead box O3 (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=974) signalling pathway. The levels of p‐AMPK, manganese‐dependent SOD (MnSOD) and http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=2979 were significantly decreased in model groups compared with control groups, with the lowest values after reoxygenation for 60 or 180 min in IEC‐6 cells and at reperfusion for 90 or 180 min in animals. The levels of p‐FoxO3α were markedly increased in model groups compared with control groups with the peak values at reoxygenation. for 60 min in IEC‐6 cells and at reperfusion for 90 min in animals (Figure 5B). Thus, miR‐351‐5p could regulate the extent of oxidative stress during II/R injury by targeting Sirt6.
Figure 5.

miR‐351‐5p aggravates oxidative stress by inhibiting Sirt6. (A) The expression of Sirt6 in IEC‐6 cells, rats and mice based on immunofluorescence staining (200× magnification, n = 5). (B) Levels of the proteins, Sirt6, AMPK, p‐AMPK, FoxO3α, p‐FoxO3α, MnSOD and catalase (CAT) in vitro and in vivo (n = 5). Data are presented as mean ± SD (n = 5).* P < 0.05, significantly different from control group. # P < 0.05, significantly different from 60 min reoxygenation or 90 min reperfusion group.
miR‐351‐5p regulates Sirt6 signalling to alter apoptosis
As shown in Figure 6A–C, the activity of the Sirt6/Bcl2‐associated X (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=910) signalling pathway was assessed. Levels of http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=910, http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1318, apoptotic peptidase activating factor 1, cleaved‐http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1619 and cleaved‐http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1625 were markedly increased in model groups compared with control groups with the peak values at reoxygenation. for 60 min in IEC‐6 cells and at reperfusion for 90 min in animals. Therefore, miR‐351‐5p was able to regulate apoptosis during II/R injury, by targeting Sirt6.
Figure 6.

miR‐351‐5p aggravates apoptosis by inhibiting Sirt6. (A) The expression of the proteins, Bax, cytochrome c (Cytc), apoptotic peptidase activating factor 1 (Apaf‐1), cleaved‐caspase 3 and cleaved‐caspase 9 in IEC‐6 cells. (B) Levels of the proteins Bax, Cytc, Apaf‐1, cleaved‐caspase 3 and cleaved‐caspase 9 in mice. (C) The levels of the proteins Bax, Cytc, Apaf‐1, cleaved‐caspase 3 and cleaved‐caspase 9 in rats. Data are presented as mean ± SD (n = 5).* P < 0.05, significantly different from control group. # P < 0.05, significantly different from 60 min reoxygenation or 90 min reperfusion group.
Effects of over‐expression or blockade of miR‐351‐5p in vitro
The results obtained so far prompted us to explore whether miR‐351‐5p aggravated II/R injury through repressing MAPK13 and Sirt6 signals, in our model system. We, therefore, tested the effects of over‐expression or blockade of miR‐351‐5p in vitro. The results showed that the cell viability was significantly decreased after transfection with mimic compared with H/R group and the cell damage was clearly suppressed after transfection with inhibitor, compared with H/R group (Figure 7A). The expression level of miR‐351‐5p was markedly increased after transfection with mimic and decreased after transfection with inhibitor compared with H/R group (Figure 7B). In addition, the extent of oxidative stress, inflammation and apoptosis in IEC‐6 cells was increased after transfection with mimic and reduced after transfection with inhibitor, compared with H/R group (Figure 7C–E). Levels of the proteins associated with MAPK13 and Sirt6 signalling are shown in Figure 7F–G. After transfection with mimic, the levels of MAPK13, Sirt6 and MnSOD were significantly decreased, and levels of NF‐κB (p65) and cleaved‐caspase 3 were increased, compared with H/R group. The levels of MAPK13, Sirt6 and MnSOD were markedly increased, and the expression levels of NF‐κB (p65) and cleaved‐caspase 3 were significantly decreased after transfection with inhibitor, compared with H/R group.
Figure 7.
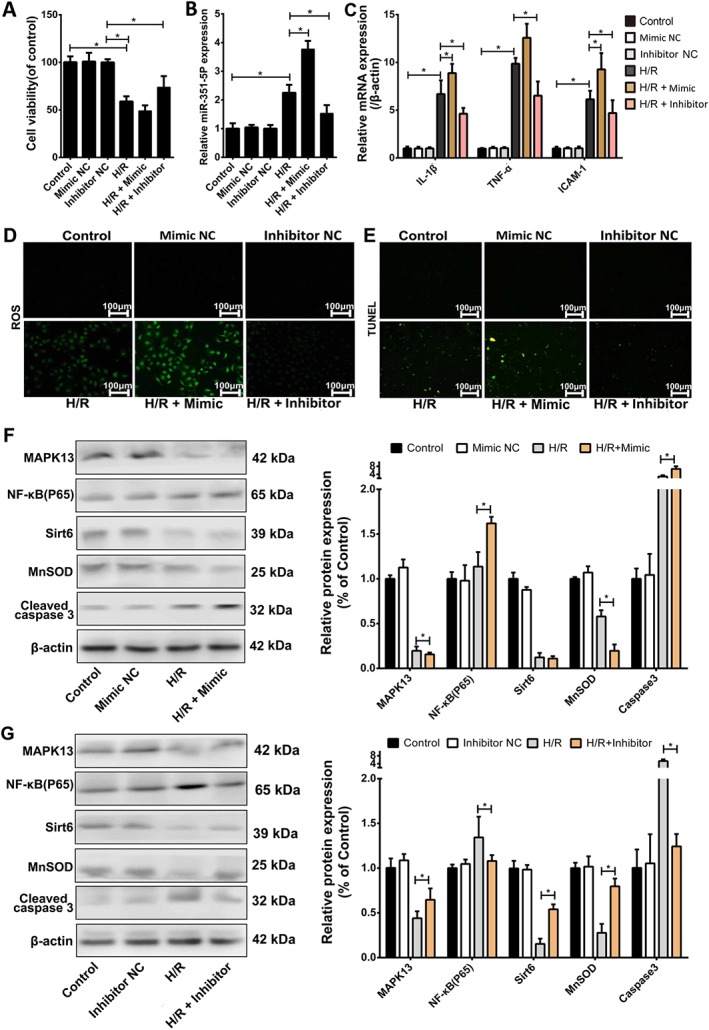
Effects of over‐expression or blockade of miR‐351‐5p on H/R injury in IEC‐6 cells. (A) Cell viabilities of IEC‐6 cells with or without transfection of miR‐351‐5p mimic and inhibitor in each group (n = 5). (B) Levels of miR‐351‐5p in IEC‐6 cells with or without transfection of miR‐351‐5p mimic and inhibitor in each group (n = 5). (C) The mRNA levels of IL‐1β, TNF‐α and ICAM‐1 in IEC‐6 cells after mimic or inhibitor of miR‐351‐5p (n = 5). (D–E) The levels of ROS and cell apoptosis in IEC‐6 cells after mimic or inhibitor of miR‐351‐5p (n = 5). (F–G) Levels of the proteins associated with miR‐351‐5p/MAPK13 and miR‐351‐5p/Sirt6 signal pathways after over‐expression or blockade of miR‐351‐5p in vitro (n = 5). All the data are expressed as the mean ± SD. * P < 0.05, significantly different as indicated.
Effects of over‐expression or blockade of miR‐351‐5p in vivo
As shown in Figure 8A, the intestinal injury was significantly reduced after transfection with antagomir and obviously increased after transfection with agomir based on the results of H&E staining and Chiu' score in mice. The level of miR‐351‐5p was markedly increased after transfection with agomir and decreased after transfection with antagomir compared with II/R group (Figure 8B). The degrees of apoptosis, inflammation and oxidative stress were increased after transfection with agomir and reduced after transfection with antagomir compared with II/R group (Figure 8C–E). Levels of the proteins associated with MAPK13 and Sirt6 signals were similar to the results in IEC‐6 cells (Figure 8F–G). In summary, our results demonstrated that miR‐351‐5p could aggravate II/R injury through inhibition of MAPK13 and Sirt6 signals in vivo.
Figure 8.
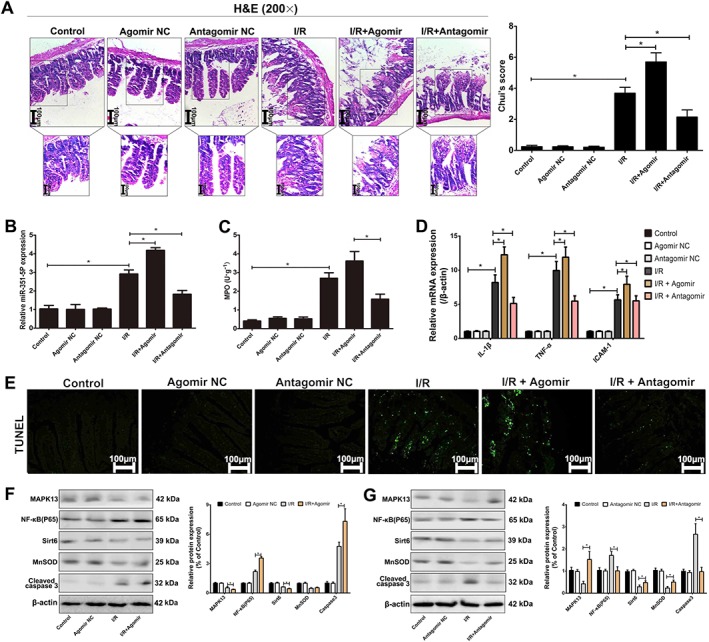
Effects of agomir or antagomir of miR‐351‐5p on II/R injury in mice. (A) Acute intestinal damage in mice assessed by H&E staining (200× magnification). (B) The expression of miR‐351‐5p in mice with or without transfection of miR‐351‐5p agomir and antagomir in each group (n = 5). (C) The levels of MPO in intestine with or without transfection of miR‐351‐5p agomir and antagomir in each group (n = 5). (D) The mRNA for IL‐1β, TNF‐α and ICAM‐1 in mice after agomir or antagomir of miR‐351‐5p (n = 5). (E) Levels of cell apoptosis in mice after agomir or antagomir of miR‐351‐5p. (F–G) Expression of the proteins associated with miR‐351‐5p/MAPK13 and miR‐351‐5p/Sirt6 signal pathways after agomir or antagomir of miR‐351‐5p in mice (n = 5). All the data are expressed as the mean ± SD. * P < 0.05, significantly different as indicated.
Discussion
II/R can cause SIRS, distant organ damage and MODS. However, the molecular mechanisms of II/R injury are not fully understood. As far as we know, the extent of II/R injury is largely dependent on the reperfusion phase (Emily and Sherry, 2014; Michael and Sherry, 2015), and our results also indicated that the injury reached its peak at 60 min of reoxygenation in IEC‐6 cells and at 90 min of reperfusion in rats and mice. The damage was slightly reduced when the times of reoxygenation and reperfusion were longer than 60 and 90 min respectively. As mentioned earlier, oxidative stress, inflammation and apoptosis play important roles in the occurrence and development of intestinal I/R (Pope et al., 2010; Pantazi et al., 2013; Taha et al., 2014). ROS and TUNEL staining are often used to evaluate oxidative stress and apoptosis. MDA and MPO are regularly considered as indices of oxidative stress. Levels of T‐SOD represent the antioxidative ability of the whole body (Wang et al., 2013). Pro‐inflammatory chemokines and cytokines including IL‐1β, TNF‐α and ICAM‐1 reflect inflammatory response (Liu et al., 2015). In the present work, the conditions of oxidative stress, inflammation and apoptosis were clearly found in H/R injury in vitro and II/R damage in vivo.
In the past decade, miRNAs have emerged as significant regulators of several pathologies and have been reported to participate in the process of II/R injury. In this study, miR‐351‐5p was selected as the target miRNA to investigate the molecular mechanisms and drug targets associated with II/R injury based on microarray assays. Our results showed that miR‐351‐5p in the model groups was notably decreased, compared with the control groups in vitro and in vivo. The GO and Pathway assays revealed the basic biological functions of miR‐351‐5p. In addition, we found that MAPK13 and Sirt6 showed putative binding sites with miR‐351‐5p based on Dual‐Luciferase Reporter assay, which confirmed that MAPK13 and Sirt6 are the target genes of miR‐351‐5p. Moreover, the protein levels of MAPK13 and Sirt6 were significantly suppressed by miR‐351‐5p. Interestingly, some studies have demonstrated that MAPK13 and Sirt6 can regulate oxidant stress, inflammation and apoptosis (Risco et al., 2012; Ikemura et al., 2013; Zhong et al., 2014; Zur et al., 2015). Therefore, miR‐351‐5p may play important roles in regulating oxidant stress, inflammation and apoptosis, thus controlling II/R damage, by targeting MAPK13 and Sirt6.
The p38 MAPKs contain four family members including p38α (MAPK14), p38β (MAPK11), p38γ (Mapk12) and p38δ (MAPK13). Among them, MAPK13 plays a key role in diabetes through regulating insulin secretion and survival of pancreatic beta cells (Cuenda and Rousseau, 2007). Furthermore, studies in knockout mice have shown that MAPK13 contributes to chronic inflammatory conditions (Alevy et al., 2012). In addition, MAPK13 can inhibit PKD1 activation and decrease NF‐κB transcription to release inflammatory factors (Schindler et al., 2009; Song et al., 2009; Ittner et al., 2012). To further investigate whether miR‐351‐5p aggravated II/R injury through repressing the MAPK13/PKD1 signalling pathway, we used miR‐351‐5p knockdown and over‐expression, in vivo and in vitro. The levels of inflammation were decreased after transfection with miR‐351‐5p antagomir compared with H/R group in cells and II/R group in mice, and the effects were achieved by reducing the expression levels of miR‐351‐5p to activate MAPK13 signal pathway. Thus, miR‐351‐5p regulated inflammation during II/R injury by targeting MAPK13.
Sirt6, a member of the mammalian sirtuins family, can be considered as a NAD+‐dependent deacylase and mono‐ADP‐ribosyltransferase of both acetyl groups and long‐chain fatty acyl groups (Lerrer et al., 2015). Sirt6 plays important roles in several cellular processes including transcription, telomere integrity, genome instability, inflammation, DNA repair, metabolism, ageing and cancer (Wang et al., 2017). In addition, Sirt6 can reduce apoptosis by binding to a specific site of Bax promoter and activate FoxO3α directly or through AMPK activation to increase the antioxidant capacity of the body (Ran et al., 2016; Wang et al., 2016). To further investigate whether miR‐351‐5p aggravated II/R injury through repressing Sirt6/AMPK/FoxO3α and Sirt6/Bax signalling, we used miR‐351‐5p knockdown and over‐expression in vivo and in vitro. The results showed that the levels of oxidant stress and cell apoptosis were decreased after transfection with miR‐351‐5p antagomir compared with the H/R group in cells and II/R group in mice, and these effects were produced by reducing the levels of miR‐351‐5p to activate the Sirt6 signalling pathway. Thus, miR‐351‐5p regulated oxidative stress and cell apoptosis in II/R injury by targeting Sirt6.
In summary, as shown in Figure S1, we found that miR‐351‐5p aggravated II/R injury by promoting oxidative stress, inflammation and apoptosis in the intestinal mucosa, via targeting MAPK13 and Sirt6. Furthermore, this study highlights the possible clinical relevance of miR‐351‐5p and will allow the potential of this molecule to be fully assessed. In the near future, miR‐351‐5p may be tested as a novel biomarker or a promising target for II/R injury in clinical trials.
Author contributions
Y.P.H. and J.Y.P. designed the experiments and wrote the manuscript. Y.P.H., X.F.T., X.H. and L.N.X. performed the animal and cell experiments. L.H.Y., H.J.S. and Y.Q. performed the quantitative real‐time PCR and Western blotting assays. Y.P.H. and Y.W.X. performed the gene transfection experiments. Y.P.H. and J.Y.P. edited the manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Declaration of transparency and scientific rigour
This http://onlinelibrary.wiley.com/doi/10.1111/bph.13405/abstract acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Table S1 The primers sequences of miRNAs in the present work.
Table S2 The primer sequencess used for real‐time PCR assay in the present work.
Table S3 The information of the antibodies used in the present work.
Table S4 The up‐regulated expression of miRNAs in II/R group compare with control.
Table S5 The down‐regulated expression of miRNAs in II/R group compare with control.
Figure S1 Proposed signaling mechanism of miR‐351‐5p on II/R‐induced intestinal injury. miR‐351‐5p was selected as the target miRNA by microarrays assay, and Sirt6 and MAPK13 were the target genes of miR‐351‐5p. II/R significantly down‐regulated the expression levels of Sirt6 and MAPK13 by increasing the level of miR‐351‐5p, up‐regulated the levels of p‐PKD1, NF‐κB(p65) and inflammatory factors including IL‐1β, IL‐6, TNF‐α and ICAM‐1, increased p‐FoxO3αlevel via inhibiting AMPK, reduced MnSOD and CAT levels, and increased the levels of Bax, Cytc, Apaf‐1, Cleaved‐ caspase3 and Cleaved‐caspase9.MicroRNA‐351‐5p aggravated intestinal ischemia/ reperfusion injury by promoting intestinal mucosal oxidative stress, inflammation and apoptosis via targeting MAPK13 and Sirt6.
Acknowledgements
This work was financially supported by the Key Research and Development Project of Liaoning Province (2017225090), the Project of Liaoning BaiQianWan Talents Program (2015‐65), the Special Grant for Translational Medicine, Dalian Medical University (2015004) and the Basic Scientific Research Projects of Liaoning University (No. LF2017010).
Hu, Y. , Tao, X. , Han, X. , Xu, L. , Yin, L. , Sun, H. , Qi, Y. , Xu, Y. , and Peng, J. (2018) MicroRNA‐351‐5p aggravates intestinal ischaemia/reperfusion injury through the targeting of MAPK13 and Sirtuin‐6. British Journal of Pharmacology, 175: 3594–3609. 10.1111/bph.14428.
References
- Alevy YG, Patel AC, Romero AG, Patel DA, Tucker J, Roswit WT et al (2012). IL‐13‐induced airway mucus production is attenuated by MAPK13 inhibition. J Clin Invest 122: 4555–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SP, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E et al (2017a). The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 174 (Suppl 1): S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SP, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD et al (2017b). The Concise Guide to PHARMACOLOGY 2017/18: Other proteins. Br J Pharmacol 174 (Suppl 1): S1–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga J, Prats P, Remirez D, Gonzalez R, Lopez‐Saura P, Aguiar J et al (2002). Prophylactic use of epidermal growth factor reduces ischemia/reperfusion intestinal damage. Am J Pathol 161: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brea R, Motiño O, Francés D, García‐Monzón C, Vargas J, Fernández‐Velasco M et al (2017). PGE2 induces apoptosis of hepatic stellate cells and attenuates liver fibrosis in mice by downregulating miR‐23a‐5p and miR‐28a‐5p. Biochim Biophys Acta 1864: 325–337. [DOI] [PubMed] [Google Scholar]
- Chassin C, Hempel C, Stockinger S, Dupont A, Kübler JF, Wedemeyer J et al (2012). MicroRNA‐146a‐mediated downregulation of IRAK1 protects mouse and human small intestine against ischemia/reperfusion injury. EMBO Mol Med 4: 1308–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou X, Qiao J, Bao A (2017). MiR‐142‐3p overexpression increases chemo‐sensitivity of NSCLC by inhibiting HMGB1‐mediated autophagy. Cell Physiol Biochem 41: 1370–1382. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S (2007). p38 MAP‐kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773: 1358–1375. [DOI] [PubMed] [Google Scholar]
- Curtis MJ, Alexander S, Cirino G, Docherty JR, George CH, Giembycz MA et al (2018). Experimental design and analysis and their reporting II: updated and simplified guidance for authors and peer reviewers. Brit J Pharmacol 175: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva W, dos Santos RA, Moraes KC (2016). Mir‐351‐5p contributes to the establishment of a pro‐inflammatory environment in the H9c2 cell line by repressing PTEN expression. Mol Cell Biochem 411: 363–371. [DOI] [PubMed] [Google Scholar]
- Deitch EA, Xu D, Kaise VL (2006). Role of the gut in the development of injury and shock induced SIRS and MODS: the gut‐lymph hypothesis, a review. Front Biosci 11: 520–528. [DOI] [PubMed] [Google Scholar]
- Dixit AK, Sarver AE, Yuan Z, George J, Barlass U, Cheema H et al (2016). Comprehensive analysis of microRNA signature of mouse pancreatic acini: overexpression of miR‐21‐3p in acute pancreatitis. Am J Physiol Gastrointest Liver Physiol 311: G974–G980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi AJ, Wu R, Nguyen E, Higuchi S, Wang H, Krishnasastry K et al (2007). Adrenomedullin and adrenomedullin binding protein‐1 prevent acute lung injury after gut ischemia‐reperfusion. J Am Coll Surg 205: 284–293. [DOI] [PubMed] [Google Scholar]
- Emily AS, Sherry DF (2014). Membrane lipid interactions in intestinal ischemia/reperfusion‐induced injury. Clin Immunol 153: 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith JR, Perez‐Chanona E, Yadav PN, Whistler J, Roth B, Jobin C (2012). Sa1939 intestinal epithelial cell (IEC)‐derived Mu‐Opioid signaling is protective against ischemia reperfusion (I/R) induced injury. Gastroenterology 142: 351–364. [Google Scholar]
- Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S et al (2018). The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura K, Yamamoto M, Miyazaki S, Mizutani H, Iwamoto T, Okuda M (2013). Microrna‐145 post‐transcriptionally regulates the expression and function of P‐glycoprotein in intestinal epithelial cells. Mol Pharmacol 83: 399–405. [DOI] [PubMed] [Google Scholar]
- Ittner A, Block H, Reichel CA, Varjosalo M, Gehart H, Sumara G et al (2012). Regulation of PTEN activity by p38δ‐PKD1 signaling in neutrophils confers inflammatory responses in the lung. J Exp Med 209: 2229–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2010). Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol 160: 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerrer B, Gertler AA, Cohen HY (2015). The complex role of SIRT6 in carcinogenesis. Carcinogenesis 37: 108–118. [DOI] [PubMed] [Google Scholar]
- Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H et al (2013). Exosomes mediate the cell‐to‐cell transmission of IFN‐α‐induced antiviral activity. Nat Immunol 14: 793–803. [DOI] [PubMed] [Google Scholar]
- Li S, Lee C, Song J, Lu C, Liu J, Cui Y et al (2017). Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget 8: 48145–48156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao Y, Cheng M, Qiao Y, Wang Y, Xiong W et al (2018). Suppression of microRNA‐144‐3p attenuates oxygen‐glucose deprivation/reoxygenation‐induced neuronal injury by promoting Brg1/Nrf2/ARE signaling. J Biochem Mol Toxicol 32: e22044. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhu C, Wang G, Xu R, Zhu Y (2015). Higenamine regulates Nrf2‐HO‐1‐Hmgb1 axis and attenuates intestinal ischemia–reperfusion injury in mice. Inflamm Res 64: 395–403. [DOI] [PubMed] [Google Scholar]
- Liu Z, Jiang J, Yang Q, Xiong Y, Zou D, Yang C et al (2016). MicroRNA‐682‐mediated downregulation of PTEN in intestinal epithelial cells ameliorates intestinal ischemia‐reperfusion injury. Cell Death Dis 7: e2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus M, Ernesto PC, Christian J (2013). Epithelial cell‐specific MyD88 signaling mediates ischemia/reperfusion‐induced intestinal injury independent of microbial status. Inflamm Bowel Dis 19: 2857–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JC, Lilley E (2015). Implementing guidelines on reporting research using animals (ARRIVE etc.): new requirements for publication in BJP. Br J Pharmacol 172: 3189–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael RP, Sherry DF (2015). TLR2 modulates antibodies required for intestinal ischemia/reperfusion‐induced damage and inflammation. J Immunol 194: 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazi E, Zaouali MA, Bejaoui M, Folch‐Puy E, Ben Abdennebi H, Roselló‐Catafau J (2013). Role of sirtuins in ischemia‐reperfusion injury. World J Gastroenterol 19: 7594–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope MR, Bukovnik U, Tomich JM, Fleming SD (2012). Small beta2‐glycoprotein I peptides protect from intestinal ischemia reperfusion injury. J Immunol 189: 5047–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope MR, Hoffman SM, Tomlinson S, Fleming SD (2010). Complement regulates TLR4‐mediated inflammatory responses during intestinal ischemia reperfusion. Mol Immunol 48: 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH, Zhou L et al (2016). SIRT6 over‐expression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2‐associated X protein–dependent apoptotic pathway. Clin Cancer Res 22: 3372–3382. [DOI] [PubMed] [Google Scholar]
- Risco A, del Fresno C, Mambol A, Alsina‐Beauchamp D, MacKenzie KF, Yang HT et al (2012). p38γ and p38δ kinases regulate the Toll‐like receptor4 (TLR4)‐induced cytokine production by controlling ERK1/2 protein kinase pathway activation. Proc Natl Acad Sci U S A 109: 11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay MC, Che N, Becker SN, Pouldar D, Hagopian R, Xiao X et al (2015). Regulation of NF‐κB signaling by oxidized glycerophospholipid and IL‐1β induced miRs‐21‐3p and ‐27a‐5p in human aortic endothelial cells. J Lipid Res 56: 38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler EM, Hindes A, Gribben EL, Burns CJ, Yin Y, Lin MH et al (2009). p38δ mitogen‐ activated protein kinase is essential for skin tumor development in mice. Cancer Res 69: 4648–4655. [DOI] [PubMed] [Google Scholar]
- Song J, Li J, Qiao J, Jain S, Mark Evers B, Chung DH (2009). PKD prevents H2O2‐induced apoptosis via NF‐κB and p38MAPK. Biochem Biophys Res Commun 378: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhotnik I, Brod V, Lurie M, Rahat MA, Shnizer S, Lahat N et al (2009). The effect of 100% oxygen on intestinal preservation and recovery following ischemia‐reperfusion injury in rats. Crit Care Med 37: 1054–1061. [DOI] [PubMed] [Google Scholar]
- Taha MO, Ferreira RM, Taha NS, Monteiro HP, Caricati‐Neto A, Fagundes DJ (2014). Heparin modulates the expression of genes encoding pro and anti‐apoptotic proteins in endothelial cells exposed to intestinal ischemia and reperfusion in rats. Acta Cir Bras 29: 445–449. [DOI] [PubMed] [Google Scholar]
- Wang G, Yao J, Li Z, Zu G, Feng D, Shan W et al (2016). miR‐34a‐5p Inhibition alleviates intestinal ischemia/reperfusion‐induced reactive oxygen species accumulation and apoptosis via activation of SIRT1 signaling. Antioxid Redox Signal 24: 961–973. [DOI] [PubMed] [Google Scholar]
- Wang J, Masika J, Zhou J, Wang J, Zhu M, Luo H et al (2015). Traditional Chinese medicine baicalin suppresses mESCs proliferation through inhibition of miR‐294 expression. Cell Physiol Biochem 35: 1868–1876. [DOI] [PubMed] [Google Scholar]
- Wang J, Qiao LF, Li SS (2013). Protective effect of ginsenoside rb1 against lung injury induced by intestinal ischemia‐reperfusion in rats. Molecules 18: 1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pan T, Wang H, Li L, Li J, Zhang D et al (2017). Overexpression of SIRT6 attenuates the tumorigenicity of hepatocellular carcinoma cells. Oncotarget 8: 76223–76230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tanigawa T, Tominaga K, Watanabe K, Fujiwara Y, Nobuhide Oshitani N et al (2008). T1299 ischemia‐reperfusion injury in small intestine is mediated by Toll‐like receptor 2. Gastroenterology 134: 515–526. [Google Scholar]
- Yuan YH, Wu YD, Chi BZ, Wen SH, Liang RP, Qiu JD et al (2017). Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction. Biosens Bioelectron 97: 325–331. [DOI] [PubMed] [Google Scholar]
- Zhong W, Zhu H, Sheng F, Tian Y, Zhou J, Chen Y et al (2014). Activation of the MAPK11/12/13/14 (p38 MAPK) pathway regulates the transcription of autophagy genes in response to oxidative stress induced by a novel copper complex in HeLa cells. Autophagy 10 (7): 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur R, Garcia‐Ibanez L, Nunez‐Buiza A, Aparicio N, Liappas G, Escós A et al (2015). Combined deletion of p38γ and p38δ reduces skin inflammation and protects from carcinogenesis. Oncotarget 6: 12920–12935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 The primers sequences of miRNAs in the present work.
Table S2 The primer sequencess used for real‐time PCR assay in the present work.
Table S3 The information of the antibodies used in the present work.
Table S4 The up‐regulated expression of miRNAs in II/R group compare with control.
Table S5 The down‐regulated expression of miRNAs in II/R group compare with control.
Figure S1 Proposed signaling mechanism of miR‐351‐5p on II/R‐induced intestinal injury. miR‐351‐5p was selected as the target miRNA by microarrays assay, and Sirt6 and MAPK13 were the target genes of miR‐351‐5p. II/R significantly down‐regulated the expression levels of Sirt6 and MAPK13 by increasing the level of miR‐351‐5p, up‐regulated the levels of p‐PKD1, NF‐κB(p65) and inflammatory factors including IL‐1β, IL‐6, TNF‐α and ICAM‐1, increased p‐FoxO3αlevel via inhibiting AMPK, reduced MnSOD and CAT levels, and increased the levels of Bax, Cytc, Apaf‐1, Cleaved‐ caspase3 and Cleaved‐caspase9.MicroRNA‐351‐5p aggravated intestinal ischemia/ reperfusion injury by promoting intestinal mucosal oxidative stress, inflammation and apoptosis via targeting MAPK13 and Sirt6.


