Abstract
Structural characterization of intrinsically disordered proteins (IDPs) has been a major challenge in the field of protein science due to limited capabilities to obtain full-length high-resolution structures. Native ESI-MS with top-down MS was utilized to obtain structural features of protein-ligand binding for the Parkinson’s disease-related protein, α-synuclein (αSyn), which is natively unstructured. Binding of heavy metals has been implicated in the accelerated formation of αSyn aggregation. Using high resolution Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry, native top-down MS with various fragmentation methods, including electron capture dissociation (ECD), collisional activated dissociation (CAD), and multistage tandem MS (MS3), deduced the binding sites of cobalt and manganese to the C-terminal region of the protein. Ion mobility MS (IM-MS) revealed a collapse toward compacted states of αSyn upon metal binding. The combination of native top-down MS and IM-MS provides structural information of protein-ligand interactions for intrinsically disordered proteins.
Introduction
Parkinson’s disease (PD) is a neurodegenerative disorder that results in impairment of movement function, including motor skills that affect speech and breathing [1–3]. One of the pathogenic hallmarks of PD is the deposition of α-synuclein (αSyn) protein as an amyloid, known as Lewy bodies, in dopaminergic neurons [2, 4, 5]. This pathogenic feature, called synucleinopathies, can also be linked to dementia and other symptoms such as multiple system atrophy (MSA) [6, 7]. The normal functions of αSyn in the brain remain largely unknown. Some studies have reported that αSyn is involved in lipid binding [8, 9] and promotes SNARE-mediated vesicle fusion [10]. It may play a role in neurotransmitter regulation [11–14].
αSyn is an intrinsically disordered protein (IDP), which is natively unstructured at physiological pH. Its unfolding is highly influenced by negatively charged acidic residues in the sequence, with a relatively high number found in the C-terminal region (Figure 1) [15–17]. Because of its unfolding and aggregation-prone properties, atomic resolution structures of the full-length protein are not yet available [18, 19]. Other biophysical methods, such as nuclear magnetic resonance (NMR) spectroscopy [9, 20, 21], electron paramagnetic resonance (EPR) spectroscopy [22, 23], and small-angle X-ray scattering (SAXS) [24, 25] are alternative approaches that have been used for structural characterization of αSyn. αSyn is a relatively small protein with 140 amino acid residues and consisting of three main regions: an N-terminal amphipathic region, a non-amyloid component (NAC) region, and the C-terminal tail (Figure 1) [16, 26, 27]. The N-terminal region (residue 1–60) has a helical structure; it is the region that interacts with phospholipid membranes and vesicles. Previous studies reported that the helical region that contains N-terminal acetylation in vivo promotes lipid binding and multimerization of the protein [28–31]. The central NAC region is usually referred as a toxic core, with evidence for its involvement in protein aggregation [19, 32]. Lastly, the C-terminal region, which is somewhat proline-rich and contains many acidic residues, is mainly unstructured [26, 27]. Toxic amyloidogenic forms of αSyn can be reduced by binding to small molecular weight ligands, such as dopamine, epigallocatechin gallate (EGCG) [33], curcumin [34, 35], rifampicin [36], scyllo-inositol [37], or a “molecular tweezer” (e.g., CLR01) [38, 39], through an off-pathway oligomer formation route.
Figure 1.

Polypeptide sequence of α-synuclein showing the three major regions: N-terminal helix (green), NAC (blue), and highly acidic C-terminal region (yellow). KTKEGV sequence repeats are bolded and underlined.
Divalent and trivalent heavy metals, such as aluminum, copper, cobalt, manganese, cadmium, and iron, have been shown to accelerate αSyn aggregation [40–42]. It is believed that metal binding triggers structural changes of the protein toward more compact states via charge neutralization, leading to neurodegenerative disease progression [41, 43]. Binding of copper by αSyn has been widely studied [44–49]. Copper binds to two regions of αSyn, the N-terminus and His-50, although the Cu-binding to His-50 is debated [34, 42]. Cu-binding is not found on the N-terminus for N-terminal acetylated forms of αSyn [50]. The αSyn binding to other transition metals, including cobalt, manganese, iron, and nickel are far less studied, but the available data to date suggests that cobalt and manganese bind to the acidic C-terminal tail, particularly Asp-121 and Glu-123, with lower affinity than copper [51, 52]. Co/Mn-binding to αSyn is measured to be in the mM range, whereas Cu-binding is in the μM range [53].
In this report, we demonstrate the applicability of native mass spectrometry (MS) approaches with electrospray ionization (ESI) to provide important structural information on the binding of cobalt and manganese to α-synuclein. Native ESI-MS has been demonstrated to be an effective experimental platform to interrogate the structures of proteins [54], protein-ligand interactions [55], and large protein complexes [56–58]. Structural features, such as the elucidation of metal binding sites have been revealed using top-down MS with electron capture dissociation (ECD) and collisionally activated dissociation (CAD) [57, 59]. Generally hydrophobic and weak noncovalent interactions involving ligand binding do not survive the energetic dissociation process of vibrational activation-based CAD. Previously, we have demonstrated that such weak interactions can be maintained under radical activation-based ECD conditions, allowing ligand binding sites to be elucidated [55]. But in the case of metal binding, which mainly involves charge-charge interactions, such electrostatic interactions are strengthened in the gas phase due to low dielectric constant [60], and supported by computational modeling [61]. Other ligands that interact with proteins through strong electrostatic forces, such as nucleotides (e.g., ATP), can be analyzed using CAD conditions [62, 63]. Our earlier native top-down MS studies of αSyn revealed the binding sites of weakly bound (solution Kd in the 10−3 – 10−6 M range) organic ligands using ECD [39, 55]. Herein we used both ECD and CAD to identify the sites of Mn- and Co-binding to αSyn. Despite the mM binding affinity of Co-/Mn-binding to αSyn, Co-/Mn-binding might be involved in the aggregation pathway by forming transient folded intermediates. Ion mobility mass spectrometry (IM-MS) provided additional structural and conformation information that may play a role in amyloid fibrillation [8, 64].
Experimental
Materials
Recombinant full-length α-synuclein (amino acids 1–140) and its trunctated polypeptides (amino acids 1–60, 61–140, and 96–140) were purchased from rPeptide (Bogard, GA). Proteins were further desalted by exchange with 20 mM ammonium acetate using 10 kDa MWCO Amicon centrifugal filters (MilliporeSigma; Burlington, MA). Cobalt acetate and manganese acetate were purchased from Sigma (St. Louis, MO) and resuspended in 20 mM ammonium acetate. Metals were added to the αSyn solution at the appropriate protein:metal concentration ratio. The final concentration of αSyn was kept at 10 μM in 20 mM ammonium acetate at pH 6.8. Samples were loaded into nano-ESI Au/Pd-coated borosilicate emitters (Thermo Scientific; San Jose, CA) for mass spectrometry analysis.
Mass Spectrometry
Top-down mass spectrometry experiments were performed with a Bruker Solarix 15-Tesla Fourier transform ion cyclotron resonance (FT-ICR) instrument and an Infinity ICR cell (Billerica, MA). To generate ions, the ESI voltage was set to ca. 1000 V to deliver protein sample at a flow rate of 50 nL/min. The capillary source temperature was 180 °C. The ion optic voltages were set to the following: deflector plate, 200 V; capillary exit, 180 V; funnel, 120 V; skimmer, 30 V.
For ECD-MS/MS experiments, precursor ions were isolated by the quadrupole and transferred into the FT-ICR cell for ECD fragmentation and ion detection. The estimated resolving power was set to 400,000 at m/z 400. For CAD-MS/MS, CAD was performed in the collision cell remote from the ICR cell. CAD fragments were transferred to and detected by the ICR cell. The CAD voltage was adjusted to a range between 8–15 V, depending on the isolated charge states. A supplemental skimmer voltage of up to 30 V was included for additional collisional activation to enhance ECD fragmentation. (Activated ion ECD from supplemental infrared radiation was not used.) Pseudo-MS3 experiments [65] for further fragmentation of MS2 product ions were conducted by a combination of the nozzle skimmer CAD (NS-CAD) and CAD or ECD. NS-CAD was used as the unbiased first stage MS/MS (i.e., collisional activation of all ions), followed by CAD or ECD of selected NS-CAD products as the second stage MS/MS. MS2 and MS3 fragments were manually assigned against the theoretical fragments indicated by Protein Prospector (University of California, San Francisco; http://prospector.ucsf.edu/prospector/mshome.htm) and visually displayed using MATLAB (MathWorks, Natick, MA).
Ion Mobility Mass Spectrometry
IM-MS was performed with a Waters Synapt G2 HDMS quadrupole traveling wave ion mobility (TWIM) orthogonal time-of-flight mass spectrometer (Waters; Manchester, UK). For IM-MS experiments, αSyn and αSyn-metal samples were prepared to have a final concentration of 20 μM in 20 mM ammonium acetate. CoCl2 and MnCl2 were used to prepare αSyn-Co(II) (1:8 molar ratio) and αSyn-Mn(II) (1:10 molar ratio). Analyte solutions were inserted into Au/Pd coated borosilicate emitters (Thermo Scientific) nano-ESI-MS. A nano-ESI capillary voltage of 1.3 kV, cone voltage of 20 V, and source temperature of 50 °C were used for the experiments. For MS experiments, the settings of the instrument were: trap bias, 45 V; IMS bias, 3 V; trap collision energy, 3 V; transfer collision energy, 0 V. The gas flow rates for the helium cell located before the IMS cell, and the IMS cell gas (N2) were 150 mL/min and 60 mL/min, respectively. The pressures of the helium cell and the IMS cell were 1.4 × 103 and 3.1 × 100 mbar, respectively. TWIM wave velocity and wave height were set to 500 m/s and 24 V, respectively. The experimental arrival times were converted into collision cross section values by performing calibration using denatured ubiquitin, cytochrome c, and apo-myoglobin proteins of which collision cross section values were previously reported [8, 66, 67].
Results and Discussion
ESI-MS and MS2 of Unbound α-Synuclein
A significant portion of α-synuclein is unstructured at pH 7, where a region near the C-terminus is highly flexible. The C-terminal tail contains several acidic residues, as shown in Figure 1. The positive ion native ESI mass spectrum of IDP protein αSyn revealed relatively high charging and a wide charge distribution, unlike those found for more compact, folded proteins at neutral pH. Charge states of αSyn from 6+ to 16+ were observed, with a maximum abundance observed at ca. 12+ (Figure 2 and Supplemental Figure S1), which echoes αSyn’s unfolded structure near physiological pH [36]. Peaks in the mass spectra that may represent either specific or nonspecific dimers with charging between 6+ to 9+ were also observed, but at much lower abundances relative to the monomer.
Figure 2.
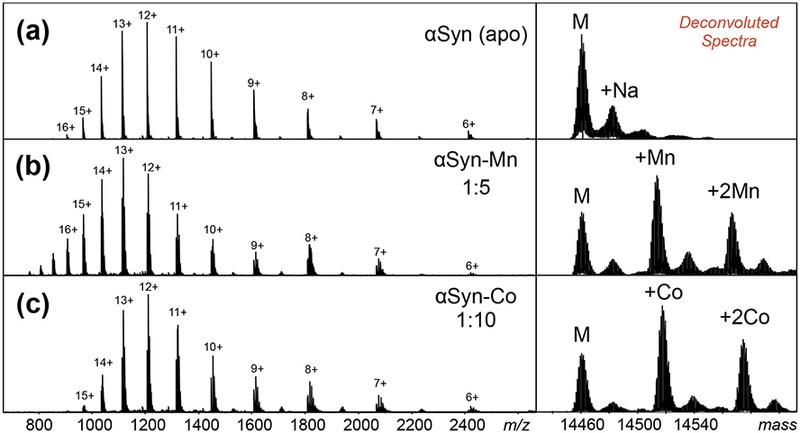
ESI-MS spectra of (a) apo-αSyn, (b) αSyn-Mn complex, and (c) αSyn-Co complex. Deconvoluted spectra were generated by Bruker DataAnalysis software using maximum entropy (MaxEnt) (right panels). Concentration ratios of αSyn:metal were optimized at 1:5 and 1:10 for manganese and cobalt, respectively.
For native top-down MS experiments, both beam-type CAD and in-cell ECD were performed. Figure 3 shows fragmentation maps from ECD and CAD. Fragmentation results are presented as a bar plot against backbone cleavage sites along the sequence. The vertical axis shows the summed charge-normalized product ion intensities resulting from top-down fragmentation of the αSyn precursors examined (12+, 13+, 14+). (No significant differences were found for the fragmentation of the individual 12+, 13+, and 14+ charged precursors.) N-terminal fragments (c-ions for ECD, or b-ions, including water and ammonia losses for CAD) are plotted above the longitudinal axis, and C-terminal fragments (z-ions for ECD, and y-ions for CAD) are plotted below the axis. ECD fragmentation was distributed throughout the protein backbone, but showed some preference toward the N-terminal end (Figure 3a). Abundant ECD products, such as c6, c9, c38, and c46 from regions where many of the positive charge sites are localized, were observed. CAD generated extensive fragmentation around the acidic C-terminal region (Figure 3b). The b116, b126, b127, and b137 product ions are among the most abundant products, most likely due to the five proline residues in the C-terminal end: Pro-108, Pro-117, Pro-120, Pro-128, and Pro-138; cleavage of the bond N-terminal to proline residues is particularly favored [68] [69]. The most abundant C-terminal product ion is y13, resulting from the cleavage of Met-126/Pro-127. Also, cleavages of bonds C-terminal to acidic residues, such as Asp, are favorable [70] and observed here as y9, y13, y14, and y21 product ions. Overall, sequence information from CAD is limited to αSyn regions mostly near the C-terminus. The results were similar to previous αSyn top-down MS studies from ion trap CAD [71] and CAD/ECD from ion trap-FT-ICR MS [55]. Backbone cleavage from ECD and CAD of αSyn reached 67% and 36% sequence coverage, respectively. Combining the results from both techniques yielded a total sequence coverage of 90%. The two fragmentation modes are complimentary to each other; combining multiple dissociation techniques, especially for native top-down MS, greatly enhances sequence information [56].
Figure 3.
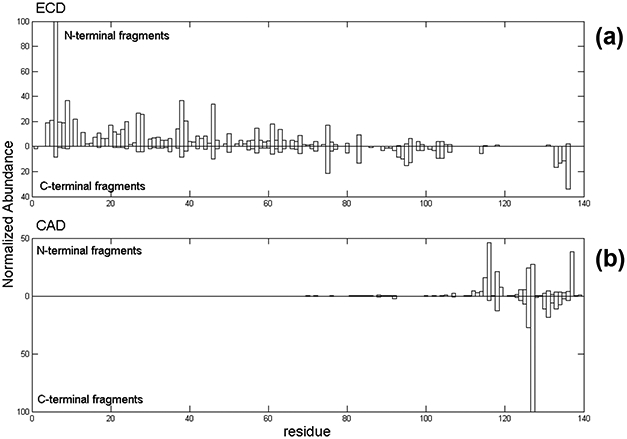
Native top-down MS fragmentation maps of α-synuclein using (a) ECD and (b) CAD.
ESI-MS of Metal-Bound α-Synuclein
Binding of cobalt and manganese to αSyn were readily observed by native ESI-MS (Figure 2). Protein:metal concentration ratios were optimized between 1:5 and 1:10, with the protein concentration kept at 10 μM. Gentle ion source conditions were used to minimize disruption of metal binding. The deconvoluted mass spectrum of unbound αSyn displayed monoisotopic and average masses at 14452 and 14460 Da, respectively. Native ESI-MS of αSyn incubated with cobalt showed a mass (monoisotopic) of 14509 Da, which corresponds to the mass of αSyn with one bound cobalt (Supplemental Table S1). A mass of 14505 Da was measured for αSyn incubated with manganese, which agrees with a single manganese bound to αSyn (Supplemental Table S2). A second bound cobalt and manganese cation was also observed, but other studies indicate that non-specific metal binding is observed for αSyn [40, 41, 51]. This work will exclusively focus on the characterization of the 1:1 protein-metal complexes.
Because αSyn contains several acidic residues, the protein is negatively charged at physiological pH (pI ~ 4.6). Binding of divalent metal cations could be expected to electrostatically neutralize some of the negative charge. It has been proposed that charge neutralization from metal binding might alter αSyn’s conformation and trigger protein fibrillation [40]. Protein charge state distributions measured by ESI-MS have been used to monitor αSyn folding induced by Cu-binding [43]. A closer examination of the ESI charge state distribution (CSD) showed little differences between the metal-free and metal-bound states, which could suggest that the global structure is not significantly changed upon metal binding. It has been widely considered that CSDs from native ESI-MS can be used to reflect a protein’s conformation [72–75], where low charge states at high m/z represent compact conformers and the extended conformers yield high charge states found at low m/z. Because αSyn is natively unstructured at pH 7, small changes in its structure due to metal binding may be difficult to produce observable changes in its CSD. Ion mobility mass spectrometry (IM-MS), however, may reflect changes in structure due to metal binding (vide infra).
Cobalt Binds to C-terminal Unstructured Tail of α-Synuclein
CAD- and ECD-MS/MS of the 12+−14+ precursor ions of the 1:1 αSyn-Co complex yielded the fragmentation maps shown in Figure 4. (An ECD mass spectrum of the 12+-charged αSyn-Co complex is shown in Supplemental Figure 2.) The ECD map revealed a sequence coverage of 79%, and that most N-terminal containing fragments up to residue 118 do not retain the Coligand, except two fragments near the C-terminal end, c131+Co and c139+Co. The majority of the C-terminal containing fragments, such as z25+Co, have cobalt bound. The only apo z-product ion is z5. The general ECD pattern of the cobalt complex appears similar to the apo-αSyn form (Figure 3), especially from the N-terminal region. The observed common fragments showing high relative abundance were c6, c9, c27, c38, and c46. The ECD data do not indicate a significant change in protein conformation upon Co-binding. The ECD fragments indicate the C-terminal tail to be the site of binding.
Figure 4.
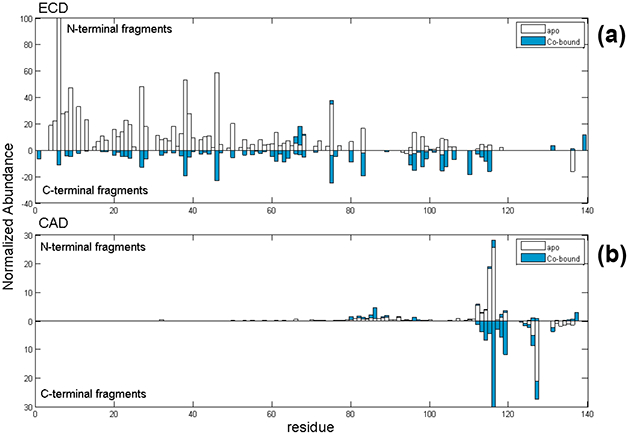
Native top-down MS fragmentation maps from (a) ECD and (b) CAD of αSyn-Co complex. Holo-products are represented in blue and apo-fragments are shown as hollow bars.
If the Co-binding site is at the C-terminus, CAD of the αSyn-cobalt complex may yield more information for metal binding due to extensive fragmentation near the C-terminus. Backbone cleavage from CAD was 61%, higher than for apo-αSyn. There are some differences evident in the CAD map for αSyn-Co compared to that for apo-αSyn (Figure 4b), and this is perhaps due to a structural change upon cobalt binding to the C-terminal region. Overall, we observed more abundant apo- and holo-y ions for the αSyn-Co complex than for apo-αSyn. The following apo-fragments were identified: b115, b116, and y127 (with low intensity). Also we observed b126+Co, b127+Co, b137+Co, y21+Co and y24+Co holo-products. With backbone cleavages from CAD and ECD, we confirmed that the cobalt-binding region is between residues 119DPDNEAYE126. Sequence coverage increased to 96% when combining data from both ECD and CAD.
Manganese and Cobalt Share the Same Binding Site to α-Synuclein
Similar to the experiments with αSyn-Co, three precursor charge states (12+, 13+, 14+) of the 1:1 αSyn-Mn complex were isolated for ECD-MS/MS and CAD-MS/MS (Figure 5); the fragmentation patterns of the Mn- and Co-bound αSyn protein are similar. ECD of the 1:1 αSyn-Mn complex produced c131+Mn and z25+Mn product ions, which are key manganese-bound fragments that pinpoint the Mn-binding site to be between residues 116–131. Sequence coverage obtained from ECD was 69%. Similar to the cobalt complex, CAD-MS/MS of the Mn-bound complex showed better sequence coverage (than by ECD) and more specific binding site identification near the C-terminus. The CAD pattern is also similar to that from the cobalt complex. CAD provided backbone cleavage efficiency of 76%, and the total sequence coverage increased to 96% when ECD and CAD data were combined. Because products b126+Mn, y21+Mn, and y24+Mn ions were observed by CAD, it is reasonable to narrow down the Mn-binding site to the region between 119DPDNEAYE126, the same region as for Co-binding. CAD generated more binding site information for manganese-binding compared to ECD because plenty of C-terminal fragments were produced.
Figure 5.
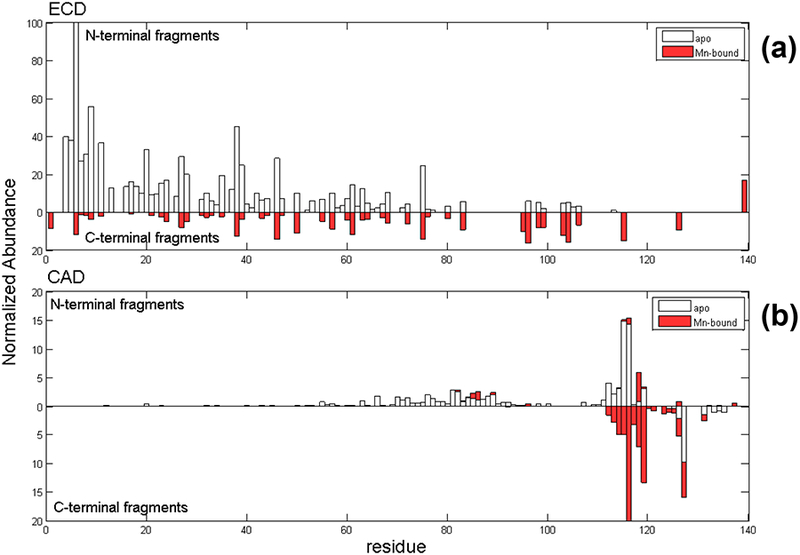
Native top-down MS fragmentation maps from (a) ECD and (b) CAD of αSyn-Mn complex. Holo-products are represented in red and apo-fragments are shown as hollow bars.
Pseudo-MS3 to Further Probe Metal-Binding of αSyn
Additional stages of MS/MS (e.g., MS3) can be useful, for example, to confirm product ion assignments or to further pinpoint the sites of modification. A “pseudo-MS3” experiment with the FT-ICR instrument was enabled to further probe the Co- and Mn-binding to αSyn. The potential on the skimmer in the atmosphere/vacuum interface was increased to fragment by CAD all ions generated by the ESI source. (This process was previously termed nozzle-skimmer dissociation (NSD) [65] and now sometimes called in-source CAD or IS-CAD.) Product ions generated by IS-CAD can be selected as precursors for further activation/dissociation, i.e., pseudo-MS3. Two product ions from each metal complex (y21+Co, y24+Co, y21+Mn, and y24+Mn) generated by IS-CAD were isolated for CAD-MS3 experiments.
MS3 fragmentation maps for the Co-bound and Mn-bound products were generated (Figure 6). Interestingly, the cobalt-binding site identified by MS3 experiments was slightly different from that by MS/MS. While the MS/MS data shows that cobalt binds to a region composed by residues 119DPDNEAYE126, the CAD-MS3 maps of y21+Co and y24+Co, originated from the same protein-metal complexes in-solution, showed in addition another binding site between 132GYQDY136 (Figures 6a, 6b), suggesting either that cobalt may have migrated from one site to another site in the gas phase, or the MS3 experiment may have picked up an additional site not detected by MS2. There are some unbound fragments observed along with metal-bound ones at the same cleavage site. For example, the Pro-128/Ser-129 cleavage to yield y24/y24+Co had more unbound fragment observed, suggesting any Co-migration to a second binding site was partial. Alternatively, structural rearrangement during in-source dissociation and/or the presence of precursors with different metal binding sites are possible explanations to be considered.
Figure 6.
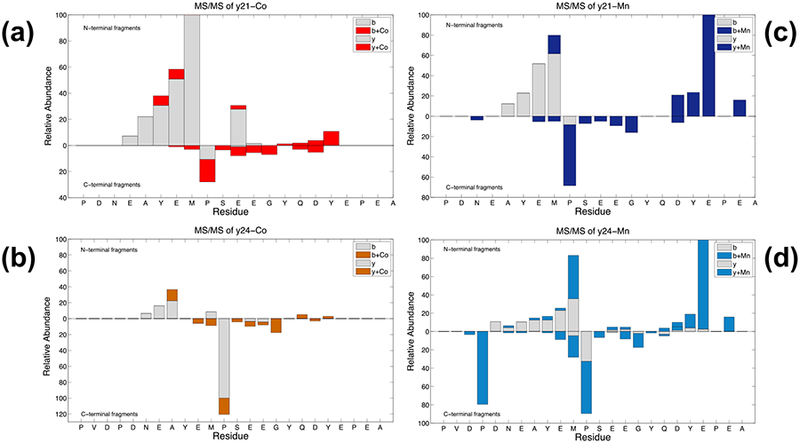
Pseudo-MS3 fragmentation maps of αSyn-Co and αSyn-Mn complexes. For cobalt-binding, (a) y21+Co and (b) y24+Co were isolated for further CAD. For Mn-binding, (c) y21+Mn and (d) y24+Mn were isolated and further underwent CAD.
The data for the Mn-complexes y21+Mn and y24+Mn showed similar results (Figures 6c, 6d), yet there were some differences compared to the Co-complexes. From the pseudo-MS3 experiments, as for Co-binding, the Mn-binding site appears to be at residues 132GYQDYE137. But there is a distinct backbone cleavage at Glu-137 (relative to the full-length sequence) that yields Mn-bound b-ions (b18+Mn from y21+Mn precursor, and b21+Mn from y24+Mn precursor). Binding of manganese may induce reconfiguration or increase stabilization around the secondary binding site, which might explain why the fragment at Glu-137/Pro-138 yield the highest abundance observed. The Pro-128/Ser-129 cleavage from y24+Mn showed more metal-bound fragment than with cobalt, suggesting the Mn-migration was more complete. The remaining product ions were similar to that found for cobalt-binding. Overall, the results from MS/MS and MS3 confirmed that cobalt and manganese have similar αSyn-binding sites.
MS/MS of Truncated αSyn Confirmed Metal Binding Sites
Aside from full-length αSyn, three truncated αSyn variants (1–60, 61–140, and 96–140) were characterized. αSyn(1–60) covers the N-terminal helical amphipathic region. αSyn(96–140) has only the C-terminal acidic region, which is mainly unfolded. αSyn(61–140) contains both the NAC and acidic regions. As expected, cobalt and manganese binds to both the 61–140 and 96–140 fragments, but not to the N-terminal 1–60 fragment (data not shown). These results confirmed the C-terminal binding regions, which were shared between the two metals. Top-down MS/MS experiments by ECD and CAD of Co/Mn-bound αSyn(61–140) and αSyn(96–140) were performed; the identified binding site was the same as for the full-length protein.
Metal-Binding Induced Structural Changes Observed by Ion Mobility Spectrometry
Ion mobility has been used to monitor changes and the molecular dynamics of biomolecule structure, such as conformational changes as a function of charge, unfolding by energetic collisions, and the stability of membrane proteins by lipid binding [64, 76–80]. In this work, ion mobility mass spectrometry (IM-MS) was used to probe for the presence of αSyn structural changes upon binding to cobalt or manganese. IM-MS was performed using a quadrupole time-of-flight (qTOF) mass spectrometer with a T-wave mobility cell. Collisional cross section (CCS) profiles derived from ion mobility of the 9+ charged αSyn is shown in Figure 7. For the apo-form, four conformations were resolved with CCSs of 1948, 2155, 2403, and 2593 Å2, with the major form (at 2593 Å2) as the most extended structure observed for this charge state. Binding of cobalt and manganese caused a significant decrease of the 2593 Å2 form relative to the three other smaller-CCS forms. However, metal binding to the C-terminal region might induce several new conformations, which could result in the peak broadening (from overlapping peaks) observed.
Figure 7.
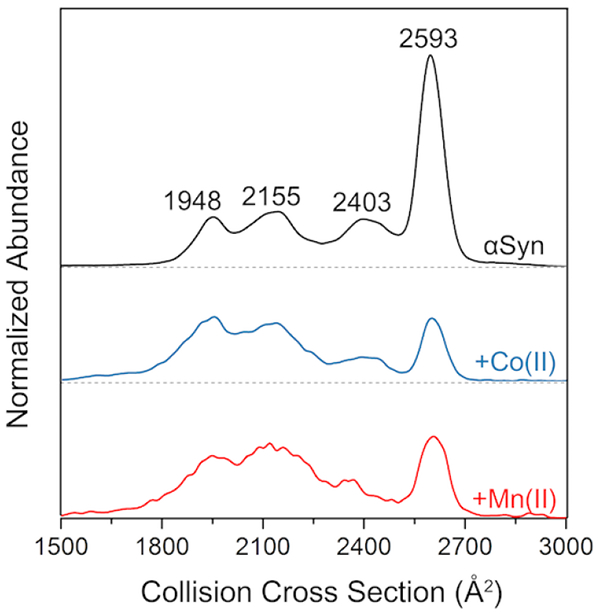
IM-MS spectra of 9+ charged ions of αSyn (apo), αSyn-Co, and αSyn-Mn. Distributions were normalized to the total area in order to reflect the difference of the proportion of the conformations. (Portions of this figure were adapted from reference [81] with permission from the publisher.)
In addition to the 9+ charge state, other charge states ranging from 7+ to 14+ were also examined by IM-MS (Figure 8). The highest charge state (14+) showed only a single conformation at high CCS values, suggesting a fully extended conformation. Lower charge states yielded more than one conformation at lower CCS towards a more compact form. Consistent with the similarity in the MS/MS data for the Co-/Mn-binding forms, the Co-/Mn-complexes exhibit a similar CCS pattern. No structural changes indicated by the IM-MS profiles were observed for the more extended 14+ charge upon Co-/Mn-binding. For charge states lower than 14+, the relative abundance (where the abundance is normalized by the total peak area) of the lower CCS conformers increased slightly with metal binding. This observation implies that Co-/Mn-binding promotes a more compact structure. This compaction is most significant for the 9+ charge, but this tendency was also observed for the higher charge states.
Figure 8.
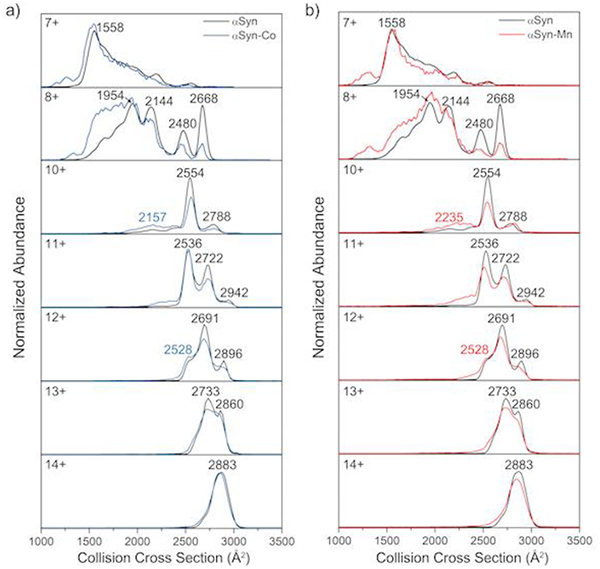
IM-MS spectra of 7+−14+ charged ions of (a) αSyn-Co and (b) αSyn-Mn overlaid with apo-αSyn profiles.
Conclusion
Native top-down mass spectrometry and ion mobility MS were used to provide in-depth structural information on the binding of divalent metal ions, Co and Mn, to α-synuclein, an intrinsically disordered protein. ECD, CAD, and pseudo-MS3 (IS-CAD/CAD-MS3) techniques were able to identify the binding sites of cobalt and manganese to the protein. Because the binding of Co and Mn to αSyn is governed by electrostatic interactions, the metal-protein interactions appeared to be highly stable in the gas phase and they survived higher-energy dissociation methods such as CAD. Sequence coverage was greatly improved by combining CAD and ECD data. Top-down MS revealed that Co and Mn share similar binding locations, with the primary and secondary binding sites, 119DPDNEAYE126 and 132GYQDY136, respectively, found at the C-terminal end. However, other weaker binding sites might be present along the polypeptide backbone. Different metals have different binding sites to αSyn and their interactions can cause unique structural changes [41, 52]. Previously, we reported the binding of Cu(II) to regions mainly near the N-terminus and residues 45–56 (wherein His-50 is located), whereas the C-terminal residues showed negligible interaction with Cu(II) [81]. We postulated that a conformational strain in the αSyn-Cu(II) complex modulates the fibrillation pathway, thereby mediating the formation of highly cell-transmissible, cytotoxic αSyn fibrils with short lengths. Mn-/Co-binding can influence protein folding, which may be important factors for neurodegenerative diseases. Newer activation/dissociation methods, such as surface induced dissociation (SID) [82] and UV photodissociation (UVPD) [83, 84], should provide additional useful tools for MS-characterization of protein-ligand complexes. Top-down MS and IM-MS are complementary biophysical tools to elucidate the structural changes induced by protein-ligand interactions.
Supplementary Material
Acknowledgments
Support from the US National Institutes of Health (R01GM103479, S10RR028893, S10OD018504 to J.A.L.), the US Department of Energy (DE-FC02-02ER63421 to J.A.L.), the Development and Promotion of Science and Technology Talents Project (DPST) and Royal Thai Government (to P.W.), the Rachadapisek Sompot Fund, Chulalongkorn University (to P.W.), the National Research Foundation of Korea (NRF) (NRF-2016R1A2B4013089 and 20100020209 to H.I.K.), Korea University Future Research Grant (to H.I.K.) and the Ministry of Science, ICT and Future Planning (CAP-15–10-KRICT to H.I.K.) are gratefully acknowledged.
References
- 1.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL: Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276, 2045–2047 (1997) [DOI] [PubMed] [Google Scholar]
- 2.Mezey E, Dehejia AM, Harta G, Suchy SF, Nussbaum RL, Brownstein MJ, Polymeropoulos MH: Alpha synuclein is present in lewy bodies in sporadic parkinson’s disease. Mol. Psych 3, 493 (1998) [DOI] [PubMed] [Google Scholar]
- 3.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K: α-Synuclein locus triplication causes Parkinson’s disease. Science 302, 841–841 (2003) [DOI] [PubMed] [Google Scholar]
- 4.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ, Jakes R, Goedert M: α-Synuclein in lewy bodies. Nature 388, 839–840 (1997) [DOI] [PubMed] [Google Scholar]
- 5.Mukaetova-Ladinska EB, McKeith IG: Pathophysiology of synuclein aggregation in lewy body disease. Mech. Ageing Dev 127, 188–202 (2006) [DOI] [PubMed] [Google Scholar]
- 6.Grazia Spillantini M, Anthony Crowther R, Jakes R, Cairns NJ, Lantos PL, Goedert M: Filamentous α-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with lewy bodies. Neurosci. Lett 251, 205–208 (1998) [DOI] [PubMed] [Google Scholar]
- 7.Wakabayashi K, Yoshimoto M, Tsuji S, Takahashi H: α-Synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci. Lett 249, 180–182 (1998) [DOI] [PubMed] [Google Scholar]
- 8.Lee SJC, Lee JW, Choi TS, Jin KS, Lee S, Ban C, Kim HI: Probing conformational change of intrinsically disordered α-synuclein to helical structures by distinctive regional interactions with lipid membranes. Anal. Chem 86, 1909–1916 (2014) [DOI] [PubMed] [Google Scholar]
- 9.Ulmer TS, Bax A: Comparison of structure and dynamics of micelle-bound human α-synuclein and Parkinson disease variants. J. Biol. Chem 280, 43179–43187 (2005) [DOI] [PubMed] [Google Scholar]
- 10.Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Sϋdhof TC: α-Synuclein promotes snare-complex assembly in vivo and in vitro. Science 329, 1663–1667 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L: Dopaminergic loss and inclusion body formation in α-synuclein mice: Implications for neurodegenerative disorders. Science 287, 1265–1269 (2000) [DOI] [PubMed] [Google Scholar]
- 12.Perez RG, Waymire JC, Lin E, Liu JJ, Guo F, Zigmond MJ: A role for α-synuclein in the regulation of dopamine biosynthesis. J. Neurosci 22, 3090–3099 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidhu A, Wersinger C, Vernier P: Α-synuclein regulation of the dopaminergic transporter: A possible role in the pathogenesis of Parkinson’s disease. FEBS Lett 565, 1–5 (2004) [DOI] [PubMed] [Google Scholar]
- 14.Illes-Toth E, Dalton CF, Smith DP: Binding of dopamine to alpha-synuclein is mediated by specific conformational states. J. Am. Soc. Mass Spectrom 24, 1346–1354 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uversky VN, Li J, Fink AL: Evidence for a partially folded intermediate in α-synuclein fibril formation. J. Biol. Chem 276, 10737–10744 (2001) [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN: A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn 21, 211–234 (2003) [DOI] [PubMed] [Google Scholar]
- 17.Sung Y. h., Eliezer D: Residual structure, backbone dynamics, and interactions within the synuclein family. J. Mol. Biol 372, 689–707 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao M, Cascio D, Sawaya MR, Eisenberg D: Structures of segments of α-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation. Protein Sci 20, 996–1004 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, Sangwan S, Guenther ,EL, Johnson LM, Zhang M, Jiang L, Arbing MA, Nannenga BL, Hattne J, Whitelegge J, Brewster AS, Messerschmidt M, Boutet S, Sauter NK, Gonen T, Eisenberg DS: Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M: Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid-state NMR. Proc. Natl. Acad. Sci. USA 102, 15871–15876 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilar M, Chou H-T, Lϋhrs T, Maji SK, Riek-Loher D, Verel R, Manning G, Stahlberg H, Riek R: The fold of α-synuclein fibrils. Proc. Natl. Acad. Sci. USA 105, 8637–8642 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jao CC, Der-Sarkissian A, Chen J, Langen R: Structure of membrane-bound α-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. USA 101, 8331–8336 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Margittai M, Chen J, Langen R: Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem 282, 24970–24979 (2007) [DOI] [PubMed] [Google Scholar]
- 24.Li J, Uversky VN, Fink AL: Conformational behavior of human α-synuclein is modulated by familial parkinson’s disease point mutations A30P and A53T. NeuroToxicol. 23, 553–567 (2002) [DOI] [PubMed] [Google Scholar]
- 25.Bernado P, Svergun DI: Structural analysis of intrinsically disordered proteins by small-angle x-ray scattering. Mol. BioSys 8, 151–167 (2012) [DOI] [PubMed] [Google Scholar]
- 26.Beyer K: α-Synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol. 112, 237–251 (2006) [DOI] [PubMed] [Google Scholar]
- 27.Bisaglia M, Mammi S, Bubacco L: Structural insights on physiological functions and pathological effects of α-synuclein. FASEB J. 23, 329–340 (2009) [DOI] [PubMed] [Google Scholar]
- 28.Bartels T, Choi JG, Selkoe DJ: α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartels T, Kim NC, Luth ES, Selkoe DJ: N-alpha-acetylation of α-synuclein increases its helical folding propensity, GM1 binding specificity and resistance to aggregation. PLoS ONE 9, e103727 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maltsev AS, Ying J, Bax A: Impact of N-terminal acetylation of α-synuclein on its random coil and lipid binding properties. Biochemistry 51, 5004–5013 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burre J, Sharma M, Sϋdhof TC: α-Synuclein assembles into higher-order multimers upon membrane binding to promote snare complex formation. Proc. Natl. Acad. Sci. USA 111, E4274–E4283 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodles AM, Guthrie DJS, Greer B, Irvine GB: Identification of the region of non-aβ component (NAC) of Alzheimer’s disease amyloid responsible for its aggregation and toxicity. J. Neurochem 78, 384–395 (2001) [DOI] [PubMed] [Google Scholar]
- 33.Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE: EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol 15, 558–566 (2008) [DOI] [PubMed] [Google Scholar]
- 34.Ahmad A, Burns CS, Fink AL, Uversky VN: Peculiarities of copper binding to alpha-synuclein. J. Biomol. Struct. Dyn 29, 825–842 (2012) [DOI] [PubMed] [Google Scholar]
- 35.Singh PK, Kotia V, Ghosh D, Mohite GM, Kumar A, Maji SK: Curcumin modulates alpha-synuclein aggregation and toxicity. ACS Chem. Neurosci 4, 393–407 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein SL, Liu D, Wyttenbach T, Bowers MT, Lee JC, Gray HB, Winkler JR: α-Synuclein: Stable compact and extended monomeric structures and ph dependence of dimer formation. J. Am. Soc. Mass Spectrom. 15, 1435–1443 (2004) [DOI] [PubMed] [Google Scholar]
- 37.Tucker WC, Edwardson JM, Bai J, Kim HJ, Martin TF, Chapman ER: Identification of synaptotagmin effectors via acute inhibition of secretion from cracked PC12 cells. J. Cell Biol 162, 199–209 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prabhudesai S, Sinha S, Attar A, Kotagiri A, Fitzmaurice AG, Lakshmanan R, Ivanova MI, Loo JA, Klarner FG, Schrader T, Stahl M, Bitan G, Bronstein JM: A novel “molecular tweezer” inhibitor of α-synuclein neurotoxicity in vitro and in vivo. Neurotherapeutics 9, 464–476 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acharya S, Safaie BM, Wongkongkathep P, Ivanova MI, Attar A, Klarner F-G, Schrader T, Loo JA, Bitan G, Lapidus LJ: Molecular basis for preventing α-synuclein aggregation by a molecular tweezer. J. Biol. Chem 289, 10727–10737 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uversky VN, Li J, Fink AL: Metal-triggered structural transformations, aggregation, and fibrillation of human α-synuclein. A possible molecular link between Parkinson’s disease and heavy metal exposure. J. Biol. Chem 276, 44284–44296 (2001) [DOI] [PubMed] [Google Scholar]
- 41.Binolfi A, Rasia RM, Bertoncini CW, Ceolin M, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO: Interaction of α-synuclein with divalent metal ions reveals key differences: A link between structure, binding specificity and fibrillation enhancement. J. Am. Chem. Soc 128, 9893–9901 (2006) [DOI] [PubMed] [Google Scholar]
- 42.Brown DR: Metal binding to alpha-synuclein peptides and its contribution to toxicity. Biochem. Biophys. Res. Commun 380, 377–381 (2009) [DOI] [PubMed] [Google Scholar]
- 43.Natalello A, Benetti F, Doglia SM, Legname G, Grandori R: Compact conformations of α-synuclein induced by alcohols and copper. Proteins 79, 611–621 (2011) [DOI] [PubMed] [Google Scholar]
- 44.Rasia RM, Bertoncini CW, Marsh D, Hoyer W, Cherny D, Zweckstetter M, Griesinger C, Jovin TM, Fernandez CO: Structural characterization of copper(II) binding to α-synuclein: Insights into the bioinorganic chemistry of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 102, 4294–4299 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung YH, Rospigliosi C, Eliezer D: Nmr mapping of copper binding sites in alpha-synuclein. Biochim. Biophys. Acta 1764, 5–12 (2006) [DOI] [PubMed] [Google Scholar]
- 46.Bharathi Rao, K.S.: Molecular understanding of copper and iron interaction with α-synuclein by fluorescence analysis. J. Mol. Neurosci 35, 273–281 (2008) [DOI] [PubMed] [Google Scholar]
- 47.Binolfi A, Lamberto GR, Duran R, Quintanar L, Bertoncini CW, Souza JM, Cervenansky C, Zweckstetter M, Griesinger C, Fernandez CO: Site-specific interactions of Cu(II) with α and β-synuclein: Bridging the molecular gap between metal binding and aggregation. J. Am. Chem. Soc 130, 11801–11812 (2008) [DOI] [PubMed] [Google Scholar]
- 48.Dudzik CG, Walter ED, Millhauser GL: Coordination features and affinity of the Cu2+ site in the α-synuclein protein of Parkinson’s disease. Biochemistry 50, 1771–1777 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dudzik CG, Walter ED, Abrams BS, Jurica MS, Millhauser GL: Coordination of copper to the membrane-bound form of α-synuclein. Biochemistry 52, 53–60 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moriarty GM, Minetti CA, Remeta DP, Baum J: A revised picture of the Cu(II)-α-synuclein complex: The role of N-terminal acetylation. Biochemistry 53, 2815–2817 (2014) [DOI] [PubMed] [Google Scholar]
- 51.Aaron S, Vladimir NU: α-Synuclein and metals In Protein Folding and Metal Ions; CRC Press, pp 169–191 (2010) [Google Scholar]
- 52.Andres B, Claudio OF: Interactions of α-synuclein with metal ions In Brain Diseases and Metalloproteins; Pan Stanford Publishing, pp 327–366 (2012) [Google Scholar]
- 53.Binolfi A, Quintanar L, Bertoncini CW, Griesinger C, Fernandez CO: Bioinorganic chemistry of copper coordination to alpha-synuclein: Relevance to parkinson’s disease. Coord. Chem. Rev 256, 2188–2201 (2012) [Google Scholar]
- 54.Han X, Jin M, Breuker K, McLafferty FW: Extending top-down mass spectrometry to proteins with masses greater than 200 kiloDaltons. Science 314, 109–112 (2006) [DOI] [PubMed] [Google Scholar]
- 55.Xie Y, Zhang J, Yin S, Loo JA: Top-down ESI-ECD-FT-ICR mass spectrometry localizes noncovalent protein-ligand binding sites. J. Am. Chem. Soc 128, 14432–14433 (2006) [DOI] [PubMed] [Google Scholar]
- 56.Li H, Wolff JJ, Van Orden SL, Loo JA: Native top-down electrospray ionization-mass spectrometry of 158 kDa protein complex by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem 86, 317–320 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA: Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J. Am. Soc. Mass Spectrom 25, 2060–2068 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Nguyen HH, Ogorzalek-Loo RR, Campuzano IDG, Loo JA: An integrated native mass spectrometry and top-down proteomics method that connects sequence to structure and function of macromolecular complexes. Nature Chem 10, 139–148 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin S, Loo JA: Top-down mass spectrometry of supercharged native protein-ligand complexes. Int. J. Mass Spectrom 300, 118–122 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woods AS, Ferre S: Amazing stability of the arginine-phosphate electrostatic interaction. J. Proteome Res 4, 1397–1402 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bartman CE, Metwally H, Konermann L: Effects of multidentate metal interactions on the structure of collisionally activated proteins: Insights from ion mobility spectrometry and molecular dynamics simulations. Anal. Chem 88, 6905–6913 (2016) [DOI] [PubMed] [Google Scholar]
- 62.Yin S, Xie Y, Loo JA: Mass spectrometry of protein-ligand complexes: Enhanced gas-phase stability of ribonuclease-nucleotide complexes. J. Am. Soc. Mass Spectrom 19, 1199–1208 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin S, Loo JA: Elucidating the site of protein-ATP binding by top-down mass spectrometry. J. Am. Soc. Mass Spectrom 21, 899–907 (2010) [DOI] [PubMed] [Google Scholar]
- 64.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT: Ion mobility analysis of molecular dynamics. Ann. Rev. Phys. Chem 65, 175–196 (2014) [DOI] [PubMed] [Google Scholar]
- 65.Loo JA, Edmonds CG, Smith RD: Primary sequence information from intact proteins by electrospray ionization tandem mass spectrometry. Science 248, 201–204 (1990) [DOI] [PubMed] [Google Scholar]
- 66.Ruotolo BT, Benesch JLP, Sandercock AM, Hyung S-J, Robinson CV: Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protocols 3, 1139–1152 (2008) [DOI] [PubMed] [Google Scholar]
- 67.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT: Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem 82, 9557–9565 (2010) [DOI] [PubMed] [Google Scholar]
- 68.Schwartz BL, Bursey MM: Some proline substituent effects in the tandem mass spectrum of protonated pentaalanine. Biol. Mass Spectrom 21, 92–96 (1992) [DOI] [PubMed] [Google Scholar]
- 69.Light-Wahl KJ, Loo JA, Edmonds CG, Smith RD, Witkowska HE, Shackleton CHL, Wu CSC: Tandem mass spectrometry of intact hemoglobin variant proteins with electrospray ionization. Biol. Mass Spectrom 22, 112–120 (1993) [DOI] [PubMed] [Google Scholar]
- 70.Yu W, Vath JE, Huberty MC, Martin SA: Identification of the facile gas-phase cleavage of the Asp-Pro and Asp-xxx peptide bonds in matrix-assisted laser desorption time-of-flight mass spectrometry. Anal. Chem 65, 3015–3023 (1993) [DOI] [PubMed] [Google Scholar]
- 71.Chanthamontri C, Liu J, McLuckey SA: Charge state dependent fragmentation of gaseous α-synuclein cations via ion trap and beam-type collisional activation. Int. J. Mass Spectrom 283, 9–16 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogorzalek Loo RR, Lakshmanan R, Loo JA: What protein charging (and supercharging) reveal about the mechanism of electrospray ionization. J. Am Soc. Mass Spectrom 25, 1675–1693 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall Z, Robinson CV: Do charge state signatures guarantee protein conformations? J. Am. Soc. Mass Spectrom 23, 1161–1168 (2012) [DOI] [PubMed] [Google Scholar]
- 74.Kaltashov IA, Abzalimov RR: Do ionic charges in ESI MS provide useful information on macromolecular structure? J. Am Soc. Mass Spectrom 19, 1239–1246 (2008) [DOI] [PubMed] [Google Scholar]
- 75.Hamdy OM, Julian RR: Reflections on charge state distributions, protein structure, and the mystical mechanism of electrospray ionization. J. Am Soc. Mass Spectrom 23, 1–6 (2012) [DOI] [PubMed] [Google Scholar]
- 76.Zhou M, Politis A, Davies RB, Liko I, Wu K-J, Stewart AG, Stock D, Robinson CV: Ion mobility-mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nature Chem. 6, 208–215 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lanucara F, Holman SW, Gray CJ, Eyers CE: The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nature Chem 6, 281–294 (2014) [DOI] [PubMed] [Google Scholar]
- 78.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV: Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV: Charge-state dependent compaction and dissociation of protein complexes: Insights from ion mobility and molecular dynamics. J. Am. Chem. Soc 134, 3429–3438 (2012) [DOI] [PubMed] [Google Scholar]
- 80.Hyung S-J, Robinson CV, Ruotolo BT: Gas-phase unfolding and disassembly reveals stability differences in ligand-bound multiprotein complexes. Chem. Biol 16, 382–390 (2009) [DOI] [PubMed] [Google Scholar]
- 81.Choi TS, Lee J, Han JY, Jung BC, Wongkongkathep P, Loo JA, Lee MJ, Kim HI: Supramolecular modulation of structural polymorphism in pathogenic α-synuclein fibrils using copper(II) coordination. Angew. Chem. Int. Ed 57, 3099–3103 (2018) [DOI] [PubMed] [Google Scholar]
- 82.Zhou M, Yan J, Romano CA, Tebo BM, Wysocki VH, Paša-Tolić L: Surface induced dissociation coupled with high resolution mass spectrometry unveils heterogeneity of a 211 kDa multicopper oxidase protein complex. J. Am Soc. Mass Spectrom 29, 723–733 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Brien JP, Li W, Zhang Y, Brodbelt JS: Characterization of native protein complexes using ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc 136, 12920–12928 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Sheng Y, McGee W, Cammarata M, Holden D, Loo JA: Structural characterization of native proteins and protein complexes by electron ionization dissociation-mass spectrometry. Anal. Chem 89, 2731–2738 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


