Abstract
Ultraviolet photodissociation or UVPD is an increasingly popular option for tandem-mass spectrometry experiments. UVPD can be carried out at many wavelengths, and it is important to understand how the results will be impacted by this choice. Here we explore the utility of 213 nm photons for initiating bond-selective fragmentation. It is found that bonds previously determined to be labile at 266 nm, including carbon-iodine and sulfur-sulfur bonds can also be cleaved with high selectivity at 213 nm. In addition, many carbon-sulfur bonds that are not subject to direct dissociation at 266 nm can be selectively fragmented at 213 nm. This capability can be used to site-specifically create alaninyl radicals that direct backbone dissociation at the radical site, creating diagnostic d-ions. Furthermore, the additional carbon-sulfur bond fragmentation capability leads to signature triplets for fragmentation of disulfide bonds. Absorption of amide bonds can enhance dissociation of nearby labile carbon-sulfur bonds and can be used for stochastic backbone fragmentation typical of UVPD experiments at shorter wavelengths. Several potential applications of the bond-selective fragmentation chemistry observed at 213 nm are discussed.
Graphical abstract
Introduction
Ultraviolet Photodissociation (UVPD) is becoming an increasingly popular choice for fragmenting ions in mass spectrometers.1 UVPD differs from other dissociation techniques such as collision-induced dissociation (CID)2,3 or electron-transfer dissociation (ETD)4 in several important ways. For example, UVPD causes dissociation through nearly simultaneous operation of two mechanisms, vibrational heating and excited-state dissociation.5 The relevant timescale for UVPD experiments can also be significantly shorter than CID or ETD, occurring within nanoseconds for experiments utilizing high-powered lasers.6 Furthermore, the requirement of absorption by a chromophore offers an orthogonal parameter, independent of mass or charge, for controlling which ions will be excited. UVPD experiments have been conducted at many wavelengths including 157, 193, 213, 266, and 355 nm.7–13 Wavelength selection significantly impacts fragmentation by modulating the ratio of fragmentation mechanisms available. For example, at 266 nm, very few direct dissociation pathways exist, and the abundance of natural chromophores in analytes is small. At 157 nm, most bonds act as chromophores, and direct dissociation pathways are expected to be more plentiful.
Previous efforts to favor bond-selective direct dissociation, which occurs due to excitation to a dissociative excited state, have primarily utilized 266 nm photons.14 Early experiments demonstrated that carbon-iodine bonds could be selectively and homolytically cleaved in peptides or proteins, leaving all other bonds intact.15 Carbon-bromine bonds can also be cleaved in this fashion, though the yield is reduced.16 Sulfur-sulfur bonds are the only native bonds in peptides or proteins that undergo direct dissociation at 266 nm.17 Carbon-sulfur bonds can be cleaved with 266 nm photons, but only if the sulfur atom is directly attached to a suitable chromophore, such as naphthalene.18,19 Importantly, all of these labile bonds can be cleaved with a high degree of selectivity even in large molecules, meaning that in many cases, a single bond is the only site of dissociation. This degree of control over fragmentation enables a variety of unique applications.
For example, disulfide bond partners from a protein can be easily identified.17 In this experiment, the protein is digested with disulfide bonds intact and then subjected to chromatographic separation and MS/MS analysis with UVPD at 266 nm. Peptide pairs bound by a disulfide bond cleave into the constituent peptides while other monomeric peptides remain intact. Bond-selective UVPD can also be used to identify UV labile post-translational modifications such as iodination of tyrosine or quinone reactions with cysteine.16 When coupled with targeted wet-chemistry modifications, bond-selective UVPD can be used to identify sites of phosphorylation by directing backbone fragmentation at the site of modification.18 All of these previous examples utilized 266 nm photons, which favor direct dissociation but also suffer from weak absorption by many chromophores.
Recently, interest in UVPD at 213 nm has increased because this wavelength is afforded by a solid-state laser, and the photon energy resides at the threshold of absorption by small chromophores such as amide bonds. In theory, this could allow UPVD at 213 nm to favor either direct dissociation or nonspecific dissociation, depending on experimental parameters (such as laser power and excitation time) or molecular composition (most critically, the chromophores available for absorption). Herein, the photochemistry of UVPD at 213 nm is explored for a variety of modified and native peptides. The propensity for bond-selective fragmentation versus undirected UVPD is examined as a function of peptide composition. The capacity for fragmenting both native and synthetically appended C-S bonds with 213 nm photons is investigated. The prospect for using bond-specific C-S bond cleavage to subsequently direct backbone fragmentation at targeted residues is evaluated. Applications of 213 nm light for bond specific cleavage of S-S bonds, which simultaneously yields C-S bond fragmentation, are explored. Advantages and disadvantages relative to similar experiments conducted at 266 nm are discussed.
Experimental Methods
Materials
Peptides RKRRQtSM, RQSVELHsPQSLPR, RGDC, RPHERNGFTVLCPKN, HCLGKWLGHPDKF, NTWTTCQSIAFPSK, and SHLVEALYLVCGERG were purchased from Anaspec (San Jose, CA). SLRRSSCFGGR and CQDSETRTFY were purchased from Abbiotec (San Diego, CA). CDPGYIGSR was purchased from Apexbio, and CGYGPKKKRKVGG was purchased from American Peptide Company (Sunnyvale, CA). GSNKGAIIGLM (Piscataway, NJ) was purchased from GenScript. DRVYIHPF was purchased from Sigma-Aldrich (St. Louis, MO). Naphthoquinone (NQ), iodoacetamide and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO). Benzoquinone (BQ), benzyl mercaptan (BM), and trifluoroacetic acid (TFA) were purchased from Alfa Aesar (Haverhill, MA). Naphthalenethiol (NT) was purchased from Fluka Analytical (Mexico City, Mexico). Iodomethane was purchased from Arcos Organics (Geel, Belgium). Chloramine-T, sodium metabisulfite, and sodium iodide were purchased from Fisher Chemical (Fairlawn, NJ). Acetonitrile (ACN) and methanol were purchased from Fisher Scientific (Waltham, MA). Water was purified by Millipore Direct-Q (Millipore, Billerica, MA). A Macrotrap holder and Macrotrap consisting of polymeric reversed-phase packing material were purchased from Michrom Bioresources, Inc (Auburn, CA).
Peptide Iodination
Peptides were iodinated using a previously published method.20 Briefly, equimolar peptide and sodium iodide were mixed with a two-fold molar excess of chloramine-T for five minutes at room temperature in water. The reaction was then quenched by the addition of 4× molar excess sodium metabisulfite. The products were then purified with a peptide microtrap, rinsed with 0.1% TFA in 90:10 H2O:ACN and eluted with 0.1%TFA in 2%:98% H2O:ACN.
Quinone Modification
Modification of peptides was carried out with either benzoquinone or naphthoquinone based on a previous procedure.16 Stocks of each quinone were prepared fresh prior to each use and stored in the dark to reduce degradation. Quinone stocks were added to peptide solutions in 0.5 to 4× excess of the peptide concentration. The reaction proceeded for 4 hours in the dark at room temperature. Following the reaction time, the solution was purified by microtrap rinsing with 0.1% TFA in 90:10 H2O:ACN and eluted in 0.1%TFA in 2%:98% H2O:ACN. The resulting solution was lyophilized, and the powder was redissolved in 50:50 H2O:MeOH in 0.1% formic acid for a final concentration of 4 µM or 10 µM for MS analysis.
Carbamidomethylation and Methylation of Free Thiols
Carbamidomethylation of cysteine was carried out by preparing a stock solution of 9 mg of iodoacetamide into 1 mL of water. To 25 µL of a 50 mM ammonium bicarbonate buffer, 10 µL of 1 mM peptide was added, followed by 6 µL of the stock iodoacetamide solution. The resulting solution was placed in the dark at room temperature for 20 minutes. The reaction mixture was lyophilized and the powder dissolved in 50:50 H2O:MeOH in 0.1% formic acid for a final concentration of 4 µM or 10 µM for MS analysis.
Methylation was carried out by adding 1µL of iodomethane to 25 µL of ammonium bicarbonate buffer and 10 µL of 1 mM peptide, followed by incubation in the dark at room temperature for 20 min. The modified peptide was purified by peptide trap as described above for MS analysis.
Preparation of Disulfide-Linked Peptides
Disulfide-linked peptides were prepared by adding 10 µL of a 1 mM peptide to 5 µL of DMSO. The reaction was mixed well and placed in a water bath at 37 °C for 18 hours. The resulting solution was lyophilized to remove DMSO and dissolved in 50:50 H2O:MeOH in 0.1% formic acid for a final concentration of 4 µM or 10 µM for MS analysis.
Dephosphorylation and Derivatization of Phosphopeptides
Peptides were dephosphorylated based on previous procedures.19 Briefly, 10 µL of 1 mM peptide stock was combined with 3 µL of saturated barium hydroxide and incubated at 60 °C for 30 min. The dephosphorylated peptides were subsequently modified through the addition of 5 µL of 50 mM naphthalenethiol (2 mg dissolved in 200 µL dioxane), or 2 µL of benzyl mercaptan at 60 °C over 4 hours. Each reagent was 0.5 to 4× excess of the peptide concentration. Purification of the peptides was carried out with a peptide trap. The resulting solution was lyophilized, and the powder was dissolved in 50:50 H2O:MeOH in 0.1% formic acid for a final concentration of 4 or 10 µM for MS analysis. Deamidation of glutamine was observed for only a single residue of the RQSVQLHsAPQSLPR peptide, and it has been previously shown that high temperature can induce deamidation.21
Photodissociation of Derivatized and Non-Derivatized Peptides
UVPD experiments were performed on an Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with a HESI II electrospray source. The HCD vacuum housing was directly modified with a quartz window to transmit the fifth harmonic (213 nm) of a diode-pumped FQSS 213-Q4 laser (Crylas, Berlin, Germany). The pulse energy was 2.5 µJ @ 1000 Hz. Photodissociation was performed by trapping ions without activation in the HCD cell for either 50, 100, or 200 ms (every millisecond equates to approximately a single pulse from the laser system), followed by mass analysis in the Orbitrap. The resolution was set to 30000. Peptides were sprayed at 4 µM concentrations at 3 µL/min with electrospray voltages set between 3 and 4 kV and the capillary inlet temperature set to 300 °C.
An LTQ linear ion trap mass spectrometer (Fisher Scientific, Waltham, MA) with a standard ESI source was utilized for fourth harmonic (266 nm) UVPD experiments. A quartz window was installed on the back plate of the LTQ for transmission of laser pulses from a flash-lamp pumped Nd:YAG Minilite laser (Continuum, Santa Clara, CA). A single pulse of the 4 mJ laser was synchronized to occur at activation step of an MS2 experiment which was triggered by the same external delay generator state above. Peptides were sprayed at 10 µM concentrations at 3 µL/min with electrospray voltages set between 3 and 4 kV and the capillary inlet temperature set to 275 °C.
An LTQ linear ion trap mass spectrometer (Fisher Scientific, Waltham, MA) with a standard ESI source with an OPO tunable laser was utilized for 213 nm UVPD experiments of disulfide-linked peptides. A quartz window was installed on the back plate of the LTQ for transmission of laser pulses from a flash-lamp pumped Nd:YAG Minilite laser (Continuum, Santa Clara, CA). Three pulses of the 0.7 mJ laser were synchronized to occur at activation step of an MS2 experiment. Peptides were sprayed at 10 µM concentrations at 3 µL/min with electrospray voltages set between 3 and 4 kV and the capillary inlet temperature set to 275 °C.
Results/Discussion
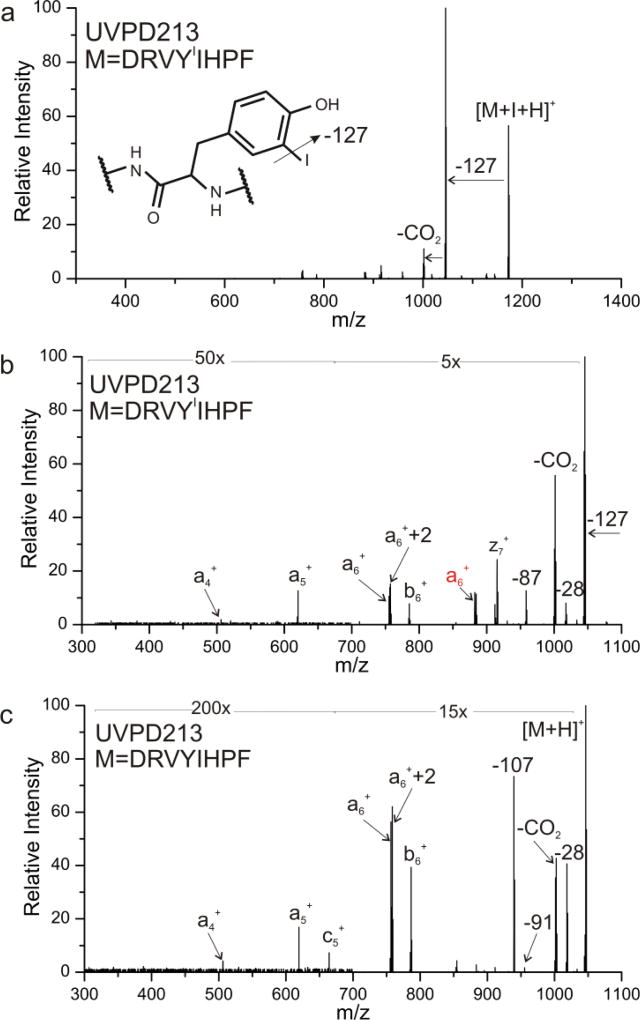
Photoactivation of DRVYIIHPF (where YI = iodotyrosine) at 213 nm produces the spectrum shown in Figure 1a. Loss of iodine yields the most abundant product ion, suggesting that bond-selective fragmentation of the C-I bond is a favorable process. In addition, a subsequent loss of CO2 is noted, as are numerous other fragments at lower intensity. These results bear similarity to those obtained previously22,23 at 266 nm, but loss of iodine is not accompanied by backbone fragmentation at the longer wavelength. Closer examination of the minor products from Figure 1a is shown in the zoomed-in view in Figure 1b. A series of a-type ions and a few other backbone fragments consistent with nonspecific UVPD are noted. Excitation of the same peptide without iodotyrosine is shown in Figure 1c, revealing a similar dissociation pattern to that found in Figure 1b with the exception of a large −107 Da peak. This loss results from direct dissociation and subsequent loss of the tyrosine side chain. This pathway is much less favorable than C-I bond dissociation, rendering it uncompetitive in the presence of iodotyrosine. These results illustrate that both bond-specific dissociation and backbone fragmentation can be produced by 213 nm light. If a favorable direct dissociation pathway is available, such as cleavage of the C-I bond in iodotyrosine, then bond-selective fragmentation will likely dominate. However, in the absence of such favorable pathways, nonspecific backbone fragmentation and side chain losses will yield the most abundant fragments.
Figure 1.
a) Photoactivation of DRVYIIHPF at 213 nm, yielding dominant loss of iodine radical. b) zoom in of minor losses from a). c) Photoactivation of unmodified DRVYIHPF at 213 nm, yielding similar nonspecific fragments.
Photodissociation of native C-S bonds
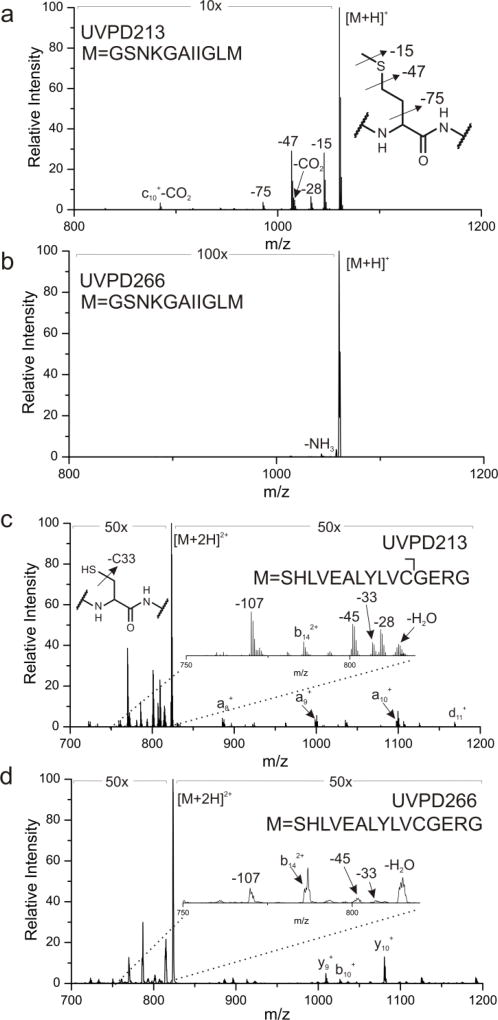
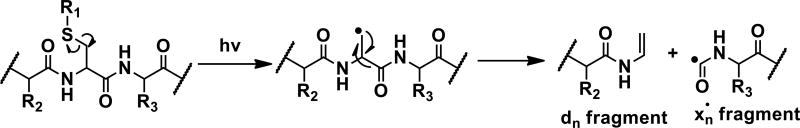
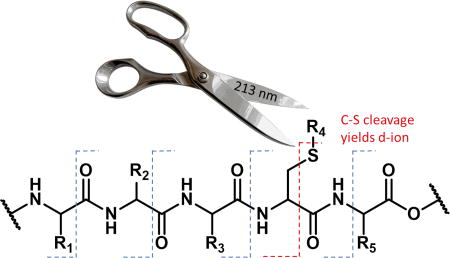
The canonical amino acids cysteine and methionine contain native C-S bonds, which are candidates for bond-selective fragmentation at 213 nm. Excitation of the methionine containing peptide [GSNKGAIIGLM+H]+ with 213 nm photons is illustrated in Figure 2a and leads to cleavage of several side chain bonds, including losses of 15 and 47 Da, corresponding to loss of methyl and thiomethyl radical, respectively. These C-S bond cleavages yield the most abundant products in Figure 2a, suggesting that bond-selective direct dissociation is occurring. In contrast, photoexcitation of the same ion with 266 nm light yields almost no fragmentation and certainly no observable C-S bond cleavage, as shown in Figure 2b. Similar experiments were conducted with [SHLVEALYLVCGERG+2H]2+, which contains a free cysteine residue. Excitation at 213 nm yields a variety of side chain losses and some backbone fragments. Importantly, the loss of 33 Da corresponds to loss of SH and fragmentation of a C-S bond. In this case, the loss of 33 Da generates an alaninyl beta radical that is poised to cleave the backbone and yield a C-terminal d-ion, as shown in Scheme 1. Indeed, a d11+ ion C-terminal to the cysteine residue is the only d-ion observed in the spectrum. Excitation of [SHLVEALYLVCGERG+2H]2+ with 266nm photons produces some dissociation, attributable primarily to absorption of the native chromophore in the side chain of tyrosine, but the loss of 33 Da is barely detectable. In addition, no d-ions are observed. These results reveal that 213 nm photons are more efficient at cleaving C-S bonds than 266 nm photons. It is likely that the dissociative excited state responsible for C-S bond cleavage is not directly accessible at 266 nm.
Figure 2.
UVPD of unmodified [GSNKGAIIGLM+H]+ at either (a) 213 nm or (b) 266 nm. Activation at 213 nm leads to dissociation of both C-S bonds of methionine. (c) 213 nm UVPD of [SHLVEALYLVCGERG+2H]2+ leads to dissociation of the single C-S bond at cysteine. (d) 266 nm activation of the same peptide leads to significantly less C-S dissociation. Relevant side chain mass losses are illustrated in the insets.
Scheme 1.
Pathway for d-ion formation.
Modification of Free Thiols
Reaction of cysteine with quinones leads to nearly quantitative coupling via Michael Addition24 chemistry, allowing for facile and specific cysteine residue labeling in peptides. If the quinone is chromophoric, then modulation of the peptide photochemistry is also possible. The results for several UVPD experiments with modified peptides are shown in Figure 3. Excitation of [RPHERNGFTVLCbqPKN + 3H]3+ (where Cbq = benzoquinone modified cysteine) with 213 nm light yields several side chains losses and backbone fragments, as illustrated in Figure 3a. Cleavage of both C-S bonds is observed, although the abundance does not appear to be enhanced significantly by the benzoquinone chromophore. Backbone fragmentation to generate the d12 ion yields similar abundance to other sites of backbone dissociation, including those at proline that are responsible for the unusual y14-2 and b12+2 ions.25 In contrast, the naphthoquinone modification leads to significant enhancement of C-S bond cleavage, as shown in Figure 3b. In this case, the majority of ions undergo rearrangement subsequent to C-S bond cleavage, leading to the abstraction of additional hydrogens from the peptide prior to quinone loss. In other words, the primary loss is observed at 192 Da instead of the expected 189 Da, but observation of the d12 ion confirms creation of the expected alaninyl radical as a transient intermediate.
Figure 3.
UVPD at 213 nm of [RPHERNGFTVLCPKN]3+ modified with either (a) BQ and (b) NQ at the cysteine residue. Red fragments retain the quinone modification. (c) UVPD at 213 nm of [NTWTTCQSIAFPSK]2+ following modification with iodoacetamide. (d) UVPD at 213 nm of [HCLGKWLGHPDKF]3+ following modification with iodomethane. Dissociation of C-S bonds is noted in the skeletal structure for each modification. Relevant side chain mass losses are illustrated in the insets.
In addition to quinone chemistry, cysteine residues are often capped with acetamide following thiol reduction in proteomics protocols.26 Photoactivation of acetamide capped NTWTTCQSIAFPSK is shown in Figure 3c, revealing surprisingly abundant C-S bond cleavage. Amide functional groups are weak chromophores at 213 nm, which may account for the enhanced fragmentation. Again, the characteristic d6 ion is generated and identifies the sequence position of the cysteine residue. Although not a commonly encountered cysteine modification, we also examined methylated cysteine in the peptide HCLGKWLGHPDKF. Methylated cysteine behaves similarly to methionine, with the exception that a modest d-ion is generated. This result is not unexpected because the cysteine side chain is one methylene unit shorter than methionine and will therefore yield an alaninyl radical following C-S bond cleavage. Comparable experiments at 266 nm yield little to no dissociation of the C-S bond.
C-S bonds formed during Identification of Phosphorylation Site Locations
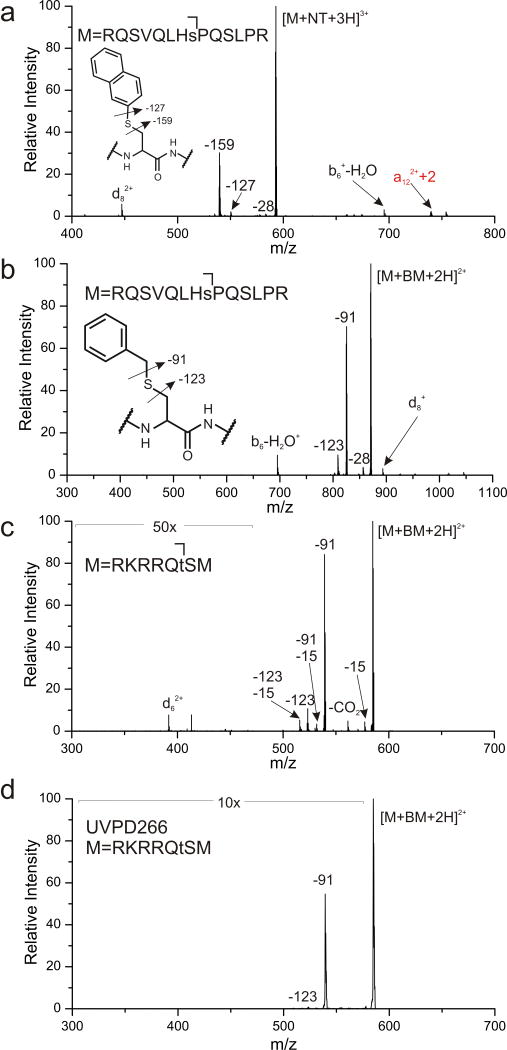
Phosphorylated serine is easily converted into dehydroalanine under basic conditions.27 Michael addition chemistry can then be used to install C-S bonds that will fragment and yield a beta radical, directing backbone dissociation and identifying the original site of phosphorylation with a characteristic d-ion. Excitation at 213 nm of RQSVQLHsAPQSLPR modified with naphthalene thiol at Ser8 (denoted with small cap s) yields cleavage of both C-S bonds and an abundant d8 ion, see Figure 4a. Unfortunately, the placement of sulfur directly adjacent to the aromatic ring makes naphthalenethiol a poor nucleophile, which leads to significantly decreased reactivity and modification yield. In contrast, spacing the sulfur out from the aromatic ring by one carbon serves to enhance nucleophilicity and reactivity. Therefore, the same modification with benzyl mercaptan proceeds much more efficiently, with yields increasing from ~20% to ~100%. However, the chromophore is also spaced away from the C-S bond that must be cleaved to yield the alaninyl radical. At 213 nm, this is not a problem, as shown in Figure 4b. A reasonable amount of the correct C-S bond is cleaved, and the d8 ion is easily detected. Similar results are obtained for the benzyl mercaptan modification of a phosphorylated threonine containing peptide, RKRRQtSM, as shown in Figure 4c. Importantly, for RKRRQtSM, the modification yield increases from ~0% to ~100% for naphthalene thiol and benzyl mercaptan, respectively. The ability to use the more reactive thiol is therefore a significant advantage for UVPD at 213 nm. This point is illustrated further in Figure 4d where photoactivation of the same peptide at 266 nm fails to produce significant yield of the −123 Da loss or a detectable d-ion.
Figure 4.
(a) UVPD at 213 nm of naphthalene thiol modified [RQSVQLHsPQSLPR]3+ leads to C-S bond dissociation. (b) UVPD at 213 nm of benzyl mercaptan modified [RQSVQLHsPQSLPR]2+ and (c) [RKRRQtSM]2+ yields loss C-S bond cleavage and d-ion formation. (d) 266 nm activation of BM modified [RQSVQLHsPQSLPR]2+.
Gas Phase Disulfide Identification
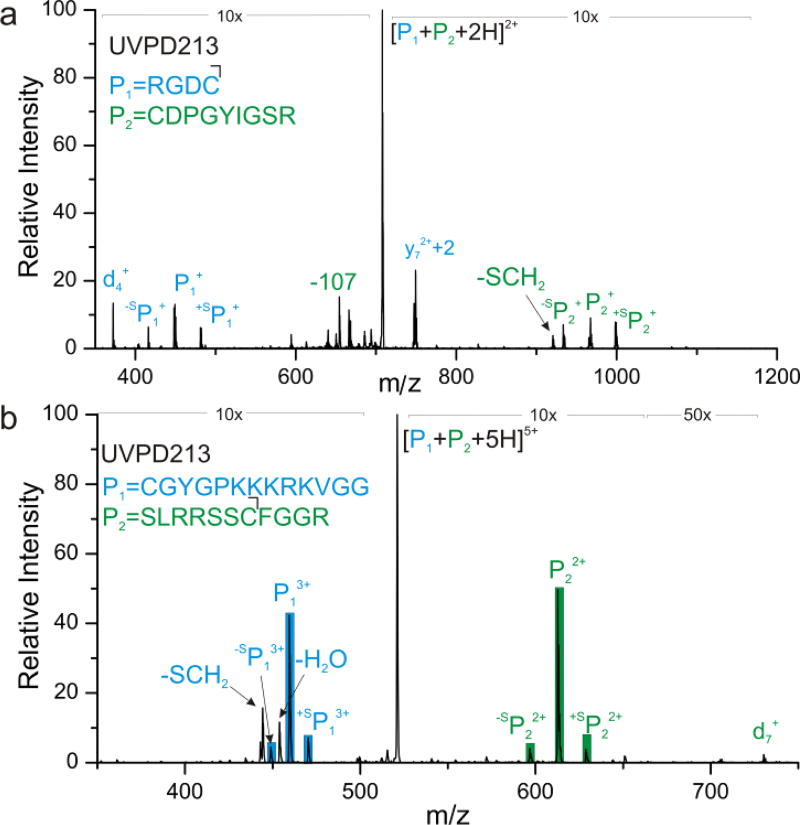
Identification of disulfide bond partners in proteins is an active area of interest. Previous results have demonstrated that 266 nm light effectively cleaves S-S bonds, enabling facile identification of peptides linked by disulfide bonds.17 Similar experiments were conducted at 213 nm and the results are shown in Figure 5. Photoactivation of disulfide-bound peptides RGDC and CDPGYIGSR yields the spectrum in Figure 5a. Similar to results obtained at 266 nm, S-S bond fracture is favorable. In addition, 213 nm light also cleaves the adjacent C-S bonds, yielding peaks at ±32 Da relative to the S-S bond dissociation products. The end result is a signature ‘triplet’ that can be used to easily identify the presence of a disulfide bond and the masses of the constituent peptides that were connected by it. In addition, several other UVPD type fragments are observed, such as the y7+2 ion, characteristic of proline. More importantly, the C-S bond fragmentations again yield beta radicals that can produce signature d-ions, identifying the location of the linking cysteine residues. Indeed, a d4 ion correctly identifies the location of cysteine in the RGDC peptide. Location of cysteine at the N-terminus prevents observation of the corresponding d-ion for CDPGYIGSR. Examination of disulfide-bound CGYGPKKKRKVGG and SLRRSSCFGGR yields similar results, as shown in Figure 5b. The abundance of S-S bond cleavage is higher for this peptide, which may be related to energy transfer from tyrosine.28 Nevertheless, significant cleavage of C-S bonds is also observed, and an easily detectable d7 ion is produced, revealing the location of the cysteine residue in SLRRSSCFGGR. Interestingly, the results shown in Figure 5 were obtained with a relatively high energy pulse (~0.4mJ, one shot), but repeated excitation at lower pulse energy (~1.5µJ/pulse) yields less prominent triplet formation (see supporting information). In particular, it appears that the β-carbon radical product generated by C-S bond cleavage is less stable than the corresponding thiyl or perthiyl radicals, and it undergoes additional losses upon repeated exposure to 213 nm light.
Figure 5.
UVPD of (a) RGDC and CDPGYIGSR (b) CGYGPKKKRKVGG and SLRRSSCFGGR each disulfide-linked. Dissociation of the disulfide bond as well as each adjacent C-S bond is identified for each peptide pair.
Conclusion
Photoactivation of peptides with 213 nm photons can be used to drive both bond-specific dissociation and traditional nonspecific UVPD. 213 nm works well for bond-selective photodissociation of labile bonds previously found to undergo direct dissociation at 266 nm. In addition, many native and non-native carbon-sulfur bonds are labile at 213 nm, including several that generate alaninyl radicals that initiate beta dissociation of the peptide backbone at the radical site. The signature d-ions generated by this mechanism can be used to site-specifically identify the location of cysteine or phosphorylated serine/threonine residues. The expanded list of labile carbon-sulfur bonds also allows for more reactive groups to be used in the derivatization of dehydroalanine, enabling quantitative conversion to a photoactive species. For disulfide bond characterization, signature triplets are generated at 213 nm due to dissociation of both the sulfur-sulfur and carbon-sulfur bonds, facilitating identification. 213 nm is a promising wavelength for the further expansion of bond-specific photodissociation and its applications for biomolecular characterization.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge assistance from John Syka, Chris Mullen, Chad Weisbrod, Jens Griep-Raming, and Jenny Brodbelt with interfacing the laser with the orbitrap. The NIH is thanked for financial support (NIGMS grant R01GM107099).
References
- 1.Brodbelt JS. Photodissociation Mass Spectrometry: New Tools for Characterization of Biological Molecules. Chem. Soc. Rev. 2014;43(8):2757. doi: 10.1039/c3cs60444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells JM, McLuckey SA. Collision-Induced Dissociation (CID) of Peptides and Proteins. Biological Mass Spectrometry. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 3.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. SPECIAL FEATURE : Mobile and Localized Protons : A Framework for Understanding Peptide Dissociation. 2000 Sep;1406:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 4.Tureček F, Julian RR. Peptide Radicals and Cation Radicals in the Gas Phase. Chem. Rev. 2013;113(8):6691–6733. doi: 10.1021/cr400043s. [DOI] [PubMed] [Google Scholar]
- 5.R Julian R. The Mechanism Behind Top-Down UVPD Experiments: Making Sense of Apparent Contradictions. J. Am. Soc. Mass Spectrom. 2017;28(9):1823–1826. doi: 10.1007/s13361-017-1721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ly T, Julian RR. Ultraviolet Photodissociation: Developments towards Applications for Mass-Spectrometry-Based Proteomics. Angew. Chemie-International Ed. 2009;48(39):7130–7137. doi: 10.1002/anie.200900613. [DOI] [PubMed] [Google Scholar]
- 7.Kim TY, Thompson MS, Reilly JP. Peptide Photodissociation at 157 Nm in a Linear Ion Trap Mass Spectrometer. Rapid Commun. Mass Spectrom. 2005;19(12):1657–1665. doi: 10.1002/rcm.1969. [DOI] [PubMed] [Google Scholar]
- 8.Morgan JW, Hettick JM, Russell DH. Peptide Sequencing by MALDI 193-Nm Photodissociation TOF MS. Biological Mass Spectrometry. 2005;402:186–209. doi: 10.1016/S0076-6879(05)02006-9. [DOI] [PubMed] [Google Scholar]
- 9.Choi KM, Yoon SH, Sun ML, Oh JY, Moon JH, Kim MS. Characteristics of Photodissociation at 193 Nm of Singly Protonated Peptides Generated by Matrix-Assisted Laser Desorption Ionization (MALDI) J. Am. Soc. Mass Spectrom. 2006;17(12):1643–1653. doi: 10.1016/j.jasms.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Morrison LJ, Brodbelt JS. 193 Nm Ultraviolet Photodissociation Mass Spectrometry of Tetrameric Protein Complexes Provides Insight into Quaternary and Secondary Protein Topology. J. Am. Chem. Soc. 2016;138(34):10849–10859. doi: 10.1021/jacs.6b03905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, Ahn W-K, Lee S, Han SY, Rhee BK, Oh H Bin. Ultraviolet Photodissociation at 266 Nm of Phosphorylated Peptide Cations. Rapid Commun. Mass Spectrom. 2009;23(23):3609–3620. doi: 10.1002/rcm.4184. [DOI] [PubMed] [Google Scholar]
- 12.Theisen A, Yan B, Brown JM, Morris M, Bellina B, Barran PE. Use of Ultraviolet Photodissociation Coupled with Ion Mobility Mass Spectrometry To Determine Structure and Sequence from Drift Time Selected Peptides and Proteins. Anal. Chem. 2016;88(20):9964–9971. doi: 10.1021/acs.analchem.6b01705. [DOI] [PubMed] [Google Scholar]
- 13.Wilson JJ, Brodbelt JS. MS/MS Simplification by 355 Nm Ultraviolet Photodissociation of Chromophore-Derivatized Peptides in 4-3 Quadrupole Ion Trap. Anal. Chem. 2007;79(20):7883–7892. doi: 10.1021/ac071241t. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks NG, Julian RR. Leveraging Ultraviolet Photodissociation and Spectroscopy to Investigate Peptide and Protein Three-Dimensional Structure with Mass Spectrometry. Analyst. 2016;141(15):4534–4540. doi: 10.1039/c6an01020b. [DOI] [PubMed] [Google Scholar]
- 15.Ly T, Julian RR. Residue-Specific Radical-Directed Dissociation of Whole Proteins in the Gas Phase. J. Am. Chem. Soc. 2008;130(1):351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 16.Diedrich JK, Julian RR. Facile Identification of Photocleavable Reactive Metabolites and Oxidative Stress Biomarkers in Proteins via Mass Spectrometry. Anal. Bioanal. Chem. 2012;403(8):2269–2277. doi: 10.1007/s00216-012-5867-0. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal A, Diedrich JK, Julian RR. Direct Elucidation of Disulfide Bond Partners Using Ultraviolet Photodissociation Mass Spectrometry. Anal. Chem. 2011;83(17):6455–6458. doi: 10.1021/ac201650v. [DOI] [PubMed] [Google Scholar]
- 18.Diedrich JK, Julian RR. Site-Specific Radical Directed Dissociation of Peptides at Phosphorylated Residues. J. Am. Chem. Soc. 2008;130(37):12212–12213. doi: 10.1021/ja8023719. [DOI] [PubMed] [Google Scholar]
- 19.Diedrich JK, Julian RR. Site-Selective Fragmentation of Peptides and Proteins at Quinone-Modified Cysteine Residues Investigated by ESI-MS. Anal. Chem. 2010;82(10):4006–4014. doi: 10.1021/ac902786q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regoeczi E. Iodine-Labeled Plasma Proteins. CRC Press; Boca Raton, FL: 1984. [Google Scholar]
- 21.Scotchler JW, Robinson AB. Deamidation of glutaminyl residues: Dependence on pH, temperature, and ionic strength. Anal. Biochem. 1974;59:319–322. doi: 10.1016/0003-2697(74)90040-2. [DOI] [PubMed] [Google Scholar]
- 22.Ly T, Julian RR. Residue-Specific Radical-Directed Dissociation of Whole Proteins in the Gas Phase. J. Am. Chem. Soc. 2008;130(1):351–358. doi: 10.1021/ja076535a. [DOI] [PubMed] [Google Scholar]
- 23.Sun Q, Nelson H, Ly T, Stoltz BM, Julian RR. Side Chain Chemistry Mediates Backbone Fragmentation in Hydrogen Deficient Peptide Radicals. J. Proteome Res. 2009;8:958–966. doi: 10.1021/pr800592t. [DOI] [PubMed] [Google Scholar]
- 24.Nair DP, Podgórski M, Chatani S, Gong T, Xi W, Fenoli CR, Bowman CN. The Thiol-Michael Addition Click Reaction: A Powerful and Widely Used Tool in Materials Chemistry. Chem. Mater. 2014;26:724–744. [Google Scholar]
- 25.Girod M, Sanader Z, Vojkovic M, Antoine R, MacAleese L, Lemoine J, Bonacic-Koutecky V, Dugourd P. UV Photodissociation of Proline-containing Peptide Ions: Insights from Molecular Dynamics. J. Am. Soc. Mass Spectrom. 2015;26:432–443. doi: 10.1007/s13361-014-1038-1. [DOI] [PubMed] [Google Scholar]
- 26.Gundry RL, White MY, Murray CI, Kane LA, Fu Q, Stanley BA, Van Eyk JE. Preparation of Proteins and Peptides for Mass Spectrometry Analysis in a Bottom-Up Proteomics Workflow. Curr. Protoc. Mol. Biol. 2009 doi: 10.1002/0471142727.mb1025s88. CHAPTER, Unit10.25-Unit10.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm C, Schröder S, Glückmann M, Beyermann M, Krause E. Derivatization of phosphorylated peptides with S- and N-nucleophiles for enhanced ionization efficiency in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:2697–2705. doi: 10.1002/rcm.1676. [DOI] [PubMed] [Google Scholar]
- 28.Hendricks NG, Julian RR. Characterizing Gaseous Peptide Structure with Action-EET and Simulated Annealing. Phys. Chem. Chem. Phys. 2015;17(39):25822–25827. doi: 10.1039/c5cp01617g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.