Abstract
The gastrointestinal tract is a complex environment in which the host immune system interacts with a diverse array of microorganisms, both symbiotic and pathogenic. As such, mobilizing a rapid and appropriate antimicrobial response depending on the nature of each stimulus is crucial for maintaining the balance between homeostasis and inflammation in the gut. Here we focus on the mechanisms by which intestinal antimicrobial peptides regulate microbial communities during dysbiosis and infection. We also discuss classes of bacterial peptides that contribute to reducing enteric pathogen outgrowth. This review aims to provide a comprehensive overview on the interplay of diverse antimicrobial responses with enteric pathogens and the gut microbiota.
Keywords: Antimicrobial peptides, microbiota, enteric pathogens, nutritional immunity, microcins, bacteriocins
1. Introduction
The mammalian intestine is faced with the constant challenge of communicating with the external environment, which include commensal microbes collectively known as the gut microbiota. Extensive co-evolution between the gut microbiota, their metabolic byproducts, and the host has resulted in a symbiotic relationship that contributes to shaping mammalian immune responses [1]. Disruptions in this homeostasis, known as dysbiosis, are associated with a plethora of disorders including inflammatory bowel disease (IBD), metabolic disorders, and cardiovascular disease [1, 2], suggesting that balanced intestinal microbiology ecology is crucial for maintaining a healthy immune system. In addition to non-infectious insults, the gut serves as a portal of entry for many food-borne bacterial pathogens. In order to successfully colonize the mammalian gut, enteric pathogenic bacteria must first encounter the commensal microbiota and establish an environmental niche, thereby disturbing the intestinal equilibrium. Thus, host responses are adapted to promote tolerance of commensals, as well as to confer protection against infection.
One of the mechanisms by which the host controls the microbiota and defends against infection with pathogens is the production of antimicrobial peptides (AMPs). AMPs are a diverse class of molecules that exert their antimicrobial activity by different mechanisms, including the disruption of bacterial membranes, and the sequestration of essential nutrients. In the gut, AMPs are produced by enterocytes and by Paneth cells, which are specialized epithelial cells that are positioned in the small intestinal crypts, in close proximity to the stem cells from which all the distinct lineage of gut epithelial cells originate. In addition, goblet cells produce mucin, which contributes to host defense by providing a physical separation between the epithelial layer and the gut microbiota. Given the importance of the gut epithelium for the maintenance of intestinal homeostasis, even slight perturbations in epithelial cell function can have drastic consequences. For example, defects in AMP responses increase host susceptibility to infection with enteric pathogens, including but not limited to, Yersinia pseudotuberculosis, Listeria monocytogenes and Citrobacter rodentium [3–6]. Moreover, individuals with IBD exhibit reduced intestinal AMP expression, which may contribute to the observed alterations in the gut microbiota [7, 8]. Thus, stringent regulation of AMP production is essential for limiting both commensal- and infection-induced dysbiosis. In addition to host-derived AMPs, peptides from bacteria have been shown to exhibit antimicrobial activity against other microbes, in some cases providing an effective weapon against pathogens.
Here we discuss the mechanisms by which intestinal AMPs promote host defense against bacterial infection, and also cover the strategies that certain enteric pathogens employ to subvert these activities. To a lesser extent, we highlight the contribution of host-derived AMPs for the maintenance of intestinal homeostasis. Equally as important are the AMP responses at other epithelial sites, including the lung, skin, and reproductive tract, which have been extensively reviewed elsewhere [9–13]. Furthermore, we describe the classes of bacterial-derived peptides that have recently been identified to restrict pathogen colonization in the intestine. We finally consider harnessing AMP activities as potential therapeutics for the treatment of gastrointestinal infections.
2. Host-derived AMPs that directly target bacteria
A major mechanism by which many AMPs mediate killing is by direct interaction with structural components of bacterial membranes, leading to pore-formation, subsequent loss of membrane potential, and eventual cell lysis [14]. One such family includes defensins, which are small, cationic peptides. The two major subfamilies, α- and β-defensins (encoded by the DEFA and DEFB genes, respectively), are classified based on structural differences in the pairing of their cysteine residues while a third group, the θ-defensins, adopt a distinct cyclical conformation [15]. In humans, protein products of six α- and ten β-defensins have been identified [16, 17], while θ-defensins have only been found in non-human primates [18].
Human α-defensins were initially identified as neutrophil protein products that exhibited antimicrobial activity, and denoted as human neutrophil peptides (hNP) 1–4 [19–21]. In addition to direct membrane disruption by these peptides, some members such as hNP-1, can interact with and sequester lipid II to inhibit bacterial cell wall synthesis as a mechanism of microbial killing [22, 23]. The only α-defensins that are produced in the human gut are human defensin 5 and 6 (HD5 and HD6), with their expression restricted to Paneth cells [24]. Importantly, HD5 and HD6 are synthesized as inactive precursors that rely on proteolysis by trypsin for their maturation and antibacterial function [25, 26]. Although in vitro studies have extensively demonstrated the antimicrobial properties of HD5 [26, 27], the contribution of this α-defensin for enteric immunity remained elusive until the development of an elegant transgenic mouse model. In particular, mice genetically modified to express HD5 in Paneth cells conferred resistance to oral challenge with the enteric pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) (Figure 1A) [28]. This protection was specific to the intestinal compartment, as HD5 became dispensable for host protection towards systemic delivery of pathogens [28]. Later studies also ascribed a role for HD5 in regulating the intestinal microbiota in the absence of infection by significantly reducing the levels of segmented filamentous bacteria (Figure 1B) [29], commensals belonging to the Clostridiales family that have important immunomodulatory properties (rev in [30]). Although HD5 and HD6 share structural similarities, HD6 exhibits negligible direct antimicrobial activity [31–33]. Instead, HD6 confers host protection by self-oligomerizing into nanonets which subsequently surround and entrap pathogens by binding to conserved surface structures (e.g. flagellin, fimbriae, or invasion apparatuses) [31]. As such, HD6 restricts S. Typhimurium infection by limiting intestinal epithelial cell invasion (Figure 1C) [31]. Importantly, this antimicrobial activity of HD6 also extends to Yersinia enterocolitica and L. monocytogenes in vitro [31, 34], suggesting a broad antimicrobial function for this peptide. Similar to HD5 and HD6, production of α-defensins in mice (more commonly referred to as cryptdins) is restricted to Paneth cells [35, 36], and requires post-translation processing by proteases for their maturation [37, 38]. Studies with cryptdins have uncovered their antimicrobial activities in vitro [39, 40], and their importance in vivo is illustrated by the finding that mice that lack mature cryptdins exhibit increased susceptibility to oral challenge with S. Typhimurium (Figure 1D) [38]. Moreover, the finding that S. Typhimurium can reduce expression of mouse cryptdins [41] further implies the contribution of these peptides for host defense in the intestinal compartment.
Figure 1. In vivo functions of intestinal antimicrobial peptides.
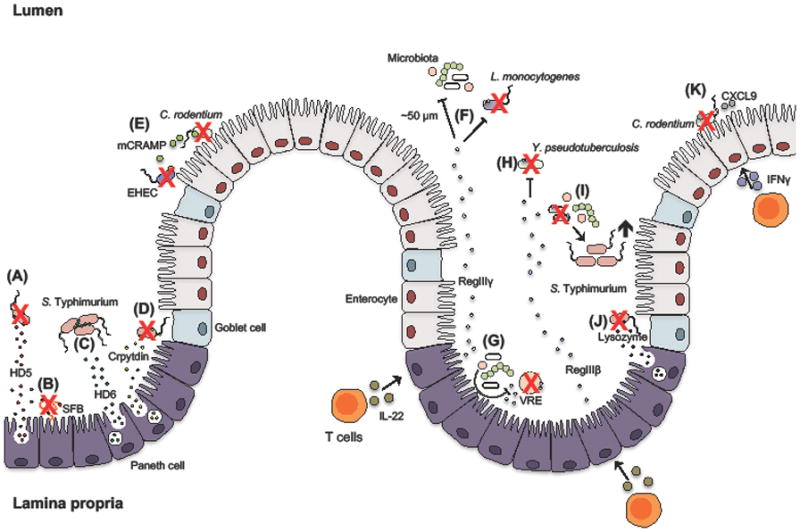
(A) Paneth cell secretion of human defensin 5 (HD5) contributes to decreasing intestinal S. Typhimurium burden and (B) also strongly reduces the levels of commensal segmented filamentous bacteria (SFB). (C) Contrary to HD5, HD6 contributes to host defense against S. Typhimurium through formation of oligomers that entrap the pathogen. (D) Furthermore, mouse α-defensins (cryptdins) promote host defense against S. Typhimurium. (E) Epithelial cell-derived murine CRAMP (mCRAMP) contributes to killing of C. rodentium and enterohemorrhagic E. coli (EHEC). Robust production of interleukin (IL)-22 induces Paneth cells to express the C-type lectins RegIIIγ and RegIIIβ. (F) RegIIIγ maintains spatial separation between the microbiota and the epithelial cell layer and also facilitates clearance of pathogenic L. monocytogenes. (G) Members of the microbiota can furthermore induce the expression of RegIIIγ, which promotes resistance to Vancomycin-Resistant Enterococcus (VRE). (H) RegIIIβ is important for host protection against enteric Y. pseudotuberculosis but can also (I) target the enteric microbiota and consequently prolong S. Typhmurium infection. (J) Secretion of lysozyme by Paneth cells promote the reduction of intestinal S. Typhimurium burden. (K) Lastly, interferon gamma (IFNγ)-inducible chemokines, such as CXCL9, mediate clearance of the mouse pathogen C. rodentium by a mechanism independent of their chemotactic ability.
In contrast to intestinal α-defensin expression by Paneth cells, β-defensins are expressed by enterocytes of both the small and large intestines [42]. In humans, the most-studied intestinal β-defensins are human β-defensins 1–4 (hBD-1, hBD-2, hBD-3, and hBD-4). Regulation of these various peptides is distinct: hBD-1 appears to be constitutively expressed [42], hBDs 2–3 are upregulated following exposure to infectious and inflammatory stimuli [43–45], and hBD4 levels are increased in colonic epithelial cells from patients with ulcerative colitis [46]. The in vitro antimicrobial activity of hBDs 2–4 against bacterial pathogens including Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pyogenes is well-established [47–50]. Interestingly, direct killing by hBD-1 under aerobic conditions is relatively poor compared to the other defensins [51]. Surprisingly, hBD-1 underwent a dramatic conformational change in anaerobic conditions, similar to the environment of the intestine, which revealed its potent antimicrobial activity against Gram-positive commensals [52]. Recent evidence shows that this form of hBD-1 can also function in a novel capacity by entrapping bacteria [53]. Similar to hBDs 1–4, murine β-defensins (mBDs) exhibit broad-spectrum antimicrobial activity in vitro [54–56], and their expression can be induced in intestinal epithelial cells [55]. Nevertheless, the in vivo requirement of mBDs for controlling intestinal pathogens remains to be determined.
A second class of cationic peptides that function by directly disrupting bacterial membranes are the cathelicidins. There have been over 30 cathelicidins identified in mammalian species (rev in [57]), including mouse mCRAMP and its human ortholog LL-37 [58, 59]. Intestinal cathelicidins are constitutively expressed by colonic epithelial cells during homeostasis [60], but may be markedly upregulated in response to enteric infection [60, 61]. Similar to α-defensins, cathelicidins are synthesized as inactive forms that require proteolytic processing; the human precursor hCAP18 is cleaved at the C-terminus to liberate the mature LL-37 peptide [62]. Notably, cathelicidin deficiency increases susceptibility to infection with enterohemorrhagic E. coli (EHEC) and C. rodentium (Figure 1E), highlighting the importance of this peptide for maintaining intestinal immunity against pathogens [6, 63]. Given the importance of cathelicidin for intestinal immunity it is perhaps unsurprising that pathogens, including the Shigella spp., are able to counteract these responses by downregulating cathecidin expression [64]. Conversely, increasing cathelicidin expression by administration of the short chain fatty acid butyrate reduces the severity of shigellosis [65].
Similar to defensins and cathelicidins, the regenerating (Reg) protein family is a group of soluble lectins that interact with structural components on the bacterial surface. RegIIIγ and its human ortholog RegIIIα (also known as HIP/PAP) are expressed by Paneth cells [66, 67] and enterocytes [68] in response to various stimuli including infection, inflammation, and Toll-like receptor signaling [4, 66, 69–71]. In homeostatic conditions, RegIIIγ regulates intestinal homeostasis by maintaining a physical separation between epithelial cells and the microbiota (Figure 1F) [72]. Another well-studied murine isoform, RegIIIβ, is often co-regulated alongside RegIIIγ [69, 70]. Like the aforementioned cationic AMPs, maturation of the Reg proteins relies on proteolytic processing, specifically trypsin-mediated removal of an N-terminal fragment [73–75]. Mechanistically, RegIII lectins are selective for Gram-positive bacteria through interaction with cell wall peptidoglycan [66, 76, 77], subsequently oligomerizing to induce pore formation [77]. Recent studies suggest that some members, such as RegIIIβ, can also target Gram-negative bacteria through interaction with surface Lipid A moieties [75, 78]. The importance of RegIIIγ and RegIIIβ for host protection is emphasized by the increased susceptibility of mice defective in these host responses to intestinal infection with L. monocytogenes (Figure 1F), and Vancomycin-Resistant Enterococcus (VRE) (Figure 1G), and Y. pseudotuberculosis (Figure 1H), [4, 5, 71]. Paradoxically, RegIIIβ deficiency can prolong enteric S. Typhimurium infection in specific cases by suppressing the recovery of a balanced microbial ecosystem (Figure 1I) [79], implying that the contribution of lectins may be distinct for different pathogens.
Another mechanism by which AMPs target pathogens is through enzymatic degradation of bacterial membranes. One such protein, lysozyme (reviewed in [80]), specifically hydrolyzes peptidoglycan linkages while a second protein, secretory phospholipase A2 (sPLA2), targets membrane phospholipids [81]. Both enzymes are produced and secreted by Paneth cells [82–84], and have a preference towards Gram-positive pathogens [85, 86]. Importantly, infection with intestinal pathogens disrupts secretory processes [87], which can interfere with AMP delivery into the intestinal lumen. Host cells subsequently employ alternative compensatory mechanisms in order to preserve intact immune responses. In particular, Paneth cells maintain their antimicrobial potency during S. Typhimurium infection by rerouting lysozyme to be secreted by secretory autophagy (Figure 1J) [88].
Other host factors that exhibit antimicrobial activity and are also expressed by intestinal cells include chemokines and members of the ribonuclease (RNase) superfamily. Chemokines are small, secreted molecules best known for their ability to orchestrate immune cell migration to various tissues. However, emerging evidence indicates that many chemokines may also function as AMPs (rev. in [89]). In particular, the interferon-inducible CXCL9, CXCL10, and CXCL11 chemokines have been shown to display antimicrobial activity against Bacillus anthracis, E. coli, and L. monocytogenes in vitro [90–93], and towards B. anthracis in vivo [94]. Mechanistically, these chemokines function either through directly disrupting bacterial membranes [92], or by targeting bacterial enzymes [93] and transporters [91, 92]. Importantly, induction of CXCL9, CXCL10, and CXCL11 by intestinal epithelial cells [95] suggests that these chemokines may play a role also in host defense against enteric pathogens. Indeed, intestinal CXCL9 is important for protection against C. rodentium infection, independently of its function in cellular recruitment (Figure 1K) [96]. The RNase angiogenin 4 (Ang4) is induced in Paneth cells following exposure to commensal microbiota [97–99]. Similar to other RNases [100, 101], Ang4 exhibits activity against both Gram-positive and Gram-negative pathogens in vitro [98, 99], although the requirement of enzymatic activity for its bactericidal function is unknown. Thus, further studies into the mechanisms by which additional chemokines and RNases mediate bacterial killing may potentially lead to the development of novel therapeutic strategies.
3. Host-derived AMPs that restrict bacterial growth by indirect mechanisms
Restriction of bacterial replication can also occur when host antimicrobial factors compete with microorganisms for essential micronutrients in a process termed nutritional immunity [102, 103]. In particular, transition metal ions are required for a myriad of biological processes, including nucleic acid and protein synthesis, precursor biosynthesis, and responses to oxidative stress [104]. Iron is among the most abundant, albeit tightly regulated transition metals in biological systems. The majority of iron in eukaryotes is stored intracellularly associated with hemoglobin, and extracellular iron is associated with the high-affinity proteins lactoferrin and transferrin [105]. Thus, iron availability to pathogens is severely restricted, and is further reduced during infection [105]. Pathogens have evolved various mechanisms to counteract iron limitation by the mammalian host; one of these mechanisms is the production and expressions of siderophores. Siderophores are low molecular weight iron-binding compounds that are produced and secreted by bacteria to aid in iron acquisition (rev. in [106]). The affinity of bacterial siderophores exceeds that of host iron-binding proteins, thus enabling pathogens to hijack iron from the host. For example, members of the Enterobactericeae family, including Salmonella spp., Escherichia spp., and Klebsiella spp. synthesize the siderophore enterobactin (also called enterochelin) [107–110], which has higher affinity for iron than host proteins like transferrin and lactoferrin [111]. To counteract bacterial siderophores, host cells produce lipocalin-2 (also known as siderocalin, 24p3, or neutrophil gelatinase-associated lipocalin, NGAL), an antimicrobial protein that sequesters enterobactin [112, 113]. In the inflamed intestine, lipocalin-2 is primarily expressed and secreted by epithelial cells following stimulation by IL-17 and IL-22 (Figure 2A) [114, 115], T cell cytokines that are highly upregulated during infectious and non-infectious colitis [69, 70, 114–116]. These cytokines also stimulate intestinal epithelial cells to produce CXC chemokines, which in turn promote the migration of neutrophils to the gut (Figure 2B) [117]. Neutrophils contain lipocalin-2 in their granules [118], and thus are an additional source of lipocalin-2 during enteric infection and inflammation [114, 119]. To overcome lipocalin-2-mediated nutritional immunity, certain pathogens produce “stealth” siderophores [120]. For instance, S. Typhimurium, Klebsiella pneumoniae, and uropathogenic E. coli synthesizes salmochelin, a C-glucosylated derivative of enterobactin, which cannot be bound by lipocalin-2 [114, 121, 122]. Salmochelin production thus enables pathogenic microbes to evade lipocalin-2-mediated iron sequestration and thrive in the host (Figure 2C) [114, 121, 123]. Evasion of lipocalin-2, however, does not appear to be a trait that is unique to pathogens. The probiotic bacterium E. coli Nissle 1917 (EcN) also produces salmochelin, as well as other stealth siderophores, which allow EcN to effectively compete with S. Typhimurium for iron in the inflamed gut [124].
Figure 2. Mechanisms of nutritional immunity in the inflamed intestine.
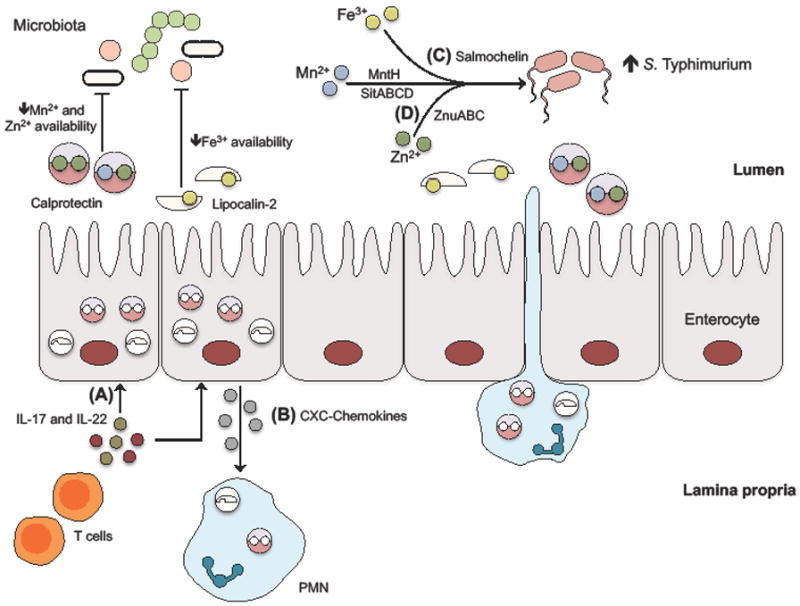
(A) During infection with enteric pathogens, high levels of interleukin (IL)-17 and IL-22 stimulate intestinal epithelial cells to express the antimicrobial protein calprotectin, which sequesters zinc and manganese, and lipocalin-2, which sequesters iron. (B) In addition, IL-17 and IL-22 also stimulate epithelial-dependent CXC-chemokine production, which recruit polymorphonucleaer cells (PMNs) to provide an additional source of lipocalin-2 and calprotectin. Nutrient-limitation by these two antimicrobial proteins restricts growth of the commensal microbiota. (C) However, successful pathogens like S. Typhimurium can overcome iron restriction by producing salmochelin, a “stealth” siderophore that is not bound by lipocalin-2. (D) Moreover, S. Typhimurium encodes high-affinity ZnuABC, MntH, SitABCD transporters to acquire zinc and manganese, which counteracts calprotectin-dependent sequestration of these metals.
Zinc and manganese are other important transition metal nutrients, although relatively little is known in comparison to iron regarding their importance in host-microbe interactions. Many bacteria can utilize these ions as cofactors for carbon metabolism, and also for the detoxification of reactive oxygen and nitrogen species [104, 125]. Uptake of these nutrients is mediated by specialized zinc and manganese import systems, including the transporters ZnuABC for zinc and MntH for manganese (rev. in [126, 127]). Importantly, defects in these transport systems are detrimental for bacteria under nutrient-limiting conditions, both in vitro and in vivo [128–132]. Given the requirement of these micronutrients for bacterial growth, host cells employ mechanisms to chelate zinc and manganese to reduce availability of these cofactors during infection. One such host protein, calprotectin, is a complex of the two calcium-binding proteins S100A8 and S100A9 [133]. Formation of the calprotectin heterodimer introduces two metal binding sites; site I is capable of binding to zinc and manganese with high affinity while site II only binds to zinc [134, 135]. In anaerobic, highly reducing conditions, site I also binds to ferric iron [136], although the relevance of this interaction in vivo remains unknown. The contribution of calprotectin for promoting host defense against S. aureus highlights the importance of zinc and manganese sequestration during infection [137]. However, pathogens are able to evade calprotectin-mediated metal sequestration to cause disease. Specifically, expression of zinc transporters facilitates Acinetobacter baumannii lung colonization and systemic dissemination by overcoming calprotectin-dependent zinc sequestration. [138]. Furthermore, S. aureus employs the MntABC and MntH transporters to counteract calprotectin-dependent manganese sequestration [130]. During intestinal infection, calprotectin is upregulated by epithelial cells in response to IL-17 and IL-22 and is also released into the intestinal lumen by translocating neutrophils (Figure 2A) [139], likely associated with neutrophil extracellular traps [140]. Notably, the measurement of calprotectin as a prognostic and diagnostic marker in patients with IBD illustrates its abundance during enteric inflammation [141]. Therefore, enteric pathogens must also evade calprotectin-mediated metal sequestration in the intestinal compartment. Indeed, S. Typhimurium is able to resist zinc limitation by expressing the high-affinity transporter ZnuABC, which enables the pathogen to outcompete the microbiota in the inflamed gut (Figure 2D) [139]. Similarly, acquisition of manganese by the MntH and SitABCD transporters is essential for S. Typhimurium to evade neutrophil-dependent oxidative stress and calprotectin, and promote the pathogen survival in the inflamed intestine (Figure 2D) [142].
Whether metal nutrients availability and nutritional immunity modulate the enteric microbiota is relatively understudied. However, recent work has uncovered the contribution of micronutrients and metal-binding proteins for the maintenance of a balanced microbiota. For example, an excess of dietary zinc reduces the microbial diversity in the gut, which in turn exacerbates Clostridium difficile-associated disease [143]. Moreover, during intestinal inflammation, IL-22-dependent induction of lipocalin-2 and calprotectin leads to a suppression of commensal Enterobacteriaceae, and enhances S. Typhimurium intestinal colonization [69]. Lipocalin-2 has also recently been shown to limit the replication of commensal microbes during severe colitis and tumorigenesis [144]. Abrogating lipocalin-2 in the context of IL-10-deficiency exacerbates the severity of colitis and tumorigenesis, which could be ameliorated by antibiotic administration [144, 145]. Further analyses revealed an enrichment of the intestinal facultative pathogenic Alistipes spp. in Lcn2 −/−Il10 −/− mice [144], implying that inflammation and iron sequestration limits the replication of some members of the microbiota. Additional studies will be necessary to further investigate the contribution of other host antimicrobial proteins in modulating the composition of the gut microbiota in the healthy and in the inflamed gut.
4. Bacterial-derived AMPs
Similar to eukaryotic hosts, certain bacterial species have evolved to produce AMPs, a mechanism that confers a competitive advantage in various ecological niches, including the mammalian intestine. Bacteriocins are a group of ribosomally synthesized AMPs that have diverse functions, ranging from targeting of the bacterial membrane to interfering with DNA, RNA, and protein metabolism (rev. in [146]). These molecules are often produced by probiotic strains to facilitate their colonization in complex microbial communities. The activity of these peptides is dependent on their export and subsequent interaction with the sensitive cell. Notably, bacteriocin gene clusters are often co-regulated with genes encoding for cognate immunity proteins to provide the producing strain with protection against self-killing (rev. in [147]).
Bacteriocins from Gram-positive bacteria are divided by the presence (class I) or absence (class II) of post-translational modifications, and are termed lantibiotics and nonlantibiotics, respectively [148]. Class II bacteriocins are further divided into four subgroups: class IIa are peptides that typically exhibit the greatest activity against Listeria, class IIb consists of two-peptide bacteriocins, class IIc bacteriocins adopt a cyclic conformation, and class IId is comprised of all remaining bacteriocins that are unable to be grouped based on their structure or activity [147]. The potency of bacteriocins against clinically relevant targets has been well documented [149–151], and the generally accepted view is that these peptides are most effective against Gram-positive pathogens. Mechanistically, bacteriocins have been shown to exert their bactericidal effect by either targeting cell wall precursors [152] or by inducing pore formation following interaction with transmembrane receptors [153]. With respect to the mammalian intestine, the Abp118 bacteriocin from probiotic Lactobacillus salivarius strain UCC118 reduces levels of L. monocytogenes (Figure 3A) [154] while thuricin CD of Bacillus thuringiensis DPC 6431 diminishes C. difficile burden (Figure 3B) [155]. The therapeutic potential of these bacterial peptides was also demonstrated when an Enterococcus faecalis strain engineered to ectopically express bacteriocins reduced intestinal colonization of VRE, without disruption to the indigenous microbiota (Figure 3C) [156]. Interestingly, bacteriocin production can also be a characteristic of virulent strains. Epidemic L. monocytogenes isolates produce the bacteriocin Listeriolysin S (LLS), which modulates the host microbiota to provide a more favorable niche for pathogen growth in the intestine (Figure 3D) [157].
Figure 3. Activity of bacterial-derived antimicrobial peptides in the intestine.
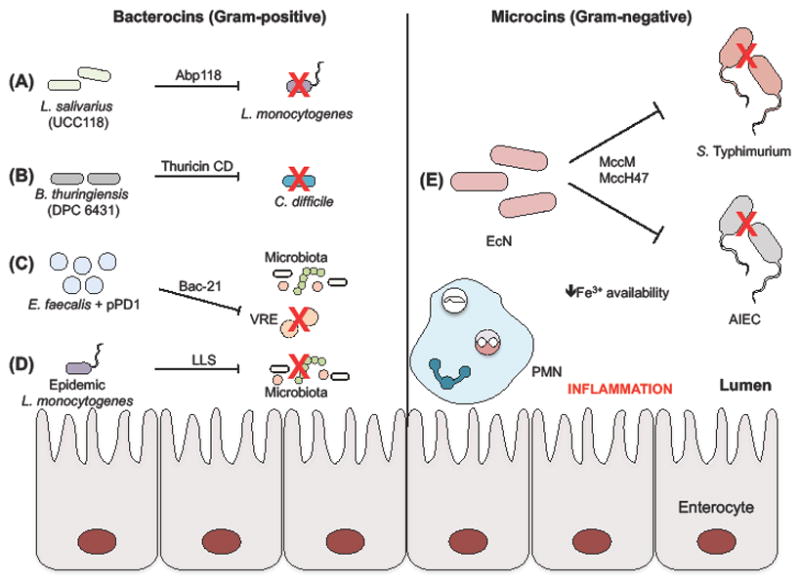
Production of bacterocins from Gram-positive bacteria is typically a characteristic of probiotic strains. (A) Specifically, Abp118 of L. salivarius strain UCC118 is able to target pathogenic L. monocytogenes, while (B) B. thuringiensis DPC 6431 restricts C. difficile expansion through the activity of thuricin CD. (C) An E. faecalis strain ectopically expressing the bacteriocin Bac-21 is able to specifically target Vancomycin-Resistant Enterococcus (VRE) while maintaining the integrity of commensal bacteria. (D) In contrast to probiotic bacteria, some epidemic strains of the Listeria spp. produce Listeriolysin S (LLS) to promote virulence by targeting the host microbiota. Microcins are peptides from Gram-negative bacteria that are smaller than 10 kDa. (E) E. coli Nissle 1917 (EcN) synthesis of microcin M (MccM) and microcin H47 (MccH47) facilities clearance of pathogenic S. Typhimurium and adherent-invasive E. coli (AIEC) in the inflamed intestine, when iron is limited.
Bacteriocins from Gram-negative microorganisms are structurally distinct from Gram-positive bacteriocins, and are classified based on the relative peptide size, with colicins ranging between 25–80 kDa and microcins being less than 10 kDa [158]. Microcins are further subdivided into class IIa, which are not post-translationally modified, and class IIb, which carry a C-terminal modification containing an enterobactin-like moiety [159]. Class IIb microcins are also known as TonB-dependent microcins, because they require functional TonB in the target cell for killing [160]. Importantly, iron starvation can induce the production of these modified siderophores [161, 162], which target cells that express cognate siderophore receptors by a ‘Trojan horse’ mechanism [163]. The probiotic strain EcN produces two class IIb microcins, microcin M and microcin H47, which are postulated to function by this mode of action. Importantly, while studies have demonstrated that microcins can restrict growth of pathogens in vitro [164], their in vivo role has remained elusive. Recent work from our group has uncovered that microcin production enables EcN to outcompete pathogens, including S. Typhimurium and adherent-invasive E. coli, in the inflamed intestine, where iron is limited (Figure 3E) [165]. Moreover, therapeutic administration of microcin-producing EcN reduced S. Typhimurium intestinal colonization in mice, providing the first in vivo evidence of microcins for host protection [165]. Additional studies will be necessary to identify the specific bacterial targets of microcins.
5. Conclusions and Future Perspectives
Maintenance of homeostasis in the gut not only requires tolerance of the commensal microbiota, but also containment of invading pathogens. Increasing evidence indicates that an essential component of the immune response regulating this balance is the production of AMPs by intestinal epithelial cells. However, there are still many unanswered questions, including the effects of specific AMPs on the composition of the microbiota, alternative functions of known AMPs, and the identity of unknown peptides and proteins with antimicrobial activities. In addition to host-derived peptides, AMPs from probiotic bacteria are also effective in reducing pathogen colonization of the gastrointestinal tract, yet their specific targets remain enigmatic in most cases. Exploiting the natural activity of AMPs may assist in the treatment of enteric infections or to restore intestinal dysbiosis. Therefore, a more comprehensive understanding of the regulatory networks governing AMP expression and their mechanisms of action will be fundamental for their therapeutic application.
Acknowledgments
Work in the Raffatellu lab is supported by Public Health Service Grants AI114625, AI121928, AI126277, and AI126465, and by the Chiba University-UC San Diego Center for Mucosal Immunology, Allergy, and Vaccines. M.R. also holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. We thank members of the Raffatellu laboratory for their critical review of the manuscript. We would also like to apologize to our colleagues whose work could not be cited due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carding S, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol. 2012;3:310. doi: 10.3389/fimmu.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandl K, et al. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204(8):1891–900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dessein R, et al. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58(6):771–6. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- 6.Iimura M, et al. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J Immunol. 2005;174(8):4901–7. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 7.Wehkamp J, et al. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn’s disease. Mucosal Immunol. 2008;1(Suppl 1):S67–74. doi: 10.1038/mi.2008.48. [DOI] [PubMed] [Google Scholar]
- 8.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci U S A. 2005;102(50):18129–34. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bals R. Epithelial antimicrobial peptides in host defense against infection. Respir Res. 2000;1(3):141–50. doi: 10.1186/rr25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braff MH, et al. Cutaneous defense mechanisms by antimicrobial peptides. J Invest Dermatol. 2005;125(1):9–13. doi: 10.1111/j.0022-202X.2004.23587.x. [DOI] [PubMed] [Google Scholar]
- 11.Hiemstra PS, et al. Antimicrobial Peptides and Innate Lung Defenses: Role in Infectious and Noninfectious Lung Diseases and Therapeutic Applications. Chest. 2016;149(2):545–51. doi: 10.1378/chest.15-1353. [DOI] [PubMed] [Google Scholar]
- 12.Schauber J, Gallo RL. Antimicrobial peptides and the skin immune defense system. J Allergy Clin Immunol. 2008;122(2):261–6. doi: 10.1016/j.jaci.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yarbrough VL, Winkle S, Herbst-Kralovetz MM. Antimicrobial peptides in the female reproductive tract: a critical component of the mucosal immune barrier with physiological and clinical implications. Hum Reprod Update. 2015;21(3):353–77. doi: 10.1093/humupd/dmu065. [DOI] [PubMed] [Google Scholar]
- 14.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6(12):1543–75. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang YQ, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science. 1999;286(5439):498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 16.Cunliffe RN. Alpha-defensins in the gastrointestinal tract. Mol Immunol. 2003;40(7):463–7. doi: 10.1016/s0161-5890(03)00157-3. [DOI] [PubMed] [Google Scholar]
- 17.Pazgier M, et al. Human beta-defensins. Cell Mol Life Sci. 2006;63(11):1294–313. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole AM, et al. Retrocyclin: a primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc Natl Acad Sci U S A. 2002;99(4):1813–8. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabay JE, et al. Antibiotic proteins of human polymorphonuclear leukocytes. Proc Natl Acad Sci U S A. 1989;86(14):5610–4. doi: 10.1073/pnas.86.14.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganz T, et al. Defensins. Natural peptide antibiotics of human neutrophils. J Clin Invest. 1985;76(4):1427–35. doi: 10.1172/JCI112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilde CG, et al. Purification and characterization of human neutrophil peptide 4, a novel member of the defensin family. J Biol Chem. 1989;264(19):11200–3. [PubMed] [Google Scholar]
- 22.de Leeuw E, et al. Functional interaction of human neutrophil peptide-1 with the cell wall precursor lipid II. FEBS Lett. 2010;584(8):1543–8. doi: 10.1016/j.febslet.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varney KM, et al. Turning defense into offense: defensin mimetics as novel antibiotics targeting lipid II. PLoS Pathog. 2013;9(11):e1003732. doi: 10.1371/journal.ppat.1003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehkamp J, et al. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580(22):5344–50. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 25.Chairatana P, et al. Proteolysis Triggers Self-Assembly and Unmasks Innate Immune Function of a Human alpha-Defensin Peptide. Chem Sci. 2016;7(3):1738–1752. doi: 10.1039/c5sc04194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh D, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3(6):583–90. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 27.Porter EM, et al. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65(6):2396–401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salzman NH, et al. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422(6931):522–6. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 29.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnupf P, et al. Segmented filamentous bacteria, Th17 inducers and helpers in a hostile world. Curr Opin Microbiol. 2017;35:100–109. doi: 10.1016/j.mib.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Chu H, et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337(6093):477–81. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ericksen B, et al. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49(1):269–75. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szyk A, et al. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15(12):2749–60. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chairatana P, Nolan EM. Molecular basis for self-assembly of a human host-defense peptide that entraps bacterial pathogens. J Am Chem Soc. 2014;136(38):13267–76. doi: 10.1021/ja5057906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bry L, et al. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc Natl Acad Sci U S A. 1994;91(22):10335–9. doi: 10.1073/pnas.91.22.10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouellette AJ, et al. Developmental regulation of cryptdin, a corticostatin/defensin precursor mRNA in mouse small intestinal crypt epithelium. J Cell Biol. 1989;108(5):1687–95. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayabe T, et al. Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem. 2002;277(7):5219–28. doi: 10.1074/jbc.M109410200. [DOI] [PubMed] [Google Scholar]
- 38.Wilson CL, et al. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286(5437):113–7. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 39.Eisenhauer PB, Harwig SS, Lehrer RI. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun. 1992;60(9):3556–65. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouellette AJ, et al. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62(11):5040–7. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman NH, et al. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect Immun. 2003;71(3):1109–15. doi: 10.1128/IAI.71.3.1109-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Neil DA, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163(12):6718–24. [PubMed] [Google Scholar]
- 43.Vora P, et al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004;173(9):5398–405. doi: 10.4049/jimmunol.173.9.5398. [DOI] [PubMed] [Google Scholar]
- 44.Wehkamp J, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72(10):5750–8. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witthoft T, et al. Enhanced human beta-defensin-2 (hBD-2) expression by corticosteroids is independent of NF-kappaB in colonic epithelial cells (CaCo2) Dig Dis Sci. 2005;50(7):1252–9. doi: 10.1007/s10620-005-2768-5. [DOI] [PubMed] [Google Scholar]
- 46.Fahlgren A, et al. beta-Defensin-3 and -4 in intestinal epithelial cells display increased mRNA expression in ulcerative colitis. Clin Exp Immunol. 2004;137(2):379–85. doi: 10.1111/j.1365-2249.2004.02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia JR, et al. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001;15(10):1819–21. [PubMed] [Google Scholar]
- 48.Harder J, et al. A peptide antibiotic from human skin. Nature. 1997;387(6636):861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 49.Harder J, et al. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276(8):5707–13. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 50.Sass V, et al. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78(6):2793–800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bensch KW, et al. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 1995;368(2):331–5. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature. 2011;469(7330):419–23. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 53.Raschig J, et al. Ubiquitously expressed Human Beta Defensin 1 (hBD1) forms bacteria-entrapping nets in a redox dependent mode of action. PLoS Pathog. 2017;13(3):e1006261. doi: 10.1371/journal.ppat.1006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bals R, Goldman MJ, Wilson JM. Mouse beta-defensin 1 is a salt-sensitive antimicrobial peptide present in epithelia of the lung and urogenital tract. Infect Immun. 1998;66(3):1225–32. doi: 10.1128/iai.66.3.1225-1232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bals R, et al. Mouse beta-defensin 3 is an inducible antimicrobial peptide expressed in the epithelia of multiple organs. Infect Immun. 1999;67(7):3542–7. doi: 10.1128/iai.67.7.3542-3547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinrichsen K, et al. Mouse beta-defensin-14, an antimicrobial ortholog of human beta-defensin-3. Antimicrob Agents Chemother. 2008;52(5):1876–9. doi: 10.1128/AAC.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosciuczuk EM, et al. Cathelicidins: family of antimicrobial peptides. A review. Mol Biol Rep. 2012;39(12):10957–70. doi: 10.1007/s11033-012-1997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758(9):1408–25. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 59.Gallo RL, et al. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem. 1997;272(20):13088–93. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- 60.Hase K, et al. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun. 2002;70(2):953–63. doi: 10.1128/iai.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hase K, et al. Expression of LL-37 by human gastric epithelial cells as a potential host defense mechanism against Helicobacter pylori. Gastroenterology. 2003;125(6):1613–25. doi: 10.1053/j.gastro.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 62.Sorensen OE, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97(12):3951–9. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- 63.Chromek M, Arvidsson I, Karpman D. The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-mediated disease. PLoS One. 2012;7(10):e46476. doi: 10.1371/journal.pone.0046476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Islam D, et al. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 2001;7(2):180–5. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 65.Raqib R, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci U S A. 2006;103(24):9178–83. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cash HL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christa L, et al. HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am J Physiol. 1996;271(6 Pt 1):G993–1002. doi: 10.1152/ajpgi.1996.271.6.G993. [DOI] [PubMed] [Google Scholar]
- 68.Ogawa H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9(3):162–70. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 69.Behnsen J, et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity. 2014;40(2):262–73. doi: 10.1016/j.immuni.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–9. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 71.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Medveczky P, Szmola R, Sahin-Toth M. Proteolytic activation of human pancreatitis-associated protein is required for peptidoglycan binding and bacterial aggregation. Biochem J. 2009;420(2):335–43. doi: 10.1042/BJ20090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukherjee S, et al. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J Biol Chem. 2009;284(8):4881–8. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stelter C, et al. Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PLoS One. 2011;6(6):e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lehotzky RE, et al. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A. 2010;107(17):7722–7. doi: 10.1073/pnas.0909449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukherjee S, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505(7481):103–7. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miki T, Holst O, Hardt WD. The bactericidal activity of the C-type lectin RegIIIbeta against Gram-negative bacteria involves binding to lipid A. J Biol Chem. 2012;287(41):34844–55. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miki T, et al. The Bactericidal Lectin RegIIIbeta Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell Host Microbe. 2017;21(2):195–207. doi: 10.1016/j.chom.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 80.Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017;13(9):e1006512. doi: 10.1371/journal.ppat.1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koprivnjak T, et al. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J Biol Chem. 2002;277(49):47636–44. doi: 10.1074/jbc.M205104200. [DOI] [PubMed] [Google Scholar]
- 82.Harwig SS, et al. Bactericidal properties of murine intestinal phospholipase A2. J Clin Invest. 1995;95(2):603–10. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peeters T, Vantrappen G. The Paneth cell: a source of intestinal lysozyme. Gut. 1975;16(7):553–8. doi: 10.1136/gut.16.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vadas P, et al. Extracellular phospholipase A2 expression and inflammation: the relationship with associated disease states. J Lipid Mediat. 1993;8(1):1–30. [PubMed] [Google Scholar]
- 85.Akinbi HT, et al. Bacterial killing is enhanced by expression of lysozyme in the lungs of transgenic mice. J Immunol. 2000;165(10):5760–6. doi: 10.4049/jimmunol.165.10.5760. [DOI] [PubMed] [Google Scholar]
- 86.Qu XD, Lehrer RI. Secretory phospholipase A2 is the principal bactericide for staphylococci and other gram-positive bacteria in human tears. Infect Immun. 1998;66(6):2791–7. doi: 10.1128/iai.66.6.2791-2797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Keestra-Gounder AM, et al. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532(7599):394–7. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bel S, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357(6355):1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolf M, Moser B. Antimicrobial activities of chemokines: not just a side-effect? Front Immunol. 2012;3:213. doi: 10.3389/fimmu.2012.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cole AM, et al. Cutting edge: IFN-inducible ELR- CXC chemokines display defensin-like antimicrobial activity. J Immunol. 2001;167(2):623–7. doi: 10.4049/jimmunol.167.2.623. [DOI] [PubMed] [Google Scholar]
- 91.Crawford MA, et al. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc Natl Acad Sci U S A. 2011;108(41):17159–64. doi: 10.1073/pnas.1108495108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Margulieux KR, et al. CXCL10 Acts as a Bifunctional Antimicrobial Molecule against Bacillus anthracis. MBio. 2016;7(3) doi: 10.1128/mBio.00334-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schutte KM, et al. Escherichia coli Pyruvate Dehydrogenase Complex Is an Important Component of CXCL10-Mediated Antimicrobial Activity. Infect Immun. 2015;84(1):320–8. doi: 10.1128/IAI.00552-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crawford MA, et al. Interferon-inducible CXC chemokines directly contribute to host defense against inhalational anthrax in a murine model of infection. PLoS Pathog. 2010;6(11):e1001199. doi: 10.1371/journal.ppat.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dwinell MB, et al. Regulated production of interferon-inducible T-cell chemoattractants by human intestinal epithelial cells. Gastroenterology. 2001;120(1):49–59. doi: 10.1053/gast.2001.20914. [DOI] [PubMed] [Google Scholar]
- 96.Reid-Yu SA, et al. CXCL9 contributes to antimicrobial protection of the gut during citrobacter rodentium infection independent of chemokine-receptor signaling. PLoS Pathog. 2015;11(2):e1004648. doi: 10.1371/journal.ppat.1004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–4. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 98.Hooper LV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–73. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 99.Walker CR, et al. Intestinal intraepithelial lymphocyte-enterocyte crosstalk regulates production of bactericidal angiogenin 4 by Paneth cells upon microbial challenge. PLoS One. 2013;8(12):e84553. doi: 10.1371/journal.pone.0084553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Becknell B, et al. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. 2015;87(1):151–61. doi: 10.1038/ki.2014.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spencer JD, et al. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013;83(4):615–25. doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10(8):525–37. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weinberg ED. Nutritional immunity. Host’s attempt to withold iron from microbial invaders. JAMA. 1975;231(1):39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 104.Diaz-Ochoa VE, et al. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front Cell Infect Microbiol. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13(5):509–19. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270(45):26723–6. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 107.O’Brien IG, Gibson F. The structure of enterochelin and related 2,3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim Biophys Acta. 1970;215(2):393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 108.Perry RD, San Clemente CL. Siderophore synthesis in Klebsiella pneumoniae and Shigella sonnei during iron deficiency. J Bacteriol. 1979;140(3):1129–32. doi: 10.1128/jb.140.3.1129-1132.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pollack JR, Neilands JB. Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem Biophys Res Commun. 1970;38(5):989–92. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 110.Rogers HJ, et al. Production of enterochelin by Escherichia coli 0111. Biochim Biophys Acta. 1977;497(2):548–57. doi: 10.1016/0304-4165(77)90211-2. [DOI] [PubMed] [Google Scholar]
- 111.Griffiths E. Iron and Infection: Molecular, Physiological and Clinical Aspects. New York, NY: John Wiley and Sons; 1999. Iron in Biological Systems. [Google Scholar]
- 112.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–21. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 113.Goetz DH, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10(5):1033–43. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 114.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5(5):476–86. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Raffatellu M, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14(4):421–8. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zindl CL, et al. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci U S A. 2013;110(31):12768–73. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ye P, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194(4):519–27. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kjeldsen L, et al. Identification of neutrophil gelatinase-associated lipocalin as a novel matrix protein of specific granules in human neutrophils. Blood. 1994;83(3):799–807. [PubMed] [Google Scholar]
- 119.Carlson M, et al. Human neutrophil lipocalin is a unique marker of neutrophil inflammation in ulcerative colitis and proctitis. Gut. 2002;50(4):501–6. doi: 10.1136/gut.50.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abergel RJ, et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci U S A. 2006;103(49):18499–503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bachman MA, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79(8):3309–16. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hantke K, et al. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100(7):3677–82. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Crouch ML, et al. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008;67(5):971–83. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- 124.Deriu E, et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14(1):26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palmer LD, Skaar EP. Transition Metals and Virulence in Bacteria. Annu Rev Genet. 2016;50:67–91. doi: 10.1146/annurev-genet-120215-035146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hantke K. Bacterial zinc uptake and regulators. Curr Opin Microbiol. 2005;8(2):196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 127.Juttukonda LJ, Skaar EP. Manganese homeostasis and utilization in pathogenic bacteria. Mol Microbiol. 2015;97(2):216–28. doi: 10.1111/mmi.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Campoy S, et al. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect Immun. 2002;70(8):4721–5. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Davis LM, Kakuda T, DiRita VJ. A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J Bacteriol. 2009;191(5):1631–40. doi: 10.1128/JB.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kehl-Fie TE, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81(9):3395–405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim S, et al. Zinc uptake system (znuA locus) of Brucella abortus is essential for intracellular survival and virulence in mice. J Vet Med Sci. 2004;66(9):1059–63. doi: 10.1292/jvms.66.1059. [DOI] [PubMed] [Google Scholar]
- 132.Makui H, et al. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol. 2000;35(5):1065–78. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- 133.Teigelkamp S, et al. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J Biol Chem. 1991;266(20):13462–7. [PubMed] [Google Scholar]
- 134.Damo SM, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110(10):3841–6. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10(2):158–64. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakashige TG, et al. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol. 2015;11(10):765–71. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319(5865):962–5. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 138.Hood MI, et al. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 2012;8(12):e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu JZ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11(3):227–39. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Urban CF, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009;5(10):e1000639. doi: 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Konikoff MR, Denson LA. Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12(6):524–34. doi: 10.1097/00054725-200606000-00013. [DOI] [PubMed] [Google Scholar]
- 142.Diaz-Ochoa VE, et al. Salmonella Mitigates Oxidative Stress and Thrives in the Inflamed Gut by Evading Calprotectin-Mediated Manganese Sequestration. Cell Host Microbe. 2016;19(6):814–25. doi: 10.1016/j.chom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zackular JP, et al. Dietary zinc alters the microbiota and decreases resistance to Clostridium difficile infection. Nat Med. 2016;22(11):1330–1334. doi: 10.1038/nm.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moschen AR, et al. Lipocalin 2 Protects from Inflammation and Tumorigenesis Associated with Gut Microbiota Alterations. Cell Host Microbe. 2016;19(4):455–69. doi: 10.1016/j.chom.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 145.Singh V, et al. Microbiota-inducible Innate Immune, Siderophore Binding Protein Lipocalin 2 is Critical for Intestinal Homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2(4):482–498 e6. doi: 10.1016/j.jcmgh.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11(2):95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 147.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–88. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 148.Hassan M, et al. Natural antimicrobial peptides from bacteria: characteristics and potential applications to fight against antibiotic resistance. J Appl Microbiol. 2012;113(4):723–36. doi: 10.1111/j.1365-2672.2012.05338.x. [DOI] [PubMed] [Google Scholar]
- 149.Eijsink VG, et al. Comparative studies of class IIa bacteriocins of lactic acid bacteria. Appl Environ Microbiol. 1998;64(9):3275–81. doi: 10.1128/aem.64.9.3275-3281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Piper C, et al. Discovery of medically significant lantibiotics. Curr Drug Discov Technol. 2009;6(1):1–18. doi: 10.2174/157016309787581075. [DOI] [PubMed] [Google Scholar]
- 151.Sandiford S, Upton M. Identification, characterization, and recombinant expression of epidermicin NI01, a novel unmodified bacteriocin produced by Staphylococcus epidermidis that displays potent activity against Staphylococci. Antimicrob Agents Chemother. 2012;56(3):1539–47. doi: 10.1128/AAC.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brotz H, et al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol. 1998;30(2):317–27. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 153.Diep DB, et al. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci U S A. 2007;104(7):2384–9. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Corr SC, et al. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104(18):7617–21. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rea MC, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci U S A. 2010;107(20):9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526(7575):719–22. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Quereda JJ, et al. Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc Natl Acad Sci U S A. 2016;113(20):5706–11. doi: 10.1073/pnas.1523899113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang SC, et al. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front Microbiol. 2014;5:241. doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Duquesne S, et al. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J Mol Microbiol Biotechnol. 2007;13(4):200–9. doi: 10.1159/000104748. [DOI] [PubMed] [Google Scholar]
- 160.Braun V, Patzer SI, Hantke K. Ton-dependent colicins and microcins: modular design and evolution. Biochimie. 2002;84(5–6):365–80. doi: 10.1016/s0300-9084(02)01427-x. [DOI] [PubMed] [Google Scholar]
- 161.Nolan EM, Walsh CT. Investigations of the MceIJ-catalyzed posttranslational modification of the microcin E492 C-terminus: linkage of ribosomal and nonribosomal peptides to form “trojan horse” antibiotics. Biochemistry. 2008;47(35):9289–99. doi: 10.1021/bi800826j. [DOI] [PubMed] [Google Scholar]
- 162.Patzer SI, et al. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149(Pt 9):2557–70. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 163.Rebuffat S. Microcins in action: amazing defence strategies of Enterobacteria. Biochem Soc Trans. 2012;40(6):1456–62. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
- 164.Duquesne S, et al. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24(4):708–34. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 165.Sassone-Corsi M, et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540(7632):280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]


