Abstract
Multidrug-resistant Escherichia coli is one of the most important public health concern worldwide that can be transferred through the food of animal origin to human being causing serious infection. The genetic responsibility of such resistant genes (Plasmids, integrons, and transposons) can be easily transmitted from the resistant strain to another. Therefore, the main objectives of the study is the molecular characterization of the resistant Escherichia coli isolates recovered from food samples and human isolates collected from outpatient clinics, KSA especially the resistance strains against aminoglycoside resistance genes which are responsible for the resistance against gentamicin and the resistance caused β-lactamases genes. Examination of food samples revealed 120 Escherichia coli isolates (22.22%) (30 strains O26: K60, 28 strains O128: K67, 20 strains O111: K58, 18 strains O126: K58, 10 strains O55: K59, 9 strains O86: K61 and 5 strains O157: H7). All the strains were highly resistance to penicillin, amoxicillin-clavulanic and erythromycin with a percentage of 100%, while the resistance to gentamicin, ampicillin, oxytetracycline, chloramphenicol, norfloxacin, trimethoprim, and nalidixic acid were 83%, 75%, 65.3%, 55.8%, 36.5%, 30.7% and 26.9% respectively. On the other hand, 59.6% of tested strains were sensitive to ciprofloxacin. Positive amplification of 896 bp fragments specific for aacC2 genes were observed by PCR designated for the detection of the aminoglycoside resistance genes. Meanwhile, multiplex PCR designed to detect the ampicillin and amoxicillin-clavulanic acid resistant E. coli isolates revealed positive amplification of 516 bp fragments specific for BlaTEM gene with all the resistant strains to ampicillin and amoxicillin-clavulanic acid. Moreover, positive amplification of 392 bp fragments specific for BlaSHV resistant gene were observed with (60.52%) of E. coli isolate. While all the tested strains were negative for amplification of BlaOXA_1.
Keywords: Multidrug-resistant, Escherichia coli, Shiga toxogenic, Aminoglycoside resistance, β-lactamases
1. Introduction
Escherichia coli enclose diverse group of strains inhabit the human intestinal tract and formulate a part in the gut flora, in addition to that Shiga-like toxin Escherichia coli, (STEC) are of zoonotic importance foodborne pathogen as they can cause severe economic losses and serious intra and extra intestinal diseases by consumption of contaminated food like hemolytic uremic syndrome (HUS), hemorrhagic colitis (HC), food poisoning and diarrhea (Cordesmeyer et al., 2011, Kuijper et al., 2011, Newell and La Ragione, 2018). The extensive use of antimicrobial therapy to control the diseases caused by Escherichia coli may emerge Multi-resistant Escherichia coli isolates that become a worldwide public health problem (Frank et al., 2011, Jacoby et al., 2006, Ukah et al., 2018).
The incidence of urinary tract infections especially with multidrug-resistant Escherichia coli have been increased (Frank et al., 2011), particularly in women and young children and the recurrence of infection is common (Hu et al., 2008). Moreover, Escherichia coli is one of the most important uropathogen that associated with uncomplicated cystitis (Chattaway et al., 2011).
The resistance to co-trimoxazole, ampicillin, and the fluoroquinolones in some uropathogenic E. coli was suggested to be originated from food of animal origin through the horizontal transfer of virulence and antibiotic resistance genes (Manges et al., 2007, Johnson and Nolan, 2009, Ramchandani et al., 2005, Rebbah et al., 2017). Moreover, the aminoglycoside- resistance genes found in uropathogenic E. coli isolates recovered from human have been suggested to be isolated from food of animal origin which considered being a reservoir of such resistant strains (Taylor et al., 2008).
The resistance of Enterobacteriaceae to third-generation cephalosporin is mediated by TEM- and SHV-type L-lactamases. While, the most incriminated plasmid-borne beta-lactamases in amoxicillin-clavulanic acid resistance Escherichia coli isolates is TEM type and OXA-1enzymes (Ahmed et al., 2014, Hamed et al., 2017).
The objective of this study was to evaluate the resistance of E. coli strains from food samples and human isolates, collected from KSA on molecular basis specially β-lactamases resistance genes (blaTEM, blaOXA_1, and blaSHV genes) and gentamicin the resistant gene (aacC2 gene).
2. Material and methods
2.1. Samples
A total of 540 beef meat samples “fresh meat samples from the abattoirs, minced meat were collected from different retail establishment markets and abattoirs collected during the period of April 2015 and March 2016 each sample weighted approximately 100 g. 25 fresh meat samples from healthy farms continuously monitored for E. coli were collected as negative control samples.
During the period of August 2015 and December 2016, 180 strains of E. coli isolates recovered from patients suffering from food poisoning and or diarrhea were collected from outpatients clinics located in Jeddah, Medina, Riyadh, and Makah, Kingdom of Saudi Arabia as a source of human isolates. All the human isolates were identified with standard microbiological techniques, serotyping and molecular techniques.
2.2. Isolation and identification of E. coli
Isolation of E. coli, from poultry and meat product, enrichment broth were used “followed by subculturing on CLED agar, MacConkey agar and Eosin methylene blue agar (Cetinkaya et al., 2014). The recovered E. coli isolates by standard microbiological techniques were serologically identified using polyvalent and monovalent E. coli antisera.
2.3. Antimicrobial susceptibility test
The resistance to wide range of antibiotics were carried out according to the recommendations of Clinical Laboratory Standards Institute (CLSI, 2013), using the Kirby-Bauer disk diffusion method and the following antibiotics disks (nalidixic acid NA30, ampicillin AMP10, gentamicin CN10, ceftazidime CAZ30, erythromycin E15, norfloxacin NOR 10, ciprofloxacin CIP5, amoxicillin-clavulanic acid AMC 30, vancomycin VA 30, penicillin G P10, chloramphenicol C 30, oxytetracycline OT30, sulphamethoxazole-trimethoprim SXT 25.
2.4. Molecular detection and characterization of the recovered E. coli
2.4.1. Extraction of DNA of E. coli
All the recovered E. coli isolates were inoculated on 10 ml tryptic soy broth at 37 °C for 24 h and the genomic DNA was extracted after centrifugation of the overnight culture at 3000 RPM for 5 min by the same method reported by (Frank et al., 2011). 5 μl from extracted DNA were used as a template DNA for PCR.
2.4.2. Molecular detection and characterization of the recovered E. coli
2.4.2.1. Molecular detection and characterization of the E. coli and its virulence genes
Molecular characterization of the recovered E. coli were carried out by multiplex-PCR using specific primers for the detection of shiga toxin type 2 (stx2) and intimin gene (eaeA) reported by (Moussa et al., 2010). Moreover, The extracted DNA were tested by using PCR primer pairs specific for uid A gene of E. coli (Heininger et al., 1999), encoding b-glucuronidase specific for E. coli, the increased serum survival iss gene of E. coli (Yaguchi et al., 2007), aerobactin iutA of E. coli (Delicato et al., 2003) and cvaC gene (Rocha et al., 2008).
2.4.2.1.1. Molecular detection of the resistance genes
PCR for the detection of the resistant gene specific for gentamicin resistance (aacC2) were carried out for all of the recovered strains using specific primers (Aleisa et al., 2013). Moreover, multiplex PCR for the molecular detection of resistant genes specific for ampicillin and amoxicillin-clavulanic acid (BlaTEM, BlaSHV and BlaOXA_1) genes were carried out using three primer pairs (Aleisa et al., 2013).
3. Results
3.1. Standard bacteriological examination of the examined herbal samples
120 E. coli isolates (22.22%) were isolated from the examined food samples. All of the strains were typed as follows; 30 strains O26: K60, 28 strains O128: K67, 20 strains O111: K58, 18 strains O126: K58, 10 strains O55: K59, 9 strains as O86: K61 while the other 5 strains were O157: H7.
The shiga toxin type 2 (stx2) and the intimin (eaeA) genes were detected in all strains E. coli serovar O157: H7 using multiplex PCR, the incidence of such two genes in the E. coli serovar O111 were 30 (53.5%) and 15 (26.79%) for stx2 and eaeA respectively and in the E. coli serovar O128 were 22 (40.74%) and 10 (18.52%) for stx2 and eaeA respectively, while the presence stx2and eaeA decreased to 30% and 20% in O126: K58, O55: K59, and O26: K60 respectively as shown in Fig. 1. Meanwhile, stx2 and eaeA genes were absent with E. coli serovars O119, O86: K61, O78: K80 and O158: K-. It is clear from the obtained result that cvaC gene showed the highest percentage (96.9%), followed by iutA gene and iss gene (80.8% and 62.99%, respectively).
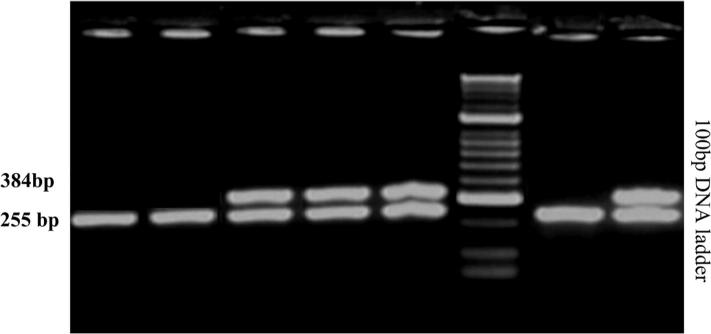
Fig. 1.
Multiplex PCR showing positive amplification of 384 bp fragments of intimin (eaeA) gene and 255 bp fragments of shiga toxin type two (stx2) genes of shiga toxogenic.
3.2. Antibiotic sensitivity test of the recovered strains of E. coli by disc diffusion method
All the recovered strains from food samples and from patients suffering from food poisoning and or diarrhea were resistant to penicillin, amoxicillin-clavulanic, and erythromycin 100%. The resistance of the recovered strains to gentamicin and ampicillin were 83% and 75% respectively, while the resistance to trimethoprim, oxytetracycline, chloramphenicol, norfloxacin, and nalidixic acid were 65.3%, 55.8%, 36.5%, 30.7% and 26.9%, respectively. On the other hand, 62.8% of tested strains were sensitive to ciprofloxacin.
3.3. Molecular detection of gentamicin-resistant gene
All the isolates recovered from food samples and from human were tested with PCR for molecular detection of the aminoglycoside resistance genes. Positive amplification of 896 bp fragments specific for aacC2 gene was observed with all the tested strains as shown in Fig. 3.
Fig. 3.
Positive amplification of 896 bp fragments of aacC2 gene of aminoglycoside resistant gene responsible to gentamicin.
3.4. Molecular detection of ampicillin and amoxicillin-clavulanic acid resistance genes by multiplex PCR
All the resistance E. coli isolates to ampicillin and amoxicillin-clavulanic were tested by multiplex PCR specific for BlaSHV (392 bp fragments), BlaTEM (516 bp fragments, and BlaOXA_1 genes. BlaTEM gene was observed in all the tested strains (100%). At the same time, positive amplification of 392 bp fragments specific BlaSHV gene was detected with 75 of the resistant E. coli isolate (62.5%). Meanwhile, all the recovered strains were negative for BlaOXA_1 gene Fig. 2.
Fig. 2.
Agarse gel electrophorsis showing amplification 392 base pair fragments specific for BiaTSHV gene and amplification of 516 base fragments specific for BiaTEM gene specific for ampicillin and amoxicillin-clavulanic acid resistance.
4. Discussion
Urinary tract infections affecting women four times more than men, and more than 50% of women will be infected at least once in their life. Multidrug-resistant E. coli, especially the extended-spectrum beta-lactamase (ESBL) E. coli is responsible for more than 80% of these infections (Ahmed et al., 2014). Recent studies found that there are genetic similarity between the ESBL E. coli isolates recovered from urinary tract infections and those isolated from chickens and food samples which indicated the ability of such strains to be transferred from food of animal origins to the human being causing serious urinary tract infections (Cetinkaya et al., 2014).
Standard Microbiological techniques for isolation and molecular characterization of E. coli revealed a total of 120 E. coli isolates (22.22%). All of the strains were typed as follows; 30 strains O26: K60, 28 strains O128: K67, 20 strains O111: K58, 18 strains O126: K58, 10 strains O55: K59, 9 strains as O86: K61 while the other 5 strains were O157: H7. Most of the recovered strains from food samples were similar to the strains recovered from serious urinary tract infections in humans (Cetinkaya et al., 2014, Yaguchi et al., 2007).
The shiga toxin type 2 (stx2) and the intimin (eaeA) genes were detected in all strains E. coli serovar O157: H7 using multiplex PCR, the incidence of such two genes in the E. coli serovar O111 were 30 (53.5%) and 15 (26.79%) for stx2 and eaeA respectively and in the E. coli serovar O128 were 22 (40.74%) and 10 (18.52%) for stx2 and eaeA respectively, while the presence stx2and eaeA decreased to 30% and 20% in O126: K58, O55: K59, and O26: K60 respectively as shown in Fig. 1. Serious complication such as Haemolytic Uremic Syndrome mostly occurs due to the infection with STEC that harboring the shiga toxin type 2 (stx2) gene than STEC that harboring the shiga toxin type 1 (stx1) (Moussa et al., 2010). The intimin (eaeA) gene which is the marker gene of enterohemorrhagic E. coli was detected in all strains of E. coli serovar O157: H7 using multiplex PCR (Kuijper et al., 2011).
All the isolates recovered from food samples and from human were tested with PCR for detection of the aminoglycoside resistance genes. Positive amplification of 896 bp fragments specific for aaC2 gene was observed with all the tested strains as shown in Fig. 3. The obtained results indicated the harboring of the gentamicin-resistant strains to the aaC2 gene responsible for aminoglycoside resistance (Ho et al., 2009). All the resistance E. coli isolates to ampicillin and amoxicillin-clavulanic were tested by multiplex PCR specific for BlaSHV (392 bp fragments), BlaTEM (516 bp fragments, and BlaOXA_1 genes. BlaTEM gene was observed in all the tested strains (100%) indicated that the predominant resistant gene for ampicillin and amoxicillin-clavulanic is the BlaTEM gene (Taylor et al., 2008). At the same time, positive amplification of 392 bp fragments specific BlaSHV gene was detected with 75 of the resistant E. coli isolate (62.5%). Meanwhile, all the recovered strains were negative for BlaOXA_1 gene Fig. 2. From this study, controlling of resistant strains should not focused only in patients but should include the environment, this raises the questions related to the classification of hospital and community-acquired infections.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ahmed O.M., Pangloli P., Hwang C.A., Zivanovic S., Wu T., D’Souza D., Draughon F.A. The occurrence of Listeria monocytogenes in retail ready-to-eat meat and poultry products related to the levels of acetate and lactate in the products. Food Control. 2014;2015(52):43–48. [Google Scholar]
- Aleisa A.M., Ashgan M.H., Alnasserallah A.A., Mahmoud M.H., Moussa I.M. Molecular detection of β-lactamases and aminoglycoside resistance genes among Escherichia coli isolates recovered from medicinal plant. A. J. M. R. 2013;7(20):2305–2310. [Google Scholar]
- Cetinkaya F., Mus T., Yibar A., Guclu N., Tavsanli H., Cibik R. Prevalence, serotype identification by multiplex polymerase chain reaction and antimicrobial resistance patterns of Listeria monocytogenesisolated from retail foods. J. Food Saf. 2014;34:42–49. [Google Scholar]
- Chattaway M.A., Dallman T., Okeke I.N., Wain J. Enteroaggregative E. coli O104 from an outbreak of HUS in Germany 2011, could it happen again. J. Infect Dev. Countries. 2011;5:425–436. doi: 10.3855/jidc.2166. [DOI] [PubMed] [Google Scholar]
- Cordesmeyer S., Peitz U., Godde N., Kasper H., Hoffmann M., Allemeyer E. Colonic ischaemia as a severe Shiga toxin/verotoxin producing Escherichia coli O104:H4 complication in a patient without hemolytic uremic syndrome, Germany. Euro Surveill. 2011;16 pii: 19895-19895. [PubMed] [Google Scholar]
- Delicato E.R., de Brito B.G., Gaziri L.C., Vidotto M.C. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 2003;94:97–103. doi: 10.1016/s0378-1135(03)00076-2. [DOI] [PubMed] [Google Scholar]
- Frank C., Werber D., Cramer J.P. Epidemic profile of Shiga-toxin–producing Escherichia coli O104:H4 outbreak in Germany preliminary report. N. Engl. J. Med. 2011 doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- Hamed O.M., Sabry M.A., Hassanain N.A., Hamza E., Hegazi A.G., Salman M.B. Occurrence of virulent and antibiotic-resistant Shiga toxin-producing Escherichia coli in some food products and human stool in Egypt. Vet World. 2017;10(10):1233–1240. doi: 10.14202/vetworld.2017.1233-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heininger A., Binder M., Schmidt S., Unertl K., Botzenhart K., Döring G. PCR and blood culture for detection of Escherichia coli bacteremia in rats. J. Clin. Microbiol. 1999;37:2479–2482. doi: 10.1128/jcm.37.8.2479-2482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.L., Yip K.S., Chow K.H., Lo J.Y., Que T.L., Yuen K.Y. Antimicrobial resistance among uropathogens that cause acute uncomplicated cystitis in women in Hong Kong: a prospective multicenter study in 2006 to 2008. Diagn. Microbiol. Infect. Dis. 2009;66:87–93. doi: 10.1016/j.diagmicrobio.2009.03.027. [DOI] [PubMed] [Google Scholar]
- Hu J.Y., Shi J.C., Chang H., Li D., Yang M., Kamagata Y. Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 2008;42(9):3415–3420. doi: 10.1021/es7026746. [DOI] [PubMed] [Google Scholar]
- Jacoby G.A., Walsh K.E., Mills D.M. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 2006;50(1178–82) doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.J., Nolan L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper E., Soonawala D., Vermont C., van Dissel J. Household transmission of hemolytic uremic syndrome associated with Escherichia coli O104:H4 in the Netherlands. Euro. Surveill. 2011;16 19897–19897. [PubMed] [Google Scholar]
- Manges A.R., Smith S.P., Lau B.J., Nuval C.J., Eisenberg J.N., Dietrich P.S., Riley L.W. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Food Borne Pathog. Dis. 2007;4:419–431. doi: 10.1089/fpd.2007.0026. [DOI] [PubMed] [Google Scholar]
- Moussa I.M., Ashgan M.H., Alwathnani H.A., Mohamed K.H.F., Al-Doss A.A. Multiplex polymerase chain reaction for detection and characterization of shiga toxigenic E. coli (STEC) Afr. J. Biotechnol. 2010;9(28):4356–4363. 12 July, 2010. [Google Scholar]
- Newell D.G., La Ragione R.M. Enterohaemorrhagic and other Shiga toxin-producing Escherichia coli (STEC): where are we now regarding diagnostics and control strategies? Transbound Emerg. Dis. 2018;25 doi: 10.1111/tbed.12789. [DOI] [PubMed] [Google Scholar]
- Ramchandani M., Manges A.R., DebRoy C., Smith S.P., Johnson J.R., Riley L.W. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin. Infect. Dis. 2005;40:251–257. doi: 10.1086/426819. [DOI] [PubMed] [Google Scholar]
- Rebbah N., Messai Y., Châtre P., Haenni M., Madec J.Y., Bakour R. Diversity of CTX-M extended-spectrum β-lactamases in Escherichia coli isolates from retail raw ground beef: first report of CTX-M-24 and CTX-M-32 in Algeria. Microb. Drug Resist. 2017 doi: 10.1089/mdr.2017.0171. [DOI] [PubMed] [Google Scholar]
- Rocha A.P., Rocha S.L.S., Lima-Rosa C.A.V., Souza G.F., Moraes H.L.S., Moraes L.B., Salle C.T.P. Genes associated with pathogenicity of avian Escherichia coli (APEC) isolated from respiratory cases of poultry. Pesq. Vet. Bras. 2008;28:183–186. [Google Scholar]
- Taylor N.M., Davies R.H., Ridley A., Clouting C., Wales A.D., Clifton-Hadley F.A. A survey of fluoroquinolone resistance in Escherichia coli and thermophilic Campylobacter spp. on poultry and pig farms in Great Britain. J. Appl. Microbiol. 2008;105:1421–1431. doi: 10.1111/j.1365-2672.2008.03877.x. [DOI] [PubMed] [Google Scholar]
- Ukah U.V., Glass M., Avery B., Daignault D., Mulvey M.R., Reid-Smith R.J., Parmley E.J., Portt A., Boerlin P., Manges A.R. Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiol. Infect. 2018;146(1):46–57. doi: 10.1017/S0950268817002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, P.A., 2013. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23.
- Yaguchi K., Ogitani T., Osawa R., Kawano M., Kokumai N., Kaneshige T., Noro T., Masubuchi K., Shimizu Y. Virulence factors of avian pathogenic Escherichia coli strains isolated from chickens with sliicolisepticemia in Japan. Avian Dis. 2007;51:656–662. doi: 10.1637/0005-2086(2007)51[656:VFOAPE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]