Abstract
Background
Oral squamous cell carcinoma (OSCC) oncogenic mechanisms still remain elusive. Herein, we proposed to understand the biological role of a newly discovered OSCC miRNA.
Methods
MiR-1297 related to OSCC was selected for this study. TaqMan qRT- PCR assay was used to profile miRNA and gene expression in 16 tumors with matched adjacent non-tumor tissues. CCK-8 assay and colony formation assay were used to demonstrate cell proliferation. Protein level was determined by western blotting. SPSS was used for statistical analyses.
Results
MiR-1297 is repressed and PTEN activated in OSCC. Moreover, the miR-1297 overexpressing cell lines displayed a decrease in cell growth rate. And, suppression of miR-1297 reversed the cell growth rate. In addition, PTEN silencing display the similar pattern as miRNA-1297 overexpression to enhance OSCC cell growth.
Conclusions
MiRNA and gene expression changes are common event in OSCC. Our results suggest miR-1297 may drive tumor progression through PTEN. And, miR-1297 could be a promising candidate for future investigation.
Keywords: MiR-1297, PTEN, Cell proliferation
1. Introduction
Oral squamous cell carcinoma (OSCC) is the 6th most common human malignancy worldwide and accounted for the 90% of oral cancers. The incidence rate is rising, especially in the younger people (Landis et al., 1999). Multi-step process driven by the accumulation of genetic or epigenetic events has become increasing clear in development of cancer (Sharma et al., 2010, Williams, 2000). Of these risk factors, the pathogenesis mechanism of miRNAs remains to be fully elucidated.
MiRNAs are naturally occurring non-coding RNAs and regulates the expression of target genes by binding to the specific sequences of the 3′ untranslated region (3′-UTR) (Zeng et al., 2005). MiRNAs have unveiled a new complexity in the gene regulatory network and played critical roles in various of biological processes (Bushati and Cohen, 2007). Many malignancies have been showed to be related to the dysfunction of miRNA expression, including oral cancer (Calin and Croce, 2006, Hui et al., 2010). Currently, the small size and low molecular weight of miRNAs made them as attractive options for therapeutic targets for cancer molecular therapy.
OSCC has become an important problem not only because of significant mortality, but also the functional defects and disfigurement often associated with its treatment (Epstein et al., 2002). In this study, we identified the expression of miR-1297 in OSCC specimens, as well as investigated the effects of miR-1297 on OSCC cell growth. And, we further discussed that miR-1297 contributed to the malignant of OSCC through targeting PTEN. Our results indicate that miR-1297 may be a new therapeutic target in OSCC and add another layer of complexity in miRNA biology.
2. Material and methods
2.1. Cell culture
The human OSCC cell line, SSC-4, was provided by American Type Cell Collection (Manassas, VA, USA). The cells were cultured in specific media according to the manufacturer and maintained in a humidified incubator at 37 °C with atmosphere of 5% (v/v) CO2 in air.
2.2. RNA isolation and quantitative real-time PCR
Total RNA was isolated from frozen tumor tissues, non-tumor tissues and cells using TRI (Molecular Research Center, Cincinnati, OH). RNA quality and quantity were determined by Nanodrop 1000 (Nanodrop; Thermos Fisher Scientific, Waltham, MA) by spectrophotometric analysis. RNAs were reverse transcribed into cDNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) after the removal of residual DNA. Quantitative real-time PCR (qRT- PCR) was performed in triplicate using the ABI 7500 system (Applied Biosystems, Foster City, CA). All the gene and miRNA specific probes are commercial available from Applied Biosystems.
2.3. miR-1297 overexpression and suppression
To evaluate the biologic roles of miR-1297, miR-1297 was stable overexpressed in SSC-4 by lentiviral system in vitro (Applied Biological Materials Inc, Richmond, BC) according to the instruction. The empty lentiviral vector (miR-NC) was used as vector control. To suppress the expression of miR-1297, miR-1297 inhibitor (anti-miR-1297) (GeneCopoeia, Guangzhou, China) assay was performed as previously described (Feng and Dong, 2015). The matched negative control is anti-miR-NC.
2.4. Western blotting analysis
15 μg of total protein extracts were separated on SDS-PAGE gels (Bio Rad Laboratories, Hercules, CA) and transferred to nitrocellulose membranes and blocked with 5% non-fat dry milk for 1 h in room temperature. After then, membranes were incubated with PTEN primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C. After being washed with TBST for 3 times, the membranes were incubated with the secondary antibody (Bio-rad) for 1 h. Band signals were visualized by the enhanced chemiluminescence kit (Pierce, Minneapolis, MN, USA). GAPDH (Abcam, Cambridge, MA) was used as internal control.
2.5. PTEN silencing
For the PTEN silencing, the lentiviral system, short hairpin RNA of PTEN lentiviral vector (shi- PTEN) and matched negative control vector (sh-NC) were commercial available from System Biosciences (Mountain View, CA). Transfection shRNA lentiviral particles were generated as previously described (Zhang et al., 2015).
2.6. Cell viability assay
Cell Counting Kit-8 (CCK-8) was used to determine the number of viable cells according to the manufacture's protocol (Promega, Madison, WI). SSC-4 cells (5000 per well) were seeded in 96-well plates in 100 μl medium. Cells were transfected with MiR-1297-mimic or miRNA control vector (miR-con) after 24 h culture. At the end of transfection, transfection medium was replaced by 100 μl fresh medium. Cells were treated with CCK-8 dye at indicated time points.
2.7. Colony formation assay
SSC-4 cells were seeded in the 6-well plates at a density of 1000 cells/well after 24 h of transfection of miR-1297 or miR-NC. Clones were fixed with methanol and stained with 2% Giemsa solution (Merck, Darmstadt, German) for 10 min after culture for 2 weeks.
2.8. Statistics
SPSS software (version 16.0.1, SPSS Inc., Chicago, IL, USA) for Windows was used for the statistical analyses. Statistical analysis data are expressed as mean ± standard deviation (N = 3). Unpaired 2 tail student t-test was performed to compare the difference between two groups. P (probability) less than .05 was considered as significant difference.
3. Results
3.1. MiR-1297 is down-regulated in tumor tissues
MiR-1297 emerged as a significant miRNA with 2.5-fold down-regulated in tumors tissues compared with the non-tumor tissues in our miRNA microarray preliminary studies (16 cases of tumors and the matched adjacent non-tumor tissues) (data not shown). By qRT-PCR, we found miR-1297 levels were dropped by 1.8-fold (p < .05) in the tumor tissues compared to non-tumor tissue samples (Fig. 1). These results suggest that miR-1297 plays a role in the progression of oral squamous carcinoma.
Fig. 1.
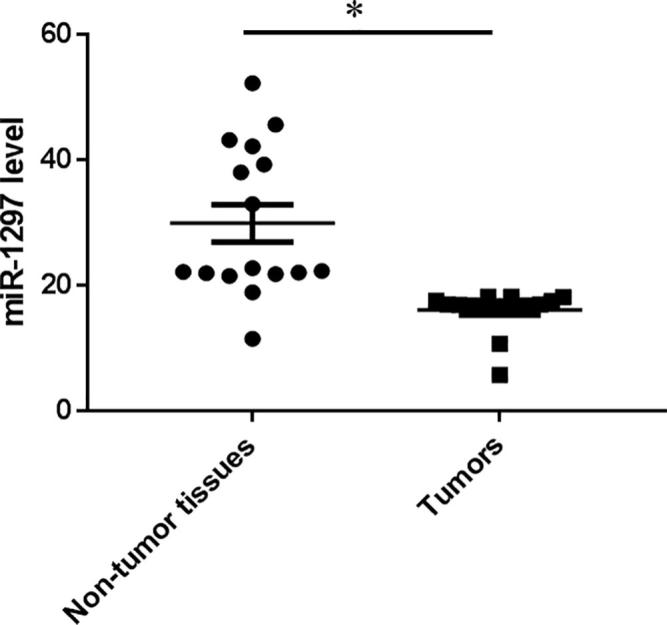
miR-1297 is down-regulated in tumors compared with non-tumor tissues. qRT-PCR examined the expression level of miR-1297 in OSCC specimens. Values represent the mean ± standard deviation, *P < .05, versus non-tumor tissues.
3.2. MiR-1297 suppresses proliferation of SSC-4 cells
To decipher the biological role of miR-1297 plays in the progression, we subsequently determined whether overexpression of miR-1297 would affect the SSC-4 cell growth. Stable transfection with lentiviral vector containing miR-1297 produced high level of miR-1297 in the SSC-4 cells (Fig. 2A). The miR-1297 overexpressing cells showed consistent decrease in cell proliferation rate compared with the miR-NC overexpressing and wild type cells, as measured by CCK-8 kit (Fig. 2B). In addition, compared with the miR-NC transfected cells, SSC-4 cells transfected with miR-1297 formed less colonies (Fig. 2C).
Fig. 2.
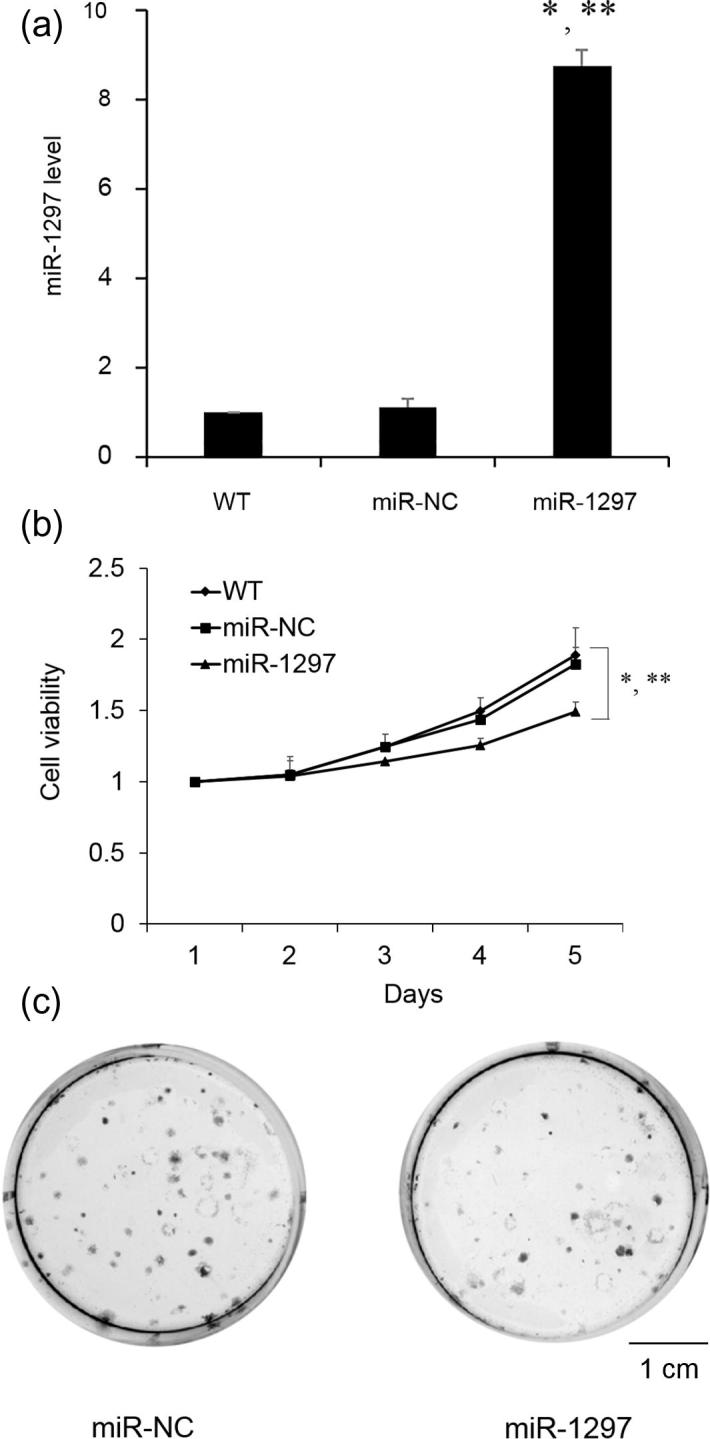
Effects of miR-1297 on SSC-4 cell proliferation. SSC-4 cells were transfected with lentiviral vector control (miR-NC) and miR-1297. (A) the expression level of miR-1297 in SSC-4 cells; (B) CCK-8 cell proliferation assay with SSC-4 cells or SSC-4 cells expressing either miR-NC or miR-1297; (C) Representative colony formation assays of SSC-4 cells expressing either miR-NC or miR-1297. Values represent the mean ± standard deviation (N = 3). *P < .05, versus WT; **P < .05, versus miR-NC. Scale bar: 1 cm.
To further confirm the suppressing function, we investigate whether block of miR-1297 would increase cell growth rate. MiR-1297 level significantly decreased after SSC-4 cells transfected with anti-miR-1297 (Fig. 3A). As shown in Fig. 3B and C, a marked increase in cell proliferation and colony numbers were observed after down-regulation of miR-1297. Our data of miR-1297 suggested its suppressing role in cell proliferation.
Fig. 3.
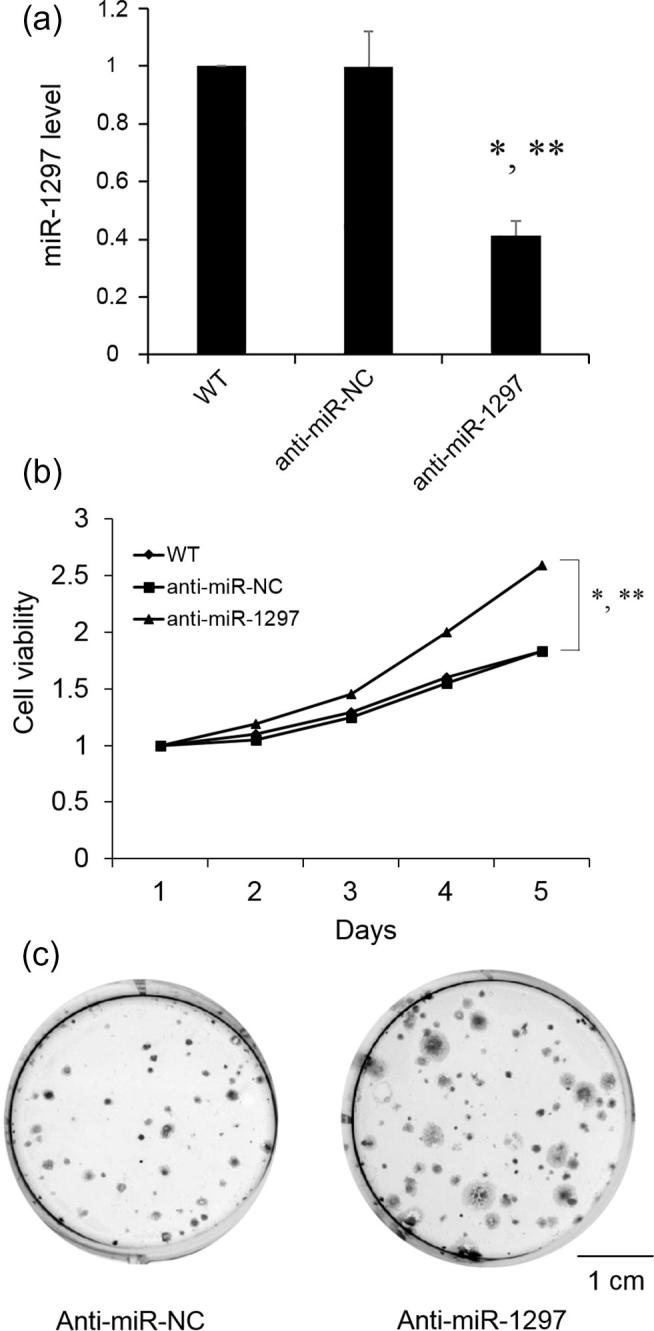
Effects of miR-1297 knockdown on SSC-4 cell proliferation. miR-1297 was knockdown in SSC-4 cells with anti-miR-1297. (A) the expression level of miR-1297 in SSC-4 cells; (B) CCK-8 cell proliferation assay with SSC-4 cells after miR-1297 knockdown; (C) Representative colony formation assays of SSC-4 cells expressing after miR-1297 knockdown. Values represent the mean ± standard deviation (N = 3). *P < .05, versus WT; **P < .05, versus anti-miR-NC. Scale bar: 1 cm.
3.3. PTEN is up-regulated in tumor tissues and repressed by miR-1297
To further clarify the cellular mechanisms underlying miR-1297 mediated cell proliferation, we used the bioinformatics tool, Targetscan, to predict target genes of miR-1297. Among these potential regulatory genes, PTEN was found to be up-regulated in the tumor tissues versus nontumor tissues by qRT- PCR (Fig. 4A). Next, we investigate the mRNA and protein levels of PTEN after miR-1297 overexpression. And, we found both the mRNA and protein levels of PTEN were significantly decreased by the increasing level of miR-1297 (Fig. 4B, C). These results suggest that both miR-1297 and PTEN are involved in the tumor progression.
Fig. 4.
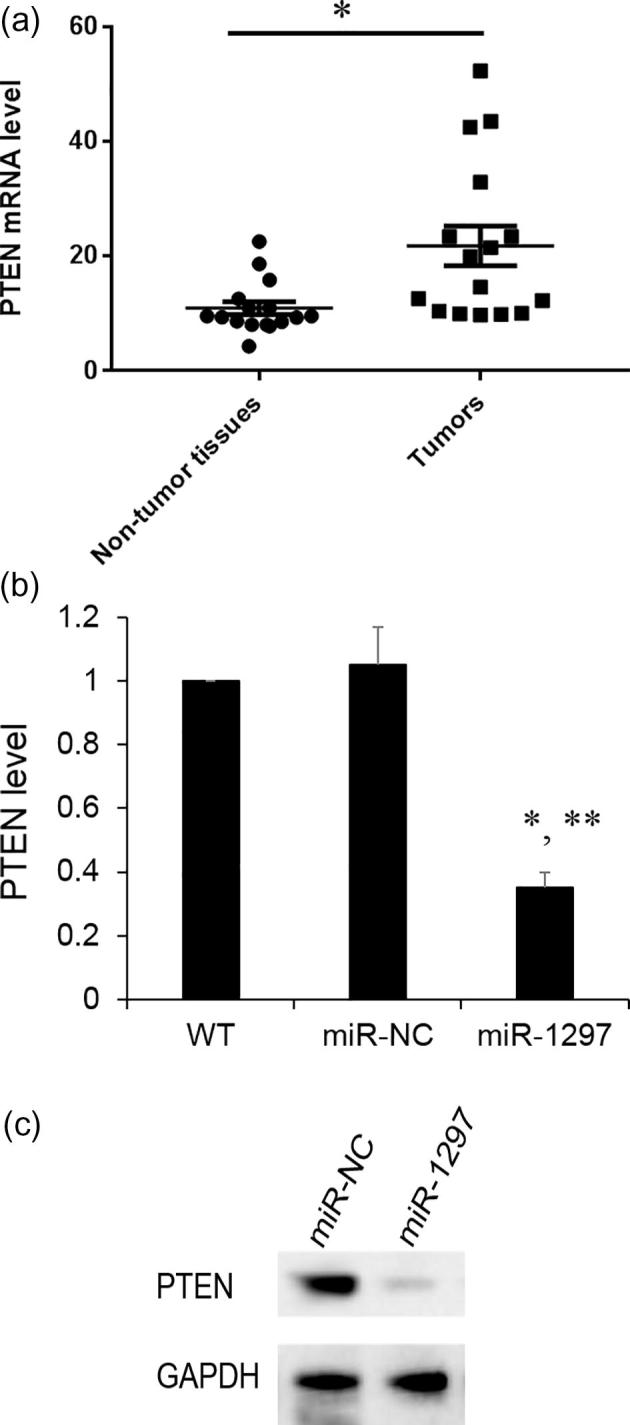
PTEN expression in tissue and SSC-4 cells. (A) PTEN mRNA expression level was determined by qRT-PCR in OSCC specimens. Values represent the mean ± standard deviation (N = 3), *P < .05, versus non-tumor tissues; (B) PTEN mRNA expression with SSC-4 cells or SSC-4 cells expressing either miR-NC or miR-1297. Results are mean ± standard deviation (N = 3). *p < .05 vs WT; **p < .05 vs miR-NC; (C) PTEN protein level was determined by western blotting. GAPDH was used as an internal control.
3.4. PTEN silencing inhibits SSC-4 cell proliferation
To assess the role of PTEN in the tumor progression, we silenced PTEN and determined whether the cell proliferation was affected. As shown in Fig. 5A, sh-PTEN significantly reduced the PTEN protein expression in SSC-4 cells. And, we also found silencing PTEN inhibits cell proliferation which is similar to miR-1297 overexpression (Fig. 5B). These data provide further evidence that miR-1297 could inhibit OSCC progression through regulation of PTEN.
Fig. 5.
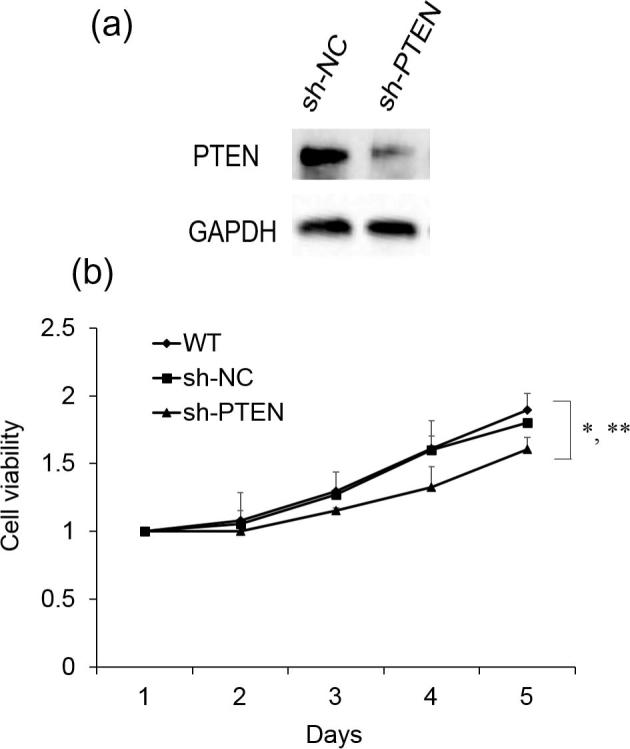
Effects of PTEN silencing on SSC-4 cell proliferation. (A) PTEN protein expression level was determined by western blotting after PTEN silencing; (B) CCK-8 cell proliferation assay with SSC-4 cells after PTEN silencing. Values represent the mean ± standard deviation (N = 3). *P < .05, versus WT; **P < .05, versus sh-NC.
4. Discussion
Accumulating evidences showed that dysregulation of miRNAs was frequently observed in a various types of cancers and plays important roles in tumor progression (Hata and Lieberman, 2015, Yates et al., 2013). The expression and biological roles of miRNAs in OSCC remain incompletely characterized and underlie the lack of effective treatment. Herein, miR-1297 expression was significantly lower in the tumor tissues than non-tumor tissues, which is consistent with a previous observation (Chen et al., 2014). And, miR-1297 was also found to suppress the SSC-4 cell growth. Additionally, miR-1297 downregulation is inversely correlated with PTEN expression in tissue samples. Furthermore, we found that PTEN expression was inhibited after miR-1297 overexpression. Moreover, under PTEN silencing, the cell growth exhibited the similar pattern as miR-1297 overexpression. Our results indicate that miR-1297 involved in the cell proliferation through PTEN.
MiRNAs are noncoding small RNAs and have been found to regulate tumor progression through targeting genes that are critical for cancers. MiR-1297 is one of the poorly understood miRNAs. The expression and functions of miR-1297 are cell and tissue specific. Recently, miR-1297 has been found to be down-regulated in colorectal cancer (Chen et al., 2014). However, miR-1297 was significantly up-regulated in Laryngeal squamous cell carcinoma (LSCC) compared with normal tissue (Li et al., 2012). In this study, we found that miR-1297 was significantly down-regulated in OSCC tumors compared with non-tumor tissues. In addition, miR-1297 overexpression has been observed to contribute to SSC-4 cell proliferation in vitro.
PTEN is a protein phosphatase and well-known as a tumor suppressor gene down-regulated in multiple types of cancers (Levine et al., 2002, Mehrian-Shai et al., 2007, Ghosh-Choudhury et al., 2009, Vinciguerra et al., 2009, Yang et al., 2010, Zhang et al., 2010, Nagy et al., 2011, Bian et al., 2012, Kong et al., 2012, Akca et al., 2013). PTEN has been found to be regulated by miR-21 in the human hepatocellular cancers (Meng et al., 2007). And, loss of PTEN in the primary prostate cancers correlates with high Gleason score and advanced stage (McMenamin et al., 1999). However, in our study, we found that PTEN is up-regulated in tumors compared with non-tumor tissues. And, PTEN expression is negatively correlated with miR-1297 expression. Furthermore, PTEN silencing inhibits the SSC-4 cell proliferation which exhibit the similar effects as miR-1297 overexpression. This leaves us with an interesting direction to follow in the future studies.
In summary, our data demonstrated that miR-1297 was down-regulated in OSCC and the overexpression of miR-1297 inhibited SSC-4 cell proliferation. Moreover, PTEN, a crucial tumor suppressor, is inversely correlated with miR-1297 expression. The identified link of miR-1297/PTEN may be useful in the understanding of OSCC progression and provide a new candidate for the therapeutic targeting in OSCC.
Conflicts of interest
None declared.
Footnotes
Peer review under responsibility of King Saud University.
References
- Akca H., Demiray A., Aslan M., Acikbas I., Tokgun O. Tumour suppressor PTEN enhanced enzyme activity of GPx, SOD and catalase by suppression of PI3K/AKT pathwayin non-small cell lung cancer cell lines. J. Enzyme Inhib. Med. Chem. 2013;28:539–544. doi: 10.3109/14756366.2011.654114. [DOI] [PubMed] [Google Scholar]
- Bian Y., Hall B., Sun Z.J., Molinolo A., Chen W., Gutkind J.S., Waes C.V., Kulkarni A.B. Loss of TGF-beta signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2012;31:3322–3332. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chen P., Wang B.L., Pan B.S., Guo W. MiR-1297 regulates the growth, migration and invasion of colorectal cancer cells by targeting cyclo-oxygenase-2. Asian Pac. J. Cancer Prev. 2014;15:9185–9190. doi: 10.7314/apjcp.2014.15.21.9185. [DOI] [PubMed] [Google Scholar]
- Epstein J.B., Zhang L., Rosin M. Advances in the diagnosis of oral premalignant and malignant lesions. J. Can. Dent. Assoc. 2002;68:617–621. [PubMed] [Google Scholar]
- Feng R., Dong L. Knockdown of microRNA-127 reverses adriamycin resistance via cell cycle arrest and apoptosis sensitization in adriamycin-resistant human glioma cells. Int. J. Clin. Exp. Pathol. 2015;8:6107–6116. [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury T., Mandal C.C., Woodruff K., St Clair P., Fernandes G., Choudhury G.G., Ghosh-Choudhury N. Fish oil targets PTEN to regulate NFkappaB for downregulation of anti-apoptotic genes in breast tumor growth. Breast Cancer Res. Treat. 2009;118:213–228. doi: 10.1007/s10549-008-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Lieberman J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 2015;8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- Hui A.B., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O'Sullivan B., Waldron J., Gullane P., Cummings B., Liu F.F. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin. Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- Kong L., Schafer G., Bu H., Zhang Y., Zhang Y., Klocker H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis. 2012;33:751–759. doi: 10.1093/carcin/bgs022. [DOI] [PubMed] [Google Scholar]
- Landis S.H., Murray T., Bolden S., Wingo P.A. Cancer statistics, 1999. CA Cancer J. Clin. 1999;49(8–31):31. doi: 10.3322/canjclin.49.1.8. [DOI] [PubMed] [Google Scholar]
- Levine R.A., Forest T., Smith C. Tumor suppressor PTEN is mutated in canine osteosarcoma cell lines and tumors. Vet. Pathol. 2002;39:372–378. doi: 10.1354/vp.39-3-372. [DOI] [PubMed] [Google Scholar]
- Li X., Wang H.L., Peng X., Zhou H.F., Wang X. miR-1297 mediates PTEN expression and contributes to cell progression in LSCC. Biochem. Biophys. Res. Commun. 2012;427:254–260. doi: 10.1016/j.bbrc.2012.09.025. [DOI] [PubMed] [Google Scholar]
- McMenamin M.E., Soung P., Perera S., Kaplan I., Loda M., Sellers W.R. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–4296. [PubMed] [Google Scholar]
- Mehrian-Shai R., Chen C.D., Shi T., Horvath S., Nelson S.F., Reichardt J.K., Sawyers C.L. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc. Natl. Acad. Sci. U S A. 2007;104:5563–5568. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R., Ganapathi S., Comeras I., Peterson C., Orloff M., Porter K., Eng C., Ringel M.D., Kloos R.T. Frequency of germline PTEN mutations in differentiated thyroid cancer. Thyroid. 2011;21:505–510. doi: 10.1089/thy.2010.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra M., Carrozzino F., Peyrou M., Carlone S., Montesano R., Benelli R., Foti M. Unsaturated fatty acids promote hepatoma proliferation and progression through downregulation of the tumor suppressor PTEN. J. Hepatol. 2009;50:1132–1141. doi: 10.1016/j.jhep.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Williams H.K. Molecular pathogenesis of oral squamous carcinoma. Mol. Pathol. 2000;53:165–172. doi: 10.1136/mp.53.4.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shao N., Luo G., Li L., Zheng L., Nilsson-Ehle P., Xu N. Mutations of PTEN gene in gliomas correlate to tumor differentiation and short-term survival rate. Anticancer Res. 2010;30:981–985. [PubMed] [Google Scholar]
- Yates L.A., Norbury C.J., Gilbert R.J. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi R., Cullen B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Huang C., Guo Y., Gou X., Hinsdale M., Lloyd P., Liu L. MicroRNA-26b modulates the NF-kappaB pathway in alveolar macrophages by regulating PTEN. J. Immunol. 2015;195:5404–5414. doi: 10.4049/jimmunol.1402933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yu D., Li X., Hu J., Gong J. Reduced expression of PTEN protein and its prognostic significance in the gastrointestinal stromal tumor. J. Huazhong Univ. Sci. Technol. Med. Sci. 2010;30:165–169. doi: 10.1007/s11596-010-0206-1. [DOI] [PubMed] [Google Scholar]


