Abstract
Advanced paternal age has very real consequences in fertility, embryogenesis, and even offspring health. Specifically, advanced paternal age has been linked to delayed time to pregnancy and in some studies even appears to be linked to a decreased likelihood of achieving a pregnancy. Epidemiological and animal model evidence also suggests that the offspring of older fathers are at an elevated risk for neuropsychiatric disease. For these reasons it is essential that we have a comprehensive understanding of what actually occurs in the gametes of the aging male. Available data suggest that there are very clear patterns of aging in the sperm epigenome that can be directly detected in DNA methylation patterns. Importantly, these alterations are so consistent that a predictive model has been successfully generated to predict an individual’s age based only on sperm DNA methylation signatures. Because this metric is the most direct way to detect aging in sperm, it is logical that these signatures may offer predictive value for the offspring abnormalities that are also correlated with advanced paternal age and as such may offer a unique opportunity to generate diagnostic tools that can identify personalized risks for each couple hoping to achieve a pregnancy. While a great deal of work still needs to be performed to understand the real diagnostic utility of sperm epigenetic marks, the potential is real and warrants further investigation particularly in the context of advanced paternal age.
Keywords: Epigenetics, aging, sperm, male fertility
Introduction
Aging is an unavoidable and unforgiving biological process with significantly disproportionate gender-based effects on human fertility (Figure 1). Females often experience an abrupt loss of fertility in their mid 30’s to early 40’s that is marked by an increased likelihood of spontaneous abortions, chromosomal defects in offspring, preterm delivery and intrauterine growth restriction (IUGR) (1). This already significant impact on female fertility is soon followed by sterility upon the onset of menopause. In stark contrast is the impact of aging on males, which has historically received far less attention, likely the result of the significantly less striking negative consequences of the aging process compared with females. Despite this, a great deal of data has recently come to light suggesting an association between advanced paternal age and neuropsychiatric disorders, among other consequences, in the offspring of aged fathers (2-5). Additional studies focusing on sperm epigenetics have found distinct and consistent alterations to the sperm epigenome associated with aging that have the potential to impact the offspring (6,7). Taken together, these findings have driven increased interest in the process of aging in fathers and the impacts of this process on both male fertility and the increased disease susceptibility in the offspring of older males.
Figure 1.
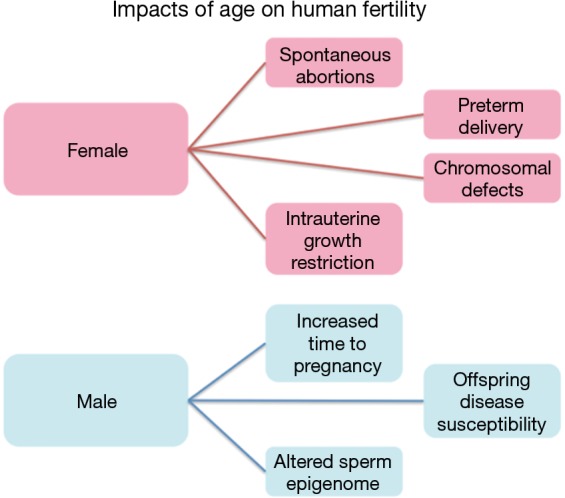
Diagram of the most prominent impacts of aging on fertility in both males and females.
Many couples necessarily weigh the risks associated with advanced maternal age in family planning decisions, while the age of the male partner has historically been viewed as a less important consideration. Based on newly available data however, it is clear that advanced paternal should be included in these considerations (2-4,8). In the future, it will be important that data regarding the impact of paternal aging be made available to be care providers and couples with aged male partners to facilitate well-informed family planning decisions in light of the data regarding advanced paternal and maternal age. Novel tools utilizing epigenetic analysis appear to be capable of identifying aging patterns directly in the gametes and in the future may serve as a tool for physicians to offer more personalized counseling with patients regarding the potential impacts of advanced paternal age (9).
Epigenetics
The epigenome is an essential and modifiable series of marks including DNA methylation, nuclear protein composition (and specifically modifications to histone protein tails), and, by some definitions, various RNA species. These epigenetic marks are competent to regulate gene expression and can be passed onto the embryo following fertilization. Because of the epigenome’s role in gene regulation, it is not surprising that each cell in the body has a distinct epigenetic profile that is tightly associated with each specific cell’s function. Human sperm are no exception to this, and in fact may represent one of the best examples of a unique cell type with a highly specialized epigenome well suited to drive the morphologically and functionally distinct attributes of this unique cell type.
In somatic cells, DNA is packaged around histones, however in sperm the majority of histones have been displaced by transition proteins during spermatogenesis. As the name indicates, these transition proteins are only in place for a short period of time before being replaced by protamine proteins 1 and 2 (P1 and P2). These protamine proteins work together in an approximately 1:1 ratio to form a toroidal structure that is 6 to 20 times more tightly compact than the nucleosome bound nucleus (10,11). The expression patterns of these proteins appear to be important for both sperm cell function and fertility. While not abnormal to observe these altered ratios in the general population, unbalanced expression of the protamines (P1 and P2) tends to be associated with abnormal sperm function, DNA damage, reduced fertilization rates, and reduced implantation rates (12,13). These findings were among the first to suggest that the nuclear protein composition of the sperm cell may have an impact beyond normal sperm function. It is important to note that not all of the sperm DNA is protamine-bound. In fact about 5% remains enriched with nucleosomes (and associated histone modifications). Thus these regions still have the capacity to affect gene activity (14). In 2009 it was demonstrated that histones are retained at specific loci important in embryonic development including: developmental gene promoters, genes encoding microRNAs, and imprinted loci (14). In addition, sites where histones are retained in the mature sperm often maintain both transcription activating and transcription silencing histone modifications within the same region. Such ‘bivalent’ histone modifications are quite important to pluripotency and are a hallmark of stem cells (14). Taken together, it appears that sperm function, embryogenesis and even early development, likely require some level of normalcy in the sperm epigenome.
DNA methylation marks are also strikingly unique in the sperm epigenome. The sperm methylome reflects and facilitates the cell’s highly specialized and unique functional role (15). Methylation marks at cytosine residues, typically found at cytosine phosphate guanine dinucleotides (CpGs), and have been shown to be capable of transcription regulation based on the presence or absence of this mark. Most notably, these marks are believed to help prevent aberrant transcription. These roles are dependent on location relative to gene architecture. One of the most distinct areas of the genome that is differentially methylated between sperm and somatic cells are imprinted regions (16). These genomic regions display a parent-of-origin-specific methylation signature such that DNA from each parent will be entirely differentially methylated (one being fully methylated and the other fully unmethylated). Thus, the methylation signature in diploid (somatic) cells when taken as a whole will be approximately 50% methylated where as in the sperm the methylation signatures at these same sites will be either fully unmethylated or fully methylated depending on the specific site. This represents only one of many distinctions that make sperm DNA methylation signatures so unique. This unique epigenetic landscape plays a key role in the many aspects of sperm function.
Delayed parenthood
A significant contributing factor to the interest in the impacts of paternal age is the trend of delayed parentage that has occurred in developed countries for many years. This trend toward delaying having children can be seen in both sexes and is believed to be a result of socioeconomic pressures and the increased frequency of divorce with subsequent remarriage (3). Though this trend in many ways is justified by increasing life expectancies in both sexes, advanced paternal age significantly affects general semen parameters, epigenetic signatures, as well as point mutations that ultimately affect fecundity and even offspring health (2,3,6-8). A very clear trend of delayed parenthood in both females and males exists in developing countries around the world. Many examples of this exist in the literature. During the 10-year span of 1993 to 2003 the percentage of 35–54 years old fathers increased from 25% of all births in Great Britain to 40%. In agreement with these data was the percent of fathers over the same period who were less than 35 years of age, which fell from 74% of total births to only 60% (17). Australian data reflects the same trend at a very similar rate. Between the years of 1988 and 2008 the average age of fathers in Australia increased by an average of approximately 3 years (18). The average age of fathers in Germany increased by 2 years over a 10-year period (3). Similar trends can be found in the United States and many other developed countries. As this trend continues there will soon be a smaller percentage of fathers <35 years of age compared to fathers >35 years of age. While the trend is justifiable, the potential impacts of delayed parenthood are real and should be thoroughly understood and considered.
Impacts of advanced paternal age
Fecundity
Though very different from the abrupt and universal cessation of fertility seen in females, a significant decline in man’s fertility and fecundity does exist and is directly correlated with age. However, the age at which this decline occurs and even the magnitude of this decline among a population of men remains poorly understood and is quite variable. In many ways this is not surprising as the decline in fertility in the male is not as abrupt as in females, but the lack of a defined point at which men are considered to be ‘aged’ (in terms of reproductive potential) can cause a level of increased uncertainty in family planning decisions. Despite this uncertainty, based on many studies it is clear that an age effect on male fecundity does exist and should be considered in family planning decisions.
In an observational study performed in the United Kingdom in 2003, Hassan et al. found that men >45 years of age had a five-fold increase in their time to pregnancy in comparison to individuals <25 years of age (19). Interestingly, when compared to males <25, men 45 and older were also 12.5 times more likely to have a time to pregnancy of greater than 2 years (19). As expected, this effect is amplified when the female member of a couple is of advanced reproductive age as well [35–39]. In these couples, men >40 were more than two times as likely to fail to conceive during a 12-month period in comparison to men <40 (20). Additionally, when taking into account unsuccessful pregnancies in the same groups, men over 40 were 3 times less likely to produce viable offspring than the younger cohort (20). Similar studies support these data and suggest an increased frequency of fetal loss, increased time to pregnancy, and decreased probability of conception with increasing paternal age (21-23). However, there are conflicting data that suggest little or no effect of paternal age when considering natural conception (24).
The impact of paternal age has also been assessed with the use of assisted reproductive techniques. In a study from France, 17,000 intrauterine insemination (IUI) cycles were analyzed and described a pregnancy rate of 12.3 for couples whose male partner was less than 30 years old. In contrast, and after adjusting for female age, this study found that when the male partner was over 30 years old the couples had significantly lower pregnancy rates with an average (25). Similarly, it was shown in 1995 by Mathieu et al. that older men (≥35 years old) had decreased rates of conception (26). It should be noted however, that these data are not without controversy. Some studies have failed to identify a paternal age effect on IUI success (27). Studies of the impact of paternal have also been conducted using in vitro fertilization (IVF) with similar controversy. Many studies suggest that pregnancy rates/outcomes in IVF cycles can be impacted by paternal age (28). In fact, some have suggested a compounded and amplified effect with couples having both partners of advanced age (29). Among the most convincing data is work using donor eggs, effectively isolating the male as the variable of interest. A significant paternal age effect was seen on pregnancy outcomes when performing a large study on IVF with donor eggs (30). However, an even more recent study, which corrected for age of the egg donor found no effect of paternal age on pregnancy outcome (31).
Offspring health
The impacts of external environments, lifestyles, and other biological processes (including aging) have been shown to impact the sperm in many ways and have also been associated with phenotypic effects in offspring. In some cases, these effects can be transmitted across multiple generations (32-34). In addition, multiple animal studies and a few human studies have observed associations between paternal exposures, sperm epigenetic status, and offspring phenotype. For example, advanced paternal age, paternal diet, exposures to stress and in utero exposures to various compounds have all been demonstrated to impact offspring phenotype and/or epigenetic profiles.
Specifically, some recent studies have suggested the existence of heritability that is uniquely propagated through the paternal germ line and believed to be a result of inheritance of DNA methylation profiles. One recent study in particular showed that in utero alcohol exposure alters proopiomelanocortin (POMC) gene promoter methylation in the sperm (35). Intriguingly, this alteration is inherited only through the paternal germ line for multiple generations. Though this study did not specifically look at the impact of aging they were able to show an interesting pattern of inheritance via DNA methylation signatures that may also play a role in aging. In support of this idea is data from a 2015 study (8) which demonstrated clear age-associated DNA methylation alterations in mouse sperm that were passed onto the offspring. These offspring were also found to have altered behavioral patterns that mimicked the patterns seen in human neuropsychiatric disorders (8). Thus, the mechanism of epigenetic inheritance provides a plausible explanation for the non-genetic paternal transmission of increased disease susceptibility from whatever source.
Of particular interest in this review is the impact of advanced paternal age on offspring. This topic has begun to receive much attention and has recently been described as the ‘paternal age effect’. In brief, this effect can be summarized by the findings of recent studies that have linked paternal aging and the prevalence of well-known neuropsychiatric disorders in offspring (5,36,37). Specifically, multiple studies have shown an increased risk of schizophrenia in the offspring of older fathers (4,5). The most notable meta-analysis found an increased relative risk (up to 1.66) in older fathers (4). Similarly, increased risk of autism has been identified through epidemiological studies in the offspring of older fathers (2). Interestingly, this increased risk of autism appears to also be detectable when assessing grand paternal age as well (38) suggesting that the risk may be compounded over multiple generations. Importantly, this neuropsychiatric/behavioral phenotype has been replicated in mouse models of paternal aging (36) where animals sired by older fathers had decreased social and exploratory behavior in the absence of influence from the father other than that provided by the sperm. One mouse study in particular showed altered behavioral patterns and DNA methylation signatures in the brain of offspring from older fathers and additionally found associated DNA methylation signature alterations in the sperm of aged fathers (8). In addition to increased incidence of neuropsychiatric disease, the offspring of older fathers also appear to have increased incidence of some forms of cancer (39-42). These intriguing findings suggest the paternal aging effect warrants study beyond what is simply seen in the affected individual. In fact, based on the available data, the study of aging should not be limited to a single generation. It appears that through epigenetic mechanisms the alterations seen in the sperm of an aged individual may be of consequence across multiple generations.
Sperm epigenetics and advanced paternal age
Despite the intriguing evidence surrounding the paternal age effect, the etiology of the increased frequency of various disorders in the offspring of aged fathers remains poorly defined, though there are likely candidates to explore. Among the most intriguing are epigenetic alterations to the sperm that are associated with age, as these alterations can accumulate over time and have the potential to impact fertility, embryogenesis and even offspring health.
In most somatic tissues throughout the body, aging alters DNA methylation profiles in consistent but quite subtle ways (43,44). Though very distinct from other cell types, it is not unreasonable to assume that similar age associated methylation alterations may take place in the sperm as well. In fact, Oakes et al. has described age associated hypermethylation at specific genomic loci in both sperm and liver tissue in male rats (44). In fact previous studies have reported that a key feature influencing the magnitude of age associated alterations to DNA methylation is the frequency at which the specific cell type of interest divides, with more frequently dividing cells displaying higher magnitude age associated DNA methylation changes (45). Thus, it is not unreasonable to assume that a great deal of methylation alteration can occur in the sperm as a result of aging similar to what is seen in other somatic tissues. Our laboratory has invested significant effort into describing the nature of age-affects in the sperm epigenome. In fact in 2014, we reported over 140 loci in sperm that consistently and predictably undergo DNA methylation changes as men age (7). The loci were first identified by evaluating DNA methylation patterns in paired sperm samples from the same men collected 10–20 years apart and subsequently validated in an independent cohort of men. The observation of systematic and predictable changes in sperm DNA methylation patterns as a function of age gave rise to the application of machine learning algorithms to develop a sperm DNA methylation-based age predicting calculator (9). Remarkably, these alterations were enriched at genes known to be associated with neuropsychiatric disease processes (specifically, bipolar disorder and schizophrenia (7). This finding is of particular interest within the context of previous reports of increased risk of neuropsychiatric disease in the offspring of older fathers. In addition, our data showed that regions where age-associated changes typically took place were enriched in sub-telomeric regions. Importantly, sub-telomeric regions have been previously shown to escape the large-scale epigenetic reprograming events following fertilization and during early sperm development suggesting that age-impacted epigenetic signals may be capable of direct transmission to the offspring (46). In further support of this, is data from murine models that have confirmed an impact of aging on the sperm methylome and have additionally shown that the age-associated methylation alterations are transmitted to offspring and impact offspring behavioral phenotypes (8).
A very striking feature associated with age associate DNA methylation change in sperm can be seen when comparing these patterns to those seen in somatic cell aging (Figure 2). Specifically, it has been reported that in somatic cells there is a global loss of DNA methylation whereas among the regions known to change with age there is a strong bias toward increases in methylation with age (47). In contrast, the opposite is observed in human sperm with global increases in methylation and a strong bias toward regional loss of methylation at the sites known to be impacted by aging. In fact, of the age-impacted regions identified in our study DNA methylation decreased with age at 140 regions and increased with age at only eight regions (7). The biological impact of this difference is very difficult to interpret (because they are not necessarily found in the same regions), but the difference between somatic and germ line aging is striking. This finding is less surprising when taken into account that sperm are known to have other age-associated modifications that defy convention (i.e., telomere length) (48-50).
Figure 2.
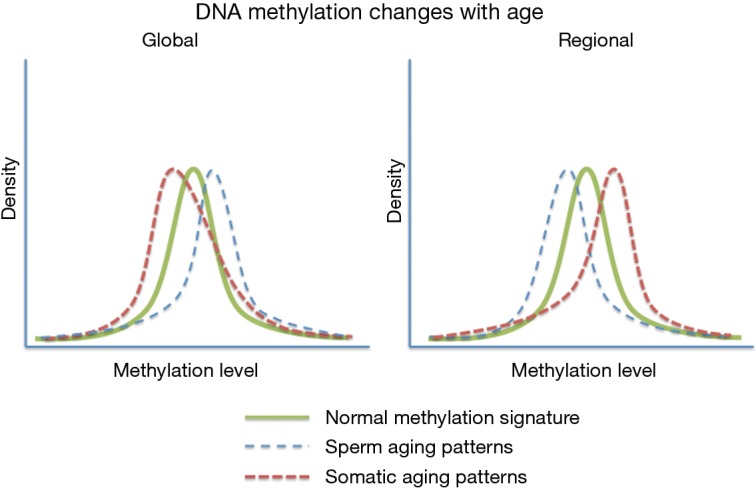
Theoretical density plots depicting the differences in methylation alterations associated with age between human sperm and somatic cells.
In addition to defining the age-associated methylation alterations that are common in sperm, our laboratory has also conducted machine learning experiments to determine the predictive nature of sperm DNA methylation signatures and aging (9). Specifically, we constructed multiple linear regression models to determine if sperm DNA methylation signatures were predictive of an individual’s chronological age. While a methylation age calculator was previously constructed by Steven Horvath in 2013 (51), this calculator was constructed for somatic tissues and the epigenetic signatures of aging are so distinct between somatic cells and sperm that the available epigenetic age calculator is entirely ineffective for age prediction based on sperm DNA methylation signatures. A total of 329 sperm samples were assessed and utilized to construct a model using DNA methylation array data. The model was trained using methylation at only 51 of the regions previously identified to be affected by age. Because of the remarkable consistency with which DNA methylation changes with age, the current model is able to predict an individual’s chronological age with up to 94% accuracy. Because it is believed that methylation alterations with age may play a role in the increased incidence of various abnormalities in the offspring of older fathers, measuring aging directly in this tissue potentially offers a great deal of insight into each individual’s risk of passing on aberrant methylation signatures. Despite the intriguing potential, a great deal of work needs yet to be performed to ensure that these patterns are truly predictive of offspring abnormalities.
Conclusions/future directions
While the data surrounding the impacts of age on epigenetic signatures in the sperm are intriguing and appear to offer significant potential utility, it is important to note that a great deal of work still needs to be performed to specifically implicate age-impacted DNA methylation signatures in abnormalities commonly seen in the offspring of older fathers. Despite this, even if these alterations were not found to be causative, but were only associated, the predictive potential of these marks is still real. With more sophisticated modeling, any prediction that could help a couple to better understand their unique risk of various pregnancy outcomes or offspring phenotypes is worth pursuing.
A key to making such prediction a reality is the completion of more focused studies using what has already been learned to guide the process. Specifically, animal model studies could be used to better understand the heritability of methylation signatures directly. In fact, new techniques may soon allow us to directly alter methylation signatures in the sperm, which would ultimately allow us to analyze the heritability of these alterations. Among the most promising of these techniques is the use of dCas9-DNMT3a fusion protein to induce DNA methylation changes in sperm (52). While this has yet to be performed in the context described above, the technology is moving rapidly enough that such an experiment is feasible in the relative near term.
An additional experiment that will be an important next step in expanding our understanding of the predictive nature of these marks is to focus on the fathers of children with already manifested disorders regardless of age. A potential design would be to recruit the fathers of children with a specific form of neuropsychiatric disease and perform sperm DNA methylation profiling on these individuals. It will be important to ensure that the selected disorder in the offspring manifests itself early in life such that an analysis of the father’s sperm if performed as close to conception as is possible. Such a study is problematic for many reasons mainly due to difficulties in the study design itself. Despite these difficulties, there is real potential to identify direct correlations between sperm DNA methylation signatures and offspring phenotypes, which would significantly augment our understanding of the predictive nature of sperm DNA methylation signatures.
Taken together, it is clear that the aging process significantly impacts the sperm epigenome. Work is still required to understand the impact of aging on histone modifications and nuclear protein composition in sperm, but the impact on sperm DNA is clear and has begun to be more thoroughly elucidated. While altered DNA methylation signatures in sperm have not been proven to escape all of the reprograming events in the early embryo, at least a portion of these marks are likely to be retained. Further, regardless of the direct impact of these alterations, the potential for prediction of the aging process has been demonstrated and as such may be useful diagnostically. Much work is still required to realize this potential, but the available data is very promising and clearly warrants further investigation.
Acknowledgements
None.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Lean SC, Derricott H, Jones RL, et al. Advanced maternal age and adverse pregnancy outcomes: A systematic review and meta-analysis. PLoS One 2017;12:e0186287. 10.1371/journal.pone.0186287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Idring S, Magnusson C, Lundberg M, et al. Parental age and the risk of autism spectrum disorders: findings from a Swedish population-based cohort. Int J Epidemiol 2014;43:107-15. 10.1093/ije/dyt262 [DOI] [PubMed] [Google Scholar]
- 3.Kuhnert B, Nieschlag E. Reproductive functions of the ageing male. Hum Reprod Update 2004;10:327-39. 10.1093/humupd/dmh030 [DOI] [PubMed] [Google Scholar]
- 4.Miller B, Messias E, Miettunen J, et al. Meta-analysis of paternal age and schizophrenia risk in male versus female offspring. Schizophr Bull 2011;37:1039-47. 10.1093/schbul/sbq011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naserbakht M, Ahmadkhaniha HR, Mokri B, et al. Advanced paternal age is a risk factor for schizophrenia in Iranians. Ann Gen Psychiatry 2011;10:15. 10.1186/1744-859X-10-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkins TG, Aston KI, Cairns BR, et al. Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril 2013;100:945-51. 10.1016/j.fertnstert.2013.05.039 [DOI] [PubMed] [Google Scholar]
- 7.Jenkins TG, Aston KI, Pflueger C, et al. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 2014;10:e1004458. 10.1371/journal.pgen.1004458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milekic MH, Xin Y, O'Donnell A, et al. Age-related sperm DNA methylation changes are transmitted to offspring and associated with abnormal behavior and dysregulated gene expression. Mol Psychiatry 2015;20:995-1001. 10.1038/mp.2014.84 [DOI] [PubMed] [Google Scholar]
- 9.Jenkins TG, Aston KI, Smith AD, et al. Paternal germ line aging: DNA methylation age prediction from human sperm. Available online: https://doi.org/ 10.1101/220764 [DOI] [PMC free article] [PubMed]
- 10.Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol 2007;8:227. 10.1186/gb-2007-8-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 1991;44:569-74. 10.1095/biolreprod44.4.569 [DOI] [PubMed] [Google Scholar]
- 12.Aoki VW, Emery BR, Liu L, et al. Protamine levels vary between individual sperm cells of infertile human males and correlate with viability and DNA integrity. J Androl 2006;27:890-8. 10.2164/jandrol.106.000703 [DOI] [PubMed] [Google Scholar]
- 13.Aoki VW, Moskovtsev SI, Willis J, et al. DNA integrity is compromised in protamine-deficient human sperm. J Androl 2005;26:741-8. 10.2164/jandrol.05063 [DOI] [PubMed] [Google Scholar]
- 14.Hammoud SS, Nix DA, Zhang H, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009;460:473-8. 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins TG, Aston KI, James ER, et al. Sperm epigenetics in the study of male fertility, offspring health, and potential clinical applications. Syst Biol Reprod Med 2017;63:69-76. 10.1080/19396368.2016.1274791 [DOI] [PubMed] [Google Scholar]
- 16.Mann JR. Imprinting in the germ line. Stem Cells 2001;19:287-94. 10.1634/stemcells.19-4-287 [DOI] [PubMed] [Google Scholar]
- 17.Bray I, Gunnell D, Davey Smith G. Advanced paternal age: how old is too old? J Epidemiol Community Health 2006;60:851-3. 10.1136/jech.2005.045179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Births, Australia, 2008. Summary of findings. Births. Australian Bureau of Statistics 2009. Available online: http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/3301.02009
- 19.Hassan MA, Killick SR. Effect of male age on fertility: evidence for the decline in male fertility with increasing age. Fertil Steril 2003;79 Suppl 3:1520-7. 10.1016/S0015-0282(03)00366-2 [DOI] [PubMed] [Google Scholar]
- 20.de la Rochebrochard E, Thonneau P. Paternal age and maternal age are risk factors for miscarriage; results of a multicentre European study. Hum Reprod 2002;17:1649-56. 10.1093/humrep/17.6.1649 [DOI] [PubMed] [Google Scholar]
- 21.Selvin S, Garfinkel J. Paternal age, maternal age and birth order and the risk of a fetal loss. Hum Biol 1976;48:223-30. [PubMed] [Google Scholar]
- 22.Ford WC, North K, Taylor H, et al. Increasing paternal age is associated with delayed conception in a large population of fertile couples: evidence for declining fecundity in older men. The ALSPAC Study Team (Avon Longitudinal Study of Pregnancy and Childhood). Hum Reprod 2000;15:1703-8. 10.1093/humrep/15.8.1703 [DOI] [PubMed] [Google Scholar]
- 23.Dunson DB, Colombo B, Baird DD. Changes with age in the level and duration of fertility in the menstrual cycle. Hum Reprod 2002;17:1399-403. 10.1093/humrep/17.5.1399 [DOI] [PubMed] [Google Scholar]
- 24.Olsen J. Subfecundity according to the age of the mother and the father. Dan Med Bull 1990;37:281-2. [PubMed] [Google Scholar]
- 25.Belloc S, Cohen-Bacrie P, Benkhalifa M, et al. Effect of maternal and paternal age on pregnancy and miscarriage rates after intrauterine insemination. Reprod Biomed Online 2008;17:392-7. 10.1016/S1472-6483(10)60223-4 [DOI] [PubMed] [Google Scholar]
- 26.Mathieu C, Ecochard R, Bied V, et al. Cumulative conception rate following intrauterine artificial insemination with husband's spermatozoa: influence of husband's age. Hum Reprod 1995;10:1090-7. 10.1093/oxfordjournals.humrep.a136100 [DOI] [PubMed] [Google Scholar]
- 27.Bellver J, Garrido N, Remohi J, et al. Influence of paternal age on assisted reproduction outcome. Reprod Biomed Online 2008;17:595-604. 10.1016/S1472-6483(10)60305-7 [DOI] [PubMed] [Google Scholar]
- 28.Klonoff-Cohen HS, Natarajan L. The effect of advancing paternal age on pregnancy and live birth rates in couples undergoing in vitro fertilization or gamete intrafallopian transfer. Am J Obstet Gynecol 2004;191:507-14. 10.1016/j.ajog.2004.01.035 [DOI] [PubMed] [Google Scholar]
- 29.de La Rochebrochard E, de Mouzon J, Thepot F, et al. Fathers over 40 and increased failure to conceive: the lessons of in vitro fertilization in France. Fertil Steril 2006;85:1420-4. 10.1016/j.fertnstert.2005.11.040 [DOI] [PubMed] [Google Scholar]
- 30.Frattarelli JL, Miller KA, Miller BT, et al. Male age negatively impacts embryo development and reproductive outcome in donor oocyte assisted reproductive technology cycles. Fertil Steril 2008;90:97-103. 10.1016/j.fertnstert.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 31.Whitcomb BW, Turzanski-Fortner R, Richter KS, et al. Contribution of male age to outcomes in assisted reproductive technologies. Fertil Steril 2011;95:147-51. 10.1016/j.fertnstert.2010.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerrero-Bosagna C, Covert TR, Haque MM, et al. Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reproductive toxicology 2012;34:694-707. 10.1016/j.reprotox.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson LM, Riffle L, Wilson R, et al. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition 2006;22:327-31. 10.1016/j.nut.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 34.Pembrey ME, Bygren LO, Kaati G, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 2006;14:159-66. 10.1038/sj.ejhg.5201538 [DOI] [PubMed] [Google Scholar]
- 35.Govorko D, Bekdash RA, Zhang C, et al. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry 2012;72:378-88. 10.1016/j.biopsych.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith RG, Kember RL, Mill J, et al. Advancing paternal age is associated with deficits in social and exploratory behaviors in the offspring: a mouse model. PLoS One 2009;4:e8456. 10.1371/journal.pone.0008456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalman C. Advanced paternal age increases risk of bipolar disorder in offspring. Evid Based Ment Health 2009;12:59. 10.1136/ebmh.12.2.59 [DOI] [PubMed] [Google Scholar]
- 38.Frans EM, Sandin S, Reichenberg A, et al. Autism risk across generations: a population-based study of advancing grandpaternal and paternal age. JAMA Psychiatry 2013;70:516-21. 10.1001/jamapsychiatry.2013.1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksuzyan S, Crespi CM, Cockburn M, et al. Birth weight and other perinatal characteristics and childhood leukemia in California. Cancer Epidemiol 2012;36:e359-65. 10.1016/j.canep.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray L, McCarron P, Bailie K, et al. Association of early life factors and acute lymphoblastic leukaemia in childhood: historical cohort study. Br J Cancer 2002;86:356-61. 10.1038/sj.bjc.6600012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemminki K, Kyyronen P, Vaittinen P. Parental age as a risk factor of childhood leukemia and brain cancer in offspring. Epidemiology 1999;10:271-5. 10.1097/00001648-199905000-00014 [DOI] [PubMed] [Google Scholar]
- 42.Yip BH, Pawitan Y, Czene K. Parental age and risk of childhood cancers: a population-based cohort study from Sweden. Int J Epidemiol 2006;35:1495-503. 10.1093/ije/dyl177 [DOI] [PubMed] [Google Scholar]
- 43.Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science 1983;220:1055-7. 10.1126/science.6844925 [DOI] [PubMed] [Google Scholar]
- 44.Oakes CC, Smiraglia DJ, Plass C, et al. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci U S A 2003;100:1775-80. 10.1073/pnas.0437971100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson RF, Atzmon G, Gheorghe C, et al. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell 2010;9:506-18. 10.1111/j.1474-9726.2010.00577.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guibert S, Forne T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 2012;22:633-41. 10.1101/gr.130997.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson B. Impact of aging on DNA methylation. Ageing Res Rev 2003;2:245-61. 10.1016/S1568-1637(03)00010-2 [DOI] [PubMed] [Google Scholar]
- 48.Unryn BM, Cook LS, Riabowol KT. Paternal age is positively linked to telomere length of children. Aging Cell 2005;4:97-101. 10.1111/j.1474-9728.2005.00144.x [DOI] [PubMed] [Google Scholar]
- 49.Njajou OT, Cawthon RM, Damcott CM, et al. Telomere length is paternally inherited and is associated with parental lifespan. Proc Natl Acad Sci U S A 2007;104:12135-9. 10.1073/pnas.0702703104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allsopp RC, Vaziri H, Patterson C, et al. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A 1992;89:10114-8. 10.1073/pnas.89.21.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stepper P, Kungulovski G, Jurkowska RZ, et al. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 2017;45:1703-13. 10.1093/nar/gkw1112 [DOI] [PMC free article] [PubMed] [Google Scholar]


