Abstract
The physiological and cellular adaptations to extreme fasting in northern elephant seals (Mirounga angustirostris, NES) are remarkable and may help to elucidate endocrine mechanisms that regulate lipid metabolism and energy homeostasis in mammals. Recent studies have highlighted the importance of thyroid hormones in the maintenance of a lipid-based metabolism during prolonged fasting in weaned NES pups. To identify additional molecular regulators of fasting, we used a transcriptomics approach to examine changes in global gene expression profiles before and after 6–8 wk of fasting in weaned NES pups. We produced a de novo assembly and identified 98 unique protein-coding genes that were differentially expressed between early and late fasting. Most of the downregulated genes were associated with lipid, carbohydrate, and protein metabolism. A number of downregulated genes were also associated with maintenance of the extracellular matrix, consistent with tissue remodeling during weight loss and the multifunctional nature of blubber tissue, which plays both metabolic and structural roles in marine mammals. Using this data set, we predict potential mechanisms by which NES pups sustain metabolism and regulate adipose stores throughout the fast, and provide a valuable resource for additional studies of extreme metabolic adaptations in mammals.
Keywords: adipose, de novo assembly, extracellular matrix, fasting, transcriptome
INTRODUCTION
Marine mammals have long been the subject of study in the field of comparative physiology because of their remarkable diving physiology and metabolic adaptations to fasting (24). Northern elephant seals (Mirounga angustirostris) (NES), in particular, have emerged as a robust marine mammal study system due to their extreme diving and fasting behavior and amenability to research handling. Adult NES forage pelagically and haul out at coastal rookeries twice a year for breeding and molting, during which they fast for a period of 1–3 mo. During fasting, NES are completely reliant on lipid mobilization from blubber and fatty acid oxidation as their primary fuel source, while maintaining extremely high metabolic rates and engaging in energy-intensive activities such as breeding and molting (5). Blubber, the specialized adipose tissue of marine mammals, is both an energy storage depot and a potent endocrine organ (8, 9, 28) and makes up nearly 50% of body mass in newly weaned NES pups (13). NES pups are nursed continuously for approximately 1 mo, weaned abruptly, and remain at their natal rookery for up to 8 wk to complete postnatal development, which includes increasing oxygen storage capacity and muscle mass (52). Due to the dependence of NES on mesopelagic prey (43), this postnatal period at the rookery requires complete fasting. This life history stage is one of the best studied in NES because of the pups’ accessibility on land, and our previous studies have examined the endocrine and metabolic adjustments that underlie this natural fasting period. Fasting seals exhibit chronically elevated triglycerides, insulin resistance, and increased thyroid hormone receptor availability, deiodinase activity, and expression of TH-mediated target genes and proteins coupled with increases in lipid metabolism and fatty acid mobilization (37–39, 41, 42, 61–64). In addition, the expression of lipid metabolic regulator genes such as peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) and uncoupling protein 2 (UCP2) is upregulated over the duration of fasting in parallel with the upregulation of TH signaling, providing an intriguing insight into natural TH-mediated reliance on lipid metabolism in seals that is not present in morbidly obese humans with similar levels of adiposity (37–39).
To date, studies of the molecular mechanisms involved in fasting metabolism and adaptation have been limited to small sets of target genes that are known to regulate adipose function in terrestrial model systems. To gain additional, broader insights into metabolic adjustments that occur during food deprivation in fasting-adapted mammals, we used a nontargeted RNA sequencing (RNA-Seq) approach to examine global gene expression profiles during fasting in energy-rich adipose tissue (blubber) in weaned NES pups. RNA-Seq has been used to profile adipose tissue transcriptome responses to short-term and intermittent fasting in mouse and human and to identify novel regulatory pathways of fasting metabolism (31, 49). Transcriptomics has also been applied to studies of adipose responses to prolonged fasting in hibernating mammals (19, 20). This approach has recently been employed in marine mammal study systems including NES (4), in which muscle and blubber transcriptome responses to a stress challenge in juveniles were examined, providing a wealth of species-specific sequence data and insights into cellular responses to stress (27, 29, 30). Profiling the blubber transcriptome during fasting in NES pups enables assessment of myriad cellular changes that occur to facilitate energy provisioning, as well as insights into potential mechanisms of avoidance of pathological consequences associated with states of insulin resistance. Our study suggests novel regulatory functions of metabolism including effects on lipid mobilization and the remodeling of the extracellular matrix (ECM) in fasting NES pups. Here for the first time, we provide a fasting-specific blubber transcriptome of northern elephant seals, revealing insights into adaptive fasting physiology that enables these mammals to survive and thrive.
METHODS
All procedures were reviewed and approved by the Institutional Animal Care and Use Committees of University of California-Merced and Sonoma State University. All work was conducted under National Marine Fisheries Service marine mammal permit #14636.
Animals.
Northern elephant seal (M. angustirostris) pups were sampled at Año Nuevo State Reserve (30 km north of Santa Cruz, CA) during their natural postweaning fast on land. Pups were tagged with hind flipper tags (Dalton Jumbo Roto-tags, Oxon, UK) and marked with black hair dye (Lady Clairol) within several days of weaning. Pups were sampled during the early (1–2 wk postweaning; n = 6) and late fasting periods (6–8 wk postweaning; n = 6) as independent cohorts. Samples were collected from two independent groups of pups. Time fasting was determined by date of flipper tagging, which occurs within several days of weaning, and confirmed by percentage of molted natal pelage. The pups were initially sedated with ~1 mg/kg Telazol (tiletamine/zolazepam HCl; Fort Dodge Laboratories, Ft. Dodge, IA) administered intramuscularly, and once the pups were immobilized, sedation was maintained with intravenous doses of ketamine and diazepam (Fort Dodge Laboratories) via an 18-gauge, 3.5-inch spinal needle inserted into the extradural spinal vein (37–39, 41, 42, 53, 54, 57, 58, 61–64).
Sample collection.
Prior to sampling, a small region in the flank of the animal near the hind flipper was cleaned with alternating wipes of isopropyl alcohol and betadine. A small (<1.5 cm) incision was made with a sterile scalpel (Sigma-Aldrich, Darmstadt, Germany), and a blubber biopsy (ca. 50–200 mg) was collected via a sterile biopsy punch needle (Henry Schein Animal Health, Dublin, OH). The biopsy samples were rinsed with cold, sterile saline, placed in cryogenic vials, and immediately frozen by immersion in liquid nitrogen (37–39, 41, 42, 53, 54, 57, 58, 61–64).
RNA isolation.
Tissue samples were stored at −80°C until extraction procedures. During RNA extraction, 80–100 mg of adipose were minced with a scalpel on ice, transferred to a glass tissue grinder, and homogenized with 1 ml of TRIzol Reagent (Life Technologies) per 100 mg of tissue. RNA was extracted according to the manufacturer’s protocol. Phenol-chloroform-isoamyl alcohol (Affymetrix) extraction was performed to remove DNase I (Roche, Indianapolis, IN). RNA concentration was quantified with the NanoDrop (Life Technologies) and Qubit RNA kit (Invitrogen).
Illumina sequencing.
Total RNA integrity was evaluated with 2100 Bioanalyzer RNA 6000 kit (Agilent). RNA samples had integrity values of 7.6–9.0. Strand-specific libraries for sequencing were prepared according to TruSeq protocol with Ribozero depletion (Human/Rat/Mouse; Illumina). Twelve libraries were sequenced (2 × 125 bp paired-end reads) on one lane with the HiSeq 2000 platform at the Otago Medical University’s (New Zealand) Genomic Limited center.
Transcriptome assembly and annotation.
Illumina HiSeq sequencing produced 41.50 ± 2.64 million reads per sample. Sequencing adapters and poor quality reads were removed with Trimmomatic before de novo assembly using Trinity v.2.1.1 with default settings (17). Reads were not abundance-normalized before assembly. Bowtie2 v2.2.7 was used to map reads back to assembly to evaluate assembly quality (33). Benchmarking Universal Single-Copy Orthologs (BUSCO) v1.22 (51) and the vertebrata ortholog database (downloaded on September 9, 2017) were used to evaluate assembly completeness. The assembly was annotated by BLASTX using DIAMOND v0.8.31 with more-sensitive option (3) and the UniProtKB/SwissProt database downloaded on August 20, 2017. The e-value threshold for significant matches was 0.001. Functional enrichment of KEGG categories in the annotated transcriptome was conducted with DAVID v6.8 with human genome as background (Homo sapiens genome, updated May 2016) and P value threshold of 0.05 for significant enrichment. The P values were adjusted for multiple hypothesis testing by the Benjamini-Hochberg method (26).
Gene expression analysis.
All gene expression analyses were conducted with the Trinity v2.4.0 pipeline. Transcript abundance was quantified with Salmon v0.8.1 (47). Differential expression analysis was performed at the gene level with the DESeq2 package [Bioconductor v3.5 (1)] in R (v3.4.1). Genes with adjusted P value <0.05 [false discovery rate (FDR) threshold = 0.05] and log2fold change of ≥1 and ≤−1 were considered as differentially expressed.
RESULTS
Blubber transcriptome assembly and annotation.
In this study, we analyzed and compared blubber transcriptomes of NES pups early and late in their postweaning fast. Blubber was collected from two separate groups of weaned pups, one group sampled 1–2 wk postweaning (“early,” n = 6), and the other group sampled 6–8 wk postweaning (“late,” n = 6). Plasma hormone levels and expression of target genes and proteins in blubber of the same animals have been previously published (38, 39).
Libraries generated from blubber of each individual (n = 12) were sequenced with Illumina HiSeq 2000, which produced 62.25 billion sequenced bases with 41.50 ± 2.64 million 125 bp paired-end reads per sample (National Center for Biotechnology Information Sequence Read Archive accession: SAMN08105288–SAMN08105299). Sequenced reads from all samples were assembled de novo into a single assembly with Trinity software after being trimmed with Trimmomatic using default Trinity settings (17). The assembly contained 1,830,330 transcripts (contigs) in 1,635,200 gene clusters, and mean and median contig lengths were 832 and 425 bp, respectively (Table 1; https://figshare.com/articles/Fasting_northern_elephant_seal_pup_blubber_transcriptome/5746227/2). The high number of transcripts is common for de novo-assembled transcriptomes (59, 66) and is likely a product of high degree of sequence polymorphism among the 12 individual seals used for the assembly. Eighty-seven percent of the reads mapped back as proper pairs to the assembly with bowtie2, indicating that the assembly accurately represented the sequenced reads (Table 1). We evaluated transcriptome completeness by calculating the percentage of BUSCOs (51) that were recovered in the assembly. The fasting elephant seal pup transcriptome contained 81.2% complete, 36.3% duplicated, and 15.5% fragmented BUSCOs from the vertebrate set of orthologs in OrthoDB. The assembly was missing only 3.2% of vertebrate BUSCOs, suggesting a fairly complete transcriptome (Table 1).
Table 1.
Transcriptome assembly statistics
| Sequenced bases | 62.25 billion |
| Reads per sample | 41.50 ± 2.64 million |
| Assembled bases | 1.09 billion |
| Trinity transcripts | 1,830,330 |
| Trinity genes | 1,635,200 |
| Mean contig length | 832 bp |
| Median contig length | 425 bp |
| Annotated transcripts | 389,363 |
| Complete vertebrate BUSCOs | 81.2% |
| Duplicated BUSCOs | 36.3% |
| Fragmented BUSCOs | 15.5% |
| Missing BUSCOs | 3.2% |
| Read alignment rate | 87.0% |
BUSCOs, Benchmarking Universal Single-Copy Orthologs, or highly conserved gene orthologs that are expected to be expressed in all vertebrates.
To identify protein-coding genes, we annotated the fasting elephant seal pup blubber transcriptome with DIAMOND BlastX with the “very sensitive” option (3) against the UniProt/SwissProt database with e-value threshold of 10−3. We identified 389,363 vertebrate homologs of elephant seal genes (Table 1, Supplementary File 1). (The online version of this article contains supplemental material.) DAVID functional annotation tool (26) was used to identify metabolic and signaling pathways (KEGG) that were enriched in the elephant seal transcriptome relative to the human genome. Thirty-seven KEGG pathways were enriched in the seal transcriptome relative to the human genome background [Benjamini-Hochberg adjusted (adj.) P < 0.05]. The top 20 significantly enriched pathways are shown in Fig. 1. The largest category was endocytosis, which contained 203 elephant seal homologs. Other significantly enriched categories of interest not shown in Fig. 1 include regulation of actin cytoskeleton (164 genes), RNA transport (135 genes), protein processing in endoplasmic reticulum (134 genes), Hippo signaling pathway (110 genes), FoxO signaling pathway (107) genes, neurotrophin signaling pathway (97 genes), glycerophospholipid metabolism (78 genes), valine, leucine, and isoleucine degradation (42 genes), and propanoate metabolism (27 genes). An additional 14 pathways were enriched at adj. P < 0.1. These include KEGG pathways of interest to the comparative physiology community such as metabolic pathways (885 genes), purine metabolism (137 genes), thyroid signaling pathway (91 genes), carbon metabolism (90 genes), glucagon signaling pathway (80 genes), TGF-β signaling pathway (69 genes), adipocytokine signaling pathway (58 genes), and biosynthesis of unsaturated fatty acids (22 genes). Lastly, we searched our annotated gene list for 168 genes in the Human Protein Atlas with elevated expression in adipose tissue compared with other tissue types. Seal homologs of 142 of these proteins were found in our transcriptome assembly, suggesting that we recovered the majority of genes enriched in adipose tissue in other mammals.
Fig. 1.
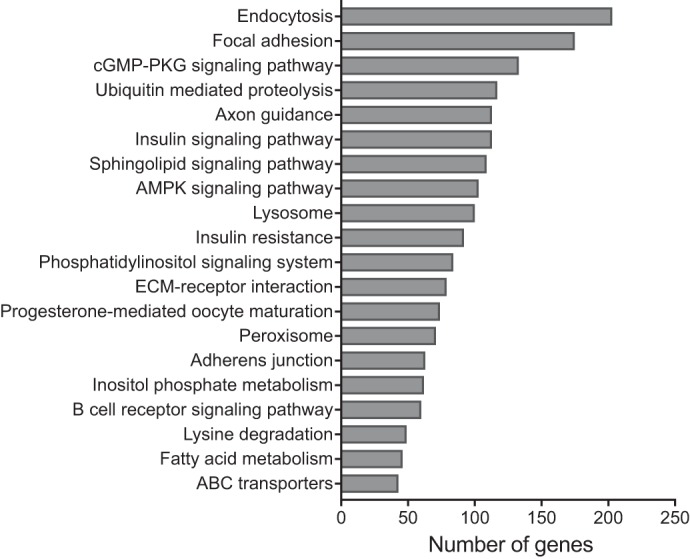
Top 20 KEGG pathways significantly enriched in the fasting elephant seal pup blubber transcriptome relative to the human genome (P < 0.05, corrected for multiple hypothesis testing with the Benjamini-Hochberg method, DAVID v6.8). Bars show number of annotated seal genes mapping to each category.
Differential gene expression analysis.
To quantify changes in gene activity in NES blubber during fasting, we conducted differential expression analysis at the gene level using Salmon and DESeq2. We initially identified 202 genes that were differentially expressed by at least twofold (adj. P value < 0.05 and FDR < 0.05) between early and late fasting in blubber of NES pups. However, a number of these had very low [transcripts per million (TPM)] counts (mean TPM < 0.1), and the gene list was subsequently filtered to remove genes with 0 TPM in two or more samples. The filtered list contains 154 differentially expressed genes (29 upregulated, 125 downregulated), of which 110 had BlastX hits to known vertebrate proteins (Fig. 2, Supplementary File 2). Some annotated genes include multiple transcript homologs; in total, there are 98 unique protein-coding genes (10 upregulated, 88 downregulated) that were differentially expressed between early and late fasting. Supplementary File 2 contains log2 fold-change, adj. P values, and individual gene expression counts (TPM) for differentially expressed genes in this study.
Fig. 2.
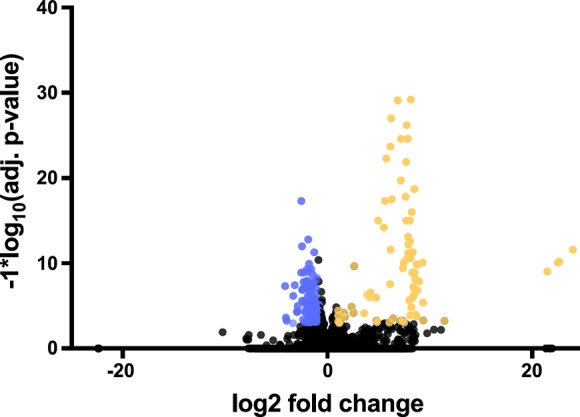
Volcano plot of blubber gene expression changes during fasting in elephant seal pups. The x-axis shows log2 fold change, and the y-axis shows significance (–log10 adj. P value). Differentially expressed genes (log2 fold change ≥|1|, adj. P value < 0.05) are shown in yellow (upregulated) and violet (downregulated).
Genes upregulated in late-fasted pups.
To infer biological functions associated with differentially expressed genes, we next conducted functional enrichment of Gene Ontology (GO) Biological Process and KEGG categories with the DAVID bioinformatics tool with human genome as background. There were no significantly enriched (adj. P > 0.05) GO categories or KEGG pathways in the upregulated gene set because of its small size. Annotated upregulated genes with the highest mean expression counts in late-fasting samples included uncharacterized protein ORF91 (1435 TPM), obscurin (62 TPM), nebulin (25 TPM), and glutathione peroxidase 3 (23 TPM). The remaining seven annotated genes in the upregulated data set had low expression counts (<20 TPM). Specific genes of interest upregulated during fasting included an adipocytokine receptor (leptin receptor), an antioxidant enzyme (glutathione peroxidase 3), an enzyme involved in amino acid degradation (aldehyde oxidase 2), and a growth factor involved in connective tissue development, energy regulation, and thermogenesis (bone morphogenic protein 8A; Tables 2 and 3, Supplementary File 2). Other upregulated genes included two putative tumor suppressors (MOB kinase activator 3B, dihydropyrimidinase-related protein 4), a GTPase involved in exocytosis of hormones (Rab3 GTPase-activating protein noncatalytic subunit), and three genes associated with skeletal muscle function (nebulin, obscurin, myopalladin). The muscle-related genes were expressed in only two of the six late-fasting samples, suggesting minor skeletal muscle contamination of blubber biopsies from these animals. The two samples did not differ from other late-fasting samples in levels of expression of downregulated and adipose-specific genes and were retained for subsequent analyses to increase statistical power.
Table 2.
Differentially expressed genes associated with lipid and carbohydrate metabolism and oxidative stress
| Transcript | Log2 FC | Uniprot ID | Gene Name | Pathway/Process |
|---|---|---|---|---|
| TRINITY_DN380627_c0_g1 | 1.73 | BMP8A_HUMAN | bone morphogenetic protein 8A (BMP8A) | brown adipose tissue thermogenesis |
| TRINITY_DN393979_c0_g1 | 1.63 | LEPR_PIG | leptin receptor (LEPR) | adipocytokine signaling pathway |
| TRINITY_DN339536_c10_g5 | 1.23 | GPX3_RAT | glutathione peroxidase 3 (GPX3) | thyroid hormone synthesis; Response to oxidative stress |
| TRINITY_DN379065_c5_g1 | −1.22 | PGM1_BOVIN | phosphoglucomutase-1 (PGM1) | pentose phosphate pathway |
| TRINITY_DN157252_c0_g1 | −1.33 | ECH1_PONAB | delta (3, 5)-delta (2, 4)-dienoyl-CoA isomerase, mitochondrial (ECH1) | fatty acid beta oxidation |
| TRINITY_DN276376_c0_g2 | −1.41 | GPX7_BOVIN | glutathione peroxidase 7 (GPX7) | thyroid hormone synthesis; lipid metabolism; response to oxidative stress |
| TRINITY_DN1029017_c0_g1 | −1.44 | TKT_HUMAN | transketolase (TKT) | pentose phosphate pathway |
| TRINITY_DN222849_c0_g2 | −1.46 | RARR2_PONAB | retinoic acid receptor responder protein 2 (RARRES2) | adipokine signaling |
| TRINITY_DN395199_c4_g12 | −1.53 | PLTP_HUMAN | phospholipid transfer protein (PTLP) | PPAR signaling pathway; Lipid transport |
| TRINITY_DN388710_c6_g4 | −1.59 | SCOT1_PIG | 3-oxoacid CoA-transferase 1 (OXCT1) | synthesis and degradation of ketone bodies |
| TRINITY_DN396178_c0_g6 | −1.60 | PXDNL_HUMAN | peroxidasin-like protein (PXDNL) | response to oxidative stress; hydrogen peroxide catabolic process |
| TRINITY_DN395661_c3_g1 | −1.76 | PCKGM_HUMAN | phosphoenolpyruvate carboxykinase 2, mitochondrial (PCK2) | glycolysis/gluco-neogenesis; PPAR signaling pathway |
| TRINITY_DN386920_c1_g3 | −1.86 | THIL_BOVIN | acetyl-CoA acetyltransferase 1 (ACAT1) | fatty acid degradation; synthesis and degradation of ketone bodies |
| TRINITY_DN292489_c1_g1 | −2.02 | IGF1_AILME | insulin-like growth factor 1 (IGF1) | PI3K-Akt, AMPK, HIF-1, FoxO signaling pathways |
| TRINITY_DN379917_c0_g4 | −2.14 | CR3L1_HUMAN | cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1) | AMPK, PI3K-Akt signaling pathways; insulin secretion; thyroid hormone synthesis |
| TRINITY_DN344610_c2_g1 | −2.25 | MOT8_HUMAN | monocarboxylate transporter 8 (SLC16A2) | thyroid hormone signaling pathway |
| TRINITY_DN129339_c0_g1 | −2.37 | APOE_ZALCA | apolipoprotein E (APOE) | cholesterol metabolic process |
| TRINITY_DN385059_c1_g1 | −2.48 | C1QT6_RAT | complement C1q tumor necrosis factor-related protein 6 (C1QTNF6) | Type I diabetes mellitus |
| TRINITY_DN390074_c3_g1 | −3.19 | PI3R4_PONAB | phosphoinositide 3-kinase regulatory subunit 4 | apelin signaling pathway; autophagy |
| TRINITY_DN4732_c2_g5 | −3.97 | ACOD_HUMAN | stearoyl-CoA desaturase (delta-9-desaturase) (SCD) | AMPK signaling pathway |
FC, fold change (late/early fasting); adjusted P < 0.05.
Table 3.
Differentially expressed genes associated with amino acid and protein metabolism and extracellular matrix remodeling
| Transcript | Log2 FC | Uniprot ID | Gene Name | Pathway/Process |
|---|---|---|---|---|
| TRINITY_DN387928_c6_g2 | 2.37 | AOXB_MACFA | aldehyde oxidase 2 (AOX2) | Val, Leu, and Ile degradation; Tyr, Trp metabolism |
| TRINITY_DN327581_c0_g1 | −1.03 | P5CR2_BOVIN | pyrroline-5-carboxylate reductase 2 (PYCR2) | Arg and Pro metabolism |
| TRINITY_DN385444_c0_g1 | −1.09 | ADA12_HUMAN | disintegrin and metalloproteinase domain-containing protein 12 (ADAM12) | metalloendopeptidase |
| TRINITY_DN392307_c1_g1 | −1.14 | MRC2_HUMAN | C-type mannose receptor 2 (MRC2) | collagen catabolic process |
| TRINITY_DN352091_c0_g1 | −1.18 | MMP23_BOVIN | matrix metalloproteinase-23 (MMP23) | metalloendopeptidase |
| TRINITY_DN252999_c0_g1 | −1.19 | CO6A3_HUMAN | collagen alpha-3(VI) chain (COL6A3) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN394004_c11_g4 | −1.26 | CO5A2_HUMAN | collagen alpha-2(V) chain (COL5A2) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN333936_c1_g2 | −1.30 | MMP14_RAT | matrix metalloproteinase-14 (MMP14) | metalloendopeptidase |
| TRINITY_DN375427_c15_g3 | −1.30 | CO4A2_HUMAN | collagen alpha-2(IV) chain (COL4A2) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN374050_c4_g1 | −1.37 | COFA1_HUMAN | collagen alpha-1(XV) chain (COL15A1) | protein digestion and absorption |
| TRINITY_DN341351_c0_g1 | −1.44 | PCOC1_HUMAN | procollagen C-endopeptidase enhancer 1 (PCOLCE) | proteolysis |
| TRINITY_DN394379_c13_g1 | −1.43 | ATS2_BOVIN | a disintegrin and metalloproteinase with thrombospondin motifs 2 (ADAMTS2) | metalloendopeptidase |
| TRINITY_DN392463_c0_g1 | −1.54 | SARDH_MOUSE | sarcosine dehydrogenase, mitochondrial (SARDH) | Gly, Ser, and Thr metabolism |
| TRINITY_DN307780_c0_g1 | −1.59 | CO6A1_MOUSE | collagen alpha-1(VI) chain (COL6A1) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN648723_c0_g3 | −1.63 | A7E3A1_BOVIN | collagen alpha-3(V) chain (COL5A3) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN288724_c0_g2 | −1.64 | SAT1_PIG | diamine acetyltransferase 1 (SAT1) | Arg and Pro metabolism |
| TRINITY_DN384694_c1_g2 | −1.72 | AL4A1_HUMAN | delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial (ALDH4A1) | Arg and Pro metabolism |
| TRINITY_DN648723_c0_g1 | −1.79 | CO5A3_HUMAN | collagen alpha-3(V) chain (COL5A3) | protein digestion and absorption |
| TRINITY_DN293757_c2_g1 | −1.88 | CO4A1_HUMAN | collagen alpha-1(IV) chain (COL4A1) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN383247_c0_g2 | −1.96 | CO6A2_HUMAN | collagen alpha-2(VI) chain (COL6A2) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN296911_c0_g1 | −1.99 | IVD_HUMAN | isovaleryl-CoA dehydrogenase, mitochondrial (IVD) | Val, Leu, and Ile degradation |
| TRINITY_DN370946_c0_g2 | −2.27 | CO5A1_HUMAN | collagen alpha-1(V) chain (COL5A1) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN392142_c1_g7 | −2.37 | CO1A2_CANLF | collagen alpha-2(I) chain (COL1A2) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN333370_c0_g1 | −2.44 | ELN_SHEEP | elastin (ELN) | protein digestion and absorption |
| TRINITY_DN394079_c2_g1 | −2.54 | CO6A6_HUMAN | collagen alpha-6(VI) chain (COL6A6) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN396844_c3_g1 | −2.56 | CO3A1_HUMAN | collagen alpha-1(III) chain (COL3A1) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN244979_c2_g2 | −2.88 | CO1A1_CANLF | collagen alpha-1(I) chain (COL1A1) | protein digestion and absorption; PI3K-Akt signaling pathway |
| TRINITY_DN154914_c0_g1 | −3.34 | KCRU_HUMAN | creatine kinase U-type, mitochondrial (CKMT1) | Arg and Pro metabolism |
| TRINITY_DN322676_c0_g1 | −4.13 | CBPZ_HUMAN | carboxypeptidase Z (CPZ) | metallocarboxy-peptidase |
FC, fold change (late/early fasting); adjusted P < 0.05.
Genes downregulated in late-fasted pups.
GO Biological Process categories enriched (adj. P < 0.05) in the gene set downregulated in late-fasted compared with early-fasted pups include collagen catabolic process, ECM organization, collagen fibril organization, cellular response to amino acid stimulus, protein heterotrimerization, skin development, cell adhesion, and skeletal system development (Fig. 3). KEGG categories enriched (adj. P < 0.05) in the downregulated gene set include protein digestion and absorption, ECM-receptor interaction, focal adhesion, PI3K-Akt signaling pathway, amoebiasis, arginine and proline metabolism, and platelet activation (Fig. 3). Downregulated genes with the highest mean expression counts in early fasting samples include SPARC (748 TPM), collagen alpha-1(I) chain (748 TPM), collagen alpha-1(III) chain (550 TPM), collagen alpha-2(I) chain (547 TPM), acyl-CoA desaturase (391 TPM), elastin (315 TPM), collagen alpha-2(I) chain (260 TPM), tetranectin (231 TPM), collagen alpha-1(IV) chain (183 TPM), EH domain-containing protein 2 (150 TPM), apolipoprotein E (145 TPM), transketolase (142 TPM), and collagen alpha-2(IV) chain (112 TPM; Supplementary File 2). An additional 24 annotated genes in the downregulated data set were expressed at TPM between 20 and 100, while the remaining 60 were expressed at TPM <20.
Fig. 3.
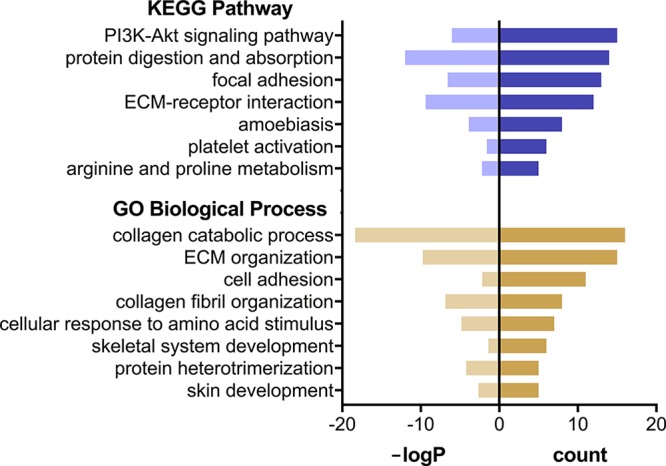
KEGG pathways (violet) and Gene Ontology (GO) Biological Process categories (yellow) overrepresented in the set of genes that were downregulated during fasting in blubber of elephant seal pups, relative to the human genome (adj. P < 0.05). −LogP, −log10(adj. P value); count, number of transcripts mapping to each process or pathway; ECM, extracellular matrix.
Many downregulated genes were associated with metabolic processes (Fig. 3, Tables 2 and 3, Supplementary File 2). They include genes involved in fatty acid synthesis (stearoyl-CoA desaturase), fatty acid oxidation (mitochondrial delta (3, 5)-delta (2, 4)-dienoyl-CoA isomerase), ketone synthesis (acetyl-CoA acetyltransferase 1 and 3-oxoacid CoA-transferase 1), lipid transport (apolipoprotein E, phospholipid transfer protein, low-density lipoprotein receptor class A domain-containing protein 2), adipocytokine signaling (retinoic acid receptor responder protein 2), insulin signaling and resistance (insulin-like growth factor 1, complement C1q tumor necrosis factor-related protein 6), thyroid hormone synthesis and signaling (monocarboxylate transporter 8, proto-oncogene tyrosine-protein kinase receptor Ret, cyclic AMP-responsive element-binding protein 3-like protein 1), protein and amino acid metabolism (mitochondrial U-type creatine kinase, diamine acetyltransferase 1, mitochondrial isovaleryl-CoA dehydrogenase, mitochondrial delta-1-pyrroline-5-carboxylate dehydrogenase, mitochondrial sarcosine dehydrogenase, follistatin-related protein 1, pyrroline-5-carboxylate reductase 2, beta-secretase 1), carbohydrate metabolism (mitochondrial phosphoenolpyruvate carboxykinase 2, phosphoglucomutase-1, transketolase), bone morphogenic protein signaling pathway (latent-transforming growth factor beta-binding protein 3, chordin-like protein 1), and oxidative stress (glutathione peroxidase 7, peroxidasin-like protein).
A large number of downregulated genes were associated with the ECM, including collagens [collagen alpha-1(I), alpha-2(I), alpha-1(III), alpha-1(VI), alpha-2(IV), alpha-1(V), alpha-2(V), alpha-3(V), alpha-1(VI), alpha-2(VI), alpha-3(VI), alpha-6(VI), alpha-1(XV)], other ECM proteins (elastin, mimecan, type II cytoskeletal 7 keratin, syndecan-3), ECM remodeling and degradation enzymes (carboxypeptidase Z, probable carboxypeptidase X1, A disintegrin and metalloproteinase with thrombospondin motifs 2, procollagen C-endopeptidase enhancer 1, matrix metalloproteinases 14 and 23, disintegrin and metalloproteinase domain-containing protein 12 serine protease HTRA3), and factors that play a role in ECM organization (coiled-coil domain-containing protein 80, SPARC; Table 3, Supplementary File 2). Therefore, the transcriptional response of blubber tissue to fasting in elephant seal pups included upregulation of genes involved in energy-regulating pathways (leptin and thyroid) and cytoskeleton organization and suppression of genes that promote catabolism of lipid, carbohydrate, and protein stores, including those that comprise the ECM.
DISCUSSION
The physiological adaptations to fasting evolved by elephant seals are remarkable and have the potential to yield insights into human and animal metabolic pathologies. In this study we characterized the first blubber transcriptome from fasting NES pups and identified 126 annotated genes that were differentially expressed in pups that have fasted for 6–8 wk relative to those that have fasted for 1–2 wk. The majority of differentially expressed genes were downregulated and were associated with metabolism, oxidative stress, and the maintenance of cellular structure and ECM (Fig. 4).
Fig. 4.
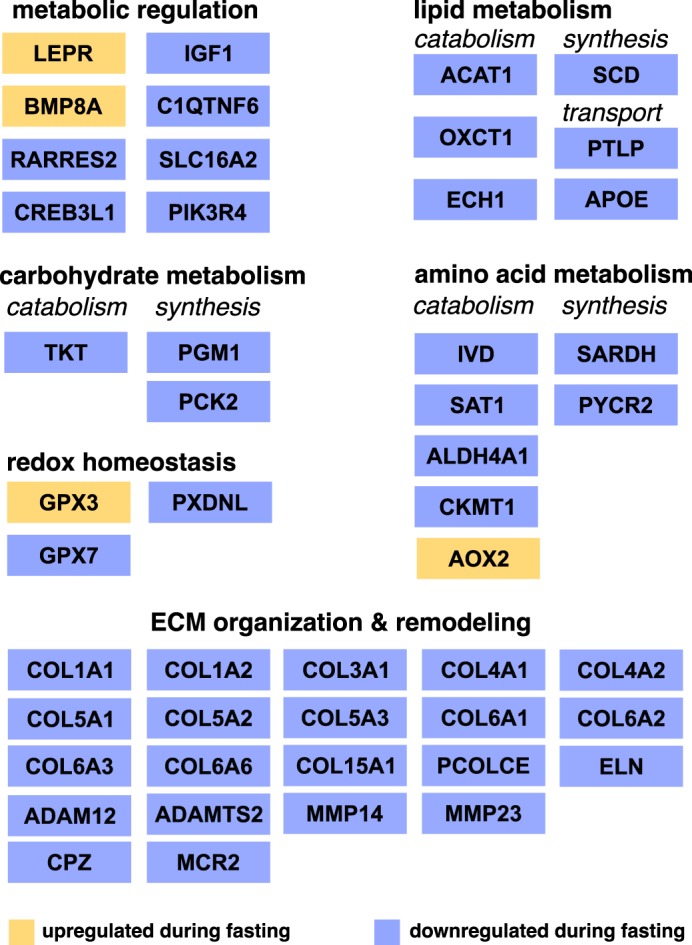
Summary diagram with differentially expressed genes grouped by their known function in metabolism in mammals. Upregulated genes are shown in yellow, downregulated genes are shown in violet.
Genes involved in lipid metabolism.
Physiological changes that have been observed during prolonged fasting in NES include increased lipolysis, elevated plasma NEFA, and a static respiratory quotient of ~0.71, demonstrating a near-strict reliance on lipid oxidation for energy (5). Our findings suggest putative mechanisms for maintenance of fasting metabolism in NES blubber during prolonged fasting. In this study, the majority of genes involved in metabolic processes were downregulated. The only metabolism-related genes that were upregulated in late-fasted pups were bone morphogenetic protein 8A (BMP8A) and leptin receptor (LEPR). BMP8A has been shown to stimulate lipolysis and brown adipose tissue thermogenesis in laboratory rodents (65). Leptin regulates adiposity via hypothalamus-mediated hunger signaling and increases triglyceride metabolism and fatty acid oxidation in adipose (2, 23). We previously showed that leptin mRNA and protein levels were unaltered in NES blubber during fasting (45). However, the approaches used in the aforementioned studies (quantitative PCR, immunoblotting) were less sensitive than RNA-Seq and were likely unable to resolve small, but potentially biologically significant changes in leptin sensitivity over fasting. Upregulation of BMP8A and LEPR over fasting is consistent with high rates of fat oxidation observed in fasting seals (5). In addition, upregulation of BMP8A suggests a potential mechanism of thermoregulatory adjustment in declining adipose stores in preparation for weaned pups’ first trip to sea (44).
Lipid metabolism enzymes that were downregulated in late-fasted pups included acetyl-CoA acetyltransferase 1 (ACAT1), 3-oxoacid CoA-transferase 1 (OXCT1), stearoyl-CoA desaturase (SCD), and delta (3, 5)-delta (2, 4)-dienoyl-CoA isomerase (ECH1). ACAT1 and OXCT1 are involved in ketone body catabolism and their downregulation is consistent with previously reported elevation in plasma ketones in NES pups during fasting (6). Dysregulation of acetyl-CoA metabolism imparts tissue-specific insulin sensitivity in obese humans (12). We propose that downregulation of ACAT1 and OXCT1 during fasting in NES pups may contribute to insulin resistance in adipose while maintaining insulin sensitivity in muscle. SCD catalyzes the rate-limiting step in the synthesis of monounsaturated fatty acids (46). Suppression of fatty acid biosynthesis and increase in beta-oxidation by downregulation of SCD is a well-known response to fasting in mammals, including hibernators (15, 20). SCD downregulation is consistent with the increase in circulating acetylarnitine previously observed in fasting NES (64). ECH1, an enzyme involved in fatty acid oxidation, was also downregulated with fasting in NES pups. While this may seem paradoxical in a fasting animal, several studies have shown that lipid oxidation is tightly regulated in fasting NES pups and that metabolic rates decline at the end of the postweaning fast to conserve energy stores in preparation for the first foraging trip (44, 52, 55).
Several downregulated genes were involved in endocrine regulation of lipid metabolism. These included the adipokines retinoic acid receptor responder protein 2 (RARRES2, also known as chemerin), complement C1q tumor necrosis factor-related protein 6 (C1QTNF6), and insulin-like growth factor 1 (IGF1); cyclic AMP-responsive element-binding protein 3-like protein 1 (CREB3L1); and the thyroid hormone transporter monocarboxylate transporter 8 (SLC16A2). RARRES2 knockdown stimulates lipolysis in 3T3-L1 adipocytes (16), deletion of C1QTNF6 increases metabolic rate and energy expenditure in obese mice (34), and plasma IGF-1, an insulin-sensitizing anabolic hormone, is downregulated during fasting in elephant seals (61). CREB3L1 was recently shown to promote adipogenesis in mice (14). Downregulation of these genes is consistent with known metabolic adjustments in fasting seals. While thyroid signaling and sensitivity increase during fasting in elephant seals (38), the TH transporter SLC16A2 is downregulated during fasting in mice (50), similar to what we observed in fasting NES pups. SLC16A2 downregulation in this study suggests that modulation of tissue-specific TH sensitivity is likely to be complex in fasting seals and warrants further investigation.
Genes involved in carbohydrate metabolism.
Downregulated genes associated with carbohydrate metabolism included the enzymes phosphoglucomutase-1 (PGM1), transketolase (TKT), and phosphoenolpyruvate carboxykinase 2 (PCK2). PGM1 catalyzes the rate-limiting step in glycogen synthesis, and its deficiency is associated with a reliance on lipid metabolism in humans (48). TKT channels excess sugar phosphates to glycolysis in the pentose phosphate pathway (PPP). Its downregulation would retain sugar phosphates in the PPP for nucleotide and NADPH recycling during fasting. PCK2 is involved in hepatic gluconeogenesis and fatty acid reesterification in adipocytes and was previously shown to decline during fasting in weaned NES pups (10). PCK2 levels are also low during winter but increase during interbout arousal in hibernating mammals (20). Downregulation of these genes would promote catabolism and metabolic substrate recycling in fasting seals, consistent with previous endocrine and metabolic studies in this species (10).
Genes involved in protein metabolism.
Several genes encoding enzymes involved in amino acid metabolism were also downregulated during fasting. These included pyrroline-5-carboxylate reductase 2 (PYCR2), sarcosine dehydrogenase (SARDH), diamine acetyltransferase 1 (SAT1), delta-1-pyrroline-5-carboxylate dehydrogenase (ALDH4A1), isovaleryl-CoA dehydrogenase (IVD), and creatine kinase (CKMT1). PYCR2 and SARDH are involved in biosynthesis of proline and glycine, respectively, while IVD is involved in leucine catabolism. ALDH4A1 is involved in proline metabolism to glutamate, linking the urea and tricarboxylic acid cycles. SAT1 catalyzes the transfer of acetyl groups from acetyl-CoA to intracellular polyamines and promotes their catabolism; SAT1 knockout mice exhibit reduced acetyl-CoA consumption and fatty acid oxidation in adipose and elevated glucose intolerance (36). Downregulation of these genes is consistent with the minimal contribution of protein stores to fasting metabolism observed in NES (10).
Genes involved in oxidative stress.
Prolonged fasting and insulin resistance are correlated with increased oxidative stress in terrestrial mammals, but minimal oxidative damage in seals due to their high antioxidant production capacity (56). Several genes that were differentially expressed during fasting were associated with oxidative stress, including aldehyde oxidase 2 (AOX2) and glutathione peroxidase 3 (GPX3), which were upregulated, and glutathione peroxidase 7 (GPX7), which was downregulated. Upregulation of GPX3 during fasting is consistent with increased antioxidant defenses observed in fasting seals (12). GPX7 is a unique member of the glutathione peroxidase family without GPX activity that is primarily associated with oxidative protein folding. Interestingly, GPX7 has been implicated in adipogenesis; mice lacking GPX7 exhibit increased fat mass and adipocyte hypertrophy (7). AOX2 is a member of the aldehyde oxidase family, which is primarily associated with xenobiotic detoxification in mammals. A recent study proposed that NADH is the endogenous substrate of AOX enzymes, which produce reactive oxygen species (ROS) as a byproduct of NADH oxidation (32). Upregulation of a ROS-producing enzyme in fasting seals is surprising given our previous work on oxidative stress tolerance in this species. However, it occurs with concomitant upregulation of the antioxidant GPX3 and highlights the complexity of regulation of oxidative status in fasting mammals, which may also be involved in regulation of adipose stores (18).
Genes involved in ECM remodeling.
One of the more novel findings of this study is that a number of highly expressed genes that were downregulated during fasting are associated with cell structure and maintenance of the ECM. Specific downregulated genes include 14 collagen isoforms, elastin, five metalloproteases (ADAMTS2, ADAM12, MMP14, MMP23, CPZ), growth factor beta binding protein 3 (LTBP3), and peroxidasin-like protein (PXDNL), which mediates collagen crosslinking. These data suggest that major alterations in structural protein production and adipose tissue remodeling occurs during postnatal development in weaned elephant seal pups. Downregulation of collagen and MMPs has also been observed in obese mice during weight loss (35). Downregulation of these genes may be a key component of the reduction in adipocyte size with fasting duration observed in fasting seals as lipid mobilization increases and fatty acids are released into circulation (10).
Conclusions
In summary, we showed that genes involved in lipid synthesis, carbohydrate recycling, amino acid metabolism, oxidative stress, and ECM remodeling are primarily downregulated in blubber during fasting in weaned NES pups. These data complement previous metabolic studies demonstrating reliance of NES on lipid metabolism, maintenance of insulin resistance, and protein sparing during fasting, as well as changes in tissue remodeling that accompany rapid weight loss. Our data highlight the complexity of metabolic regulation that sustains prolonged fasting concomitant with energy-intensive processes in fasting-adapted seals. The unique adaptation of multifunctional blubber tissue in marine mammals sustains fasting metabolism by mechanisms distinct from those observed in adipose tissue of hibernating and nonfasting adapted mammals, which involve significant metabolic suppression and increases in insulin sensitivity (19, 24). As such, this study provides some preliminary insights into maintenance of insulin resistance and adiposity in fasting-adapted mammals.
GRANTS
B. Martinez was supported by the Dennis R. Washington Graduate Achievement Scholarship and the National Institutes of Health (NIH) Minority Health and Health Disparities International Research Training Program. R. M. Ortiz was partially supported by NIH Grant K02HL-103787. Research was funded by NIH Grant R01HL-09176. This work used the Extreme Science and Engineering Discovery Environment and was supported by National Science Foundation Grant ACI-1053575.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
B.M. conceived and designed research; B.M. performed experiments; B.M., J.K., K.R., and D.E.C. analyzed data; B.M., J.K., D.E.C., N.G., and R.M.O. interpreted results of experiments; B.M. and J.K. prepared figures; B.M. drafted manuscript; B.M., J.K., K.R., D.E.C., N.G., and R.M.O. edited and revised manuscript; B.M., J.K., K.R., D.E.C., N.G., and R.M.O. approved final version of manuscript.
Supplemental Data
ACKNOWLEDGMENTS
The authors thank Derek Somo and David Ensminger for assistance with sample collection and Patrick Robinson, Daniel Costa, University of California Natural Reserve System, and the Rangers at Año Nuevo State Park for facilitating access to the study animals.
REFERENCES
- 1.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 11: R106, 2010. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan AM, Mantzoros CS. Drug Insight: the role of leptin in human physiology and pathophysiology–emerging clinical applications. Nat Clin Pract Endocrinol Metab 2: 318–327, 2006. doi: 10.1038/ncpendmet0196. [DOI] [PubMed] [Google Scholar]
- 3.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12: 59–60, 2015. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 4.Cammen KM, Andrews KR, Carroll EL, Foote AD, Humble E, Khudyakov JI, Louis M, McGowen MR, Olsen MT, Van Cise AM. Genomic methods take the plunge: recent advances in high-throughput sequencing of marine mammals. J Hered 107: 481–495, 2016. doi: 10.1093/jhered/esw044. [DOI] [PubMed] [Google Scholar]
- 5.Champagne CD, Crocker DE, Fowler MA, Houser DS. Fasting physiology of the pinnipeds: the challenges of fasting while maintaining high energy expenditure and nutrient delivery for lactation, in Comparative Physiology of Fasting, Starvation, and Food Limitation (McCue MD, editor). Berlin, Heidelberg: Springer, 2012, p. 309–336. doi: 10.1007/978-3-642-29056-5_19. [DOI] [Google Scholar]
- 6.Champagne CD, Houser DS, Crocker DE. Glucose production and substrate cycle activity in a fasting adapted animal, the northern elephant seal. J Exp Biol 208: 859–868, 2005. doi: 10.1242/jeb.01476. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y-C, Yu Y-H, Shew J-Y, Lee W-J, Hwang J-J, Chen Y-H, Chen Y-R, Wei P-C, Chuang L-M, Lee W-H. Deficiency of NPGPx, an oxidative stress sensor, leads to obesity in mice and human. EMBO Mol Med 5: 1165–1179, 2013. doi: 10.1002/emmm.201302679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe SS, Huh JY, Hwang IJ, Kim JI, Kim JB. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Front Endocrinol (Lausanne) 7: 30, 2016. doi: 10.3389/fendo.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci 9: 191–200, 2013. doi: 10.5114/aoms.2013.33181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crocker DE, Champagne CD, Fowler MA, Houser DS. Adiposity and fat metabolism in lactating and fasting northern elephant seals. Adv Nutr 5: 57–64, 2014. doi: 10.3945/an.113.004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crocker DE, Khudyakov JI, Champagne CD. Oxidative stress in northern elephant seals: Integration of omics approaches with ecological and experimental studies. Comp Biochem Physiol A Mol Integr Physiol 200: 94–103, 2016. doi: 10.1016/j.cbpa.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Noren DP. Thermoregulation of weaned northern elephant seal (Mirounga angustirostris) pups in air and water. Physiol Biochem Zool 75: 513–523, 2002. doi: 10.1086/342254. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlund A, Mejhert N, Björk C, Andersson R, Kulyté A, Åström G, Itoh M, Kawaji H, Lassmann T, Daub CO, Carninci P, Forrest ARR, Hayashizaki Y, Sandelin A, Ingelsson E, Rydén M, Laurencikiene J, Arner P, Arner E; FANTOM Consortium . Transcriptional Dynamics During Human Adipogenesis and Its Link to Adipose Morphology and Distribution. Diabetes 66: 218–230, 2017. doi: 10.2337/db16-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster DW. The role of the carnitine system in human metabolism. Ann N Y Acad Sci 1033: 1–16, 2004. doi: 10.1196/annals.1320.001. [DOI] [PubMed] [Google Scholar]
- 16.Goralski KB, McCarthy TC, Hanniman EA, Zabel BA, Butcher EC, Parlee SD, Muruganandan S, Sinal CJ. Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J Biol Chem 282: 28175–28188, 2007. doi: 10.1074/jbc.M700793200. [DOI] [PubMed] [Google Scholar]
- 17.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a genome. Nat Biotechnol 29: 644–652, 2011. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gummersbach C, Hemmrich K, Kröncke K-D, Suschek CV, Fehsel K, Pallua N. New aspects of adipogenesis: radicals and oxidative stress. Differentiation 77: 115–120, 2009. doi: 10.1016/j.diff.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Hampton M, Melvin RG, Andrews MT. Transcriptomic analysis of brown adipose tissue across the physiological extremes of natural hibernation. PLoS One 8: e85157, 2013. doi: 10.1371/journal.pone.0085157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampton M, Melvin RG, Kendall AH, Kirkpatrick BR, Peterson N, Andrews MT. Deep sequencing the transcriptome reveals seasonal adaptive mechanisms in a hibernating mammal. PLoS One 6: e27021, 2011. doi: 10.1371/journal.pone.0027021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris RBS. Direct and indirect effects of leptin on adipocyte metabolism. Biochim Biophys Acta 1842: 414–423, 2014. doi: 10.1016/j.bbadis.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houser DS, Champagne CD, Crocker DE. A non-traditional model of the metabolic syndrome: the adaptive significance of insulin resistance in fasting-adapted seals. Front Endocrinol (Lausanne) 4: 164, 2013. doi: 10.3389/fendo.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Khudyakov JI, Champagne CD, Meneghetti LM, Crocker DE. Blubber transcriptome response to acute stress axis activation involves transient changes in adipogenesis and lipolysis in a fasting-adapted marine mammal. Sci Rep 7: 42110, 2017. doi: 10.1038/srep42110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. J Clin Endocrinol Metab 89: 2548–2556, 2004. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 29.Khudyakov JI, Champagne CD, Preeyanon L, Ortiz RM, Crocker DE. Muscle transcriptome response to ACTH administration in a free-ranging marine mammal. Physiol Genomics 47: 318–330, 2015. doi: 10.1152/physiolgenomics.00030.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khudyakov JI, Preeyanon L, Champagne CD, Ortiz RM, Crocker DE. Transcriptome analysis of northern elephant seal (Mirounga angustirostris) muscle tissue provides a novel molecular resource and physiological insights. BMC Genomics 16: 64, 2015. doi: 10.1186/s12864-015-1253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K-H, Kim YH, Son JE, Lee JH, Kim S, Choe MS, Moon JH, Zhong J, Fu K, Lenglin F, Yoo J-A, Bilan PJ, Klip A, Nagy A, Kim J-R, Park JG, Hussein SMI, Doh K-O, Hui CC, Sung H-K. Intermittent fasting promotes adipose thermogenesis and metabolic homeostasis via VEGF-mediated alternative activation of macrophage. Cell Res 27: 1309–1326, 2017. doi: 10.1038/cr.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kundu TK, Velayutham M, Zweier JL. Aldehyde oxidase functions as a superoxide generating NADH oxidase: an important redox regulated pathway of cellular oxygen radical formation. Biochemistry 51: 2930–2939, 2012. doi: 10.1021/bi3000879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359, 2012. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei X, Seldin MM, Little HC, Choy N, Klonisch T, Wong GW. C1q/TNF-related protein 6 (CTRP6) links obesity to adipose tissue inflammation and insulin resistance. J Biol Chem 292: 14836–14850, 2017. doi: 10.1074/jbc.M116.766808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin D, Chun T-H, Kang L. Adipose extracellular matrix remodelling in obesity and insulin resistance. Biochem Pharmacol 119: 8–16, 2016. doi: 10.1016/j.bcp.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Perez-Leal O, Barrero C, Zahedi K, Soleimani M, Porter C, Merali S. Modulation of polyamine metabolic flux in adipose tissue alters the accumulation of body fat by affecting glucose homeostasis. Amino Acids 46: 701–715, 2014. doi: 10.1007/s00726-013-1548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez B, Ortiz RM. Thyroid hormone regulation and insulin resistance: insights from animals naturally adapted to fasting. Physiology (Bethesda) 32: 141–151, 2017. [DOI] [PubMed] [Google Scholar]
- 38.Martinez B, Scheibner M, Soñanez-Organis JG, Jaques JT, Crocker DE, Ortiz RM. Increased sensitivity of thyroid hormone-mediated signaling despite prolonged fasting. Gen Comp Endocrinol 252: 36–47, 2017. doi: 10.1016/j.ygcen.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez B, Soñanez-Organis JG, Godoy-Lugo JA, Horin LJ, Crocker DE, Ortiz RM. Thyroid hormone-stimulated increases in PGC-1α and UCP2 promote life history-specific endocrine changes and maintain a lipid-based metabolism. Am J Physiol Regul Integr Comp Physiol 312: R189–R196, 2017. doi: 10.1152/ajpregu.00395.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez B, Soñanez-Organis JG, Vázquez-Medina JP, Viscarra JA, MacKenzie DS, Crocker DE, Ortiz RM. Prolonged food deprivation increases mRNA expression of deiodinase 1 and 2, and thyroid hormone receptor β-1 in a fasting-adapted mammal. J Exp Biol 216: 4647–4654, 2013. doi: 10.1242/jeb.085290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez B, Soñanez-Organis JG, Viscarra JA, Jaques JT, MacKenzie DS, Crocker DE, Ortiz RM. Glucose delays the insulin-induced increase in thyroid hormone-mediated signaling in adipose of prolong-fasted elephant seal pups. Am J Physiol Regul Integr Comp Physiol 310: R502–R512, 2016. doi: 10.1152/ajpregu.00054.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naito Y, Costa DP, Adachi T, Robinson PW, Fowler M, Takahashi A. Unravelling the mysteries of a mesopelagic diet: a large apex predator specializes on small prey. Funct Ecol 27: 710–717, 2013. doi: 10.1111/1365-2435.12083. [DOI] [Google Scholar]
- 44.Noren DP, Crocker DE, Williams TM, Costa DP. Energy reserve utilization in northern elephant seal (Mirounga angustirostris) pups during the postweaning fast: size does matter. J Comp Physiol B 173: 443–454, 2003. doi: 10.1007/s00360-003-0353-9. [DOI] [PubMed] [Google Scholar]
- 45.Ortiz RM, Wade CE, Ortiz CL. Effects of prolonged fasting on plasma cortisol and TH in postweaned northern elephant seal pups. Am J Physiol Regul Integr Comp Physiol 280: R790–R795, 2001. doi: 10.1152/ajpregu.2001.280.3.R790. [DOI] [PubMed] [Google Scholar]
- 46.Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab 297: E28–E37, 2009. doi: 10.1152/ajpendo.90897.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419, 2017. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preisler N, Laforêt P, Echaniz-Laguna A, Ørngreen MC, Lonsdorfer-Wolf E, Doutreleau S, Geny B, Stojkovic T, Piraud M, Petit FM, Vissing J. Fat and carbohydrate metabolism during exercise in phosphoglucomutase type 1 deficiency. J Clin Endocrinol Metab 98: E1235–E1240, 2013. doi: 10.1210/jc.2013-1651. [DOI] [PubMed] [Google Scholar]
- 49.Schupp M, Chen F, Briggs ER, Rao S, Pelzmann HJ, Pessentheiner AR, Bogner-Strauss JG, Lazar MA, Baldwin D, Prokesch A. Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genomics 14: 758, 2013. doi: 10.1186/1471-2164-14-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schutkowski A, Wege N, Stangl GI, König B. Tissue-specific expression of monocarboxylate transporters during fasting in mice. PLoS One 9: e112118, 2014. doi: 10.1371/journal.pone.0112118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212, 2015. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 52.Somo DA, Ensminger DC, Sharick JT, Kanatous SB, Crocker DE. Development of Dive Capacity in Northern Elephant Seals (Mirounga angustirostris): Reduced Body Reserves at Weaning Are Associated with Elevated Body Oxygen Stores during the Postweaning Fast. Physiol Biochem Zool 88: 471–482, 2015. doi: 10.1086/682386. [DOI] [PubMed] [Google Scholar]
- 53.Soñanez-Organis JG, Vázquez-Medina JP, Crocker DE, Ortiz RM. Prolonged fasting activates hypoxia inducible factors-1α, -2α and -3α in a tissue-specific manner in northern elephant seal pups. Gene 526: 155–163, 2013. doi: 10.1016/j.gene.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki M, Vázquez-Medina JP, Viscarra JA, Soñanez-Organis JG, Crocker DE, Ortiz RM. Activation of systemic, but not local, renin-angiotensin system is associated with upregulation of TNF-α during prolonged fasting in northern elephant seal pups. J Exp Biol 216: 3215–3221, 2013. doi: 10.1242/jeb.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tift MS, Ranalli EC, Houser DS, Ortiz RM, Crocker DE. Development enhances hypometabolism in northern elephant seal pups (Mirounga angustirostris). Funct Ecol 27: 1155–1165, 2013. doi: 10.1111/1365-2435.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vázquez-Medina JP, Crocker DE, Forman HJ, Ortiz RM. Prolonged fasting does not increase oxidative damage or inflammation in postweaned northern elephant seal pups. J Exp Biol 213: 2524–2530, 2010. doi: 10.1242/jeb.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vázquez-Medina JP, Popovich I, Thorwald MA, Viscarra JA, Rodriguez R, Sonanez-Organis JG, Lam L, Peti-Peterdi J, Nakano D, Nishiyama A, Ortiz RM. Angiotensin receptor-mediated oxidative stress is associated with impaired cardiac redox signaling and mitochondrial function in insulin-resistant rats. Am J Physiol Heart Circ Physiol 305: H599–H607, 2013. doi: 10.1152/ajpheart.00101.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vázquez-Medina JP, Soñanez-Organis JG, Rodriguez R, Viscarra JA, Nishiyama A, Crocker DE, Ortiz RM. Prolonged fasting activates Nrf2 in post-weaned elephant seals. J Exp Biol 216: 2870–2878, 2013. doi: 10.1242/jeb.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vijay N, Poelstra JW, Künstner A, Wolf JBW. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol 22: 620–634, 2013. doi: 10.1111/mec.12014. [DOI] [PubMed] [Google Scholar]
- 61.Viscarra JA, Champagne CD, Crocker DE, Ortiz RM. 5'AMP-activated protein kinase activity is increased in adipose tissue of northern elephant seal pups during prolonged fasting-induced insulin resistance. J Endocrinol 209: 317–325, 2011. doi: 10.1530/JOE-11-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viscarra JA, Rodriguez R, Vazquez-Medina JP, Lee A, Tift MS, Tavoni SK, Crocker DE, Ortiz RM. Insulin and GLP-1 infusions demonstrate the onset of adipose-specific insulin resistance in a large fasting mammal: potential glucogenic role for GLP-1. Physiol Rep 1: e00023, 2013. doi: 10.1002/phy2.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viscarra JA, Vázquez-Medina JP, Crocker DE, Ortiz RM. Glut4 is upregulated despite decreased insulin signaling during prolonged fasting in northern elephant seal pups. Am J Physiol Regul Integr Comp Physiol 300: R150–R154, 2011. doi: 10.1152/ajpregu.00478.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viscarra JA, Vázquez-Medina JP, Rodriguez R, Champagne CD, Adams SH, Crocker DE, Ortiz RM. Decreased expression of adipose CD36 and FATP1 are associated with increased plasma non-esterified fatty acids during prolonged fasting in northern elephant seal pups (Mirounga angustirostris). J Exp Biol 215: 2455–2464, 2012. doi: 10.1242/jeb.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whittle AJ, Carobbio S, Martins L, Slawik M, Hondares E, Vázquez MJ, Morgan D, Csikasz RI, Gallego R, Rodriguez-Cuenca S, Dale M, Virtue S, Villarroya F, Cannon B, Rahmouni K, López M, Vidal-Puig A. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 149: 871–885, 2012. doi: 10.1016/j.cell.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Smith SA. Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genomics 14: 328, 2013. doi: 10.1186/1471-2164-14-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


