Abstract
Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) are rare autosomal recessive disorders associated with a defect in the nucleotide excision repair (NER) pathway required for the removal of DNA damage induced by UV light and distorting chemical adducts. Although progressive neurological dysfunction is one of the hallmarks of CS and of some groups of XP patients, the causative mechanisms are largely unknown. Here we show that mice lacking both the XPA (XP-group A) and CSB (CS-group B) genes in contrast to the single mutants display severe growth retardation, ataxia, and motor dysfunction during early postnatal development. Their cerebella are hypoplastic and showed impaired foliation and stunted Purkinje cell dendrites. Reduced neurogenesis and increased apoptotic cell death occur in the cerebellar external granular layer. These findings suggest that XPA and CSB have additive roles in the mouse nervous system and support a crucial role for these genes in normal brain development.
Xeroderma pigmentosum (XP) and Cockayne syndrome (CS) are rare autosomal recessive disorders associated with inherited defects in the nucleotide excision repair (NER) system. Complementation studies by using cell hybridization have revealed the existence of eight genes in XP (groups A-G and a variant) and two in CS (groups A and B) (1, 2). The clinical hallmark of both disorders is pronounced hypersensitivity of the skin to UV light. XP patients also exhibit >1,000-fold increased risk of UV-induced skin cancer and a moderately enhanced predisposition to internal tumors. About 20% of XP patients develop progressive neurological dysfunction. XP-A patients usually exhibit the most severe symptoms of XP with both skin symptoms and central nervous system abnormalities. Many XP-A individuals exhibit manifestations from birth or early in life and correspond to the clinical category of DeSanctis–Cacchione syndrome with progressive neurologic degeneration, such as microcephaly, mental deterioration, choreoathetosis, cerebellar ataxia, spasticity, sensorineuronal deafness, and peripheral neuropathy. CS patients exhibit a combination of photosensitivity, postnatal growth failure, and severe early onset neurological abnormalities (i.e., microcephaly, mental retardation, limb ataxia, and abnormal gait, retinal pigmentation, sensorineural hearing loss, and peripheral and central myelinopathy), but they do not develop skin cancer (1, 3). Although the neuropathological features of XP and CS patients show similarities and the association of XP with CS has been extensively discussed (1, 4), the main cause(s) of those symptoms are still largely unknown.
Several mouse models for XP and CS have been established by gene targeting (5–10). They reliably reflect the NER defect but do not always represent a complete phenocopy of the corresponding disorder. For example, in XPA-deficient mice, susceptibility to UV- and chemical carcinogen-induced skin cancer is greatly increased, but animals fail to develop clearly detectable neurological abnormalities (5, 6). CSB-deficient mice exhibit most of the CS repair characteristics, but in contrast to human CS, show increased susceptibility to UV- and chemically induced skin cancer, develop normally, and show only a mild neurological phenotype (8). The mechanisms underlying the difference in neurological phenotypes between XPA- and CSB-deficient mice and the corresponding human patients remain unknown.
Here we show that animals lacking both XPA and CSB genes (XPA−/−CSB−/− mice) display growth retardation and abnormal behavior, features that closely resemble symptoms observed in human XP-A and/or CS-B patients at early postnatal stage. This observation may provide new insight into the physiological roles of NER genes in the nervous system and the pathogenesis of XP and CS diseases.
Materials and Methods
Mice.
XPA+/− (genetic background: CBA/C57BL6/CD-1) (6) and CSB+/− (genetic background: C57BL6/129ola) (8) mice were crossed to generate XPA+/−CSB+/− animals, that were intercrossed to give XPA+/−CSB−/− and XPA−/−CSB+/− mice. Matings between XPA+/−CSB−/− and XPA−/−CSB+/− animals yielded the XPA+/−CSB+/−, XPA−/−CSB+/−, XPA+/−CSB−/−, and XPA−/−CSB−/− littermate mice. Mouse genotyping was performed as previously described (6, 11).
Immunohistochemistry.
Histological analysis of the cerebellum in mice lacking XPA and/or CSB was performed by using double heterozygous (XPA+/−CSB+/−) as control. Mice were anesthetized with chloroform, and brains were removed and fixed with 4% paraformaldehyde. Brain sections (10 μm) were cut by cryostat and processed for histochemical staining with cresyl violet. Alternatively, sections were processed for Purkinje cell (PC)-specific immunohistochemical staining by using an anticalbindin D antibody diluted 1:500 (Sigma). Signals were visualized by using a Vectastain avidin-biotin peroxydase complex (ABC) kit (Vector Laboratories). The length of the PC dendritic tree was determined by measuring the thickness of the molecular layer of six randomly chosen fields in the middle of midsagittal section of lobules I/II, III, IX, and X per cerebellum stained with anticalbindin D antibody. Five to eight animals of each genotype were examined.
BrdUrd Labeling and Terminal Deoxynucleotidyl Transferase-Mediated dUTP-Biotin Nick End-Labeling (TUNEL) Staining.
To examine the proliferative ability of neuroblasts in the cerebellum, mice were injected with BrdUrd (50 mg/kg in saline) and anesthetized 1.5 h after injection. BrdUrd incorporated into the cells was immunohistochemically detected by using a BrdUrd staining Kit (Oncogene Research Products). Cells undergoing apoptosis were identified by the terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling (TUNEL) method, as previously described (12).
Results
XPA−/−CSB−/− Mice Display Abnormal Behavior at an Early Postnatal Age.
XPA−/− and CSB−/− single mutant mice (6, 8) did not display severe developmental and neurological abnormalities. In marked contrast, XPA−/−CSB−/− mice (obtained from XPA+/−CSB−/− × XPA−/−CSB+/− crossings) revealed developmental defects starting from an early postnatal age and died around postnatal day (P) 21 (life span 20.95 ± 0.81, n = 20). XPA−/−CSB−/− mice were born at nearly expected Mendelian frequency [XPA+/−CSB+/− 39 (29.5%), XPA−/−CSB+/− 27 (20.5%), XPA+/−CS−/− 38 (28.8%) and XPA−/−CSB−/− 28 (21.2%), n = 132 newborns in 17 litters] and were indistinguishable from other littermates in overall appearance and size at birth; poor development, low body weight, and small body size became apparent during early postnatal life. In contrast, XPA+/−CSB+/− offspring was indistinguishable from wild-type mice in development, size, and behavior (M.Y. and G.T.J.H., unpublished observations) and was therefore used as a control throughout the experiments.
Signs of ataxia progressively appeared in the XPA−/−CSB−/− mice as early as around 1 week after birth and became clearly evident around 2 weeks. XPA−/−CSB−/− mice sat with their hind limbs spread widely to obtain balance (Fig. 1A) and walked with a wide gait and tremors (data not shown). When XPA−/−CSB−/− mice were suspended by their tails (a treatment that causes wild-type and XPA+/−CSB+/− control mice to extend and shake their hindlimbs), animals kept their hind limbs often in a crossed or clasped position, indicating the loss of coordinated movements of legs (Fig. 1B). Furthermore, XPA−/−CSB−/− mice showed waltzing locomotion, a sign indicative for inner ear defect (Fig. 1C), and often fell down (Fig. 1D). XPA+/−CSB+/− control mice walked carefully, often stopping and resting (data not shown). These features strongly suggest that the neurological abnormalities displayed by XPA−/−CSB−/− mice originate from cerebellar and vestibular dysfunctions. To further examine the effects of XPA and CSB gene defects on the central nervous system, we focused on the cerebellar phenotype of XPA−/−CSB−/− mice.
Figure 1.
Motor dysfunction and abnormal behavior in XPA−/−CSB−/− mice. (A) Compared with control littermates of XPA+/−CSB+/− (Left), XPA−/−CSB−/− mice (Right) had to spread their hind limbs to maintain balance (P14). (B) When the mice were suspended by their tails, the XPA−/−CSB−/− mice (Right) often crossed or clasped their hind limbs, whereas XPA+/−CSB+/− mice (Left) usually extended and shook their hind limbs. XPA−/−CSB−/− mice frequently performed waltzing (C 1–4) or lost their balance and fell (D 1–4).
Cerebellar Morphological Defects in XPA−/−CSB−/− Mice.
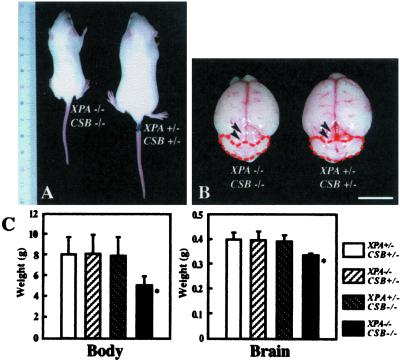
XPA−/−CSB−/− mice showed growth retardation from an early postnatal stage (Fig. 2A). Compared with XPA+/−CSB+/− control mice, XPA−/−CSB−/− mice showed 40 and 20% reduction in body and brain weight at P14, respectively (n = 20, P < 0.01), whereas those of XPA−/−CSB+/− (n = 26) and XPA+/−CSB−/− (n = 22) mice were comparable to the control (n = 35) (Fig. 2 B and C). We also examined several organs (the colon, lung, pancreas, small intestine, spleen, stomach, testis, and thymus) of XPA−/−CSB−/− mice. Although they were apparently smaller in size in comparison with those of the control, no obvious histological abnormalities were found (data not shown). Further examinations will be necessary to elucidate the principal cause of the growth retardation and premature death of XPA−/− CSB −/− mice.
Figure 2.
Comparison of the body and brain of each genotypic mouse. (A) XPA−/−CSB−/− mice show growth retardation from an early postnatal stage (P14). (B) The gross view of the brain of XPA−/−CSB−/− versus XPA+/−CSB+/− littermates. Comparison to XPA+/−CSB+/− control, XPA−/−CSB−/− cerebellum was remarkably smaller (red interrupted line), so the colliculi were more exposed (black arrowheads). (Bar = 10 mm.) (C) Averaged body and brain weight of each genotype at P14. Values are mean ± SD derived from mice of each genotype. *, P < 0.01, against XPA+/−CSB+/−.
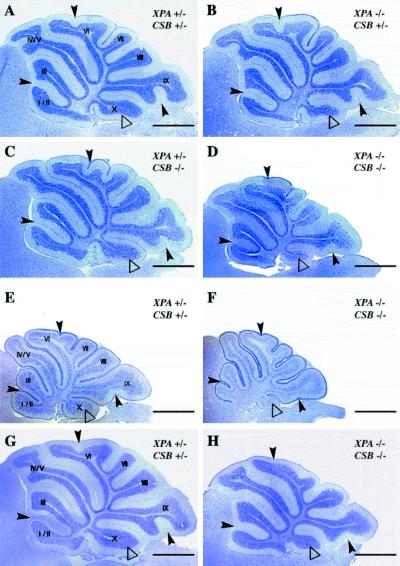
Although slight reduction in size and weight was observed, the gross appearance of the brain of XPA−/−CSB−/− mice seemed normal (e.g., the cerebrum and olfactory bulbs). However, the cerebellum of the XPA−/−CSB−/− mice was strikingly small in comparison to that of XPA+/−CSB+/− mice (Fig. 2B). These abnormalities were not observed at birth but became apparent around P8 (data not shown), suggesting that cell depletion takes place during early postnatal development. Analysis of cresyl violet-stained sagittal sections of cerebella derived from XPA−/−CSB−/− mice confirmed a gross reduction in the size of the cerebellum. In comparison to XPA+/−CSB+/− mice (n = 6), XPA−/−CSB−/− animals showed impaired foliation and sulcus formation throughout the cerebellum, which appeared more prominent at lobules I/II, VI, IX, and X (n = 6) at P14 (Fig. 3 A and D). In contrast, cerebella of XPA−/−CSB+/− (n = 6) and XPA+/−CSB−/− (n = 11) mice did not show any irregularities and were indistinguishable from those of control XPA+/−CSB+/− mice (Fig. 3 B and C). The abnormalities of XPA−/−CSB−/− mice were not simply caused by a delay in cerebellar folding, because they were already observed at P8 (n = 6 of each XPA+/−CSB+/− and XPA−/−CSB−/−) (Fig. 3 E and F) and did not disappear even at P20 when cell proliferation and migration of neuronal precursors in the mouse cerebellum is completed (n = 5 of XPA+/−CSB+/− and n = 6 of XPA−/−CSB−/−) (Fig. 3 G and H).
Figure 3.
Histological abnormalities of cerebellum in XPA−/−CSB−/− mice. Sagittal cerebellar sections of P14 (A–D), P8 (E and F), and P20 (G and H) XPA+/−CSB+/− (A, E, and G), XPA−/−CSB+/− (B), XPA+/−CSB−/− (C), and XPA−/−CSB−/− (D, F, and H) mice. XPA−/−CSB−/− mice (D) showed impaired cerebellar foliation pattern and some fissures (black arrowheads), and the tenth lobule (Flocculonodular lobe, white triangles) appeared to be small compared with other genotypes (A–C). (Bar = 1 mm.)
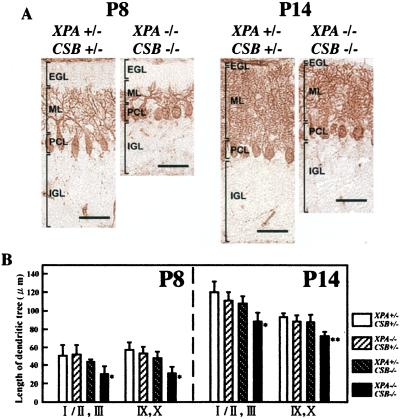
To further examine the morphological abnormalities in the cerebellum of XPA−/−CSB−/− mice, we stained PCs with anticalbindin D antibody, a specific marker for this cell type (Fig. 4). The structural organization of the external granular layer (EGL), molecular layer (ML), and internal granular layer (IGL) formed in an orderly fashion. However, the thickness of these cerebellar layers was strikingly reduced, and stunted PC dendrites were observed in XPA−/−CSB−/− mice in comparison to those of control XPA+/−CSB+/− mice (Fig. 4A). Moreover, size measurements of dendritic trees of PCs in XPA−/−CSB−/− mice both at P8 (n = 8) and at P14 (n = 6) revealed a significant reduction in length compared with those of control animals (n = 8 at P8 and n = 5 at P14) (Fig. 4B). These results suggest that PC dendritic differentiation is impaired in XPA−/−CSB−/− mice. In contrast, the cerebellar layer formation and the length of dendritic trees of PCs were not affected in XPA−/−CSB+/− (n = 7 at both P8 and P14) and XPA+/−CSB−/− (n = 7 at P8 and n = 6 at P14) mice (data not shown, Fig. 4B). These results indicate that the abnormal cerebellar histogenesis takes place during early postnatal development in XPA−/−CSB−/− mice.
Figure 4.
Immunohistochemical staining of cerebellar PCs with anticalbindin antibody. (A) PC morphologies of XPA+/−CSB+/− and XPA−/−CSB−/− mice at P8 (Left) and P14 (Right). EGL, external granule cell layer; PCL, PC layer; IGL, internal granule cell layer; ML, molecular layer. (Bar = 50 μm.) (B) Measures of the length of the dendritic tree of PCs in lobules I/II and III, and IX/X of each genotypic mice cerebellum at P8 and P14. Values are mean ± SD for six midsagittal sections derived from five to eight mice of each genotype. *, P < 0.001, and **, P < 0.0001, against XPA+/−CSB+/−.
Reduced Neurogenesis in the EGL in XPA−/−CSB−/− Cerebellum.
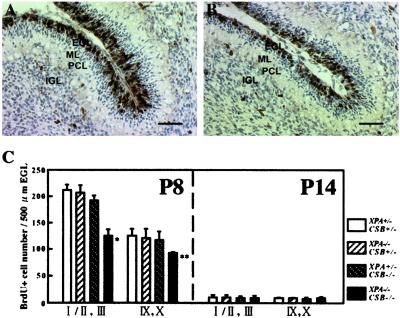
During postnatal development, the cerebellar morphogenesis largely depends on the proliferation and migration of granular neuron precursors in the EGL (13–15). Thus, we next examined the proliferative capacity of cells in the EGL by using BrdUrd incorporation in dividing cells followed by immunohistochemistry. At P8, when neurogenesis in cerebellar granule cells in the EGL reaches its maximum, the thickness of the BrdUrd-positive cell region in the EGL was reduced in XPA−/−CSB−/− animals (n = 5) in comparison with XPA+/−CSB+/− mice (n = 4) (Fig. 5 A and B). A reduction in the number of BrdUrd-positive cell was observed on the anterior (lobule I/II and lobule III) side and, to a lesser extent, on the posterior (lobules IX and X) side (Fig. 5C). We could not find a significant difference in the number of BrdUrd-positive cell between XPA−/−CSB+/− (n = 3) or XPA+/−CSB−/− (n = 5) mice and XPA+/−CSB+/− mice (Fig. 5C). At P14, BrdUrd-positive cells had almost disappeared from the EGL, and there was no longer a significant difference in the numbers among the genotypes (n = 3 for each genotype examined) (Fig. 5C). At P20, we could find few proliferating cells in the EGL of XPA−/−CSB−/− mice as well as littermates of the other genotypes (data not shown). These results demonstrate that the XPA and CSB gene defects do not delay the disappearance of the EGL but rather impair neurogenesis in the EGL.
Figure 5.
Reduced neurogenesis in XPA−/−CSB−/− cerebellur EGL. Compared with the EGL of XPA+/−CSB+/− cerebellum (A), proliferating cells (black) were obviously decreased in number, and the row of cells disordered in XPA−/−CSB−/− cerebellum at P8 (B). (Bar = 50 μm.) (C) A comparison of proliferative ability of cells in the EGL of lobules I/II and III, and IX/X of each genotypes at P8 and P14. Values are mean ± SD for six sections derived from three to five mice of each genotype. *, P < 0.002, and **, P < 0.02, against XPA+/−CSB+/−.
Accelerated Apoptosis in the EGL in XPA−/−CSB−/− Cerebellum.
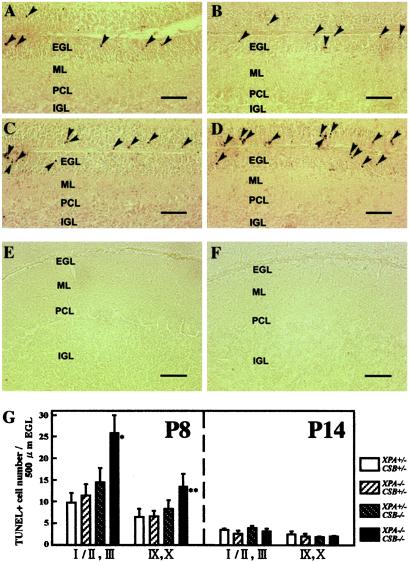
To examine whether neuronal cell death is accelerated in the developing cerebellum of XPA−/−CSB−/− mice, we performed a TUNEL staining to detect cells dying from apoptosis. At P8, a significant increase in the number of TUNEL-positive cells in the EGL of XPA−/−CSB−/− (n = 5) mice was observed when compared with that of control XPA+/−CSB+/− (n = 5) mice (Fig. 6 A, D, and G). Although we noted a slight increase in the number of TUNEL-positive cells in the EGL of XPA+/−CSB−/− (n = 5) mice (Fig. 6 C and G), the difference with XPA−/−CSB+/− (n = 3) (Fig. 6 B and G) and XPA+/−CSB+/− mice was not significant. Moreover, we could find hardly any TUNEL-positive cells in the PC layer, molecular layer, and internal granular layer in all genotypes (Fig. 6 A–D, data not shown). At P14, neither EGL nor other layers contained many TUNEL-positive cells in both XPA−/−CSB−/− and XPA+/−CSB+/− mice (Fig. 6 E–G). These results indicate that a combined XPA and CSB gene defect preferentially accelerates apoptotic cell death in the EGL and suggest that, together with the impaired neurogenesis in the EGL, this may associate with the reduced size of the XPA−/−CSB−/− cerebellum.
Figure 6.
Accelerated apoptosis in XPA−/−CSB−/− cerebellar EGL. (A–D) The cerebellum of each genotypic mouse at P8 was stained by TUNEL. The number of TUNEL-positive cells (arrowhead) was significantly higher in XPA−/−CSB−/− EGL (D) than XPA+/−CSB+/− EGL (A) but comparable between XPA−/−CSB+/− (B) and XPA+/−CSB−/− (C). At P14, only a few TUNEL-positive cells remained in the EGL of both XPA+/−CSB+/− (E) and XPA−/−CSB−/− (F). (Bar = 50 μm.) (G) A comparison of the number of TUNEL-positive cells in the EGL in lobules I/II and III, and IX/X of each genotypes at P8 and P14. Values are mean ± SD for six sections derived from three to five mice of each genotype. *, P < 0.004, and **, P < 0.03, against XPA+/−CSB+/−.
Discussion
In this study, we have shown that XPA−/−CSB−/− mice exhibit distinct neurological symptoms resembling those of human XP-A and CS-B patients, and that the cerebella of these animals display abnormal cerebellar histogenesis and PC differentiation. Moreover, immunohistochemical examinations revealed decreased neurogenesis and increased apoptosis within the EGL. Our results provide in vivo evidence that a NER gene defect can prevent neural cell proliferation and affect survival during development, which may trigger various neurological abnormalities.
The cerebellum is one of the best characterized regions of the brain with respect to development. It undergoes dramatic developmental changes during the first 3 weeks of postnatal life in the mouse (13). During this period, neuroblasts proliferate in the EGL and differentiate into cerebellar granular cells that migrate toward the internal granular layer. This leads to a dramatic increase in the volume of the cerebellum (over 1,000-fold) and to the formation of the mature cerebellar structure, including deep fissures and folia. In contrast to the granular cells, PCs have already stopped proliferating at birth and then (P3–P28) extend their dendrites into the molecular cell layer, where they form synapses with parallel fibers and climbing fibers from granule cells and inferior olivary nucleus cells, respectively (14, 15). In the present study, histological analysis revealed an apparent reduction in size and an impaired foliation pattern in the cerebellum of XPA−/−CSB −/− mice (Fig. 3). Interestingly, lobule X was strikingly smaller than in the cerebella of mice with other genotypes. Lobule X has a profound functional connection with the vestibular system that controls equilibrium, upright stance, gait, and the auditory system (15), suggesting that the vestibular neurological phenotypes of XPA−/−CSB−/− mice might originate from this structural abnormality. Alternatively, it is possible that the defect of the vestibular system impairs in turn the normal cerebellar development in XPA−/−CSB−/− mice.
It has been shown that proliferation and apoptosis of neuronal cells in the EGL play a crucial role in postnatal cerebellar histogenesis in the mouse (14–16). Given the impaired proliferation and accelerated apoptotic cell death in the EGL of XPA−/−CSB−/− mice, the abnormal histogenesis of the cerebellum in XPA−/−CSB−/− mouse might result from hypocellularity of the EGL, which in turn affects the cerebellar layer structure and foliation. On the other hand, in comparison to control XPA+/−CSB+/− mice, the number of PCs in the cerebellum of XPA−/−CSB−/− animals at birth is not significantly changed (M.M. and I.N., unpublished observations), and apoptosis in the PC layer does not increase in later postnatal life (Fig. 6). These suggest that PC degeneration does not predominantly contribute to the neurological symptoms of XPA−/−CSB−/− mice. Rather, PCs are the only afferent output cells in the cerebellar cortex; thus, abnormal morphology of PC dendrites in which afferent inputs are integrated may primarily relate to the motor dysfunction of XPA−/−CSB−/− mice.
Unlike XPA and CSB single mutant mice, XPA−/−CSB−/− animals die before weaning and exhibit severe neurological symptoms during early postnatal development. Which molecular mechanism underlies these neurological abnormalities in XPA−/−CSB−/− mice? The findings that, in XPA−/−CSB−/− mice, the mild neurological phenotype of CSB−/− mice (8) seems to be dramatically enhanced when XPA is additionally inactivated suggest that CSB and XPA have at least in part an additive role in the developing mouse brain, and that mutations in these genes cause abnormal neuronal development and brain dysfunction.
One explanation is that accumulation of DNA damage is enhanced in XPA−/−CSB−/− mice in comparison with XPA and CSB single mutant animals. Both XPA and CSB gene products are involved in NER, which deals mainly with severely distorting DNA injuries, including bulky nucleotide adducts and intrastrand DNA crosslinks (17). NER includes two damage-sensing pathways: one for the entire genome, global genome NER, and one focusing on the transcribed strand of active genes, designated transcription-coupled NER. XPA binds to the damaged site as one of the DNA damage recognition/verification factors required for both NER pathways, whereas CSB acts specifically in transcription-coupled repair (18, 19). In addition, recent studies have shown that XPA is required for removal of oxygen free radical-induced 5′, 8-purine cyclodeoxynucleosides lesions (20, 21), and that CSB is required for the transcription-coupled base excision repair of 8-oxoGuanine, a highly mutagenic oxidative DNA lesion (22). On the basis of these results, it could be hypothesized that in the absence of certain repair (sub) pathways, increased levels of unrepaired or misrepaired (endogenous) DNA damage can lead to abnormal neuronal proliferation and survival in XPA−/−CSB−/− mice. This may dramatically worsen the phenotypes of single mutant mice. Because the brain is thought to be one of the organs most sensitive to oxidative stress, oxidative DNA damage is a strong candidate. In fact, recent studies have suggested that oxidative stress and disturbed glutamate transport (a hallmark of the excite neurotoxicity) may be involved in cerebellar neurodegeneration of XP-A and CS patients (23). Furthermore, Nouspikel and Hanawalt (24) showed that global genome repair rather than transcription-coupled repair of UV-induced DNA lesions was significantly impaired in terminally differentiated human hNT neurons, whereas both repair systems are active in undifferentiated NT2 cells. To elucidate the role of XPA and CSB in the development and aging of the nervous system, it would therefore be of interest to examine DNA repair in neuronal cells of XPA−/−CSB−/− mice as well as young and aged single mutant animals of various types of DNA lesions.
Another possibility is a direct or indirect involvement of XPA and CSB gene products in neural gene expression. The key example for the connection between NER and transcription is the presence of the XPB and XPD proteins in the transcription/repair factor TFIIH, involved in opening of the DNA helix during the transcription initiation step or as a preparatory step to the damage excision reaction (25). Both XPA and CSB proteins form a NER complex with TFIIH (2, 26, 27). Moreover, the CSB protein is found in a complex with RNA polymerase II (28) and enhances transcription elongation in vitro (29, 30). These findings open the possibility that the defects of XPA and CSB gene defects directly affect the level of neuronal gene expression. Taken together, if the transcription and/or DNA repair machinery is substantially impaired, the neuronal developmental process may be disrupted, resulting in inappropriate premature neuronal differentiation and apoptosis. At this point, it should be further clarified whether XPA and CSB have a similar redundant function that prevents the onset of a severe phenotype in the single mutant mice or whether defects in the XPA and CSB gene act synergistically. In the latter scenario, it is envisaged that the potential transcription (elongation) defect, caused by the CSB deficiency, is further hampered by the increased levels of endogenously produced transcription-blocking DNA lesions, originating from the lack of global genome repair by the XPA defect.
Interestingly, XPG−/− mice display postnatal growth failure and undergo premature death before weaning and as such strongly resemble XPA−/−CSB−/− mice (9). Moreover, Sun et al. recently reported PC degeneration and abnormal dendritic arborization were observed at P20 in XPG−/− mice (31). XPG interacts with various NER proteins (including XPA, CSB, and TFIIH) and acts as a structure-specific 3′ DNA endonuclease, cleaving the damaged strand at the 3′-side of the lesion. In addition, XPG takes part in transcription-coupled repair of oxidative DNA damage caused by 8-oxoGuanine and thymine glycol in human cells as well as CSB and TFIIH (22, 32). These results suggest that the mechanisms underlying the severe and comparable phenotype of XPG−/− and XPA−/−CSB−/− mice may be very similar.
The relationship between neurological function and the normal repertoire of DNA-damage responses has only recently been firmly established. Compelling examples are mouse models with deficiencies in XRCC2 (thought to be central to homologous recombination repair) (33), XRCC4, Ligase IV, Ku70, and Ku80 (all involved in nonhomologous DNA double-strand break repair) (34–36), and DNA polymerase β (which functions in DNA base excision repair) (37). Furthermore, mutations in the ataxia-telangiectasia-mutated (ATM) gene have been shown to rescue neuronal apoptosis in Ligase IV mutant mice, indicating that DNA damage checkpoint proteins are involved (38, 39). In addition to defects in different types of DNA repair pathways, these animals exhibit strikingly similar neurological phenotypes, that is, defective neurogenesis associated with extensive apoptosis in the developing nervous system. Here, by using XPA−/−CSB−/− mice, we have shown that deficiencies in NER genes can also cause accelerated apoptosis in developing neuronal tissues. Apart from the connection between human NER gene deficiencies and neurodegeneration, our present findings further support the idea that DNA repair processes play a crucial role in normal development and maintenance of the nervous system (40).
The molecular basis for the difference in neurological phenotypes between human XP-A and CS-B diseases and the corresponding mouse models remains unknown. In XP-A and CS patients, postnatal cerebellar neurodegeneration and abnormal dendritic morphology of PC have been reported (41, 42). Moreover, in the developing normal human cerebellum, the EGL appears between the ninth and eleventh gestational weeks and persists until the end of the first postnatal year (43), whereas abnormal motor activity, gait disturbance, and cerebellar ataxia start at early postnatal age in XP-A and CS patients, and in some CS patients the signs are observed even at birth (3). Taken together, these results led us to hypothesize that, similar to the severe and early onset cerebellar abnormalities in XPA−/−CSB−/− mice, the severe forms of XP-A and CS-B may be caused by the synergy of impaired neurogenesis, cell survival, and PC differentiation in development. In contrast, in the less severe forms, neurons that escape from cell death and are not severely damaged in the developmental stage might become the cause of neurological abnormalities at later stages of life. In this respect, it is interesting to note that the ATM protein has been suggested to act as a neural survival checkpoint component during neurogenesis (40, 44). Although the neuropathological analysis of the developing cerebellum of XP-A and CS-B patients remains to be clarified, our findings raise the possibility that at least some early onset neurological abnormalities in XP-A and/or CS-B patients are caused by neurodevelopmental defects rather than by neurodegeneration later in life.
Acknowledgments
We thank S. Yamagishi for technical support and Drs. Y. Nakamura and M. Hayashi for helpful advice about cerebellar anatomy and human XP and CS neuropathology. This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture, Japan (Grant 08280101), Nissan Science Foundation (to Y.E. and H.H.), the Netherlands Organization for Scientific Research NWO (“Diseases of the Elderly”), the National Institutes of Health (Grant AG17242–02), the Dutch Cancer Society (EUR 98–1774), and the European Community (QLRT-1999–02002) (to G.T.J.H. and J.H.J.H.).
Abbreviations
- XP
xeroderma pigmentosum
- CS
Cockayne syndrome
- NER
nucleotide excision repair
- PC
Purkinje cell
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling
- Pn
postnatal day n
- EGL
external granular layer
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12860.
References
- 1.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 2001. pp. 677–703. [Google Scholar]
- 2.de Laat W L, Jaspers N G J, Hoeijmakers H J. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 3.Nance M A, Berry S A. Am J Med Genet. 1992;42:68–82. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 4.Greenhaw G A, Hebert A, Duke-Woodside M E, Butler I J, Hecht J T, Cleaver J E, Thomas G H, Horton W A. Am J Hum Genet. 1992;50:677–689. [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries A, van Oostrom C T, Hofhuis F M, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J, van Steeg H, Verbeek S J. Nature (London) 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 6.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, et al. Nature (London) 1995;377:165–168. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 7.Sands A T, Abuin A, Sanchez A, Conti C J, Bradley A. Nature (London) 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- 8.van der Horst G T, van Steeg H, Berg R J, van Gool A J, de Wit J, Weeda G, Morreau H, Beems R B, van Kreijl C F, de Gruijl F R, et al. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 9.Harada Y N, Shiomi N, Koike M, Ikawa M, Okabe M, Hirota S, Kitamura Y, Kitagawa M, Matsunaga T, Nikaido O, Shiomi T. Mol Cell Biol. 1999;19:2366–2372. doi: 10.1128/mcb.19.3.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer J, de Wit J, van Steeg H, Berg R J, Morreau H, Visser P, Lehmann A R, Duran M, Hoeijmakers J H, Weeda G. Mol Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 11.Berg R J, Rebel H, van der Horst G T, van Kranen H J, Mullenders L H, van Vloten W A, de Gruijl F R. Cancer Res. 2000;60:2858–2863. [PubMed] [Google Scholar]
- 12.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldowitz D, Hamre K. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 14.Hatten M, Heintz N. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 15.Voogd J, Glickstein M. Trends Neurosci. 1998;21:370–375. doi: 10.1016/s0166-2236(98)01318-6. [DOI] [PubMed] [Google Scholar]
- 16.Kuida K, Zheng T S, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell R A. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 17.Citterio E, Vermeulen W, Hoeijmakers J H. Cell. 2000;101:447–450. doi: 10.1016/s0092-8674(00)80854-5. [DOI] [PubMed] [Google Scholar]
- 18.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 19.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston R E, Hoeijmakers J H, Vermeulen W. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuraoka I, Bender C, Romieu A, Cadet J, Wood R D, Lindahl T. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. . (First Published April 4, 2000; 10.1073/pnas.070471597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks P J, Wise D S, Berry D A, Kosmoski J V, Smerdon M J, Somers R L, Mackie H, Spoonde A Y, Ackerman E J, Coleman K, et al. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 22.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi M, Itoh M, Araki S, Kumada S, Shioda K, Tamagawa K, Mizutani T, Morimatsu Y, Minagawa M, Oda M. J Neuropathol Exp Neurol. 2001;60:350–356. doi: 10.1093/jnen/60.4.350. [DOI] [PubMed] [Google Scholar]
- 24.Nouspikel T, Hanawalt P C. Mol Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoeijmakers J H, Egly J M, Vermeulen W. Curr Opin Genet Dev. 1996;6:26–33. doi: 10.1016/s0959-437x(96)90006-4. [DOI] [PubMed] [Google Scholar]
- 26.Park C H, Mu D, Reardon J T, Sancar A. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 27.Frit P, Bergmann E, Egly J M. Biochimie. 1999;81:27–38. doi: 10.1016/s0300-9084(99)80035-2. [DOI] [PubMed] [Google Scholar]
- 28.van Gool A J, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly J M, Bootsma D, Hoeijmakers J H. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selby C P, Sancar A. Proc Natl Acad Sci USA. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tantin D, Kansal A, Carey M. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X Z, Harada Y N, Takahashi S, Shiomi N, Shiomi T. J Neurosci Res. 2001;64:348–354. doi: 10.1002/jnr.1085. [DOI] [PubMed] [Google Scholar]
- 32.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 33.Deans B, Griffin C S, Maconochie M, Thacker J. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes D E, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 36.Gu Y, Sekiguchi J, Gao Y, Dikkes P, Frank K, Ferguson D, Hasty P, Chun J, Alt F W. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Barnes D E, Lindahl T, McKinnon P J. Genes Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekiguchi J, Ferguson D O, Chen H T, Yang E M, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, Alt F W. Proc Natl Acad Sci USA. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. . (First Published March 6, 2001; 10.1073/pnas.051632098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rolig R L, McKinnon P J. Trends Neurosci. 2000;23:417–424. doi: 10.1016/s0166-2236(00)01625-8. [DOI] [PubMed] [Google Scholar]
- 41.Kohji T, Hayashi M, Shioda K, Minagawa M, Morimatsu Y, Tamagawa K, Oda M. Neurosci Lett. 1998;243:133–136. doi: 10.1016/s0304-3940(98)00109-8. [DOI] [PubMed] [Google Scholar]
- 42.Soffer D, Grotsky H W, Rapin I, Suzuki K. Ann Neurol. 1979;6:340–348. doi: 10.1002/ana.410060407. [DOI] [PubMed] [Google Scholar]
- 43.Rakic P, Sidman R L. J Comp Neurol. 1970;139:473–500. doi: 10.1002/cne.901390407. [DOI] [PubMed] [Google Scholar]
- 44.Herzog K H, Chong M J, Kapsetaki M, Morgan J I, McKinnon P J. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]