Abstract
Recent studies have reported hundreds of genes linked to Alzheimer’s Disease (AD). However, many of these candidate genes may be not identified in different studies when analyses were replicated. Moreover, results could be controversial. Here, we proposed a computational workflow to curate and evaluate AD related genes. The method integrates large scale literature knowledge data and gene expression data that were acquired from postmortem human brain regions (AD case/control: 31/32 and 22/8). Pathway Enrichment, Sub-Network Enrichment, and Gene-Gene Interaction analysis were conducted to study the pathogenic profile of the candidate genes, with 4 metrics proposed and validated for each gene. By using our approach, a scalable AD genetic database was developed, including AD related genes, pathways, diseases and info of supporting references. The AD case/control classification supported the effectiveness of the 4 proposed metrics, which successfully identified 21 well-studied AD genes (i.g. TGFB1, CTNNB1, APP, IL1B, PSEN1, PTGS2, IL6, VEGFA, SOD1, AKT1, CDK5, TNF, GSK3B, TP53, CCL2, BDNF, NGF, IGF1, SIRT1, AGER and TLR) and highlighted one recently reported AD gene (i.g. ITGB1). The computational biology approach and the AD database developed in this study provide a valuable resource which may facilitate the understanding of the AD genetic profile.
Keywords: Alzheimer’s disease, ResNet database, Pathway enrichment analysis, Sub-network enrichment analysis, Gene-gene interaction analysis
1. Introduction
Alzheimer's disease (AD) is a chronic neurodegenerative disease that usually onsets slowly and progresses more rapidly over time (Burns and Iliffe, 2009). It is the leading cause of dementia, beginning with impaired memory, and most often onsets in people over 65 years of age (Mendez 2012). The global prevalence of AD as of 2015 was estimated to be as high as 48 million people worldwide (World Health Organization, 2015). Although the cause of most Alzheimer's cases largely remains unknown, about 70% of the risk is believed to come from a large network of genes (Ballard et al., 2011). As such, researches into the causes of AD are currently being explored.
In recent years, an increased number of genetic researches have been conducted revealing over a thousand altered genes linked to AD. For example, increased GSK3B activity and decreased phosphorylation of the gene have been repeatedly observed in AD cases (Cole et al., 2007, Koedam et al., 2013, Xu et al., 2016). Significantly increased expression levels of TP53, PTGS2 and TGFB1 were suggested by many independent studies to be associated with AD (Cenini et al., 2008, Lanni et al., 2012, Ramalho et al., 2008, Yoo et al., 2008, Wang et al., 2014, Luo et al., 2006). Observations from these previous studies are valuable in studying the genetic basis of the pathogenic development of the disease.
However, approximately one third of these AD-gene linkages were reported once with no further replication, and over 60% were supported by no more than three citations. Moreover, most of these studies had small sample sizes that were more susceptible to noise. Additionally, due to the variation in data collection and processing approaches, results from different studies were not always consistent. Meanwhile, there are dozens of new AD risk genes being reported every year, posing an increased need for further validation of these candidate genes to AD. While biological experiments were effective towards this validation task, they could be very costly. To address this issue, we propose a computational biology approach for a systematic evaluation of these AD candidate genes.
In recent years, Pathway Studio ResNet relation data have been widely used to study modeled relationships between proteins, genes, complexes, cells, tissues and diseases (http://pathwaystudio.gousinfo.com/Mendeley.html). In this study, we integrated large scale AD related ResNet literature knowledge data, independent gene expression data and related pathway/network information to study the functional profile of a large gene pool that has been reported to be linked to AD. The purpose of the study is to provide an easy-update computational evaluation workflow, through which an AD genetic database (AD_GD) could be generated to present a weighted landscape view of the genetic basis underlying the pathogenic development of AD. Our results support the hypothesis that AD candidate genes are functionally linked to each other, forming a large genetic network to regulate the pathogenic development of AD through multiple pathways.
2. Materials and methods
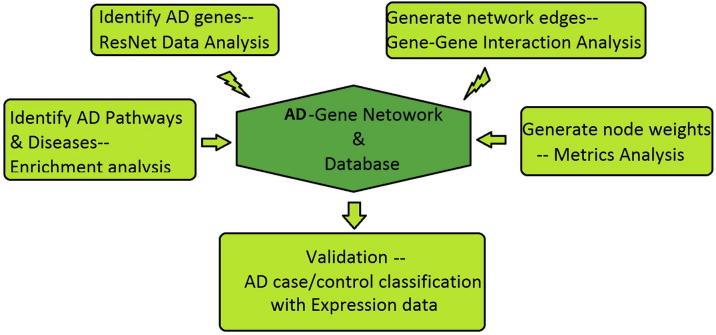
Fig. 1 presents the diagram of the proposed computational gene marker evaluation system, with detailed descriptions in the following sub-sections. Using our approach, a genetic database (AD_GD) was developed and deposited into an open source ‘Bioinformatics Database’ online available at http://database.gousinfo.com, including 1699 genes (with metric scores), 151 pathways and 114 diseases that are linked to AD. Also included in AD_GD are information of 27000+ supporting references for AD-gene relationships, including the titles and relevant sentences where the relations were identified. The AD_GD database is scalable and will be updated monthly or upon request using our approach.
Fig. 1.
Diagram for the integrative computational marker evaluation approach for AD. First, literature based analysis were conducted to identify the AD related genes, then Gene-Gene Interaction Analysis, Enrichment analysis, and Metrics analysis were conducted on these gene and results were saved in the AD database. Finally, AD case/control classification were conducted to test the effectiveness of the identified genes, using gene expression datasets.
2.1. ResNet literature knowledge data
ResNet relation data (AD-Gene) were acquired from the Pathway Studio ResNet® Mammalian database (http://pathwaystudio.gousinfo.com/ResNetDatabase.html) updated November 2016. The ResNet® Mammalian database are a group of real-time updated literature knowledge databases, including curated signaling, cellular processes and metabolic pathways, ontologies and annotations, as well as molecular interactions and functional relationships (http://pathwaystudio.gousinfo.com/ResNetDatabase.html). Modeled relation data are extracted from the 41M+ references covering entire PubMed abstracts and Elsevier and third party full text journals. The ResNet database employs an automated natural language processing-based information extraction system, MedScan, with precision of over 91% (Daraselia et al., 2004). Each relationship within the database is supported with one or more references. By far, Pathway Studio ResNet Databases is the largest database among known competitors in the field (Lorenzi et al., 2014).
2.2. Enrichment and gene-gene interaction analysis
Pathway enrichment analysis (PEA) and sub-network enrichment analysis (SNEA) (http://pathwaystudio.gousinfo.com/SNEA.pdf) was conducted using Pathway Studio to identify genetic pathways and diseases potentially linked to AD (Sivachenko et al., 2007). Furthermore, a pathway based gene-gene interaction (GGI) analysis was conducted to generate weighted edges/linkage between genes. The weight of an edge is the number of pathways where both nodes were included.
2.3. Metrics analysis
For the gene network built through the aforementioned steps, 4 metrics were proposed for each node/gene, including 2 literature based metric scores (RScore and AScore), and 2 enrichment based metric scores (PScore and SScore). The logic is that, a gene is likely linked to AD if it satisfies one or more of the following conditions: the gene has been frequently observed in independent studies to be associated with AD (high RScore), plays roles within multiple pathways associated with AD (high PScore), and demonstrates strong functional linkage to many of other genes associated with AD (high SSCore). Additionally, an AScore was proposed to present the history of each AD-gene relation. The detailed definitions of the proposed metrics are described as follows.
2.3.1. Two literature metrics
The reference score (RScore) of a gene is defined as the reference number underlying a gene-disease relationship, as shown in Eq. (1).
| (1) |
The age score (AScore) of a gene is defined as the earliest publication age of a gene-disease relationship, as shown in Eq. (2).
| (2) |
where n is the total number of references supporting a gene-disease relation, and
| (3) |
2.3.2. Two enrichment metrics
We define a network significance score (SScore) of a node as Freeman's formalized node degree centrality (Freeman, 2012), as defined in Eq. (4).
| (4) |
where n is the total number of nodes, and j represents all other nodes within the network; x is the adjacency matrix, in which the cell x is 1 if the jth node is connected with the current node, other with is 0.
Given a disease is associated with a set of genetic pathways we define the PScore for the gene as Eq. (5).
| (5) |
2.4. Validation using independent gene expression data
We hypothesized that significant AD related genes should contribute to distinguishing AD patients from healthy controls. To validate the effectiveness of the selected genes and the proposed metrics, we performed a Euclidean distance-based multivariate classification (Wang et al., 2015) on two independent gene expression data sets (NCBI GEO: GSE29378 and GSE28146), followed by a leave-one-out (LOO) cross validation, using the overall gene set and the sub-sets selected by different scores as tentative markers. In each run of LOO, gene expression data of one subject is used for testing and the rest for training. A permutation of 5000 runs was then conducted to test the hypothesis that a randomly selected gene set in same size can reach equal or higher classification accuracy (CR).
For dataset GSE29378, RNA expression profile of 64 subjects (AD case/control: 31/32) were obtained from 60 μm sections of frozen human hippocampus using scalpel dissection. Of the 1699 genes evaluated, 1605 were included in the database. For GSE28146, brain sections from a total of 30 subjects were analyzed (case/control: 22/8), with 1621 out of 1699 genes included.
3. Results
3.1. AD genes for evaluation
AD-Gene literature knowledge data analysis identified 1699 AD genes, supported by 27128 scientific articles (AD_GD → Related Genes and AD_GD → References for Disease-Gene Relation). A scalable genetic database, AD_GD, was developed through our study, which is online available at ‘Bioinformatics Database’ (http://database.gousinfo.com).
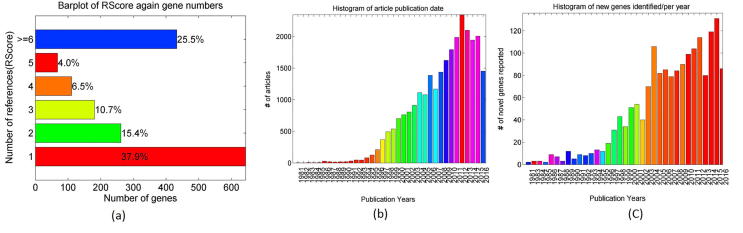
Of the 1699 genes, 644 (37.90%) have been reported with one reference (RScore = 1), 262 (15.42%) with 2, 181 (10.65%) with 3, and 433 (25.49%) with more than 5 references, as shown in Supplementary Fig. S1(a). Publication date statistics of the 27128 supporting references are presented in Supplementary Fig. S1(b), with novel genes reported in each year (Supplementary Fig. S1(c)). To note, these articles have an average publication age of only 7.4 years, indicating that most of the articles were published in recent years.
3.2. Enrichment analysis results
PEA showed that, 1453 out of 1669 genes got significantly enriched within 151 AD candidate pathways/gene sets (p-values < 1e−15, q = 0.001 for FDR; AD_GD → Related Pathways). Not surprisingly, aging (GO: 0016280; overlap 140 genes; q-value = 2.28E−84) is the top one enriched gene group. In addition, 11 pathways/groups were related to the neuronal system (619 unique genes) (Gong and Lippa, 2010, Marcello et al., 2012), 10 to cell growth and proliferation (441 unique genes) (Hohman et al., 2015, Li and Yao, 2013), 8 to cell apoptosis (452 unique genes) (Behl, 2000), 5 to protein phosphorylation (246 unique genes) (Shapiro et al., 1991), 2 to protein kinase (209 unique genes) (Martin et al., 2013), 3 to brain function/development (99 unique genes) (Llorente-Vizcaíno and Cejudo-Bolívar, 2001), and 2 to immune system (268 unique genes) (Heneka et al., 2001). Due to lack of space, we only present the top 10 pathways enriched in Table 1 (p-value ≤ 2.6e−55, including 755 out of 1699 genes).
Table 1.
Top 10 Molecular function pathways/groups enriched by 1669 genes reported.
| Pathway/gene set name | GO ID | # of Entities | Overlap | q-value | Jaccard similarity |
|---|---|---|---|---|---|
| Aging | 0016280 | 254 | 140 | 2.28E−84 | 0.077 |
| Neuronal cell body | 0043025 | 466 | 172 | 4.7E−72 | 0.086 |
| Neuron projection | 0043005 | 378 | 153 | 2.8E−70 | 0.079 |
| Response to lipopolysaccharide | 0032496 | 252 | 126 | 4.9E−69 | 0.069 |
| Response to hypoxia | 0001666 | 259 | 127 | 8.11E−68 | 0.069 |
| Response to organic cyclic compound | 0014070 | 253 | 122 | 1.79E−64 | 0.067 |
| Response to ethanol | 0017036 | 161 | 94 | 4.21E−59 | 0.053 |
| Negative regulation of apoptotic process | 0006916 | 650 | 187 | 1.21E−56 | 0.086 |
| Perinuclear region of cytoplasm | 0048471 | 688 | 188 | 2.11E−55 | 0.085 |
| Axon | 0030424 | 318 | 125 | 2.56E−55 | 0.066 |
For each gene set, the p-value was calculated using Fisher’s-Exact test against the hypothesis that a randomly selected gene group of same size (1669) can generate a same or higher overlap with the corresponding gene set (q = 0.001 for FDR correction). The Jaccard similarity (js) is a statistic used for comparing the similarity and diversity of sample sets, which is defined by where A and B are two sample sets.
A SNEA was also performed to identify the pathogenic significance of the reported genes to other disorders which are potentially related to AD. Interestingly, besides neuropathic related diseases (e.g., Parkinson's disease and Schizophrenia) and some types of cancers (e.g., breast cancer), AD seems to share major gene overlaps with many blood related diseases (e.g., diabetes mellitus, obesity, type 2 diabetes, atherosclerosis, ischemia, myocardial infarction, hyperglycemia and stroke). The full list of 113 disease related sub-networks enriched with p-value < 1e−150 (q = 0.001 for FDR; 1625 out of 1669 genes enriched; see AD_GD → Related Diseases).
3.3. GGI results
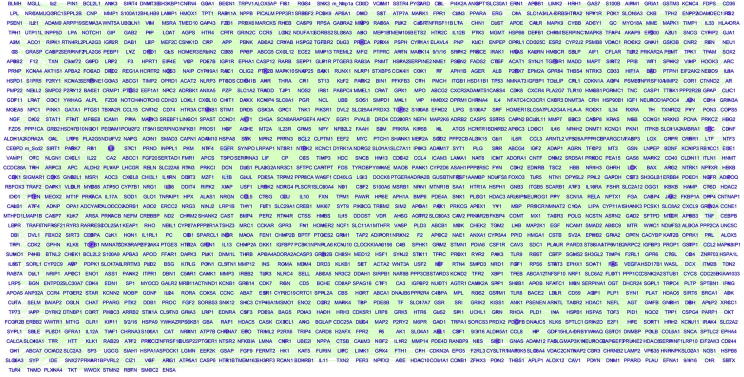
Fig. 2 presents the genetic network for AD, which was built through GGI analysis. The nodes of the network are 1453 out of 1669 genes that were enriched within the 151 AD target pathways. There were 319206 edges within the network, the weight of which are the numbers of pathways shared by the corresponding pair of nodes. The average node strength (sum of the number of genes directly connected) of the network was 219.69, and the node strength for the 246 unconnected genes was signed with 0.
Fig. 2.
Gene-gene interaction network for AD. The network contains 1453 out of 1669 genes AD target genes that enriched within the 151 AD target pathways. The weight of an edge between two nodes is the number of pathways shared by both nodes. The larger the size of a node, the larger the number of AD candidate pathways including the gene (high PScore); the brighter the color, the larger number of AD candidate genes associated with gene (high SScore). 216 out of 1669 genes were not included in the network as they were not enriched within the top 151 AD candidate pathways.
Along with GGI, SScore and PScore were calculated for each gene (AD_GD → Related Genes). The value of a PScore represents how many AD candidate pathways involve the gene, and an SScore represents how strong of a gene associated with other genes within the network.
3.4. Validation results
We hypothesized that, if our selected gene set (1669 genes) and the top genes selected by the proposed metric scores are significant to the pathogenesis of AD, they should lead to significant higher classification accuracy comparing to randomly selected genes. To test the hypothesis, classification and LOO cross validation were conducted on two independent public RNA expression dataset (NCBI GEO: GSE29378 and GSE28146), followed by a permutation test of 5000 runs.
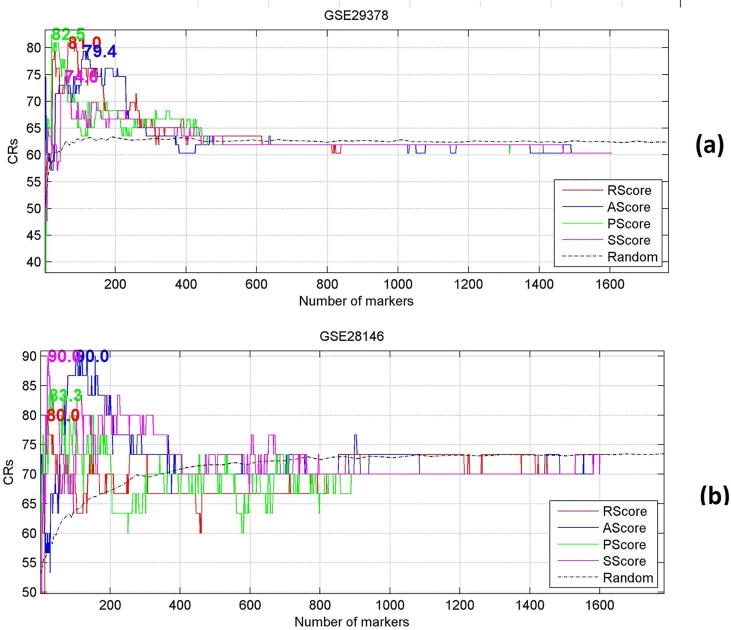
For the LOO cross validation, the 1669 genes were first ranked by different metric scores, then the top n (n = 1, 2 …) genes were used as input variables for classification and LOO cross validation. Table 2 and Fig. 3 presents the results with the maximum classification ratios (CRs) marked at the position of corresponding number of genes.
Table 2.
Permutation test on top genes corresponding to highest CRs.
Fig. 3.
Validation of different metrics through a LOO cross validation. (a) Results from GSE29378; (b) Results from GSE28146. Mean of CRs by randomly selected genes are displayed in a dash-grey line (Legend: Random). The maximum CR by different metrics are presented at the corresponding positions.
From Fig. 3 we see that the top genes selected by different scores (in descending order) can lead to the highest classification accuracies, which are significantly higher than the average CRs of randomly selected gene set in same size, while adding more genes with lower score may not necessarily lead to improved CRs.
3.5. Cross metrics analysis
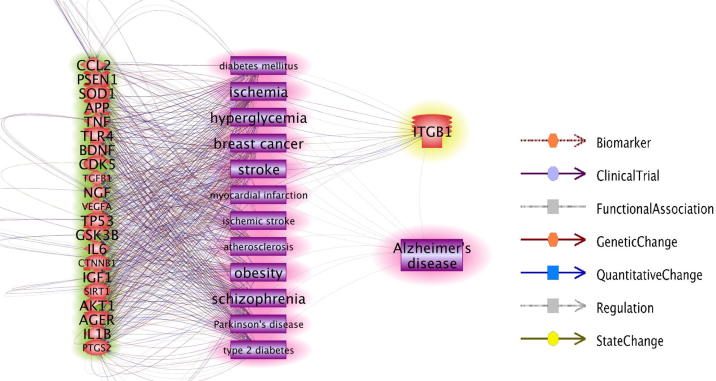
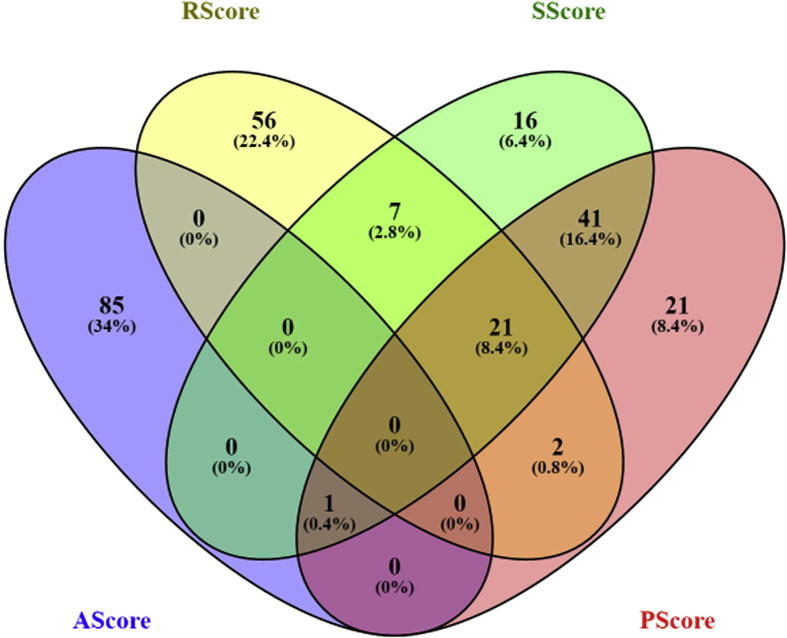
Results from AD case/control classification (Table 2 and Fig. 3) showed that, the top genes selected using each of the four proposed metrics led to significantly higher CR compared to randomly selected gene sets, demonstrating the effectiveness of the proposed metrics. Therefore, it is worthy to study the overlaps among these top genes. Cross metrics analysis of the top 5% (86 genes, corresponding to the number of genes reported this years, 2016) of 1699 genes selected using different scores showed that (see Veen diagram at Supplementary Fig. S2), there was strong overlap between PScore group and SScore group (63/86). Among these 63 genes, 21 were also identified to be within RScore group, including TGFB1, CTNNB1, APP, IL1B, PSEN1, PTGS2, IL6, VEGFA, SOD1, AKT1, CDK5, TNF, GSK3B, TP53, CCL2, BDNF, NGF, IGF1, SIRT1, AGER and TLR4, with RScore = 542 ± 16 refrences, PScore = 29 ± 6 pathways, SScore = 969 ± 85 connected genes. Network analysis using Pathway Studio showed that, these 21 genes also demonstrated strong correlation with the top disorders that are linked to AD (Fig. 4, highlighted in red). The genes related to these diseases present significant overlap with these genes linked to AD (see AD_GD → Related Diseases). On the other hand, only one gene, ITGB1, was identified to be the overlap of AScore, PScore and SScore groups (Fig. 4, highlighted in yellow), which also linked to several other diseases (e.g., diabetes, stroke and breast cancer) that are genetically linked to AD.
Fig. 4.
AD genes selected by cross metrics analysis and their relation with other diseases. The 21 genes that were overlap in RScore, PScore and SScore groups are highlighted in green; Gene ITGB1 that was the overlap in AScore, PScore and SScore groups and is highlighted in yellow. The network was built using the ‘network building’ module of Pathway Studio.
4. Discussion
Recent studies proposed over a thousand of AD risk genes with dozens of novel targets identified each year. However, over half of these AD target genes were lack of replication and results were not always consistent, posing an increasing need of a systematic evaluate approach to test the significance of these genes as a network to AD. In this study, we integrated large scale literature knowledge data, gene expression data and related pathways and disease-sub networks to evaluate 1669 AD candidate genes. Four metric scores have been proposed and validated. A scalable genetic database, AD_GD, was developed through our study, which is online available at ‘Bioinformatics Database’ (http://database.gousinfo.com).
PEA results showed that most genes within the network (1453 out of 1699) were significantly enriched (FDR corrected p-value < 1e−15) in the pathways previously implicated with AD, including pathways/groups related to aging, neuronal system pathways, cell growth and proliferation, cell apoptosis, protein phosphorylation brain function/development and immune system (Gong and Lippa, 2010, Marcello et al., 2012, Hohman et al., 2015, Li and Yao, 2013, Behl, 2000, Shapiro et al., 1991, Martin et al., 2013, Llorente-Vizcaíno and Cejudo-Bolívar, 2001, Heneka et al., 2001). These observations support the hypothesis that, most of the AD target genes are functionally linked to each other and play roles within multiple pathways associated with AD.
In addition to PEA, we performed a SNEA, which can provide high levels of confidence when interpreting experimentally-derived genetic data against the background of previously published results (http://pathwaystudio.gousinfo.com/SNEA.pdf). SNEA results demonstrated that over 95% (1625) of the 1669 AD-genes were as well identified as causal genes for other disorders that were linked to AD (AD_GD → Related Diseases).
For a quantitative measure of the significance of these 1669 AD candidate genes, we proposed 4 metrics: (1) publication frequency (RScore), (2) novelties (AScore), (3) Number of associated AD candidate pathways (PScore), and (4) Network centrality (SScore). We hypothesized that if a gene satisfies one or more of the following conditions, it has high possibility to be linked to AD: The gene is frequently identified by independent studies to be linked to AD (high RScore), plays roles within multiple AD pathways (high PScore), and is functionally linked to multiple AD genes (high SScore).
The effectiveness of our 4 proposed metrics were supported by the AD case/control classification study using two independent gene expression data sets (GSE29378 and GSE28146). Results of the LOO cross validation and permutation process showed that, the top genes by the 4 proposed metrics can lead to significantly higher classification ratio than using randomly selected gene sets (Table 2 and Fig. 3). While using the identified gene set as a whole (1605 and 1621 out of 1669 for GSE29378 and GSE28146, respectively) showed no significant efficiency in terms of AD prediction (permutation p-value > 0.9; see Table 2), suggesting the necessity of using our network metrics for further analysis of the candidate AD genes when dealing with specific experiment data. Notably, for each score, the number of top genes corresponded to the maximum CRs for the two data sets were different. This may reflect the group-wise variation in terms of sample size (63 vs. 30) and clinical parameter dissimilarities (e.g., age, gender). The difference may be also caused by the unique variation of different subjects’ genome in case of AD.
Cross metrics analysis showed that 21 genes were overlapped within RScore, SScore and PScore groups (Fig. 4, highlighted in green). These genes were frequently identified by different studies to be linked to AD (RScore = 542 ± 16 references), play roles with in multiple AD candidate pathways (PScore = 29 ± 6 pathways), and demonstrate strong network centrality (SScore = 969 ± 85 direct gene connections). Therefore, our results suggest that they are among the top AD risk genes that likely pose biological significance with the disease. As a matter of fact, these genes were also identified to play role within many other disorders that were linked to AD, such as diabetes mellitus, obesity, Parkinson's disease, Schizophrenia, and breast cancer (Fig. 4). These results support the effectiveness of the proposed metric scores in the identification of top genes for AD.
Additionally, there was one newly reported gene, ITGB1 (AScore = 1), also demonstrated high SScore and PScore (Fig. 4, highlighted in yellow). Although ITGB1 were not frequently replicated in their association with AD (RScore = 1 reference), and presented less relationships with other AD related mental disorders, it demonstrated high interaction with other genes within the genetic network (SScore = 1050 directly connected genes) and play role within multiple pathways implicated with AD (PScore = 28 pathways). Therefore, our study suggests that it may be worthy of further study. In fact, activation of integrin β1 (ITGB1) has been reported to regulate the synthesis of enterovirus 71-induced and NADPH oxidase-driven reactive oxygen species (ROS) (Tung et al., 2011), which are closely involved in pathogeneses of AD (Aliev et al., 2003). It also revealed that adhesion of HeLa cells to β1 integrin clustering can increase the release of arachidonic acid (Xu and Clark, 1997) that has therapeutic functions against AD (Huang and Cheung, 2011). These findings support our observation that ITGB1 may play roles for pathogenic development of AD, demonstrating the effectiveness of our proposed PScore and SScore in identifying novel genes for the disease. To note that, although in this study we evaluated 1669 known AD candidate genes acquired from ResNet database, which already received literature support for their association with AD, our proposed PScore and SScore can be applied to any given genes and therefore could be used for evaluation and discovery of novel target genes for AD.
The genetic database built through our approach, namely AD_GD, is scalable and can be automatically updated using the computational workflow proposed in this study. Any novel AD-gene relationships can be added to update the database. Moreover, further network analysis with more experiment data may extract additional meaningful features that can be added into our proposed system to gain improved evaluation of existing and/or novel AD genes.
To our knowledge, this is the first study integrating large scale literature knowledge data, experiment data and related pathway/network data for a systematical evaluation of AD candidate genes. The computational biology approach of this study provides a comprehensive weighted genetic network and genetic database for AD, which may help in the evaluation and prioritization of AD genes for further study in the field.
Acknowledgments
Acknowledgement
We would like to thank Mckenzie Ritter for her contribution to the English writing of this manuscript. This work was partly funded by National Nature Science Foundation of China (81130024 and 81630030, Tao Li), National Key Technology R & D Program of the Ministry of Science and Technology of China during the 12th Five-Year Plan (2012BAI01B06, Tao Li), National Key Research and Development Program of the Ministry of Science and Technology of China (2016YFC0904300, Tao Li), Science and Technology Department of Sichuan Province of China (2017SZ0049, Zhe Li; 2018ZR0216, Zhenzhen Xiong), and Health and Family Planning Commission of Sichuan Province of China (17PJ080, Zhe Li). The Fundamental Research Funds for The Central Universities of China (2017SCU11072, Zhe Li).
Conflict of interest
The author HC is with Elsevier Inc., the company that owns the software Pathway Studio used in this study.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.sjbs.2018.05.019.
Appendix A. Supplementary material
Supplementary Figure S1.
Relations between the AD candidate genes and the underlying references. (a) Number of genes with different number of supporting references; (b) number of articles by year; (c) number of novelty genes identified in each year.
Supplementary Figure S2.
Veen diagram of the top 5% out of the 1699 genes selected using different scores.
References
- Aliev G., Obrenovich M.E., Smith M.A., Perry G. Hypoperfusion, mitochondria failure, oxidative stress, and alzheimer disease. J. Biomed. Biotechnol. 2003;2003(3):162–163. doi: 10.1155/S1110724303305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., Jones E. Alzheimer's disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- Behl C. Apoptosis and Alzheimer's disease. J. Neural Transm. (Vienna) 2000;107(11):1325–1344. doi: 10.1007/s007020070021. [DOI] [PubMed] [Google Scholar]
- Burns A., Iliffe S. Alzheimer's disease. BMJ. 2009;338:b158. doi: 10.1136/bmj.b158. [DOI] [PubMed] [Google Scholar]
- Cenini G., Sultana R., Memo M., Butterfield D.A. Effects of oxidative and nitrosative stress in brain on p53 proapoptotic protein in amnestic mild cognitive impairment and Alzheimer disease. Free Radic. Biol. Med. 2008;45(1):81–85. doi: 10.1016/j.freeradbiomed.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A.R., Astell A., Green C., Sutherland C. Molecular connexions between dementia and diabetes. Neurosci. Biobehav. Rev. 2007;31(7):1046–1063. doi: 10.1016/j.neubiorev.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Daraselia N., Yuryev A., Egorov S., Novichkova S., Nikitin A., Mazo I. Extracting human protein interactions from MEDLINE using a full-sentence parser. Bioinformatics. 2004;20:604–611. doi: 10.1093/bioinformatics/btg452. [DOI] [PubMed] [Google Scholar]
- Freeman L.C. Centrality in social networks conceptual clarification. Soc. Netw. 2012;1:215–239. [Google Scholar]
- Gong Y., Lippa C.F. Review: disruption of the postsynaptic density in Alzheimer's disease and other neurodegenerative dementias. Am J Alzheimers Dis Other Demen. 2010;25(7):547–555. doi: 10.1177/1533317510382893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M.T., Golenbock D.T., Latz E., 2001. Innate immunity in Alzheimer's disease. Nat Immunol. 2015;16(3):229–236. [DOI] [PubMed]
- Hohman T.J., Bell S.P., Jefferson A.L. The role of vascular endothelial growth factor in neurodegeneration and cognitive decline: exploring interactions with biomarkers of Alzheimer disease. JAMA Neurol. 2015;72(5):520–529. doi: 10.1001/jamaneurol.2014.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.J., Cheung P.C. +UVA treatment increases the degree of unsaturation in microalgal fatty acids and total carotenoid content in Nitzschia closterium (Bacillariophyceae) and Isochrysis zhangjiangensis (Chrysophyceae) Food Chem. 2011;129(3):783–791. doi: 10.1016/j.foodchem.2011.05.021. [DOI] [PubMed] [Google Scholar]
- Koedam E.L., van der Vlies A.E., van der Flier W.M., Verwey N.A., Koene T., Scheltens P. Cognitive correlates of cerebrospinal fluid biomarkers in frontotemporal dementia. Alzheimers Dement. 2013;9(3):269–275. doi: 10.1016/j.jalz.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Lanni C., Racchi M., Memo M., Govoni S., Uberti D. p53 at the crossroads between cancer and neurodegeneration. Free Radic. Biol. Med. 2012;52(9):1727–1733. doi: 10.1016/j.freeradbiomed.2012.02.034. [DOI] [PubMed] [Google Scholar]
- Li J.S., Yao Z.X. Modulation of FGF receptor signaling as an intervention and potential therapy for myelin breakdown in Alzheimer's disease. Med. Hypotheses. 2013;80(4):341–344. doi: 10.1016/j.mehy.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Llorente-Vizcaíno A., Cejudo-Bolívar J.C. Memories and Alzheimer's disease. Rev. Neurol. 2001;32(12):1163–1172. [PubMed] [Google Scholar]
- Lorenzi P.L., Claerhout S., Mills G.B., Weinstein J.N. A curated census of autophagy-modulating proteins and small molecules: candidate targets for cancer therapy. Autophagy. 2014;10:1316–1326. doi: 10.4161/auto.28773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Lin A.H., Masliah E., Wyss-Coray T. Bioluminescence imaging of Smad signaling in living mice shows correlation with excitotoxic neurodegeneration. Proc. Natl. Acad. Sci. USA. 2006;103(48):18326–18331. doi: 10.1073/pnas.0605077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcello E., Epis R., Saraceno C., Di Luca M. Synaptic dysfunction in Alzheimer's disease. Adv. Exp. Med. Biol. 2012;970:573–601. doi: 10.1007/978-3-7091-0932-8_25. [DOI] [PubMed] [Google Scholar]
- Martin L., Latypova X., Wilson C.M., Magnaudeix A., Perrin M.L., Yardin C. Tau protein kinases: involvement in Alzheimer's disease. Ageing Res. Rev. 2013;12(1):289–309. doi: 10.1016/j.arr.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Mendez M.F. Early-onset Alzheimer's disease: nonamnestic subtypes and type 2 AD. Arch. Med. Res. 2012;43(8):677–685. doi: 10.1016/j.arcmed.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho R.M., Viana R.J., Low W.C., Steer C.J., Rodrigues C.M. Bile acids and apoptosis modulation: an emerging role in experimental Alzheimer's disease. Trends Mol. Med. 2008;14(2):54–62. doi: 10.1016/j.molmed.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Shapiro I.P., Masliah E., Saitoh T. Altered protein tyrosine phosphorylation in Alzheimer's disease. J. Neurochem. 1991;56(4):1154–1162. doi: 10.1111/j.1471-4159.1991.tb11405.x. [DOI] [PubMed] [Google Scholar]
- Sivachenko A.Y., Yuryev A., Daraselia N., Mazo I. Molecular networks in microarray analysis. J. Bioinform. Comput. Biol. 2007;5:429–456. doi: 10.1142/s0219720007002795. [DOI] [PubMed] [Google Scholar]
- Tung W.H., Hsieh H.L., Lee I.T., Yang C.M. Enterovirus 71 induces integrin β1/EGFR-Rac1-dependent oxidative stress in SK-N-SH cells: role of HO-1/CO in viral replication. J. Cell. Physiol. 2011;226(12):3316–3329. doi: 10.1002/jcp.22677. [DOI] [PubMed] [Google Scholar]
- Wang J., Cao H., Liao Y., Liu W., Tan L., Tang Y. Three dysconnectivity patterns in treatment-resistant schizophrenia patients and their unaffected siblings. Neuroimage Clin. 2015;8:95–103. doi: 10.1016/j.nicl.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pang T., Hafko R., Benicky J., Sanchez-Lemus E., Saavedra J.M. Telmisartan ameliorates glutamate-induced neurotoxicity: roles of AT(1) receptor blockade and PPARγ activation. Neuropharmacology. 2014;79:249–261. doi: 10.1016/j.neuropharm.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Dementia Fact sheet No. 362. March 2015. Archived from the original on 18 March 2015. Retrieved 13 January 2016.
- Xu J., Clark R.A. A three-dimensional collagen lattice induces protein kinase C-zeta activity: role in alpha2 integrin and collagenase mRNA expression. J. Cell Biol. 1997;136(2):473–483. doi: 10.1083/jcb.136.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.P., Yang S.L., Zhao S., Zheng C.H., Li H.H., Zhang Y. Biomarkers for early diagnostic of mild cognitive impairment in type-2 diabetes patients: a multicentre, retrospective, nested case-control study. EBioMedicine. 2016;5:105–113. doi: 10.1016/j.ebiom.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.J., Cho I.H., Park M., Cho E., Cho S.C., Kim B.N. Association between PTGS2 polymorphism and autism spectrum disorders in Korean trios. Neurosci. Res. 2008;62(1):66–69. doi: 10.1016/j.neures.2008.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.