Abstract
Increasing evidence suggests that perturbations in the intestinal microbiota in early infancy are implicated in the pathogenesis of food allergy (FA); existing evidence on the structure and composition of the intestinal microbiota in human beings with FA is limited and conflicting. The main object of the study was to compare the faecal microbiota between healthy and cow’s milk allergy (CMA) infants at the baseline immediately after the diagnosis, and to evaluate the changes in the faecal microbiota after 6 months of treatment of CMA infants with hypoallergenic formula (HF), compared with healthy children fed on standard milk formulae. Sixty infants younger than 4 months of age with challenge-proven CMA and 60 healthy age-matched children were investigated in this prospective case - control follow-up study. Faecal samples were collected at baseline and at 6 months of follow-up, microbial diversity and composition were characterized by high-throughput 16S rRNA sequencing. The average age (±SD) of the infants at inclusion was 2.9 ± 1.0 months. Children with CMA have lower gut microbiota diversity and an elevated Enterobacteriaceae to Bacteroidaceae (E/B ratio) in early infancy compared with healthy children (115.8 vs. 0.8, P = 0.0002). After 6 months of treatment with HF, CMA infants had a higher Lactobacillaceae (6.3% vs. 0.5%, P = 0.04) and lower Bifidobacteriaceae (0.3% vs. 8.2%, P = 0.03) and Ruminococcaceae (1.5% vs. 10.5%, P = 0.03) abundance compared with control children. Conclusion: Low gut microbiota diversity and an elevated E/B ratio in early infancy may contribute to the development of FA, including CMA. A strict elimination diet may weaken FA by reducing E/B ratio and promoting a gut microbiota that would benefit the acquisition of oral tolerance.
Keywords: Cow’s milk allergy, Infants, Gut microbiota, Diversity, 16S rRNA sequencing
1. Introduction
It has been estimated that the human gut is populated with up to 100 trillion microbes (Ley et al., 2006). Rough estimates are that the microbiota (previously termed flora or microflora) contain on the order of 150-fold more genes than are encoded in the human genome (Qin et al., 2010). A growing body of evidence suggests that gut microbiota plays an essential role in host health by processing energy from food, protecting intestinal epithelial cells from injury, and promoting local and systemic immunity (Hooper et al., 2012, Mazmanian et al., 2005), which has been extensively studied in recent years using culture-independent molecular methods. The next-generation DNA sequencing technologies, including high throughput 454 pyrosequencing, provide a large number of sequencing reads in a single run, resulting in a large sampling depth and the detection of low-abundance taxa. Thus, the results of studies using high-throughput sequencing technologies have revolutionized our understanding of the gut microbiota in healthy and disease conditions.
An increase in the prevalence of allergic diseases has been noted over the past few decades and 2–15% of the population worldwide is estimated to suffer from asthma, which can affect a high percent of children in some countries (Bousquet et al., 2004). A rising prevalence of food hypersensitivity and severe allergic reactions to food has also been reported. Cow’s milk allergy (CMA) is the most prevalent type of food allergy in childhood, with an incidence of 2–3% in the first year of life. An elimination diet that avoids the offending cow’s milk protein is the only available treatment (De Greef et al., 2012, Fiocchi et al., 2010). Current guidelines suggest the use of extensively hydrolyzed formula (eHF) as first-line dietary management of CMA in infancy and an amino acid-based formula (AAF) for management of complex CMA or when an eHF is not tolerated (De Greef et al., 2012), and they collectively known as hypoallergenic formula (HF).
So far a few studies (Azad et al., 2015, Bunyavanich et al., 2016, Ling et al., 2014, Thompson-Chagoyan et al., 2011, Thompson-Chagoyan et al., 2010) have assessed the fecal microbiota for infants with food allergy and/or hypersensitivity and postulated a possible association between allergy and an altered microbiota pattern. Only one Spanish cohort study (Thompson-Chagoyan et al., 2010) compared the faecal microbiota between healthy and CMA infants at the baseline, and evaluated the changes in the faecal microbiota after a period of treatment, but using a conventional bacterial culture method.
With this background, we performed high-throughput sequencing for V3-V4 hypervariable regions of the 16S rRNA gene from gut faecal material to compare faecal microbiota between healthy and challenge-proven CMA infants at baseline, immediately after the diagnosis and after 6 months of treatment with HF, compared with healthy children fed on standard milk formulae.
2. Methods
2.1. Subjects and study design
Sixty infants diagnosed with CMA from the Gastroenterology and Immunology clinics at the Children’s Hospital of Fudan University were recruited between January 2014 and September 2015. The children were less than 4 months of age and received no human milk. CMA (IgE or non-IgE-mediated) were confirmed by family allergic history, feeding history, clinical manifestation (acute severe reaction after cow’s milk ingestion combined with either cow’s milk-specific IgE > 5 kU/l or with SPT wheal diameter ≥ 3 mm, no confirmed history of cow’s milk protein reaction but with either SPT wheal diameter ≥ 6 mm, or cow’s milk associated allergic eosinophilic gastroenteritis or other non-IgE-mediated symptoms) and positive result of double-blind placebo-controlled food challenge with cow’s milk, of which the food challenge test was the main method to make definite diagnosis (Koletzko et al., 2012). The choice of an eHF or AAF was determined by the pediatrician in charge according to the severity of the infant’s symptoms, and a switch from eHF to AAF was considered if an eHF was not tolerated. The control group comprised 60 healthy age-matched infants recruited from our health care clinic.
These children had been exclusively breast fed until introduced to milk formulae at the same age as the paired CMA infants. The recommendations on solid food introduction were given to all the recruited children from 4 to 6 months old. The following exclusion criteria were established: allergic rhinitis; asthma; use of antibiotics, probiotics, prebiotics, or synbiotics in the previous month; and known active bacterial, fungal, and/or viral infection(s).
Written informed consent was obtained from the children’s parents. The study was approved by the Ethics Committee of the Children’s Hospital of Fudan University (Children’s Hospital of Fudan University Ethics Protocol 2013–033).
2.2. Sample collection and DNA extraction
At baseline and at 6 months of follow-up, faecal samples were collected at the hospital or by the parent at home. Samples were refrigerated during transport and stored at −80 °C until analysis.
DNA was extracted from 300 mg of feces using a QIAamp DNA stool mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The amount of DNA was determined using a Qubit® 2.0 Fluorometer (Life Technologies, USA); the integrity and size were checked by 1.5% agarose gel electrophoresis containing 0.5 mg/ml ethidium bromide. All DNA was stored at −20 °C until further analysis.
2.3. PCR and pyrosequencing
The bacterial genomic DNA was amplified with 343F (5′- TACGGRAGGCAGCAG-3′) and 798R (5′- AGGGTATCTAATCCT-3′) primers specific for the V3-V4 hypervariable regions of the 16S rRNA gene (Nossa et al., 2010). Barcodes that allowed sample multiplexing during pyrosequencing were incorporated between the 454 FLX Titanium adapter and the 5′ end of the forward primer. The thermocycling steps were as follows: 95 °C for 5 min, followed by 20 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 10 min. Each PCR reaction was performed in a 50 μL system, and the products were extracted with the QIAquick gel extraction kit (Qiagen) and quantified on a NanoDrop 2000C spectrophotometer and Qubit® 2.0 Fluorometer (Life technologies). Pooled PCR amplicons were subjected to paired-end sequencing by Illumina MiSeq. Using a QIIME pipeline (v 1.6.0, qiime.org), forward and reverse reads were assembled using PandaSeq for a final length of 144 bp (unassemblable sequences discarded), demultiplexed and filtered against the GREENGENES reference database (v12.10) to remove all sequences with <60% similarity. Remaining sequences were clustered with Usearch61 at 97% sequence similarity against the GREENGENES database (closed-picking algorithm), and taxonomic assignment was achieved using the RDP classifier constrained by GREENGENES. Operational taxonomic units (OTUs) with overall relative abundance below 0.0001 were excluded from subsequent analyses.
2.4. Bioinformatics and statistical analysis
Using default settings in QIIME, OTU relative abundance was summarized at the phylum, family and genus levels of taxonomy. The Shannon diversity index was used to measure the biodiversity in samples. Briefly, it is a test that takes into account the number of species and the evenness of the species, typically with a value between 1.5 and 3.5. It was calculated as -∑ log (pi) pi, where pi denotes the frequency of OTU i, and differences in this index were tested with the Mann-Whitney U test by using R software (http://www.r-project.org/). Microbiota community differences between samples (beta diversity) were tested by permutational multivariate analysis of variance (PERMANOVA) comparison of unweighted UNIFRAC (Lozupone and Knight, 2005) distance matrices, with 500 permutations. Median relative abundance of dominant taxa were compared by nonparametric Kruskal–Wallis test and Spearman rank correlation. As gut microbiota coexist in functional communities, ratios of specific taxa are commonly evaluated. We evaluated the ratio of Enterobacteriaceae to Bacteroidaceae (E/B ratio) as a measure of gut microbiota maturity as Proteobacteria (mainly Enterobacteriaceae) are prevalent in the early gut microbiota, while Bacteroidetes (mainly Bacteroidaceae) become dominant as the community matures towards an adult-like profile (Matamoros et al., 2013).
3. Results
3.1. Demographic and clinical data
The average age (±SD) of the infants at inclusion was 2.9 ± 1.0 months. The baseline characteristics of the enrolled infants are presented in Table 1. The symptoms that occurred most often in infants due to CMA were atopic eczema (61%) and gastrointestinal symptoms (25%), all of which were resolved within 4 weeks of HF feeding. Infants in both groups were introduced to solid foods at the same time as follows: cereal first, at ∼4.5 months; fruits and vegetables at ∼5.5 months; and meats at ∼7.0 months of age. After the 6-month follow-up, 47 (78.3%) infants with CMA were eHF-fed, and 13 (21.7%) were AAF-fed.
Table 1.
Descriptive data of children included in the study.
| Infants withCMA (n = 60) |
Control (n = 60) |
|
|---|---|---|
| Gender, male n (%) | 33 (55.0) | 31 (51.7) |
| Asian/others, (%) | 100.0/0 | 100.0/0 |
| Mode of delivery | ||
| Vaginal | 47 | 49 |
| Cesarean | 13 | 11 |
| Exclusive breast-feeding, mo | 2.6 ± 0.1 | 2.4 ± 0.1 |
| Age at study entry, mo | 3.0 ± 0.9 | 2.8 ± 1.2 |
| Weight at study entry, kg | 6.1 ± 0.6 | 6.4 ± 0.9 |
| Length at study entry, cm | 61.0 ± 2.8 | 61.8 ± 2.6 |
There were no significant differences between groups for all variables.
3.2. Faecal microbiota
Infants with CMA had a lower diversity of the total microbiota and the bacterial phylum Bacteriodetes and its genus Bacteroides at baseline than healthy controls (Table 2). After the 6-month follow-up, the diversity of the phylum Bacteriodetes was still significantly reduced in the CMA infants (Table 2). Furthermore, these phyla and genera differed significantly between CMA and healthy infants with repeated-measures ANOVA including the two sampling time points (baseline and 6 months: P = .045 for the total microbiota, P = .04 for Bacteroidetes, P = .03 for Bacteroides species).
Table 2.
Shannon diversity index of the total microbiota, dominant phyla, and significant genera in stool samples obtained at baseline and after 6 months of follow-up in healthy and CMA infants.
| Infants with CMA (n = 60) Median (IQR) |
Control (n = 60) Median (IQR) |
p value* | |
|---|---|---|---|
| Baseline | |||
| Total microbiota | 1.46 (1.17–1.67) | 1.70 (1.52–2.17) | 0.005 |
| Actinobacteria | 0.39 (0.14–0.47) | 0.41(0.19–0.68) | 0.35 |
| Firmicutes | 0.58 (0.37–1.21) | 0.60 (0.46–0.99) | 0.63 |
| Bacteroidetes | 0.05 (0.00–0.37) | 0.47 (0.07–0.61) | 0.03 |
| Bacteroides species | 0.01 (0.00–0.28) | 0.43 (0.09–0.51) | 0.01 |
| Proteobacteria | 0.19 (0.06–0.38) | 0.27 (0.15–0.35) | 0.34 |
| Six months | |||
| Total microbiota | 2.89 (2.23–3.32) | 2.72 (2.20–3.27) | 0.66 |
| Actinobacteria | 0.23 (0.13–0.42) | 0.19 (0.04–0.38) | 0.53 |
| Firmicutes | 2.02 (1.70–2.59) | 1.85 (1.48–2.40) | 0.17 |
| Bacteroidetes | 0.15 (0.02–0.37) | 0.51 (0.11–0.65) | 0.02 |
| Proteobacteria | 0.04 (0.01–0.07) | 0.07 (0.04–0.13) | 0.06 |
CMA, Cow’s milk allergy; IQR, Interquartile range.
Mann-Whitney U test.
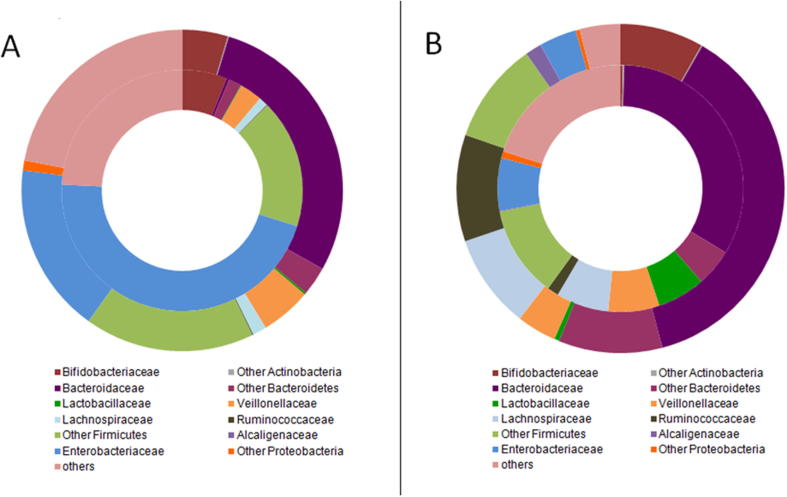
Gut microbiota composition at baseline and at 6 months differed between CMA and control infants (Table 3 and Fig. 1). Significant overall community differences at the OUT level of taxonomy were detected by PERMANOVA at baseline (PseudoF = 1.48, P = .03) and 6 months (PseudoF = 1.51, P = .04). Among dominant microbial families, Enterobacteriaceae was substantially and significantly overrepresented among CMA infants at baseline (median relative abundance 46.0% vs. 17.1%, P = .004). Conversely, Bacteroidaceae was underrepresented (0.4% vs. 28.4%, P = .01 at baseline). Given these differences, we compared the ratio of Enterobacteriaceae to Bacteroidaceae (E/B ratio) between CMA and control infants, finding a significant difference at baseline (115.8 vs. 0.8, P = .0002). After the 6-month follow-up, there was no further significant difference in neither relative abundance of Enterobacteriaceae, Bacteroidaceae nor E/B ratio between the two groups; however, the CMA infants tended to have a higher relative abundance of Lactobacillaceae (6.3% vs. 0.5%, P = .04) and lower relative abundance of Bifidobacteriaceae (0.3% vs. 8.2%, P = .03) and Ruminococcaceae (1.5% vs. 10.5%, P = .03) compared with healthy controls. At both sampling times, relative abundance of class Clostridia was comparable among allergic and non-allergic infants (P > .50, data not shown).
Table 3.
Mean of the relative abundance of dominant phyla and families in stool samples obtained at baseline and after 6 months of follow-up in healthy and CMA infants.
| Dominant taxa* | Baseline |
Six months |
||||||
|---|---|---|---|---|---|---|---|---|
| Infants with CMA (n = 60) Median (IQR) |
Control (n = 60) Median (IQR) |
p value | FDRp | Infants with CMA (n = 60) Median (IQR) |
Control (n = 60) Median (IQR) |
p value | FDRp | |
| Actinobacteria | 6.0 (0.3–22.2) | 4.7 (1.2–12.8) | 0.69 | 0.85 | 0.5 (0.0–1.7) | 8.3 (3.6–14.6) | 0.04 | 0.05 |
| Bifidobacteriaceae | 6.0 (0.0–22.1) | 4.6 (1.0–12.3) | 0.79 | 0.94 | 0.3 (0.0–1.6) | 8.2 (3.6–14.5) | 0.03 | 0.04 |
| Bacteroidetes | 2.1 (0.3–21.8) | 31.3 (0.4–61.3) | 0.02 | 0.05 | 38.1 (12.4–52.6) | 47.8 (40.0–61.3) | 0.09 | 0.25 |
| Bacteroidaceae | 0.4 (0.1–7.5) | 28.4 (0.1–57.8) | 0.01 | 0.04 | 33.2 (10.5–47.0) | 37.6 (29.5–55.8) | 0.09 | 0.25 |
| Firmicutes | 21.7 (13.3–36.4) | 23.9 (7.9–50.1) | 0.56 | 0.95 | 33.4 (25.6–57.1) | 34.2 (26.1–43.6) | 0.58 | 0.67 |
| Lactobacillaceae | 0.1 (0.0–2.1) | 0.2 (0.3–3.8) | 0.58 | 0.69 | 6.3 (0.6–11.2) | 0.5 (0.1–4.8) | 0.04 | 0.04 |
| Veillonellaceae | 3.0 (0.7–13.9) | 5.1 (0.9–17.7) | 0.51 | 0.80 | 6.7 (0.7–15.3) | 3.9 (1.5–9.2) | 0.51 | 0.55 |
| Lachnospiraceae | 1.2 (0.1–3.0) | 1.4 (0.1–7.5) | 0.44 | 0.69 | 6.9 (0.5–19.0) | 9.3 (3.9–20.5) | 0.33 | 0.57 |
| Ruminococcaceae | 0.1 (0.0–2.6) | 0.1 (0.0–2.2) | 0.87 | 0.87 | 1.5 (1.2–7.6) | 10.5 (2.4–13.9) | 0.03 | 0.03 |
| Proteobacteria | 46.0 (27.0–78.9) | 18.1 (8.7–37.0) | 0.006 | 0.01 | 7.9 (3.0–20.1) | 5.7 (2.5–8.6) | 0.21 | 0.33 |
| Alcaligenaceae | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.86 | 0.96 | 0.1 (0.0–2.3) | 1.6 (0.0–2.9) | 0.27 | 0.37 |
| Enterobacteriaceae | 46.0 (27.0–78.1) | 17.1 (6.6–35.9) | 0.004 | 0.01 | 6.9 (2.3–7.6) | 3.7 (0.3–5.2) | 0.09 | 0.22 |
| E/B Ratio | 115.8 (10.3–299.0) | 0.8 (0.2–154.4) | 0.0002 | 0.003 | 0.22 (0.07–16.69) | 0.10 (0.01–0.19) | 0.06 | 0.11 |
CMA, Cow’s milk allergy; IQR, Interquartile range; E/B, Enterobacteriacea/Bacteroidaceae; FDR, false discovery rate.
Dominant taxa have overall median relative abundance >1% at baseline and/or 6 months; phyla are in plain text and families are italicized. Comparisons by nonparametric Kruskal–Wallis test with FDR correction for multiple testing.
Fig. 1.
Mean relative abundance of dominant families (those with overall median relative abundance >1% at either sampling time). A: Baseline; B: After 6 months of follow-up. The outer circle represents gut microbiota of healthy infants, and the inner circle represents gut microbiota of infants with cow’s milk allergy (CMA).
4. Discussion
Bacterial diversity is widely considered to be an important factor in determining the stability of gut ecology to perturbation (Millar et al., 2003). The predominance of some bacteria (e.g. bifidobacteria) is beneficial, whereas the predominance of others (e.g. enterobacteriaceae and some pseudomonadaceae and clostridia species) is detrimental, and the balance among the populations of these micro-organisms maintains the health of the host. In speaking of allergic diseases, it is debated whether low diversity of the gut microbiota in early childhood is more important than the prevalence of specific bacterial taxa when trying to explain why the prevalence of allergic disease is increasing in relatively affluent countries. The underlying rationale is that the gut immune system reacts to exposure to new bacterial antigens and repeated exposure would enhance the development of immune regulation. Although this theory emerged more than two decades ago (Holt, 1995), there are still only a few studies relating this diversity with allergy, likely because of methodological limitations. A new generation of powerful noncultivation-based microbiologic methods has now made it possible to analyze the total microbiota down to the genus level, even in large cohorts (Azad et al., 2015, Turnbaugh et al., 2009). Previously uncultivated bacteria can now be detected, and there is no need to decide what bacteria to analyze in advance. Thus the assessment can be made without prejudice. This will allow for more comprehensive knowledge of the intestinal microbiota and its effects on the immune system.
So far by using the high-throughput 16S-based sequencing to study the early-life gut microbiome in infants with CMA is rare. Bunyavanich et al. (2016) characterized the gut microbiota of 226 children aged 3 to 16 months with CMA and followed these children up to age 8 years; they found that enrichment of Clostridia and Firmicutes as well as a trend of increased total bacterial diversity in the infant gut microbiome at age 3 to 6 months was associated with milk allergy resolution by age 8 years. In our current study, infants with CMA showed a lower diversity of the total microbiota at baseline than healthy controls, thus further support the hypothesis that low microbial diversity early in life is associated with an increased risk for CMA. Besides, we also showed that the differences in diversity were attributed to specific bacterial phyla and genera, possibly because the sensitivity of our analyses was higher. The low diversity of the phylum Bacteroidetes and its genus Bacteroides in infants with CMA is consistent with the results of previous studies reporting low levels of these bacteria to be associated both with allergic disease (Watanebe et al., 2003) and factors associated with allergic disease, such as a Western lifestyle (De Filippo et al., 2010) and cesarean section (Jakobsson et al., 2014). Bacteroides species have also been demonstrated to have anti-inflammatory properties. Thus Bacteroides fragilis prevented the induction of colitis through suppression of the proinflammatory cytokines TNF and IL-23 in an experimental colitis model (Mazmanian et al., 2008) and also mediated a conversion from CD4+ T cells into IL-10–producing forkhead box protein 3 regulatory T cells during commensal colonization, eliciting mucosal tolerance in another murine model (Round and Mazmanian, 2010). Furthermore, Bacteroides thetaiotaomicron modulates the expression of a large quantity of genes involved in mucosal barrier reinforcement (Hooper et al., 2001).
In terms of gut microbiota composition, our present study demonstrated that infants with CMA had a higher baseline abundance of Enterobacteriaceae and lower abundance of Bacteroidaceae as compared with paired healthy controls. The association of CMA with Enterobacteriaceae and Bacteroidaceae was particularly evident when the ratio of these taxa was evaluated. The E/B ratio could be considered an indicator of gut microbiota maturity, as normal development involves early colonization by facultative anaerobes (predominantly Enterobacteriaceae), which deplete initial oxygen supplies to create a favourable environment for subsequent colonization by anerobes including Bacteroidaceae (Davis et al., 2017). Thus, the E/B ratio is expected to decline with age as relative proportions of Enterobacteriaceae decline and Bacteroidaceae become more dominant, reflecting maturation toward an adult-like gut microbiota (Bergström et al., 2014, Yatsunenko et al., 2012). Our finding that the E/B ratio is elevated among CMA infants suggest that delayed maturation of the gut microbiota may be a predictor of CMA. After 6 months of nutritional intervention, we indentified a significant increase in the relative abundance of Lactobacillaceae and a parallel reduction in Bifidobacteriaceae and Ruminococcaceae, and a disappearance of the difference in the abundance of Enterobacteriaceae and Bacteroidaceae between allergic and healthy infants observed at baseline. Our findings are partially consistent with evidence from Thompson-Chagoyan et al.(2010), by using bacterial culture, they found CMA infants had a higher count and proportion of lactobacilli, a lower count and proportion of bifidobacteria and lower proportions of enterobacteria and yeasts after 6 months of eHF feeding. All of these changes may be attributable to the almost complete avoidance of cow’s milk proteins during the 6 months of treatment, when the CMA infants received a HF (eHF or AAF) formula. Nevertheless, eHF does not completely eliminate cow’s milk antigens (Høst and Halken, 2004) and an AFF-feeding infants may intake at least some degree of cow’s milk proteins from solid food or other oral nutrition supplementation, the constant administration of these antigens favors the acquisition of oral tolerance (Longo et al., 2008), as observed in chronic intestinal parasitosis (Thompson-Chagoyán et al., 2005). This tolerance is reflected in the reduction in specific immunoglobulin concentrations in the serum of the CMA infants at 6 months of follow-up. This was accompanied by a major decrease in the amount of Enterobacteriaceae and Bifidobacteriaceae and an increase in Lactobacillaceae, with a significant improvement in or disappearance of clinical symptoms. Besides, we were the first to find a decreased relative abundance of Ruminococcaceae in faecal samples of CMA infants at 6 months of follow-up, as compared with that in matched controls. It is reported that members of these families stimulate the production and degradation of mucin, which is required to maintain an intact gut microbiota–mucin barrier (McGuckin et al., 2011), the association of decline in Ruminococcaceae abundance and the clinical outcome of CMA need to be further investigated.
Our results should be interpreted with caution, as with all faecal microbiota studies, it is necessary to consider that microbiota colonizing the gut mucosa may not be accurately reflected by the communities observed in stool, although Centanni et al. (2013) have recently reported that (contrary to adults) the phylogenetic structures of faecal and enterocyte-associated microbiota are remarkably similar in infants.
In conclusion, our study showed that low gut microbiota diversity and an elevated E/B ratio in early infancy may contribute to the development of food allergy, including CMA. The observed differences in phylum and families between cases and controls may provide insight into the link between the microbiome and food allergy outcomes in childhood. Further studies are needed to confirm these findings.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Children’s Hospital of Fudan University (Children’s Hospital of Fudan University Ethics Protocol 2013–033). Written informed consent was obtained from the children’s parents.
Acknowledgments
Acknowledgements
The authors would like to acknowledge all the staff of Tong An Pharmacy of Children’s Hospital of Fudan University for registering the potential patients for us; we also want to express our gratitude to Shanghai Hanyu Bio-Technology Limited to provide bacteria sequencing for us.
Author’s contributions
PD and XX conceived the study and participated in its design and coordination; PD, JJF, DYY and YJL performed the recruitment, anthropometric measurements and sample collection; PD performed the DNA extraction and bioinformatics analyses; PD collected the data and performed the statistical analysis; PD wrote the manuscript; XX revised the manuscript; and all authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (to Ping Dong) (grant number 81302436).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Ping Dong, Email: dongpingis@163.com.
Jing-jing Feng, Email: 13788944690@163.com.
Dong-yong Yan, Email: yandongyong_88@aliyun.com.
Yu-jing Lyu, Email: 504877260@qq.com.
Xiu Xu, Email: xuxiu@fudan.edu.cn.
References
- Azad M.B., Konya T., Guttman D.S., Field C.J., Sears M.R., HayGlass K.T., Mandhane P.J., Turvey S.E., Subbarao P., Becker A.B. Infant gut microbiota and food sensitization: associations in the first year of life. Clin. Exp. Allergy. 2015;45:632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- Bousquet J., Ansotegui I.J., van Ree R., Burney P.G., Zuberbier T., van Cauwenberge P. European Union meets the challenge of the growing importance of allergy and asthma in Europe. Allergy. 2004;59:1–4. doi: 10.1111/j.1398-9995.2004.00425.x. [DOI] [PubMed] [Google Scholar]
- Bunyavanich S., Shen N., Grishin A., Wood R., Burks W., Dawson P., Jones S.M., Leung D.Y.M., Sampson H., Sicherer S., Clemente J.C. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016;138:1122–1130. doi: 10.1016/j.jaci.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A., Skov T.H., Bahl M.I., Roager H.M., Christensen L.B., Ejlerskov K.T., Mølgaard C., Michaelsen K.F., Licht T.R. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014;80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanni M., Turroni S., Consolandi C., Rampelli S., Peano C., Severgnini M., Biagi E., Caredda G., De Bellis G., Brigidi P., Candela M. The enterocyte-associated intestinal microbiota of breast-fed infants and adults responds differently to a TNF-alpha-mediated proinflammatory stimulus. PLoS One. 2013;8:e81762. doi: 10.1371/journal.pone.0081762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greef E., Hauser B., Devreker T., Veereman-Wauters G., Vandenplas Y. Diagnosis and management of cow’s milk protein allergy in infants. World J. Pediatr. 2012;8:19–24. doi: 10.1007/s12519-012-0332-x. [DOI] [PubMed] [Google Scholar]
- De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J.B., Massart, S., Collini, S., Pieraccini, G., Lionetti, P., 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A.107, 14691–14696. [DOI] [PMC free article] [PubMed]
- Davis E.C., Wang M., Donovan S.M. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut. Microbes. 2017;8:143–171. doi: 10.1080/19490976.2016.1278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiocchi A., Brozek J., Schünemann H., Bahna S.L., von Berg A., Beyer K., Bozzola M., Bradsher J., Compalati E., Ebisawa M. World allergy organization (WAO) diagnosis and rationale for action against cow’s milk allergy (DRACMA) guidelines. Pediatr. Allergy Immunol. 2010;21:1–125. doi: 10.1111/j.1399-3038.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Wong M.H., Thelin A., Hansson L., Falk P.G., Gordon J.I. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Holt P.G. Postnatal maturation of immune competence during infancy and childhood. Pediatr. Allergy Immunol. 1995;6:59–70. doi: 10.1111/j.1399-3038.1995.tb00261.x. [DOI] [PubMed] [Google Scholar]
- Høst A., Halken S. Hypoallergenic formulas-when, to whom and how long: after more than 15 years we know the right indication. Allergy. 2004;59:45–52. doi: 10.1111/j.1398-9995.2004.00574.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson H.E., Abrahamsson T.R., Jenmalm M.C., Harris K., Quince C., Jernberg C., Björkstén B., Engstrand L., Andersson A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- Koletzko S., Niggemann B., Arato A., Dias J.A., Heuschkel R., Husby S., Mearin M.L., Papadopoulou A., Ruemmele F.M., Staiano A. Diagnostic approach and management of cow’s-milk protein allergy in infants and children: ESPGHAN GI Committee practical guidelines. J. Pediatr. Gastroenterol Nutr. 2012;55:221–229. doi: 10.1097/MPG.0b013e31825c9482. [DOI] [PubMed] [Google Scholar]
- Ley R.E., Peterson D.A., Gordon J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ling Z., Li Z., Liu X., Cheng Y., Luo Y., Tong X., Yuan L., Wang Y., Sun J., Li L., Xiang C. Altered fecal microbiota composition associated with food allergy in infants. Appl. Environ. Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo G., Barbi E., Berti I., Meneghetti R., Pittalis A., Ronfani L., Ventura A. Specific oral tolerance induction in children with very severe cow’s milk induced reactions. J Allergy Clin Immunol. 2008;121:343–347. doi: 10.1016/j.jaci.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Liu C.H., Tzianabos A.O., Kasper D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Matamoros S., Gras-Leguen C., Le Vacon F., Potel G., de La Cochetiere M.F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013;21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Millar, M., Wilks, M., Costeloe, K., 2003. Probiotics for preterm infants? Arch Dis Child Fetal Neonatal Ed.88, F354–358. [DOI] [PMC free article] [PubMed]
- Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- McGuckin M.A., Lindén S.K., Sutton P., Florin T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Nossa C.W., Oberdorf W.E., Yang L., Aas J.A., Paster B.J., Desantis T.Z., Brodie E.L., Malamud D., Poles M.A., Pei Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010;16:4135–4144. doi: 10.3748/wjg.v16.i33.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. Inducible Foxp31 regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Chagoyan O.C., Vieites J.M., Maldonado J., Edwards C., Gil A. Changes in faecal microbiota of infants with cow’s milk protein allergy - a Spanish prospective case - control 6-month follow-up study. Pediatr. Allergy Immunol. 2010;21:e394–400. doi: 10.1111/j.1399-3038.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- Thompson-Chagoyan O.C., Fallani M., Maldonado J., Vieites J.M., Khanna S., Edwards C., Doré J., Gil A. Faecal microbiota and short-chain fatty acid levels in faeces from infants with cow’s milk protein allergy. Int. Arch. Allergy Immunol. 2011;156:325–332. doi: 10.1159/000323893. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Chagoyán O.C., Maldonado J., Gil A. Aetiology of inflammatory bowel disease (IBD): role of intestinal microbiota and gut-associated lymphoid tissue immune response. Clin. Nutr. 2005;24:339–352. doi: 10.1016/j.clnu.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Watanebe S., Narisawa Y., Arase S., Okamatsu H., Ikenaga T., Tajiri Y., Kumemura M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J. Allergy Clin. Immunol. 2003;111:587–591. doi: 10.1067/mai.2003.105. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]