Abstract
In this paper, we propose a new hybrid method based on Correlation-based feature selection method and Artificial Bee Colony algorithm,namely Co-ABC to select a small number of relevant genes for accurate classification of gene expression profile. The Co-ABC consists of three stages which are fully cooperated: The first stage aims to filter noisy and redundant genes in high dimensionality domains by applying Correlation-based feature Selection (CFS) filter method. In the second stage, Artificial Bee Colony (ABC) algorithm is used to select the informative and meaningful genes. In the third stage, we adopt a Support Vector Machine (SVM) algorithm as classifier using the preselected genes form second stage. The overall performance of our proposed Co-ABC algorithm was evaluated using six gene expression profile for binary and multi-class cancer datasets. In addition, in order to proof the efficiency of our proposed Co-ABC algorithm, we compare it with previously known related methods. Two of these methods was re-implemented for the sake of a fair comparison using the same parameters. These two methods are: Co-GA, which is CFS combined with a genetic algorithm GA. The second one named Co-PSO, which is CFS combined with a particle swarm optimization algorithm PSO. The experimental results shows that the proposed Co-ABC algorithm acquire the accurate classification performance using small number of predictive genes. This proofs that Co-ABC is a efficient approach for biomarker gene discovery using cancer gene expression profile.
Keywords: Gene expression profile, Gene selection method, CFS, Cancer classification, Artificial bee colony, ABC, Correlation-based feature selection
1. Introduction
Gene expression profile or microarray data can be used to identify which genes are being expressed in a given cell type at a particular time and under particular conditions. This very helpful to compare the gene expression in two different cell types or tissue samples. Therefore, we can select the most informative and predictive genes that are responsible for causing a specific disease or cancer (Alshamlan et al., 2013, Alba et al., 2007). However, gene expression profile is conceded as high dimensional dataset. In other word, it suffers from the curse of dimensionality, the small number of samples, and the level of irrelevant and noise genes, all of which makes the classification task for a given sample more challenging (Alshamlan et al., 2013, Ghorai et al., 2010, Sheng-Bo et al., 2006).
In this paper, we developed a new gene selection method to select the smallest subset of informative genes that are most predictive to its relative class using a classification model. In addition our new algorithm aim to determine the genes that contribute the most to cancer diagnosis, which would assist in drug discovery and early diagnosis, and increase the classifier’s ability to classify new samples accurately.
The artificial bee colony (ABC) algorithm is an effective meta-heuristic algorithm that was invented in 2005 by Karaboga (2005). ABC algorithm was inspired by the social life of bees and is used to look for an optimal solution in numerical optimization problems (Karaboga, 2005). Becouse its simplicity and ease of implementation, the ABC algorithm; it has been widely applied in many optimization applications such as protein tertiary structures (Bahamish et al., 2009), digital IIR filters (Karaboga, 2009), artificial neural networks (Karaboga and Akay, 2005) and others. However, the ABC algorithm suffer from major critical problems, which is shared and similar to other evolutionary algorithms. Especially in computational efficiency, when it is applied to high dimensional dataset such as gene expression profile.
In our previous researches, we applied ABC algorithm with support vector machine (SVM) classifier in order to generate new wrapper feature selection method, which is named ABC-SVM (Alshamlan et al., 2016). Also, we developed a new hybrid feature selection algorithm based on ABC, called mRMR-ABC (Alshamlan et al., 2015). In mRMR -ABC, we successfully combined minimum Redundancy Maximum Relevance (mRMR) filter algorithm with ABC algorithm in order to select informative genes that are minimum redundancy for other genes and maximum relevancy for specific cancer classe. This hybrid mRMR-ABC gene selection algorithm offers a good tradeoff between computationally effective and feature dependencies. However, we did not get high classification accuracy with some of microarray dataset.
In order to solve this problem and further improve the performance of the ABC algorithm. In this paper, we propose combined a Correlation-based Feature Selection (CFS) filtering method, as a preprocessing stage, with ABC algorithm. It worth mentioning that CFS can be effectively combined with other feature selectors, such as wrappers. This can be done to find a very compact subset from candidate features at lower expense. Chuang et al. (2011) proposed a novel hybrid gene selection method by combining the CFS and TGA methods. The experimental results for both binary and multi-class cancer microarray datasets show that the proposed method reduced the dimensionality of microarray datasets by illuminating the redundant genes and achieved high classification accuracy.
In addition, Yang et al. (2008) proposed an improved binary particle swarm optimization (IBPSO), which further developed the standard BPSO. The filter methods applied in this study were information gain (IG) and correlation-based feature selection (CFS). The authors used the Weka software package (Quinlan, 1986) to determine the information value of each feature and to sort the features in accordance with their information gain value. The wrapper method that was adopted in this study was IBPSO. The authors evaluated the performance of these hybrid methods using a leukemia dataset. The experimental results indicated that CFS with IBPSO achieved a minimum number of selected genes, while IG with IBPSO produced high classification accuracy.
In our proposed algorithm, which is named Co-ABC, we select the genes that have maximal correlation between genes and class and have minimal correlation between gene to gene. This step will reduced the dimensional of microarray dataset, because we will identify and select the relevant and informative genes only. After that, we applied ABC algorithm for those selected genes in order to select small number of predictive genes. Then, we will measure the efficiency of the selected genes using a support vector machine (SVM) as a classifier. We used an SVM classifier because its displayed substantial benefits when compared to other classification approaches (Alshamlan et al., 2014). In addition, it addresses this problem by mapping the input space into a high-dimensional feature (gene) space. After that, it generates a linear classification decision to classify the initial dataset (microarray dataset) with a maximum margin hyperplane. An SVM is more efficient, very accurate, and faster than other machine learning methods, such as Neural Networks (NN) and k-Nearest Neighbour (K-NN) classifiers when they applied with gene expression profile (Wang and Gotoh, 2009).
The Co-ABC algorithm is tested using six gene expression profile for binary and multi-class cancer datasets. Also, it compared with our previous proposed algorithms ABC-SVM (Alshamlan et al., 2016), and mRMR-ABC (Alshamlan et al., 2015). In addition, in order to proof the efficiency of our proposed Co-ABC algorithm, we compare it with previously known related methods. Two of these methods was re-implemented for the sake of a fair comparison using the same parameters. These two methods are: Co-GA, which is CFS combined with a genetic algorithm GA. The second one named Co-PSO, which is CFS combined with a particle swarm optimization algorithm PSO. Furthermore, Co-ABC was compared with other related and recently published algorithms. The experimental results show improvements in both the number of selected informative genes and cancer classification accuracy.
The rest of this paper is organized as follows: Section 2 provides a description of proposed Co-ABC algorithm. Section 3 outlines the experimental setup and provides results. Finally, Section 4 concludes our paper.
2. The proposed Co-ABC algorithm
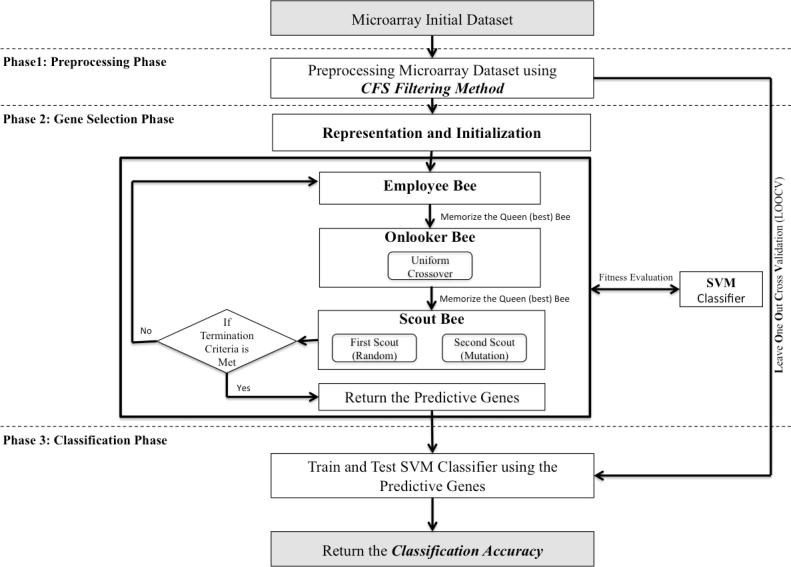
In this section, we present the proposed Co-ABC algorithm for elect the highly informative genes from the cancer gene expression profile. As shown in Fig. 1, Co-ABC consists of three main phases: preprocessing phase, gene selection phase, and classification phase. In the following sub section, we introduce the function of each phase.
Fig. 1.
The main phases and steps of the proposed Co-ABC algorithm.
2.1. Preprocessing phase: Correlation-based Feature Selection (CFS) filter method
Correlation-based feature selection (CFS) scores (and ranks) the worth of subsets of features according to a correlation-based heuristic evaluation function, rather than scoring (and ranking) individual features (Yvan et al., 2007). As microarray feature (genes) space is usually huge, CFS uses a best-first-search heuristic that takes into account the usefulness of individual features for predicting the class. Therefore, CFS selects the subset that has maximal correlation to the class, and minimal correlation between features (Yvan et al., 2007). CFS first calculates a matrix of (feature to class) and (feature to feature) correlations from the training data. Then, a score for the subset of features assigned by the heuristic is calculated using Eq. (1).
| (1) |
where is the heuristic merit of a feature subset S containing k features, is the average correlation between features and class, and is the average correlation between features.
CFS starts from the empty set of features and then the subset with the highest Merit found during the search will be selected. In our problem, genes which have correlation with specific cancer classes should be selected using the CFS method. As microarray feature (genes) space is usually huge, CFS uses a best-first-search heuristic that takes into account the usefulness of individual features for predicting the class.
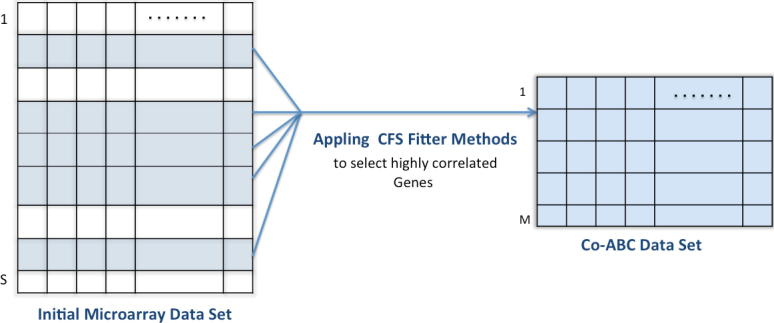
The main purpose of applying the CFS gene selection method is to find the highly correlated subset of genes from initial microarray dataset. As illustrate in Fig. 2, the initial gene expression profile is preprocessed using the CFS filtering method. Each gene is evaluated and sorted based one CFS criteria as explained previously in this section. The highly correlated genes that give high classification accuracy with an SVM classifier will selected to create a new subset named the CFS dataset. Suppose the initial microarray dataset contains S genes, as shown in Fig. 2. After applying CFS filter method, the number of genes will be reduced to m genes that have the maximal correlation between genes to class and minimal correlation between gene to gene.
Fig. 2.
Co-ABC dataset which is contains the highly correlated genes m selected by the CFS filter approach.
Our main objective is to maximize the classification accuracy and reduce the number of informative genes. Hence, we adopted the CFS filter methods as a preprocessing step for ABC algorithm to enhance the speed and classification accuracy performance of the search. In addition, in order to eliminate the irrelevant genes and filter the noisy genes, which is reduces the computational complexity for the ABC algorithm and SVM classifier as well.
2.2. Gene selection phase: Artificial Bee Colony (ABC) algorithm
In this phase, we apply ABC algorithm to elect and identify the most informative and predictive genes from an CFS dataset that achieve the highest classification accuracy with an SVM classifier.The ABC algorithm is a meta-heuristic evolutionary algorithm that simulates the search for food in a group of bees. An ABC algorithm is implemented as presented in our prevues research article (Alshamlan et al., 2015).
This section will present briefly the main step of ABC algorithm and how can we apply them in gene selection for gene expression data analysis, which is already explained in more detail in our prevues research article (Alshamlan et al., 2015).
2.2.1. Employed bee step
In this step, the employee bees looking around the solutions (food resources) at in order to find the best genes index at the new location . We determined the new gene index using following equation (Xiang and An, 2013):
| (2) |
where is the new gene index (the location vector in the artificial bees), is the current gene index (the location vector of the bee), is a right random number in , and the SN is the number of artificial bees, which considers as a solution in our problem. is a random number that is distributed randomly between . The selection of random numbers from the CFS dataset index is done using the following equation (Xiang and An, 2013):
| (3) |
where and are the high (up) limit and the low (down) limit of the variable respectively, , and . While, is the random selected numbers between . When the new selected indes of the gene is determined, the optimization of it must be computed based on the fitness function. In this problem, our fitness value identified based on the solution classification accuracy using an SVM classifier. If the new fitness value is greaterr than the fitness value acquired thus far, then the bee moves to the new solution (i.e,food source) leaving the old one; otherwise it stay in the old one.
2.2.2. Onlooker bee step
After the employed bees complete there task, looking for the best solutions, they shared the information with onlooker bees. Then, an onlooker bee selects the genes depending on their winning probability value that is very similar to roulette wheel selection in genetic algorithm (GA) as follows: the possibility of selecting the particular solution (food source) by the onlooker bees is calculated using the following equation:
| (4) |
2.2.3. Scout bee step
Each employee or onlooker bee will search for best genes (solution) for a certain number and limited number of cycles. If the fitness value for any employee or looked bee does not improve, then that particular bee becomes a scout bee. In other word, a solution which could not be improved through “limit” number of trials becomes a scout bee. A scout bee select an index of genes randomly from CFS dataset (search space).
It is worth mentioning that the ABC algorithm suffer from some critical issues, especially in computational and speed efficiency, when it is applied on huge and high dimensional dataset such as a cancer gene expression profile. This motivates us to solve these difficulties and further enhance the performance of the ABC algorithm by proposing a hybrid technique between the ABC algorithm and mRMR filter gene selection approach, namely, the mRMR-ABC algorithm. In the following subsection, we explain the mRMR algorithm when applied to our problem.
2.3. Classification phase: Support Vector Machine (SVM) classifier
Support vector machines (SVM) is a classification algorithm that belongs to a new generation of learning system based on recent advances in statistical learning theory (Vapnik, 1998). A SVM is very effective classification algorithm that showed a good performance in a variety of computational biological classification tasks. It worth mentioning that SVM-based classifiers are becoming increasingly popular classifiers for gene expression profile. Support vector machine is very helpful in cancer diagnostic models, where the number of feature, which is genes in our problem, is so large in relative to the sample size. Because, it has the ability to be fitted with all genes and at the same time with stable performance when using the full set of genes (Alonso et al., 2012, Huerta et al., 2006, Lee and Leu, 2011, Mukherjee, 2003). Also, SVM is aim is to identify the hyperplane that is separating the feature with the largest margin (distance between itself and the closest samples from each classes). Generally, the effective SVM classifier seeks to a trade-off between increasing the margin and decreasing the number of errors.
In our proposed Co-ABC algorithm, we use the informative and predictive genes that are predicted from the second phase to train the SVM classifier. Then, the SVM is applied again to classify the testing gene expression dataset and restore the classification accuracy.
The main steps for the proposed Co-ABC algorithm illustrated in Fig. 1. In addition, the pseudo code for the proposed Co-ABC algorithm is demonstrated in Algorithm 1.
Algorithm 1
Co-ABC algorithm
1: Select the maximum relevant genes subset using CFS filter method that achieve highly classification accuracy with SVM Classifier from initial microarray dataset. 2: ABC parameters setting, include maximum cycles, bee colony size and limited trail. 3: Initialize ABC food sources randomly. 4: Food sources quality evaluation using fitness calculation, which is SVM classification accuracy. 5: 6: WhileDo 7: Generate new employed bees (new candidate solutions) 8: New solution quality evaluation using fitness calculation. 9: Adopt greedy selection approche. 10: Determine the probability values by using fitness values. 11: Generate new onlooker bees (new candidate solutions) using the probability of food source. 12: New solution quality evaluation using fitness calculation. 13: Adopt greedy selection process. 14: Identify abandoned solutions and produce new solutions randomly using scout bee. 15: Identify and save the best solution found so far. 16: 17: End While 18: Generate and return best solution (predictive and biomarker genes). 19: Train the SVM classifier algorithm using generated biomarker genes. 20: Classify gene expression profile using SVM classifier. 21: Calculate the classification accuracy
3. Experimental setup and results
3.1. Experiential setup
In this section, we evaluated the overall performance of the Co-ABC algorithm using six more useful benchmark binary and multi-class microarray cancer datasets, which we used to evaluate our previously proposed algorithms ABC-SVM (Alshamlan et al., 2016), and mRMR-ABC (Alshamlan et al., 2015). The binary-class microarray datasets are: colon (Alon et al., 1999), leukemia (Golub et al., 1999), and lung (Beer et al., 2002). Where the multi-class microarray datasets are: SRBCT (Khan et al., 2001), lymphoma (Alizadeh et al., 2000), and leukemia (Armstrong et al., 2001). Table 2 shows a detailed description of these six popular cancer microarray datasets with illustration of number of classes, number of samples, number of genes, and a brief description of each dataset construction.
Table 2.
The cancer microarray datasets statistical values.
| Microarray datasets | No of classes | No of samples | No of genes | Description |
|---|---|---|---|---|
| Colon Alon et al. (1999) | 2 | 62 | 2000 | 40 cancer samples and 22 normal samples |
| Leukemia1 Golub et al. (1999) | 2 | 72 | 7129 | 25 AML samples and 47 ALL samples |
| Lung Beer et al. (2002) | 2 | 96 | 7129 | 86 cancer samples and 10 normal samples |
| SRBCT Khan et al. (2001) | 4 | 83 | 2308 | 29 EWS cancer samples, 18 NB cancer samples, 11 BL cancer samples, and 25 RMS cancer samples |
| Lymphoma Alizadeh et al. (2000) | 3 | 62 | 4026 | 42 DLBCL cancer samples, 9 FL cancer samples, and 11 B-CLL cancer samples |
| Leukemia2 Armstrong et al. (2001) | 3 | 72 | 7129 | 28 AML sample, 24 ALL sample, and 20 MLL samples |
In order to make fair evaluation and comparison, we applied same control parameters that have been used for our previous proposed algorithms. Table 1 shows the control parameters for the Co-ABC algorithm applied in this research. The first control parameter is the bee colony size or population size. The second one is the maximum cycle or maximum number of generations. The third control parameter is the number of runs, we use it a stopping criteria. The fourth control parameter is the non-improved limit, which is means the number of iterations allowed when the food source is not enhanced (i.e, exhausted). If the food source(bee) exceeds this limit value, it will become as scout bee.
Table 1.
The control parameters for Co-ABC algorithm.
| Control parameter | Value |
|---|---|
| 80 | |
| 100 | |
| 30 | |
| 5 |
In this research paper, we tested the performance of the proposed Co-ABC algorithm by comparing it with our previously proposed algorithms ABC-SVM (Alshamlan et al., 2016), and mRMR-ABC (Alshamlan et al., 2015) using two parameters: the first one is the classification accuracy and the second is the number of predictive and informative genes that have been applied in cancer classification task. Moreover, we adopt leave-one-out cross-validation (LOOCV) (Ng et al., 1997) in order to evaluate the performance of Co-ABC and the existing methods in the literature. We applied LOOCV in this study, because it is more suitable to our research problem, and it has the capability to prevent and reduce the “overfitting” problem (Ng et al., 1997).
3.2. Experimental results
In this section, we illustrate and evaluate the results that are generated from Co-ABC algorithm. First, we applied CFS filter method to select the highly correlated genes that acquire accurate classification result using SVM classifier. From Table 3, we can observe that the most correlated 80 genes from leukemia1 dataset achieve 100% accuracy. For colon cancer dataset, we can generate 91.94% classification accuracy by 25 genes. While, in the lung cancer dataset, we got 100% with 71 genes and 110 correlated genes to achieve the same performance percentage for the SRBCT dataset. Also, by selecting 184 highly correlated genes from the lymphoma cancer dataset and 103 correlated genes from the leukemia2 cancer dataset, we generated 100% as classification performance.
Table 3.
The CFS with an SVM classification performance.
| Microarray datasets | Number of genes | Classification accuracy |
|---|---|---|
| Colon | 25 | 91.94% |
| Leukemia1 | 80 | 100% |
| Lung | 71 | 100% |
| SRBCT | 110 | 100% |
| Lymphoma | 184 | 100% |
| Leukemia2 | 103 | 100% |
When we compared the classification accuracy performance of CFS with mRMR (Alshamlan et al., 2015) using SVM classifier. We noted that the CFS achieved same classification accuracy used small number of genes exempt the colon and lymphoma datasets. In colon dataset, CFS achieved lass classification accuracy than mRMR, which is 91.94%. While in lymphoma dataset, CFS used more genes to generate 100% classification accuracy.
After employing CFS filter method, we used these highly correlated genes as input in the ABC algorithm to select the predictive and informative genes from these correlated ones. In addition, we compare the performance of the proposed Co-ABC algorithm with the previously proposed ABC-based algorithm, ABC-SVM (Alshamlan et al., 2016), and mRMR-ABC (Alshamlan et al., 2015), with the similar number of selected genes for all six benchmark gene expression microarray datasets. The performance comparison for binary-class cancer dataset, which are colon, leukemia1, and lung are presented in Table 4, Table 5, Table 6, respectively. While, Table 7, Table 8, Table 9, respectively, show the performance comparison for multi-class cancer datasets, which are SRBCT, lymphoma, and leukemia2. As demonstrated on these tables, we noted that Co-ABC algorithm performs better than mRMR-ABC algorithm and ABC-SVM algorithm for all cancer datasets (binary or multi- classes) with different number of predictive genes. In addition, in order to make our experiments statistically validated, we apply each experiment 30 times for all cancer dataset. Furthermore, the best, worst, and average classification accuracies results of the 30 independent runs are computed to evaluate the performance of Co-ABC algorithm. The values obtained for classification accuracy are further analyzed using the Kruskal–Wallis test (Kruskal and Wallis, 1952) for statistical significance. The p-values obtained for classification accuracy and discovered informative genes is 0.0063, which indicates the statistical significance of the results.
Table 4.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for Colon cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 3 | 90.32% | 90.16% | 88.71% | 88.71% | 87.50% | 85.48% | 87.10% | 85.91% | 83.87% |
| 4 | 91.94% | 91.34% | 90.32% | 90.23% | 88.27% | 87.10% | 87.10% | 86.71% | 85.48% |
| 5 | 91.94% | 91.94% | 91.94% | 91.94% | 89.50% | 87.10% | 90.32% | 87.98% | 85.48% |
| 6 | 93.55% | 92.42% | 91.94% | 91.94% | 90.12% | 87.10% | 90.32% | 88.44% | 85.48% |
| 7 | 95.16% | 93.55% | 91.94% | 93.55% | 91.64% | 88.81% | 91.94% | 90.20% | 88.81% |
| 8 | 95.16% | 94.25% | 93.55% | 93.55% | 91.80% | 88.81% | 91.94% | 90.61% | 88.81% |
| 9 | 96.77% | 94.62% | 93.55% | 93.55% | 92.11% | 90.16% | 91.94% | 90.95% | 88.81% |
| 10 | 96.77% | 94.68% | 93.55% | 93.55% | 92.74% | 90.16% | 93.55% | 91.31% | 88.81% |
| 15 | 95.16% | 94.95% | 93.55% | 96.77% | 93.60% | 91.93% | 93.55% | 91.38% | 90.32% |
| 20 | 95.16% | 93.44% | 91.94% | 96.77% | 94.17% | 91.93% | 95.61% | 92.44% | 90.32% |
Table 5.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for Leukemia1 cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 2 | 97.22% | 97.22% | 97.22% | 91.66% | 89.63% | 81.94% | 87.5% | 86.45% | 81.94% |
| 3 | 100% | 99.58% | 98.61% | 93.05% | 90.37% | 83.33% | 88.88% | 89.82% | 83.33% |
| 4 | 100% | 100% | 100% | 94.44% | 91.29% | 86.11% | 88.8% | 91.15% | 83.33% |
| 14 | 100% | 100% | 100% | 100% | 95.83% | 93.05% | 93.05% | 92.51% | 88.88% |
Table 6.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for Lung cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 2 | 97.91% | 97.91% | 97.91% | 96.87% | 95.83% | 93.75% | 88.54% | 87.5% | 84.37% |
| 3 | 100% | 100% | 100% | 97.91% | 96.31% | 93.75% | 89.58% | 88.54% | 84.37% |
| 4 | 100% | 100% | 100% | 98.95% | 97.91% | 96.87% | 91.66% | 89.58% | 87.5% |
| 8 | 100% | 100% | 100% | 100% | 98.95% | 96.87% | 97.91% | 93.75% | 91.66% |
Table 7.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for SRBCT cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 2 | 77.11% | 77.03 % | 75.90 % | 75.90% | 71.08% | 68.67% | 72.28% | 69.87% | 67.46% |
| 3 | 89.16% | 86.51% | 83.13% | 85.54% | 79.51% | 71.08% | 73.34% | 71.08% | 68.67% |
| 4 | 100% | 95.82% | 92.77% | 87.95% | 84.33% | 77.10% | 84.33% | 81.92% | 77.10% |
| 5 | 100% | 98.43% | 96.38% | 91.56% | 86.74% | 84.33% | 87.95% | 84.33% | 77.10% |
| 10 | 100% | 98.43% | 96.38% | 100% | 96.30% | 92.77% | 95.36% | 91.56% | 89.15% |
Table 8.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for Lymphoma cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 2 | 100% | 99.1% | 98.48% | 86.36% | 86.36% | 86.36% | 86.36% | 86.36% | 86.36% |
| 3 | 100% | 100% | 100% | 93.93% | 90.90% | 86.36% | 89.39% | 87.87% | 86.36% |
| 5 | 100% | 100% | 100% | 100% | 96.96% | 93.93% | 96.96% | 92.42% | 90.90% |
Table 9.
The classification performance of the Co-ABC algorithm with comparison to mRMR-ABC algorithm and ABC-SVM algorithm for Leukaemia2 cancer dataset.
| Number of genes | Classification accuracy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Co-ABC |
mRMR-ABC |
ABC-SVM |
|||||||
| Best | Mean | Worst | Best | Mean | Worst | Best | Mean | Worst | |
| 2 | 87.5% | 87.5% | 87.5% | 84.72% | 84.72% | 84.72% | 84.72% | 84.72% | 84.72% |
| 3 | 97.22% | 96.32% | 95.83% | 87.5% | 86.11% | 84.72% | 86.11% | 85.23% | 84.72% |
| 4 | 98.61% | 96.81% | 95.83% | 90.27% | 87.5% | 84.72% | 87.5% | 86.11% | 84.72% |
| 5 | 98.61% | 98.19% | 97.22% | 90.27% | 88.88% | 86.11% | 87.5% | 86.45% | 84.72% |
| 6 | 100% | 99.21% | 98.61% | 94.44% | 90.27% | 87.5% | 90.27% | 88.88% | 86.11% |
| 20 | 100% | 99.21% | 98.61% | 100% | 96.12% | 95.83% | 97.22% | 93.15% | 91.66% |
In order to sake of a fair comparison using the same parameters. In this research, we re-implement two related evolutionary based algorithm: Co-GA, which is CFS combined with a genetic algorithm GA. The second one named Co-PSO, which is CFS combined with a particle swarm optimization algorithm PSO. Furthermore, we compared Co-ABC with recently published related algorithms. Notably, all these algorithms that are under comparison have been combined with the support vector machine (SVM) for classification task.
Table 10 present the performance comparison results of the Co-ABC algorithm and other related algorithms. Compared with the mRMR-ABC algorithm, the mRMR-ABC algorithm selected 15 genes to achieve 96.77% classification accuracy. In contrast, the Co-ABC algorithm achieves same classification accuracy using only 9 genes.
Table 10.
The classification performance of the related algorithms under comparison for six cancer gene expression profile Numbers between parentheses means the numbers of informative genes that has been used in classification task.
| Algorithms | Colon | Leukemia1 | Lung | SRBCT | Lymphoma | Leukemia2 |
|---|---|---|---|---|---|---|
| Co-ABC | 96.77(9) | 100(3) | 100(2) | 100(4) | 100(2) | 100(6) |
| CFS-GA | 90.32(8) | 100(24) | 100(20) | 100(38) | 100(17) | 100(36) |
| CFS-PSO | 91.94(7) | 100(15) | 100(5) | 100(35) | ||
| mRMR-ABC Alshamlan et al. (2015) | 96.77(15) | 100(14) | 100(8) | 100(10) | 100(5) | 100(20) |
| ABC-SVM Alshamlan et al. (2016) | 95.61(20) | 93.05(14) | 97.91(8) | 95.36(10) | 96.96(5) | 97.22(20) |
| PSO Qi et al. (2007) | 85.48(20) | 94.44(23) | ||||
| PSO Javad and Giveki (2013) | 87.01(2000) | 93.06 (7129) | ||||
| mRMR-PSOJavad et al. (2012) | 90.32(10) | 100(18) | ||||
| GADPLee and Leu (2011) | 100(6) | |||||
| mRMR-GA Amine et al. (2009) | 100(15) | 95(5) | ||||
| ESVM Huang and Chang (2007) | 95.75(7) | 98.75(6) | ||||
| MLHD-GA Huang et al. (2007) | 97.1(10) | 100(11) | 100(6) | 100(9) | ||
| CFS-IBPSO Yang et al. (2008) | 100(6) | 98.57(41) | ||||
| GA Peng et al. (2003) | 93.55(12) | |||||
| mAnt Yu et al. (2009) | 91.5(8) | 100(7) |
For the Leukemia1 dataset, Co-ABC generate very accurate calcification, which is 100% using only three informative genes. As observed in Table 10, there are three algorithms acquired 100% accuracy result, however, their informative genes are greater. The mRMR-ABC algorithm got 100% classification accuracy using 14 genes. Also, the mRMR-PSO algorithm proposed by Javad et al. (2012) acquired 100% classification percentage with 18 informative genes. And, the Co-GA algorithm select 24 genes to get 100% classification accuracy.
For the Lung dataset, the Co-ABC algorithm make a superior improvement by select only two predictive genes to achieve 100% classification result. This accuracy was achieved by other algorithms, which are CFS-GA, mRMR-ABC, and mRMR-GA algorithm proposed by Amine et al. (2009), however they using greater number of selected genes.
In SRBCT dataset, the MLHD-GA algorithm developed by Huang et al. (2007) got 100% classification result with 11 predictive genes. Also, the mRMR-ABC algorithm used 10 predictive genes and acquires 100% classification accuracy. In addition, the CFS-GA and CFS-PSO achieve 100% classification accuracy using greater number of selected genes. By contrast, our proposed algorithm select only four informative genes to achieve 100% classification accuracy.
For the Lymphoma cancer dataset, there are many proposed algorithms achieve 100%, the Co-ABC algorithm identified a fewer number of informative genes. The Co-ABC selected only two genes to achieve 100% classification accuracy, which is obviously improved the other existing result so far. For the Leukemia2 dataset, the MLHD-GA algorithm developed by Huang et al. (2007), select 9 genes to achieve 100% classification accuracy. While our previous mRMR-ABC algorithm used 20 genes to got 100% classification accuracy. In contrast, our proposed Co-ABC achieve a greatest improvement in number of selected genes demand. It select only 6 genes to acquire 100% classification performance.
Computational complexity is an important aspect in algorithm assessment. Therefore, we tested and compared the time and memory complexity of Co-ABC with proposed bio-inspired meta-heuristics algorithms, ABC-SVM, and mRMR-ABC. Table 11 shows the average runtime in seconds for the Cot-ABC algorithm and other algorithms under comparison; the Co-ABC algorithm has a faster execution time. Table 12 shows the average memory space in virtual machine in giga bites for the Cot-ABC algorithm and other algorithms under comparison. The Co-ABC algorithm used approximately similar memory space as mRMR-ABC algorithm, and less than ABC-SVM algorithm.
Table 11.
Average runtime (in s) for the Co-ABC algorithm and other classification algorithms.
| Algorithms | Preprocessing time | Average classification time | Total |
|---|---|---|---|
| Co-ABC with SVM | 24.37 s | 40.22 s | 64.59 s |
| mRMR-ABC with SVM | 25.17 s | 72.13 s | 97.3 s |
| ABC with SVM | 0.0 s | 134.74 s | 134.74 s |
Table 12.
Average memory space (in GB) for the Co-ABC algorithm and other classification algorithms.
| Algorithms | Memory space |
|---|---|
| Co-ABC with SVM | 1.56 GB |
| mRMR-ABC with SVM | 1.58 GB |
| ABC with SVM | 1.89 GB |
It is worth noted that the Co-ABC algorithm generally outperforms the previously reported results, it generates the highest classification accuracy and the lowest average of selected genes when evaluated using all six cancer datasets, as compared to the original ABC algorithm under the same cross-validation approach. In comparison between CFS and mRMR filter methods when combined with ABC algorithm, the CFS elects less number of genes than mRMR with relatively improved in classification accuracy. This implies that CFS method is able to improve the classification accuracy and it is an effective tool for identifying the highly correlated genes and omitting the non-relevant and noisy genes. Therefore, we can conclude that Co-ABC is a promising method for biomarker gene discovery using gene expression profile.
The explanation of the highly correlative and informative genes that achieve highest classification accuracy for all microarray datasets using Co-ABC algorithm have been reported in Table 13.
Table 13.
The highly correlative and informative genes that achieve highest classification accuracy for six cancer gene expression profile using Co-ABC algorithm.
| Datasets | Predictive genes | Accuracy |
|---|---|---|
| Colon | Gene 625, Gene1562, Gene576, Gene1328, Gene1917, Gene 1772, Gene682, Gene1200, Gene1671 | 96.77% |
| Leukemia1 | S50223_at, U05259_rna1_at, M23197_at | 100% |
| Lung | X64559_at, U19247_rna1_s_at | 100% |
| SRBCT | Gene123, Gene742, Gene1954, Gene1003 | 100% |
| Lymphoma | Gene2403X, Gene3519X | 100% |
| Leukemia2 | L47738_at, X00274_at, X58072_at, X95735_at, D63880_at, U48251_at | 100% |
Furthermore, in order to evaluate the performance of Co-ABC algorithm result more accurately. Table 14 present the precision and sensitivity of Co-ABC algorithm with selected informative genes. Precision is the positive predictive value or the fraction of the positive predictions that are actually positive. While the specificity is the true negative rate or the proportion of negatives that are correctly identified.
| (5) |
| (6) |
Table 14.
The precision and sensitivity of Co-ABC algorithm using selected informative genes.
| Datasets | Precision | Sensitivity |
|---|---|---|
| Colon | 97.04% | 96.77% |
| Leukemia1 | 100% | 100% |
| Lung | 100% | 100% |
| SRBCT | 100% | 100% |
| Lymphoma | 100% | 100% |
| Leukemia2 | 100% | 100% |
It worth noting, that one of the CFS filters method limitation, it selects only the highly correlated genes. In some dataset such as colon datasets, the gene expression not correlated with each other. For this reason, the CFS select only few number of genes and loos some predictive and important genes. This is definitely effect the classification accuracy. While in other datasets such as lymphoma dataset, there are many correlated genes. In this case, the CFS retrieves many related genes as presented in Table 3. Therefore, we recommend addressing this limitation in feature work.
4. Conclusion
In this research, we developed a new hybrid feature selection approach, called Co-ABC algorithm. In our proposed Co-ABC algorithm, we adopted the CFS filter method as a preprocessing step to the ABC algorithm to enhance the search speed and classification performance. In addition, in order to eliminate the unimportant and filter the noisy genes and reduces the computational complexity for the ABC algorithm with SVM as classifier. The major aim for adopting the CFS filter method is to find the highly correlated subset of genes from initial microarray dataset. Extensive experiments and comparisons were conducted using six binary and multi-class microarray cancer gene expression profile. The results showed that the proposed Co-ABC algorithm perfumes better than related algorithms. Moreover, the Co-ABC algorithm generates greater classification accuracy performance with fewer average of informative genes when tested using all six cancer datasets as compared to the original ABC algorithm under the same cross-validation technique (LOOCV). Also, when we compared the performance of CFS and mRMR when they combined with ABC algorithm, the CFS identify less number of predictive genes than mRMR with relatively greater classification accuracy performance. This implies that CFS method is able to improve the classification accuracy and it is an effective tool for identifying the highly correlated genes and omitting the non-relevant and noisy genes. Therefore, we can conclude that Co-ABC is a promising method for biomarker gene discovery using gene expression profile
Acknowledgment
This research project was supported by a grant from the Research Center of the Center for Female Scientific and Medical Colleges Deanship of Scientific Research, King Saud University.
Footnotes
Peer review under responsibility of King Saud University.
References
- Alba, E., Garcia-Nieto, J., L.J.L., Talbi, E., Gene selection in cancer classification using pso/svm and ga/svm hybrid algorithms. In: IEEE Congress on Evolutionary Computation, 2007. CEC 2007, pp. 284–290.
- Alizadeh A., Eisen M., Davis M., Rosenwald A., Boldrick J., Sabet T., Powell Y., Yang L., Marti G., Moore T., Hudson J., Lu L., Lewis D., Tibshirani R., Sherlock G., Chan W., Greiner T., Weisenburger D., Armitage J., Warnke R., Levy R., Wilson W., Grever M., Byrd J., Botstein D., Brown P., Staudt L. Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Alon U., Barkai N., Notterman D., Gish K., Ybarra S., Mack D., Levine A. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. 1999;96(12):6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Moro-Sancho I., Simon-Hurtado A., Varela-Arrabal R. Microarray gene expression classification with few genes: criteria to combine attribute selection and classification methods. Exp. Syst. Appl. 2012;39(8):7270–7280. [Google Scholar]
- Alshamlan H.M., Badr G.H., Alohali Y. Proceedings of the World Congress on Engineering. vol. 2. 2013. A study of cancer microarray gene expression profile: objectives and approaches; pp. 1–6. [Google Scholar]
- Alshamlan, H., Badr, G., Alohali, Y., 2014. A comparative study of cancer classification methods using microarray gene expression profile 285, 389–398.
- Alshamlan, H., Badr, G., Alohali, Y., 2015. mrmr-abc: a hybrid gene selection algorithm for microarray cancer classification. [DOI] [PMC free article] [PubMed]
- Alshamlan H.M., Badr G.H., Alohali Y.A. Abc-svm: artificial bee colony and svm method for microarray gene selection and multi class cancer classification. Int. J. Mach. Learn. Comput. 2016;6(3):184. [Google Scholar]
- Amine, A., El Akadi,A., El Ouardighi, A., Aboutajdine, D., 2009. A new gene selection approach based on minimum redundancy-maximum relevance (mrmr) and genetic algorithm (ga). In: IEEE/ACS International Conference on Computer Systems and Applications, 2009. AICCSA 2009, pp. 69–75.
- Armstrong S.A., Staunton J.E., Silverman L.B., Pieters R., den Boer M.L., Minden M.D., Sallan S.E., Lander E.S., Golub T.R., Korsmeyer S.J. Mll translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2001;30(1):41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Bahamish, H.A.A., Abdullah, R., Salam, R.A., 2009. Protein Tertiary Structure Prediction Using Artificial Bee Colony Algorithm. Paper presented at the Modelling & Simulation, 2009. AMS '09. Third Asia International Conference on Modelling & Simulation.
- Beer D.G., Kardia S.L., Huang C.-C., Giordano T.J., Levin A.M., Misek D.E., Lin L., Chen G., Gharib T.G., Thomas D.G. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Chuang L.-Y., Yang C.-H., Wu K.-C., Yang C.-H. A hybrid feature selection method for dna microarray data. Comput. Biol. Med. 2011;41(4):228–237. doi: 10.1016/j.compbiomed.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Ghorai, S., Mukherjee, A., Sengupta, S., Dutta, P., 2010. Multicategory cancer classification from gene expression data by multiclass nppc ensemble. In: 2010 International Conference on Systems in Medicine and Biology (ICSMB), pp. 4–48.
- Golub T., Slonim D., Tamayo P., Huard C., Gaasenbeek M., Mesirov J., Coller L., Downing J., Caligiuri M., Bloomfield C., Lander E. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Huang H.-L., Chang F.-L. Esvm: Evolutionary support vector machine for automatic feature selection and classification of microarray data. Biosystems. 2007;90(2):516–528. doi: 10.1016/j.biosystems.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Huang H.-L., Lee C.-C., Ho S.-Y. Selecting a minimal number of relevant genes from microarray data to design accurate tissue classifiers. Biosystems. 2007;90(1):78–86. doi: 10.1016/j.biosystems.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Huerta E., Duval B., kao Hao J. EvoWorkshops 2006. vol. 3907. Springer; 2006. A hybrid ga/svm approach for gene selection and classification of microarray data; pp. 34–44. (LNCS). [Google Scholar]
- Javad A.M., Giveki D. Automatic detection of erythemato-squamous diseases using pso-svm based on association rules. Eng. Appl. Artif. Intell. 2013;26(1):603–608. [Google Scholar]
- Javad A.M., Mohammad H.S., Rezghi M. A novel weighted support vector machine based on particle swarm optimization for gene selection and tumor classification. Comput. Math. Meth. Med. 2012;2012:7. doi: 10.1155/2012/320698. Article ID: 320698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaboga, D., 2005. An Idea Based on Honey Bee Swarm for Numerical Optimization. Tech. rep., Technical Erciyes university, engineering faculty, computer engineering department.
- Karaboga N. A new design method based on artificial bee colony algorithm for digital IIR filters. J. Franklin Inst. 2009;346(4):328–348. [Google Scholar]
- Karaboga, D., Akay, B.B., 2005. An artificial bee colony (abc) algorithm on training artificial neural networks (Technical Report TR06): Erciyes University, Engineering Faculty, Computer Engineering Department.
- Khan J., Wei J.S., Ringner M., Saal L.H., Ladanyi M., Westermann F., Berthold F., Schwab M., Antonescu C.R., Peterson C. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 2001;7(6):673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruskal W.H., Wallis W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952;47(260):583–621. [Google Scholar]
- Lee C.-P., Leu Y. A novel hybrid feature selection method for microarray data analysis. Appl. Soft Comput. 2011;11(1):208–213. [Google Scholar]
- Mukherjee, S., 2003. Chapter 9. Classifying microarray data using support vector machines. In: Of Scientists from the University of Pennsylvania School of Medicine and the School of Engineering and Applied Science, Kluwer Academic Publishers.
- Ng, A.Y., 1997. Preventing overfitting of cross-validation data. In: ICML, vol. 97, pp. 245–253.
- Peng S., Xu Q., Ling X.B., Peng X., Du W., Chen L. Molecular classification of cancer types from microarray data using the combination of genetic algorithms and support vector machines. FEBS Lett. 2003;555(2):358–362. doi: 10.1016/s0014-5793(03)01275-4. [DOI] [PubMed] [Google Scholar]
- Qi S., Shi W.-M., Wei K., Ye B.-X. A combination of modified particle swarm optimization algorithm and support vector machine for gene selection and tumor classification. Adv. Comput. Sci. 2007;71(4):157–162. doi: 10.1016/j.talanta.2006.07.047. [DOI] [PubMed] [Google Scholar]
- Quinlan J.R. Induction of decision trees. Mach. Learn. 1986;1(1):81–106. [Google Scholar]
- Sheng-Bo G., Michael L., Ming L. Proceedings of the 2006 International Conference on Computational Intelligence and Bioinformatics - Volume Part III, ICIC’06. Springer-Verlag; 2006. Gene selection based on mutual information for the classification of multi-class cancer; pp. 454–463. [Google Scholar]
- Vapnik V.N. Wiley; 1998. Statistical Learning Theory. [DOI] [PubMed] [Google Scholar]
- Wang X., Gotoh O. Microarray-based cancer prediction using soft computing approach. Cancer Inform. 2009;7:123–139. doi: 10.4137/cin.s2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W.-L., An M.-Q. An efficient and robust artificial bee colony algorithm for numerical optimization. Comput. Oper. Res. 2013;40(5):1256–1265. [Google Scholar]
- Yang C.-S., Chuang L.-Y., Ke C.-H., Yang C.-H. A hybrid feature selection method for microarray classification. Int. J. Comput. Sci. 2008;35:285–290. [Google Scholar]
- Yu H., Gu G., Liu H., Shen J., Zhao J. A modified ant colony optimization algorithm for tumor marker gene selection. Genom. Proteom. Bioinform. 2009;7(4):200–208. doi: 10.1016/S1672-0229(08)60050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan S., aki I., Pedro L. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]