We describe here the first known peptidase displaying exclusive activity toward N-terminal glycine residues. This work indicates a specific role for intracellular glycyl peptidases in deep sea hyperthermophilic archaeal metabolism. These observations also provide critical evidence for the use of these archaeal extremozymes for biotechnological applications.
KEYWORDS: archaea, hyperthermophiles, peptidases
ABSTRACT
The TET peptidases are large self-compartmentalized complexes that form dodecameric particles. These metallopeptidases, members of the M42 family, are widely distributed in prokaryotes. Three different versions of TET complexes, with different substrate specificities, were found to coexist in the cytosol of the hyperthermophilic archaeon Pyrococcus horikoshii. In the present work, we identified a novel type of TET complex that we named PhTET4. The recombinant PhTET4 enzyme was found to self-assemble as a tetrahedral edifice similar to other TET complexes. We determined PhTET4 substrate specificity using a broad range of monoacyl chromogenic and fluorogenic compounds. High-performance liquid chromatographic peptide degradation assays were also performed. These experiments demonstrated that PhTET4 is a strict glycyl aminopeptidase, devoid of amidolytic activity toward other types of amino acids. The catalytic efficiency of PhTET4 was studied under various conditions. The protein was found to be a hyperthermophilic alkaline aminopeptidase. Interestingly, unlike other peptidases from the same family, it was activated only by nickel ions.
IMPORTANCE We describe here the first known peptidase displaying exclusive activity toward N-terminal glycine residues. This work indicates a specific role for intracellular glycyl peptidases in deep sea hyperthermophilic archaeal metabolism. These observations also provide critical evidence for the use of these archaeal extremozymes for biotechnological applications.
INTRODUCTION
In all cell types, metalloaminopeptidases play crucial roles in energy metabolism, protein maturation and degradation, and the regulation of biologically active peptides by removing the N-terminal amino acid from proteins and oligopeptides (1, 2). In most cases, they operate after the action of an endoprotease and their activity is limited to small peptides. Several aminopeptidases were found to self-assemble as large dodecameric particles (for a review, see reference 3). These 0.5-MDa molecular machines were first discovered in archaea and were named TET due to their peculiar tetrahedral shape (4). The 13-nm hollow dodecahedrons enclose 12 active sites distributed in four funnel-shaped chambers located at the apices of the particle and four large access holes, formed by the junction of six subunits, situated at the facets (3). This organization strongly distinguishes TET from the other cytosolic compartmentalized peptidases, which mostly adopt a barrel-shaped architecture. Biochemical and structural studies of TET peptidases have accumulated over the past 10 years (3); they revealed that TET dodecamers represent a common scaffold for an efficient system for polypeptide capture and processing (5). In all TET complexes, the aminopeptidase activity is based on cocatalytic dinuclear metal active sites belonging to the M18 or M42 peptidase family, according to the MEROPS classification system (6). Bound peptides are cleaved through a common mechanism involving a water molecule and a glutamate residue (7). The nature of the metal occupying the bimetallic active site has been shown to modulate TET enzymatic activity, and Co2+ ions appear to be the best activators for almost all archaeal and bacterial M42 TET complexes (8–13).
TET machines are widespread and are found in the three life domains. Interestingly, in prokaryotes, one to four different types of TET complexes can coexist in the cytosol, depending on the cell type. These enzymes can be categorized according to their preferences for the chemical structure of the N-terminal amino acid residues present in the polypeptide chains. Three main categories have been identified so far, i.e., glutamyl/aspartyl aminopeptidases, lysine aminopeptidases, and leucine aminopeptidases; the latter exhibit broader specificities. In eukarya, M18 TET complexes displayed aspartyl aminopeptidase activity (14, 15). In bacteria, M42 TET peptidases from Clostridium thermocellum and Thermotoga maritima were assigned as leucyl aminopeptidases (12). Two M42 enzymes from the pathogens Streptococcus pneumoniae and Mycoplasma hyopneumoniae displayed glutamyl-aminopeptidase activities (11, 16). In archaea, the common TET structural scaffold can harbor disparate functions. The unique TET complex observed in the halophilic archaeon Haloarcula marismortui displayed broader specificity, with a preference for neutral and basic residues (4). Pyrococcus horikoshii and related hyperthermophilic Thermococcales species are distinguished by the fact that they possess three different TET complexes, namely, PhTET1, a glutamyl/aspartyl aminopeptidase (9, 17), PhTET2, a leucyl aminopeptidase with broad activity against neutral amino acids (8), and PhTET3, a lysyl aminopeptidase with a clear preference for positively charged residues (10). The analysis of their activities on synthetic peptides of different sizes and compositions, using reverse-phase high-performance liquid chromatography (HPLC), indicated that the TET peptidases degraded oligopeptides in a sequential manner and displayed strict aminopeptidase behavior.
During a search for novel peptidase genes from the genome sequence of P. horikoshii, we found the PH0737 gene, which encodes a new protein displaying about ∼20% sequence identity with the three M42 self-compartmentalized peptidases, PhTET1, PhTET2, and PhTET3, that coexist in the cytosol of Thermococcales species. The biophysical and biochemical characterizations presented in this work revealed that the PH0737 gene product self-assembled as a dodecameric complex with a macromolecular shape similar to that of the previously described TET complexes. Functional assays with monoacyl and peptide substrates revealed that the gene product displayed aminopeptidase activity; therefore, the protein was named PhTET4. To understand the biological function of this novel TET complex, we conducted biochemical characterizations of its optimal enzymatic conditions, preferred substrate, and metal cofactor specificities. Those studies revealed that, unlike other M42 aminopeptidases, PhTET4 is activated by nickel ions and is a strict glycyl aminopeptidase (GAP).
RESULTS
The PH0737 gene product displays homology with the large dodecameric TET peptidases from P. horikoshii.
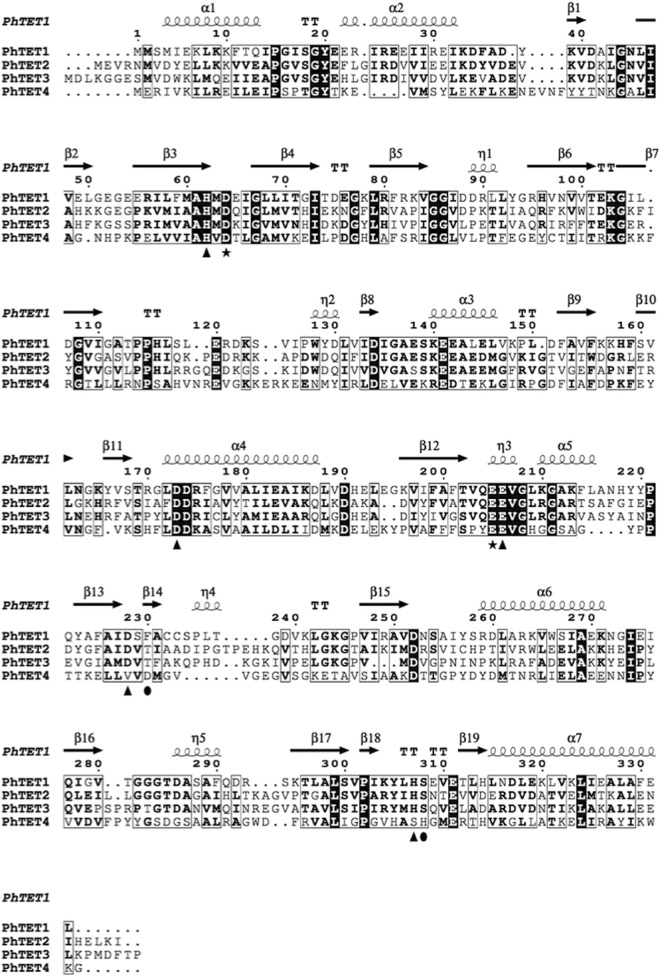
An analysis of the genomes of Thermococcales species revealed the existence of a conserved gene coding for an unassigned peptidase of the MH clan in the MEROPS database (http://merops.sanger.ac.uk). The protein encoded by PH0737 shares 20.6%, 22.5%, and 22% sequence identity with the characterized aminopeptidases PhTET1, PhTET2, and PhTET3, respectively (Fig. 1). The residues involved in the coordination of metal ions in the M42 peptidase family are well conserved among PhTET1, PhTET2, PhTET3, and the new protein encoded by the PH0737 gene (6). Interestingly, two regions (the catalytic domain and the dimerization domain) that confer the ability of M42 peptidases to form large multimers are also present in the sequence of PH0737 (9). These observations suggested that the product of the PH0737 gene could represent a paralogue to the three other TET peptidase complexes.
FIG 1.
Multiple-sequence alignment of the four P. horikoshii PhTETs, i.e., PhTET1, PhTET2, PhTET3, and PhTET4, as determined by ESPript 3.0 (39). The numbering and the secondary structural elements are those of PhTET1 (PDB code 2WYR) (9). Triangles and the star highlight the metal-binding and active residues, respectively. Most likely, a shift occurred for the last two putative metal-binding residues of PhTET4, Asp231 and His311 (highlighted by ovals), in comparison with the conserved positions of PhTET1, PhTET2, and PhTET3.
PH0737 gene product oligomerization state.
Recombinant PH0737 protein was produced in Escherichia coli. The cellular extract was clarified by heat shock precipitation, and the recombinant protein was purified by ion-exchange and size exclusion chromatography. In the final step, PH0737 was eluted in the same exclusion volume as the homologous 480-kDa complex PhTET2 (Fig. 2A). Negative-stain electron microscopy was used to investigate the molecular architecture of the PH0737 protein complex. The images showed that the protein forms hollow triangular particles of homogeneous size (Fig. 2B), with dimensions and shapes similar to those of the P. horikoshii TET2 complex. This indicated that the PH0737 protein self-assembled as a 12-subunit complex similar to the ∼500-kDa TET dodecameric complexes (9, 10, 18). Therefore, the PH0737 protein was named PhTET4.
FIG 2.
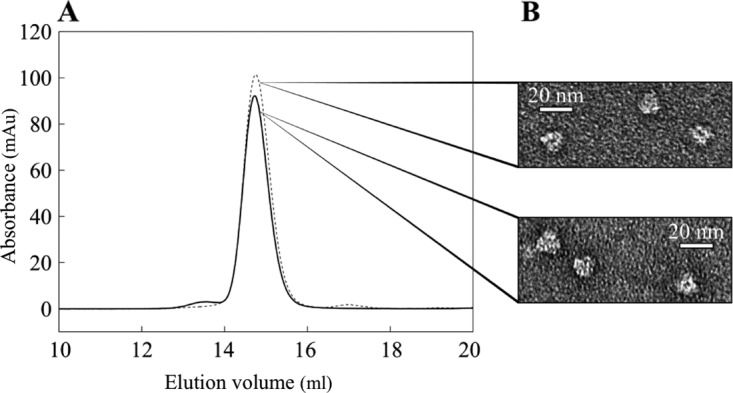
Oligomeric state of PhTET4 in solution. (A) Gel filtration analysis of PhTET2 and PhTET4 dodecameric complexes on a Superose 6 column. Dashed line, PhTET2 dodecamer; solid line, PhTET4 dodecamer. The recombinant protein PhTET4 eluted in the same volume as the 450-kDa PhTET2 dodecameric complex. (B) PhTET2 (top) and PhTET4 (bottom) observed by negative-stain electron microscopy. Homogeneous populations of hollow triangular particles are observed at a magnification of ×49,000 for both PhTET2 and PhTET4 samples.
PhTET4 is a nickel-activated peptidase.
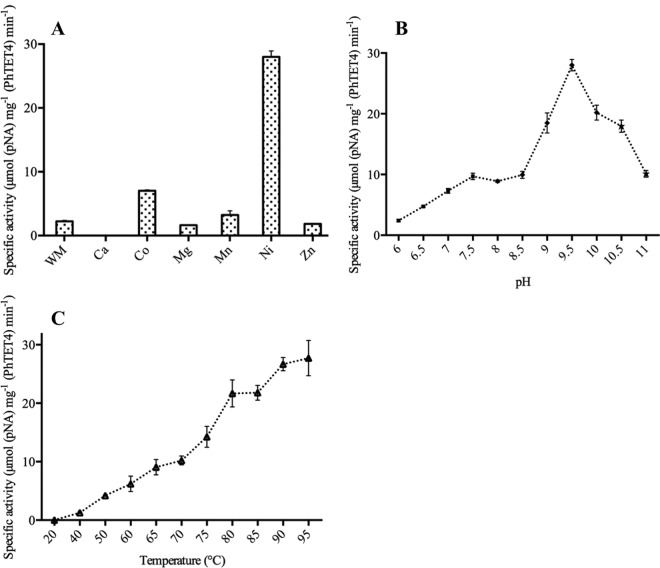
To determine the functional identity of PhTET4, we tested its amidolytic activity toward the 20 amino acids by using a broad array of chromogenic p-nitroaniline (pNA)-conjugated or fluorogenic 7-amino-4-methylcoumarin (AMC)-conjugated aminoacyl compounds. Since the three TET peptidases from P. horikoshii were found to be hyperthermophilic cobalt-activated aminopeptidases, we first assayed PhTET4 activity in the presence of 0.1 mM CoCl2 as an enzyme cofactor, at 80°C and under identical buffer conditions, i.e., 5 mM substrate, 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 150 mM KCl (pH 7.5). Surprisingly, no hydrolysis was observed with any tested substrate with the notable exception of Gly-pNA, toward which PhTET4 exhibited weak catalytic activity. Since metal cofactors have been shown to be essential for controlling the activity and oligomeric state of the various TET edifices characterized so far (19), we tested the influence of several metal ions on PhTET4 GAP activity (Fig. 3A). We determined that Ni2+ had an important stimulating effect on PhTET4 cleavage activity, with 12 times greater activity than in control assays in which no metallic ion was added to the reaction mixture. Co2+ and Mn2+ also stimulated PhTET4 activity but did so less efficiently than Ni2+ (3-fold and 1.4-fold activation, respectively). Interestingly, Zn2+, Ca2+, and Mg2+ were found to inhibit PhTET4 hydrolytic activity, and total inhibition was observed in the presence of Ca2+ ions. This is the first time that Ni2+ ions have been described as essential activating cofactors for an aminopeptidase from the M42 family.
FIG 3.
Effects of metals, pH, and temperature on amidolytic activity of PhTET4. (A) Variation of PhTET4 specific activity in the absence (without metal [WM]) or presence of several metal ions. (B) PhTET4 specific activity as a function of pH (■, PIPES; ◆, CHES; ★, CAPS). (C) PhTET4 specific activity in response to temperature variations.
PhTET4 is a hyperthermophilic alkaline peptidase.
In order to determine the influence of pH on PhTET4 enzymatic behavior, the amidolytic activity against Gly-pNA was measured between pH 6 and pH 11, at 80°C (Fig. 3B). The optimal activity was found at pH 9.5. Interestingly, a significant percentage of activity was maintained at elevated pH. These experiments revealed that, in contrast to the other three PhTET enzymes, which display narrower pH optima at ∼7.5, PhTET4 can be defined as an alkaline peptidase (8–10).
PhTET4 activity was studied at different temperatures ranging from 20°C to 95°C. PhTET4 enzymatic activity increased with temperature, with a maximal activity in the measurable temperature range being observed at 95°C (Fig. 3C). Thus, PhTET4 displays a high level of hyperthermophilic behavior, comparable to that reported for the three other PhTET aminopeptidases present in Pyrococcus horikoshii cells (8–10).
PhTET4 is a strict glycyl aminopeptidase.
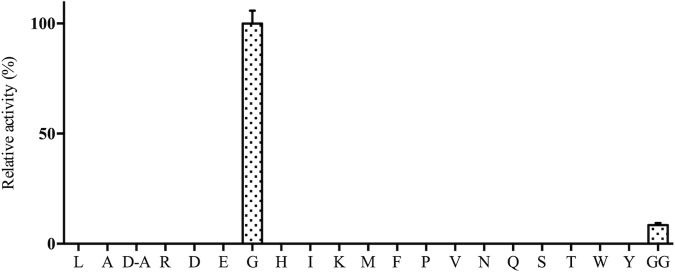
The initial characterization of PhTET4 aminopeptidase activity indicated that the enzyme displayed narrow substrate specificity, with a strong preference for glycine residues. In order to confirm this finding, the experiments were repeated in the presence of nickel and under the optimal temperature and pH conditions defined above (0.1 mM NiCl2, pH 9.5, and 85°C). The results showed unambiguously that the enzyme acted only on Gly-pNA (Fig. 4). No hydrolytic activity toward any other amino acids could be detected, even with long incubation times. We also tried to investigate whether PhTET4 exhibited high d-stereospecificity with d-alanine, as shown for Aspergillus oryzae glycine aminopeptidase (20). For this, d-Ala-pNA was used as the chromogenic substrate under optimal activity conditions (0.1 mM NiCl2, pH 9.5, and 85°C). The experiment showed that PhTET4 was unable to cleave alanine residues in the d-conformation, thus demonstrating that PhTET4 was devoid of d-stereospecificity.
FIG 4.
PhTET4 as a strict GAP. PhTET4 activity was assayed with different synthetic chromogenic and fluorogenic compounds. L, Leu-pNA; A, l-Ala-pNA; D-A, d-Ala-pNA; R, Arg-pNA; D, Asp-pNA; E, Glu-pNA; G, Gly-pNA; H, His-pNA; I, Ile-pNA; K, Lys-pNA; M, Met-pNA; F, Phe-pNA; P, Pro-pNA; V, Val-pNA; N, Asn-AMC; Q, Gln-AMC; S, Ser-AMC; T, Thr-AMC; W, Trp-AMC; Y, Tyr-AMC; GG, Gly-Gly-pNA. These substrates are levorotatory compounds except for d-Ala. Activities are expressed as a percentage of the enzymatic activity measured with Gly-pNA, which was given a value of 100%. The amidolytic activity of PhTET4 against (H-Cys-pNA)2 was not measured because it precipitated under our enzymatic conditions.
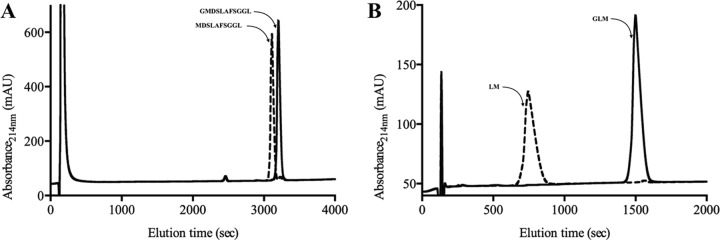
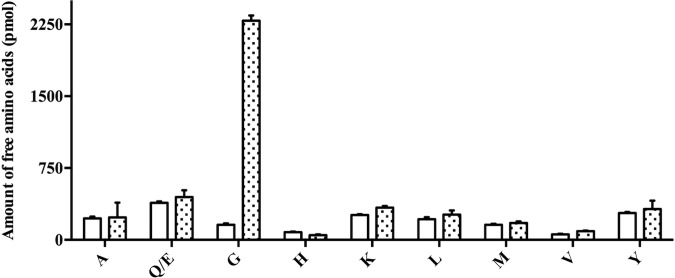
For aminopeptidases, the catalytic activities and specificities can be affected by the length and N-terminal amino acid composition of the peptide substrates (21–25). Consequently, we tested whether PhTET4 maintains its narrow specificity toward glycine residues in a peptide context. For this, we measured the capacity of PhTET4 to cleave the N-terminal residues of the following peptides: GI, GL, GLM, GMDSLAFSGGL, and LGG. After incubation of PhTET4 with the peptide substrates under optimal activity conditions, the reaction products were separated by reverse-phase HPLC and identified by N-terminal sequencing. These experiments showed that no enzymatic activity was detected against peptides that did not start with glycine even if this residue was present in the P1′ position, as demonstrated with the LGG tripeptide. The HPLC profiles for the degradation of the GLM and GMDSLAFSGGL peptides are shown in Fig. 5. The sequences of the detected accumulating peptides were determined, and the results clearly demonstrated that PhTET4 did not exert amidolytic activity beyond the N-terminal glycine in a peptide context. To assess whether PhTET4 could sequentially process several glycine residues in a peptide sequence, we tested the enzymatic activity against the chromogenic peptide Gly-Gly-pNA under the same conditions as described above. PhTET4 displayed significant activity against this substrate but corresponding to only 10% of the total activity exhibited in the presence of the monoacyl compound Gly-pNA (as shown in Fig. 4). To further demonstrate the narrow PhTET4 specificity, the enzyme was incubated with three different complex peptide mixtures obtained from the hydrolysis of casein or whey proteins, as described in Materials and Methods. The amino acids released after incubation were measured in the supernatants. No amidolytic activities were detected using these hydrolysates unless a synthetic peptide of known composition containing a N-terminal glycine (GISEQFKTNVLREYA) was added to the peptide mixtures. The experiment performed with the whey protein hydrolysate (WPH) is presented in Fig. 6. Taken together, these experiments clearly mark PhTET4 as an aminopeptidase that is strictly specialized in the hydrolysis of N-terminal glycine residues.
FIG 5.
Chromatographic profiles of peptide degradation by PhTET4. The sequences of peptides are indicated. In the presence of PhTET4 (dashed lines), only the glycine residue was removed from the original peptide (solid lines). (A) Degradation profile of the GMDSLAFSGGL peptide. (B) Degradation profile of the GLM peptide.
FIG 6.
Relative amounts of free amino acids generated from the hydrolysis of a complex peptide mixture by PhTET4. A WPH was prepared and supplemented with GISEQFKTNVLREYA. The concentrations of the free amino acids produced from the peptide mixture in the absence (white bars) or presence (dotted bars) of PhTET4 are indicated. Only the amino acids that were detected in the blank and in the assay are shown. A, Ala; Q/E, Gln or Glu; G, Gly; H, His; K, Lys; L, Leu; M, Met; V, Val; Y, Tyr.
Effects of amastatin and bestatin on PhTET4 activity.
Amastatin and bestatin represent typical aminopeptidase and leucyl aminopeptidase inhibitors, respectively. The addition of 0.2 mM amastatin and 1 mM bestatin reduced the amidolytic activity of PhTET4 against Gly-pNA to 78% and 73%, respectively, of the activity detected in the control experiment. These experiments showed that these inhibitors had moderate effects on PhTET4, compared to those reported for the other TET peptidases, especially PhTET3.
DISCUSSION
Pyrococcus horikoshii is a deep sea hyperthermophilic archaeon that was isolated from a hydrothermal vent, at a depth of 1,395 m (26). Like other hyperthermophilic organisms, its genome encodes numerous peptidases (27). In particular, three different versions of the ∼500-kDa dodecameric M42 TET peptidase were found to coexist in its cytosol (8–10). Here, we characterized a fourth member of the TET family in the same organism, which we named PhTET4. Despite weak sequence similarity with other PhTET enzymes, the protein complex was found to exhibit the same typical tetrahedral architecture as the other TET dodecamers characterized previously in archaea, bacteria, and eukarya; however, its biochemical characterization revealed important and unsuspected properties that clearly distinguished PhTET4 from all known TET complexes.
The dinuclear metal site is involved both in the stability of the dodecameric edifice and in the fine-tuning of TET catalytic activity (19). The M42 canonical residues involved in metal ion binding and catalysis are well conserved in the PhTET4 sequence, which suggests that PhTET4 has a similar catalytic mechanism, compared to other members of the M42 family. Extended X-ray absorption fine structure (EXAFS) experiments have shown that the dinuclear metal center of TET can be occupied by Zn2+, Co2+, and Mn2+ ions (10, 15, 18). Co2+ and Mn2+ ions, when added to the reaction mixture, strongly activated the amidolytic activities of previously characterized archaeal, bacterial, and eukaryotic TET aminopeptidases (8–13). In archaea, Co2+ was the main activating metal for the three TET enzymes from P. horikoshii, while Mn2+ activated the DkamTET enzyme from Desulfurococcus kamchatkensis (28). Here we report for the first time an aminopeptidase from the M42 family being strongly activated by nickel ions. This is an interesting finding, since nickel is rarely the main activator in metallo-aminopeptidases. To our knowledge, this has been reported only for methionine aminopeptidases, such as methionine aminopeptidase A from Mycobacterium tuberculosis (29).
The most striking feature of PhTET4 revealed in this work is its exclusive amidolytic activity against glycine residues, which justified categorizing this novel M42 peptidase as a GAP. Examples of GAP enzymes are rare. While aminopeptidases from different families are capable of hydrolyzing glycine residues, their activities remain weak in comparison with their preferred substrates. To our knowledge, only three aminopeptidases have been found to exhibit clear preferences for glycine residues; the first is a Zn-dependent metallopeptidase from the M61 family secreted by the Gram-negative bacterium Sphingomonas capsulata (30), the second is a eukaryotic S12-family serine peptidase found in the cytosol of Actinomucor oryzae (20), and the third is the cytosolic GAP of Actinomucor elegans (31), for which the residues implicated in the enzymatic mechanism are still ambiguous. Therefore, PhTET4 is the first GAP enzyme identified in archaea. Moreover, we showed that it displayed interesting hyperthermophilic and alkaliphilic properties. It is also noteworthy that, unlike PhTET4, the bacterial and eukaryotic TET enzymes did not display strict specificity for glycine residues. PhTET4 is thus the first known aminopeptidase displaying apparently exclusive activity toward N-terminal glycine residues.
Glycine is a difficult residue for hydrolysis, due to the flexibility that it introduces in the polypeptide chains and the absence of a side chain. Thus, by specifically cleaving glycine from peptide N termini, PhTET4 could be employed to improve the degradation of proteins and peptides in industrial processes. Furthermore, glycine is a taste enhancer that is often used to modify the palatability of food preparations (32). Because PhTET4 maintains significant activity and stability in a broad temperature range, the enzymatic release of glycine by PhTET4 could be employed to enhance positive tastes (e.g., sweetness) or to mask unpleasant tastes (e.g., saltiness) in many preparations, such as fermented foods or seasonings (33, 34).
The biological significance of the specialized TET aminopeptidase is still not well understood. In the case of Thermococcales, the presence of different TET complexes displaying distinct substrate preferences can be interpreted in different ways. As already suggested, the TET complex may act cooperatively to hydrolyze efficiently intracellular polypeptides arising from ATP-dependent protein degradation machines such as the 20S proteasome or Lon (10, 18). In the case of heterotrophic archaea, such as P. horikoshii, a combination of streamlined aminopeptidase activities could also provide an advantage to hydrolyze peptides imported from the extracellular environment as a carbon source (10). In this context, GAP activity may represent an advantage under physiologically extreme temperature conditions that may provide additional flexibility. However, the presence of GAP enzymes cannot be a crucial condition for living at high temperatures, as other hyperthermophilic strains contain only one type of TET. However, there are also arguments supporting a specific physiological role for PhTET4 in P. horikoshii. This was already proposed for PhTET1, as this enzymatic complex specializes in the hydrolysis of aspartate and glutamate and is not accumulated in cells at the same time as PhTET2 and PhTET3 dodecamers (35). Moreover, the hetero-oligomeric complex PhTET2-PhTET3 is capable of hydrolyzing a wide variety of substrates, including glycine, without the help of PhTET4 (35). This raises the possibility that PhTET4 GAP activity could be devoted to peculiar cellular functions. Indeed, glycine represents a thermoprotectant for enzymes and is also an important precursor residue for the synthesis of osmoprotectants such as trimethylglycine (36). In addition, glycine is a precursor for glutathione, which is an important antioxidant antimetal detoxification precursor in eukaryotic cells (37), and for glyoxylate, which, when metabolized, generates oxalate and formate, two antioxidant molecules (38). P. horikoshii lives under strictly anaerobic conditions, but the high temperatures and the presence of heavy metals are sources of free radicals, which are responsible for intense DNA damage. Therefore, it is tempting to speculate that PhTET4 could be involved in stress responses under certain conditions encountered by the archaea.
MATERIALS AND METHODS
Recombinant protein expression and purification.
The gene coding for the PH0737 protein (GenBank accession no. O58468) was cloned in the pET-41c vector by GeneCust Europe (Luxembourg). Recombinant protein was overexpressed in the Escherichia coli BL21(DE3)-RIL strain for 4 h at 37°C with induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in 1 liter of lysogeny broth. The cell pellet was stored at −80°C until use. The cells were resuspended in 25 ml of 50 mM Tris-HCl (pH 8)–90 mM NaCl–0.1% Triton X-100, supplemented with 6.25 mg of lysozyme (Euromedex), 1.25 mg of DNase I (grade II; Roche), 5 mg of RNase (Roche), 25 mg of Pefabloc SC (Roche), and 0.25 ml of 2 M MgSO4. Cells were disrupted by sonication at 30 W at 4°C, with five on/off cycles of 30 s each, and then were heated at 75°C for 20 min to eliminate most mesophilic proteins of the host strain. The lysate was clarified by centrifugation at 17,000 × g for 1 h at 4°C with a JA20 rotor (Beckman), and the supernatant was loaded on a Resource Q column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5)–100 mM NaCl. The 30-ml flowthrough fraction was retained and diluted with 20 ml of Tris-HCl (pH 7.5) to a final concentration of 105 mM NaCl. The new supernatant volume was loaded a second time on the Resource Q column. After the column was washed with 20 mM Tris-HCl (pH 7.5)–100 mM NaCl, bound proteins were eluted with a linear salt gradient (154 to 290 mM NaCl). The protein-containing fractions were pooled and loaded on a Superdex 200 10/300 GL size exclusion column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5)–150 mM NaCl. The peak fractions were combined, concentrated with an Amicon Ultra 30-kDa-cutoff filter, and stored at 4°C. The purity of the prepared protein was checked by SDS-PAGE. Three milligrams of recombinant protein was produced from 1 liter of E. coli culture.
PH0737 oligomeric state characterization.
To evaluate the oligomeric state of PH0737, the purified protein was loaded on a Superose 6 size exclusion column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.5)–100 mM NaCl. The column was calibrated using a protein molecular size standard (GE Healthcare). The homologous 480-kDa PhTET2 dodecameric complex was purified as described previously (8) and was used as a standard to assess the oligomeric state of PH0737.
PH0737 negative-stain electron microscopy.
After the size exclusion chromatography step, 4 μl of PH0737 at 0.05 mg/ml was deposited onto carbon-coated 400-mesh copper grids. The samples were stained using 2% uranyl acetate and air dried. Images were obtained under low-dose conditions in a T12FEI electron microscope working at 120 kV, with a nominal magnification of ×40,000, using an Orius SC1000 charge-coupled device (CCD) camera.
Determination of PhTET4 substrate specificity with synthetic chromogenic and fluorogenic compounds.
The hydrolytic activity of PhTET4 was determined by using different aminoacyl-pNA and aminoacyl-AMC conjugates (Bachem), as described in reference 8. The substrate stock solutions (X-pNA and X-AMC) were solubilized in 100% dimethyl sulfoxide (DMSO) (final concentration, 40 mM). Reactions were initiated by addition of the enzyme (final concentration, 4 μg/ml) to 400 μl of prewarmed mixture containing 5 mM chromogenic or fluorogenic substrate, 50 mM PIPES, 150 mM KCl, and 0.1 mM NiCl2 (pH 7.5). Incubations were performed at 85°C. To avoid water evaporation, the total volume was covered by a layer of mineral oil. Catalytic activities were monitored for 10 min by measuring the absorbance of released pNA at 405 nm or measuring the fluorescence of AMC using excitation and emission wavelengths of 355 and 460 nm, respectively. Three replicates and two enzyme blanks were assayed for each experimental point.
Effects of metal cations on PhTET4 activity.
The effects of various metal cations on PhTET4 activity were assayed by using CaCl2, CoCl2, MgCl2, MnCl2, NiCl2, and ZnCl2. The metal cations were incubated at final concentrations of 0.1 mM in the reaction volume containing 4 μg/ml PhTET4, 50 mM N-cyclohexyl-2-aminoethanesulfonic acid (CHES), 150 mM KCl, and 5 mM Gly-pNA (pH 9.5). The reaction was monitored by measuring the absorbance of released pNA at 405 nm for 10 min at 80°C. Three replicates and two enzyme blanks were assayed for each experimental point.
Determination of PhTET4 optimal pH.
The effect of pH on PhTET4 enzymatic activity was studied by using different buffers, as follows: PIPES, pH 6 to 7.5; CHES, pH 8.2 to 10; N-cyclohexyl-3-aminopropanesulfonic acid (CAPS), pH 10.5 to 11. All buffers were used at a final concentration of 50 mM in the presence of 4 μg/ml PhTET4, 150 mM KCl, 0.1 mM NiCl2, and 5 mM Gly-pNA, at 80°C. The incubation was performed for 10 min by measuring the absorbance of released pNA at 405 nm. Three replicates and two enzyme blanks were assayed for each experimental point.
PhTET4 optimal temperature.
The temperature effects on the enzymatic activity of PhTET4 were measured in a range from 20°C to 95°C. In all cases, 4 μg/ml PhTET4 was incubated for 10 min with 50 mM CHES, 150 mM KCl, 0.1 mM NiCl2, and 5 mM Gly-pNA (pH 9.5). The enzyme activity was assessed as described previously. Three replicates and two enzyme blanks were assayed for each experimental point.
Study of PhTET4 activity with synthetic peptides.
PhTET4 cleavage activity was assayed with synthetic peptides of various sizes and compositions. In these experiments, PhTET4 (final concentration, 6 μg/ml) was added to a prewarmed mixture of 50 mM CHES, 150 mM KCl, and 0.1 mM NiCl2 (pH 9.5) containing 3 mM GI, GL, GLM, GMDSLAFSGGL, or LGG peptides. To avoid water evaporation, 20 μl of mineral oil was added on top of the total volume. The reaction incubation was performed at 85°C for 6 min. Aliquots of 80 μl were removed and added to 220 μl of 2% acetonitrile-0.1% trifluoroacetic acid (TFA). Proteins were removed by centrifugation at 15,000 × g for 15 min. One hundred microliters of the supernatant was retrieved and injected onto a Nova-Pak C18 column (4 μm, 3.9 by 300 mm; Waters) in a HPLC purification system (PerkinElmer) equilibrated with 2% acetonitrile-0.1% TFA. The elution of peptide products was achieved with a linear acetonitrile gradient of 2% to 33.2% and was monitored by measurement of the absorbance at 214 nm. Chromatographic runs were carried out at room temperature. The separated fragments were collected and subjected to N-terminal sequence analysis as described in reference 8.
Study of PhTET4 activity with complex peptide mixtures.
The PhTET4 N-terminal specificity with a complex mixture of peptides was tested against a WPH (enzymatically hydrolyzed in the laboratory) and a casein hydrolysate (chemically hydrolyzed) from Sigma. Whey proteins were obtained from Sigma. The protein solution was incubated with thermolysin (Sigma), trypsin (Sigma), and chymotrypsin (Sigma) at 100 μg/ml for 2 h at 40°C, with constant stirring. The three enzymes were then inactivated by incubation of the solution at 100°C for 15 min. The WPH was collected after centrifugation at 15,000 × g for 10 min.
The hydrolytic activity of PhTET4 was then assayed against the casein hydrolysate, the WPH, and the WPH supplemented with the synthetic peptide GISEQFKTNVLREYA at a concentration of 0.5 mM. The hydrolysate pH was adjusted to 9.5 at the working temperature, and the mixture was supplemented with 0.1 mM NiCl2. Reactions were initiated by addition of the enzyme (0.1 mg/ml) in a final volume of 100 μl. Incubations were performed at 70°C for 2 h, with constant stirring. Reactions were stopped by addition of 100 μl of cold acetonitrile, and the samples were placed at 4°C. After protein precipitation with acetonitrile-TFA, 250 μl of supernatant was dried under vacuum. Solids were then dissolved in the same volume of Beckman analysis buffer, and the generation of free amino acids from the peptide mixture was determined with a model 7300 Beckman amino acid analyzer, using the standard sodium citrate elution buffer system. The free amino acids generated from the peptide mixture hydrolysis were determined as described previously (8). Four replicates (samples and enzyme blanks) were assayed for each experimental point.
PhTET4 inhibition.
The capacities of EDTA, bestatin, and amastatin to inhibit PhTET4 activity were studied. These inhibitors were added to PhTET4 reaction mixtures at 5 mM, 1 mM, and 0.2 mM, respectively, under optimal conditions (50 mM CHES, 150 mM KCl, 0.1 mM NiCl2, and 5 mM Gly-pNA [pH 9.5], at 85°C). PhTET4 activity was measured as described above. Three replicates and two enzyme blanks were assayed for each experimental point.
ACKNOWLEDGMENTS
We acknowledge financial support from the Agence Nationale de la Recherche (grant Archelyse ANR-12-BSV8-0019-0). This work was supported by the French National Research Agency in the framework of the Investissements d'Avenir program (grant ANR-15-IDEX-02). H.B. was supported by the CEA Ph.D. program IRTELIS. This work used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (grant ANR-10-INSB-05-02) and GRAL (grant ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology.
We thank D. Fenel from the Partnership for Structural Biology-Institute de Biologie Structurale for the negative-stain electron microscopy. We thank J. P. Andrieu from the Partnership for Structural Biology-Institute de Biologie Structurale for the N-terminal sequencing.
REFERENCES
- 1.Lowther WT, Matthews BW. 2002. Metalloaminopeptidases: common functional themes in disparate structural surroundings. Chem Rev 102:4581–4608. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- 2.Gonzales T, Robert-Baudouy J. 1996. Bacterial aminopeptidases: properties and functions. FEMS Microbiol Rev 18:319–344. doi: 10.1111/j.1574-6976.1996.tb00247.x. [DOI] [PubMed] [Google Scholar]
- 3.Appolaire A, Colombo M, Basbous H, Gabel F, Girard E, Franzetti B. 2016. TET peptidases: a family of tetrahedral complexes conserved in prokaryotes. Biochimie 122:188–196. doi: 10.1016/j.biochi.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Franzetti B, Schoehn G, Hernandez JF, Jaquinod M, Ruigrok RW, Zaccai G. 2002. Tetrahedral aminopeptidase: a novel large protease complex from archaea. EMBO J 21:2132–2138. doi: 10.1093/emboj/21.9.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appolaire A, Rosenbaum E, Dura MA, Colombo M, Marty V, Savoye MN, Godfroy A, Schoehn G, Girard E, Gabel F, Franzetti B. 2013. Pyrococcus horikoshii TET2 peptidase assembling process and associated functional regulation. J Biol Chem 288:22542–22554. doi: 10.1074/jbc.M113.450189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawlings ND, Barrett AJ, Bateman A. 2012. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 40:D343–D350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomis-Ruth FX. 2008. Structure and mechanism of metallocarboxypeptidases. Crit Rev Biochem Mol Biol 43:319–345. doi: 10.1080/10409230802376375. [DOI] [PubMed] [Google Scholar]
- 8.Dura MA, Receveur-Brechot V, Andrieu JP, Ebel C, Schoehn G, Roussel A, Franzetti B. 2005. Characterization of a TET-like aminopeptidase complex from the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemistry 44:3477–3486. doi: 10.1021/bi047736j. [DOI] [PubMed] [Google Scholar]
- 9.Schoehn G, Vellieux FM, Asuncion Dura M, Receveur-Brechot V, Fabry CM, Ruigrok RW, Ebel C, Roussel A, Franzetti B. 2006. An archaeal peptidase assembles into two different quaternary structures: a tetrahedron and a giant octahedron. J Biol Chem 281:36327–36337. doi: 10.1074/jbc.M604417200. [DOI] [PubMed] [Google Scholar]
- 10.Dura MA, Rosenbaum E, Larabi A, Gabel F, Vellieux FM, Franzetti B. 2009. The structural and biochemical characterizations of a novel TET peptidase complex from Pyrococcus horikoshii reveal an integrated peptide degradation system in hyperthermophilic archaea. Mol Microbiol 72:26–40. doi: 10.1111/j.1365-2958.2009.06600.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, San BH, Moh SH, Park H, Kim DY, Lee S, Kim KK. 2010. Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease. Biochem Biophys Res Commun 391:431–436. doi: 10.1016/j.bbrc.2009.11.075. [DOI] [PubMed] [Google Scholar]
- 12.Dutoit R, Brandt N, Legrain C, Bauvois C. 2012. Functional characterization of two M42 aminopeptidases erroneously annotated as cellulases. PLoS One 7:e50639. doi: 10.1371/journal.pone.0050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen DD, Pandian R, Kim D, Ha SC, Yoon HJ, Kim KS, Yun KH, Kim JH, Kim KK. 2014. Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa. Biochem Biophys Res Commun 447:101–107. doi: 10.1016/j.bbrc.2014.03.109. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Farquhar ER, Chance MR, Palczewski K, Kiser PD. 2012. Insights into substrate specificity and metal activation of mammalian tetrahedral aspartyl aminopeptidase. J Biol Chem 287:13356–13370. doi: 10.1074/jbc.M112.347518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaikuad A, Pilka ES, Riso AD, Delft FV, Kavanagh KL, Venien-Bryan C, Oppermann U, Yue WW. 2012. Structure of human aspartyl aminopeptidase complexed with substrate analogue: insight into catalytic mechanism, substrate specificity and M18 peptidase family. BMC Struct Biol 12:14. doi: 10.1186/1472-6807-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson MW, Buchtmann KA, Jenkins C, Tacchi JL, Raymond BB, To J, Chowdhury PR, Woolley LK, Labbate M, Turnbull L, Whitchurch CB, Padula MP, Djordjevic SP. 2013. MHJ_0125 is an M42 glutamyl aminopeptidase that moonlights as a multifunctional adhesin on the surface of Mycoplasma hyopneumoniae. Open Biol 3:130017. doi: 10.1098/rsob.130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dura MA, Franzetti B. 2013. PhTET1 aminopeptidase, p 1638–1645. In Rawlings ND, Salvesen G (ed), Handbook of proteolytic enzymes, 3rd ed Academic Press, Oxford, United Kingdom. [Google Scholar]
- 18.Borissenko L, Groll M. 2005. Crystal structure of TET protease reveals complementary protein degradation pathways in prokaryotes. J Mol Biol 346:1207–1219. doi: 10.1016/j.jmb.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 19.Colombo M, Girard E, Franzetti B. 2016. Tuned by metals: the TET peptidase activity is controlled by 3 metal binding sites. Sci Rep 6:20876. doi: 10.1038/srep20876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marui J, Matsushita-Morita M, Tada S, Hattori R, Suzuki S, Amano H, Ishida H, Yamagata Y, Takeuchi M, Kusumoto K. 2012. Enzymatic properties of the glycine d-alanine aminopeptidase of Aspergillus oryzae and its activity profiles in liquid-cultured mycelia and solid-state rice culture (rice koji). Appl Microbiol Biotechnol 93:655–669. doi: 10.1007/s00253-011-3610-y. [DOI] [PubMed] [Google Scholar]
- 21.Singh H, Kalnitsky G. 1980. α-N-Benzoylarginine-β-naphthylamide hydrolase, an aminoendopeptidase from rabbit lung. J Biol Chem 255:369–374. [PubMed] [Google Scholar]
- 22.Koldamova RP, Lefterov IM, Gadjeva VG, Lazo JS. 1998. Essential binding and functional domains of human bleomycin hydrolase. Biochemistry 37:2282–2290. doi: 10.1021/bi9722204. [DOI] [PubMed] [Google Scholar]
- 23.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. 1999. A giant protease with potential to substitute for some functions of the proteasome. Science 283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 24.Turk D, Janjic V, Stern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B. 2001. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J 20:6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandu D, Nandi D. 2003. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 149:3437–3447. doi: 10.1099/mic.0.26518-0. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. 1998. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles 2:123–130. doi: 10.1007/s007920050051. [DOI] [PubMed] [Google Scholar]
- 27.Ward DE, Shockley KR, Chang LS, Levy RD, Michel JK, Conners SB, Kelly RM. 2002. Proteolysis in hyperthermophilic microorganisms. Archaea 1:63–74. doi: 10.1155/2002/503191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrova TE, Slutskaya ES, Boyko KM, Sokolova OS, Rakitina TV, Korzhenevskiy DA, Gorbacheva MA, Bezsudnova EY, Popov VO. 2015. Structure of the dodecamer of the aminopeptidase APDkam598 from the archaeon Desulfurococcus kamchatkensis. Acta Crystallogr F Struct Biol Commun 71:277–285. doi: 10.1107/S2053230X15000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayanan SS, Nampoothiri KM. 2012. Biochemical characterization of recombinant methionine aminopeptidases (MAPs) from Mycobacterium tuberculosis H37Rv. Mol Cell Biochem 365:191–202. doi: 10.1007/s11010-012-1260-8. [DOI] [PubMed] [Google Scholar]
- 30.Byun T, Tang M, Sloma A, Brown KM, Marumoto C, Fujii M, Blinkovsky AM. 2001. Aminopeptidase from Sphingomonas capsulata. J Biol Chem 276:17902–17907. doi: 10.1074/jbc.M010608200. [DOI] [PubMed] [Google Scholar]
- 31.Ito K, Ma X, Azmi N, Huang HS, Fujii M, Yoshimoto T. 2003. Novel aminopeptidase specific for glycine from Actinomucor elegans. Biosci Biotechnol Biochem 67:83–88. doi: 10.1271/bbb.67.83. [DOI] [PubMed] [Google Scholar]
- 32.Kawai M, Okiyama A, Ueda Y. 2002. Taste enhancements between various amino acids and IMP. Chem Senses 27:739–745. doi: 10.1093/chemse/27.8.739. [DOI] [PubMed] [Google Scholar]
- 33.Wada A, Isobe Y, Yamaguchi S, Yamaoka R, Ozaki M. 2001. Taste-enhancing effects of glycine on the sweetness of glucose: a gustatory aspect of symbiosis between the ant, Camponotus japonicus, and the larvae of the lycaenid butterfly, Niphanda fusca. Chem Senses 26:983–992. doi: 10.1093/chemse/26.8.983. [DOI] [PubMed] [Google Scholar]
- 34.Lioe HN, Apriyantono A, Takara K, Wada K, Naoki H, Yasuda M. 2004. Low molecular weight compounds responsible for savory taste of Indonesian soy sauce. J Agric Food Chem 52:5950–5956. doi: 10.1021/jf049230d. [DOI] [PubMed] [Google Scholar]
- 35.Appolaire A, Dura MA, Ferruit M, Andrieu JP, Godfroy A, Gribaldo S, Franzetti B. 2014. The TET2 and TET3 aminopeptidases from Pyrococcus horikoshii form a hetero-subunit peptidasome with enhanced peptide destruction properties. Mol Microbiol 94:803–814. doi: 10.1111/mmi.12775. [DOI] [PubMed] [Google Scholar]
- 36.Canovas D, Vargas C, Csonka LN, Ventosa A, Nieto JJ. 1996. Osmoprotectants in Halomonas elongata: high-affinity betaine transport system and choline-betaine pathway. J Bacteriol 178:7221–7226. doi: 10.1128/jb.178.24.7221-7226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briat JF, Lebrun M. 1999. Plant responses to metal toxicity. C R Acad Sci III 322:43–54. doi: 10.1016/S0764-4469(99)80016-X. [DOI] [PubMed] [Google Scholar]
- 38.Alhasawi A, Castonguay Z, Appanna ND, Auger C, Appanna VD. 2015. Glycine metabolism and anti-oxidative defence mechanisms in Pseudomonas fluorescens. Microbiol Res 171:26–31. doi: 10.1016/j.micres.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]