Abstract
Most members of the large family of ATP-Binding Cassette (ABC) proteins function as membrane transporters. However, the most evolutionarily conserved group, the ABCE protein subfamily, comprises soluble proteins that were initially denoted RNase L inhibitor (RLI) proteins. ABCE proteins are present in all eukaryotes and archaea and are encoded by a single gene in most genomes, or by two genes in a few cases. Functional analysis of ABCE genes, primarily in Saccharomyces cerevisiae, has shown that ABCE proteins have essential functions as part of the translational apparatus. In this review, we summarize the current understanding of ABCE protein function in ribosome biogenesis and recycling, with a particular focus on their known and proposed developmental roles in different species. The ABCE proteins might represent another class of factors contributing to the role of the ribosome in gene expression regulation.
Keywords: ABCE, ribosome, translation, development, RLI
ABC protein structure, function, and classification
The ATP-Binding Cassette (ABC) proteins, which are present in all living organisms, constitute one of the largest known protein families. Actually, in prokaryotes, the ABC genes constitute 1–3% of the genome (Tomii and Kanehisa, 1998). The Saccharomyces cerevisiae and human genomes encode 30 and 48 ABC proteins, respectively (Dean et al., 2001; Paumi et al., 2009; Vasiliou et al., 2009). By contrast, in plants like Arabidopsis thaliana, there are more than 100 genes encoding ABC proteins (Verrier et al., 2008). Such multiplication and functional diversification of ABC proteins is consistent with the sessile nature of plants and their adaptation to changing terrestrial environments, as well as with the history of whole-genome duplications in plant evolution (Hwang et al., 2016).
Most ABC proteins transport solutes across cell membranes. These solutes, referred to as allocrites (Holland and Blight, 1999), range from small inorganic and organic molecules to large organic compounds. ABC transporters contain two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs), otherwise known as ATP-binding cassettes, which are hallmarks of the ABC family. TMDs and NBDs can be contained together in a unique full-sized protein, as commonly found in eukaryotes, or can be separated into individual peptides (subunits), as observed in prokaryotes. ABC proteins can also occur as homo- or heterodimers formed by half-sized proteins, which contain one TMD and one NBD or consist only of fused NBDs (Hyde et al., 1990). TMDs are responsible for allocrite specificity and have polyphyletic origins. TMDs belonging to a specific transporter subtype display similar membrane topologies among distant species. Each TMD typically comprises 6–10 α-helices that span the cell membrane, thus generating a pore that is accessible from the cytoplasm or from the extracellular space (Wang et al., 2009; Zheng et al., 2013).
In contrast to TMDs, NBDs are monophyletic with conserved sequences and structures (Higgins et al., 1986). The NBD regions that display the highest level of conservation are those that function specifically in ATP binding and hydrolysis, namely the Walker A and Walker B motifs; the LSGGQ signature or ABC motif; and the A-, D-, H-, and Q-loops, named after the conserved residues at their N- or C-termini (ter Beek et al., 2014). NBDs are arranged in a head-to-tail orientation, which allows them to bind two ATP molecules. These ATP molecules interact with motifs from both NBDs in a sandwich-like manner, bringing NBDs together in a closed conformation. ATP cleavage (hydrolysis) relaxes this conformation and drives the transport cycle by producing coupled conformational changes in the TMDs that recognize and translocate the allocrite across the membrane. Whether both NBDs stay in contact (continuous contact models) or totally separate (NBDs separation models) after ATP hydrolysis remains a matter of active debate. However, since there are several ABC transporter subtypes, it is reasonable to assume that more than one transport mechanism exists (Shi and Barna, 2015).
To facilitate ABC protein recognition and comparison among different species, a standardized nomenclature was proposed for all eukaryotes (Dean and Annilo, 2005; Verrier et al., 2008; Paumi et al., 2009; Xie et al., 2012; Dermauw and Van Leeuwen, 2014). Mammalian ABC proteins were divided into seven subfamilies (ABCA to ABCG) based on NBD sequence similarity (Dean et al., 2001). Two additional subfamilies were later proposed for non-mammalian ABC proteins, specifically the ABCH subfamily, which is found in insects and osteichthyes (Dean and Annilo, 2005), and the ABCI subfamily, which is found exclusively in plants and contains prokaryotic-like ABC proteins, among others (Verrier et al., 2008). ABCA, ABCB, ABCC, ABCD, ABCG, and ABCH proteins function as transporters (Figure 1A; Hopfner, 2016).
Figure 1.
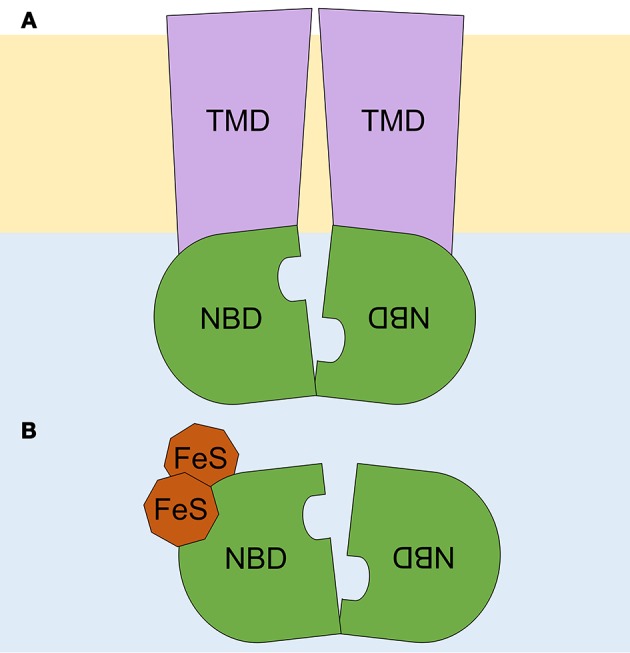
Schematic representation of ABC proteins. (A) An ABC transporter, and (B) an ABCE soluble protein. A lipid bilayer is depicted in pale orange, and the cytoplasm, in pale blue. Transmembrane domains (TMD) are shown in purple and nucleotide binding domains (NBD), arranged in a head-to-tail orientation, in green. The two diamagnetic [4Fe-4S]2+ clusters are depicted in brown. Adapted from Hopfner (2016).
By contrast, soluble ABC proteins belong to the ABCE and ABCF subfamilies, which lack TMDs but have the two NBDs in a single peptide chain (Figure 1B). ABCF proteins function in translation, as exemplified by eukaryotic elongation factor 3 (eEF3) (Andersen et al., 2006), or in chromosome segregation and DNA repair, as is the case for the Structural Maintenance of Chromosomes (SMC) proteins and the SMC-like protein Rad50 (Hirano, 2002). Here, we focus on the ABCE subfamily, which includes single-copy genes that, although initially thought to be specific to mammals (Bisbal et al., 1995), are found in both eukaryotes and archaea.
The ABCE subfamily of ABC proteins
ABCE proteins display the highest level of conservation among the ABC subfamilies. For example, the yeast ABCE1/Rli1 shares 68 and 43% sequence identity with its human and archaeal (Sulfolobus solfataricus) orthologs, respectively (Kispal et al., 2005; Barthelme et al., 2007). The ABCE subfamily is represented in most genomes by a single-copy, essential ABCE1 gene (Kerr, 2004). However, two ABCE paralogs, specifically ABCE1/RLI1 and ABCE2/RLI2, exist in plants such as Arabidopsis thaliana and Oryza sativa (Sarmiento et al., 2006; Verrier et al., 2008), and in animals such as catfish (Liu et al., 2013) and the mosquitoes Anopheles gambiae, Aedes aegypti, and Culex pipiens quinquefasciatus (Lu et al., 2016).
Loss-of-function of ABCE1 genes, either via null alleles or RNAi suppression, is associated with a lethal phenotype in all studied species, and hypomorphic ABCE1 alleles result in slow-growth phenotypes (Amsterdam et al., 2004; Dong et al., 2004; Estévez et al., 2004; Zhao et al., 2004; Coelho et al., 2005a; Kispal et al., 2005; Chen et al., 2006; Barthelme et al., 2007; Broehan et al., 2013; Kougioumoutzi et al., 2013; Table 1). Conversely, in Saccharomyces cerevisiae, RLI1 overexpression leading to accumulation of either the wild-type protein or mutated versions disrupted in conserved residues or lacking entire conserved domains caused a dominant negative effect on growth (Dong et al., 2004; Khoshnevis et al., 2010).
Table 1.
Mutations affecting ABCE genes in different species.
| Organism | Gene name | Loss of function caused by | Phenotype | References |
|---|---|---|---|---|
| Drosophila melanogaster | pixie | Strong hypomorphic alleles | Recessive lethal | Coelho et al., 2005a,b |
| Weak hypomorphic alleles | Slow growth; disproportionate organ sizes; slender bristles; eye roughening | Coelho et al., 2005a,b | ||
| Caenorhabditis elegans | abce-1 | RNAi | Slow growth; embryonic lethality | Kamath et al., 2003; Zhao et al., 2004 |
| Danio rerio | abce1 | Retroviral insertional allele | Small head and eyes; underdeveloped liver and gut; pericardial edema; lethal at 5 days post-fertilization | Amsterdam et al., 2004 |
| Xenopus laevis | abce1 | Antisense ABCE1 morpholino oligonucleotides | Arrested growth at the gastrula stage | Chen et al., 2006 |
| Cardamine hirsuta | SIL3; ChRLI2 | Hypomorphic allele | Reduced growth; small and simple leaves; delayed leaf initiation; reduced auxin signaling; reduced cell proliferation; high rates of endoreplication | Kougioumoutzi et al., 2013 |
| Nicotiana benthamiana | RLIh | Virus-induced gene silencing | Reduced growth; distorted leaves; whitened veins; reduced cell size and number | Petersen et al., 2004 |
Determination of the crystal structure of archaeal ABCE1 (aABCE1) proteins (Karcher et al., 2005, 2008; Barthelme et al., 2011) showed that these proteins contain four conserved domains: (1–2) the two NBDs that are present in all ABC proteins; (3) a hinge region proposed to facilitate NBD orientation and function as a pivot point in the tweezer-like power stroke of NBDs following ATP binding (Karcher et al., 2005); and (4) an iron-sulfur (FeS) binding domain with eight cysteine residues that coordinate two diamagnetic [4Fe-4S]2+ clusters present at the ABCE protein N-terminal region (Figure 1B; Barthelme et al., 2007; Karcher et al., 2008). The latter domain plays an essential role in ABCE protein function (Kispal et al., 2005; Yarunin et al., 2005; Barthelme et al., 2007, 2011; Alhebshi et al., 2012). The high-level conservation among all ABCE amino acid sequences, in particular within their four conserved domains, allows to deduce the structure of eukaryotic ABCE orthologs based on that of aABCE1 (Karcher et al., 2005, 2008). Moreover, cryoelectron microscopy showed that Pyrococcus furiosus aABCE1 and yeast Rli1 associate similarly with ribosomes (Becker et al., 2012; Preis et al., 2014).
The function of the yeast ABCE1 protein Rli1 has been well characterized. Furthermore, based on the sequence conservation, ABCE1 protein function is likely to be well conserved among different organisms. Yeast Rli1 triggers the dissociation of ribosomes during different processes related to translation, as described in further detail below (Figure 2). Additional roles have been proposed for ABCE proteins in higher eukaryotes. For example, ABCE1 participates in the assembly of immature HIV-1 capsids in mammals (Zimmerman et al., 2002; Dooher and Lingappa, 2004), and ABCE proteins act as endogenous suppressors of RNA silencing in plants and humans (Sarmiento et al., 2006; Kärblane et al., 2015).
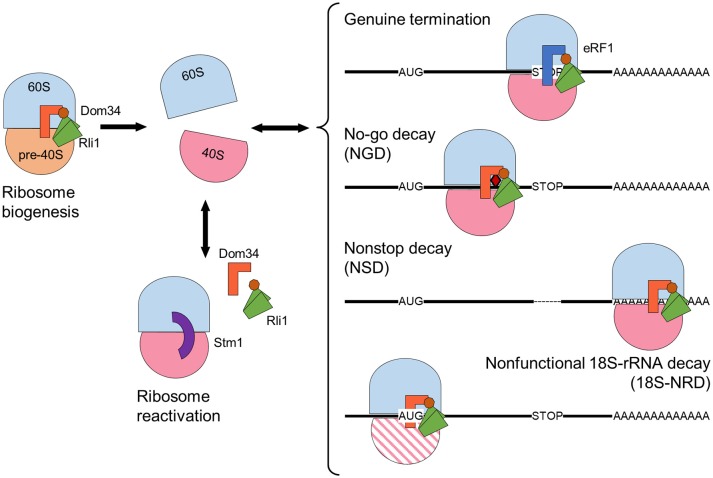
Figure 2.
Rli1 ribosome-dissociation activity has essential functions in diverse cellular processes, including ribosome maturation and release of stalled ribosomes. Schematic representation of a maturing ribosome formed by 60S (blue) and pre-40S (orange) subunits. Mature 40S subunits are depicted in pink. A faulty 40S subunit is represented by pink parallel lines. Class I release factors Dom34 (orange) and eRF1 (dark blue) dissociate ribosomes together with Rli1, whose NBDs are represented in green, and the FeS domain in brown. The “clamping” factor Stm1 is depicted in purple. The no-go decay mRNA contains a secondary structure represented by a red rhombus. The non-stop mRNA lacks an in-frame stop codon, depicted by a dashed line. Arrows indicate ribosome association-dissociation flow.
Yeast ABCE1/Rli1 functions in ribosome biogenesis and recycling
The Saccharomyces cerevisiae genome encodes 30 ABC proteins, including one member of the ABCE subfamily, Rli1. RLI1 encodes a canonical ABCE protein and contains two NBDs arranged in a head-to-tail manner (thus allowing the binding of two ATP molecules), and a FeS domain (Barthelme et al., 2007). The Drosophila melanogaster Rli1 ortholog, Pixie, localizes exclusively in the cytoplasm (Coelho et al., 2005a), whereas yeast Rli1 localizes in the cytoplasm and in the nucleus (Dong et al., 2004; Kispal et al., 2005; Yarunin et al., 2005).
Yeast Rli1 associates with eukaryotic translation initiation factors, 40S ribosomal subunits, 80S ribosomes, and polysomes. Suppression of Rli1 strongly reduces polysome size and abundance, as well as translation rates (Dong et al., 2004; Kispal et al., 2005; Yarunin et al., 2005; Shoemaker and Green, 2011). These observations suggested that Rli1 participates in translation initiation by promoting assembly of the preinitiation complex. Equivalent observations were made for human ABCE1 (Chen et al., 2006) and Drosophila Pixie (Andersen and Leevers, 2007). Additionally, ABCE proteins in Trypanosoma brucei (Estévez et al., 2004) and Caenorhabditis elegans (Zhao et al., 2004) were also implicated in translation. A role for yeast Rli1 and mammalian ABCE1 in translation initiation was confirmed following analysis of the 48S initiation and the post-splitting complexes (Heuer et al., 2017; Mancera-Martínez et al., 2017).
The role of Rli1 as a ribosome biogenesis factor is supported by the nuclear accumulation of 40S and 60S ribosome subunits when Rli1 function is compromised (Kispal et al., 2005; Yarunin et al., 2005). Nevertheless, the most well-known Rli1 function is that of ribosome disassembly (Figure 2). Indeed, Rli1 facilitates ribosomal subunit dissociation through its direct interaction with the class I release factor Supressor 45 (Sup45), otherwise known as eukaryotic release factor 1 (eRF1) or its paralog Duplication of Multilocus region 34 (Dom34). Such interaction was confirmed by affinity pull-down, coimmunoprecipitation, and yeast two-hybrid analyses (Khoshnevis et al., 2010; Shoemaker and Green, 2011).
Yeast Rli1 participates in ribosome recycling
Translation termination occurs when the stop codon of an mRNA enters the A site of the associated ribosome, after which the tRNA-mimicking eRF1 recognizes the stop codon via codon-anticodon recognition using its conserved NIKS (Asn-Ile-Lys-Ser) motif and occupies the A site of the ribosome (Song et al., 2000). Following this, the GTP bound to the eRF1-linked class II release factor Sup35/eRF3 is hydrolyzed, thus dissociating the post-termination complex.
During stop codon recognition, eRF3 has been proposed to facilitate the interaction between eRF1 and the ribosome. This function is thought to resemble delivery of aminoacylated tRNA to the ribosome A site during peptide elongation, which is performed by the eRF3 paralog eEF1α (Inagaki et al., 2003; Salas-Marco and Bedwell, 2004; des Georges et al., 2014). In an alternative scenario, DEAD-box protein 5 (Dbp5), an RNA helicase that participates in mRNA export, recruits eRF3·GTP to the ribosome following eRF1 stop codon recognition (Gross et al., 2007). Regardless of the exact protein-protein interactions, eRF3 must break down GTP and leave the ribosome to allow Rli1 binding (Shoemaker and Green, 2011; Preis et al., 2014).
The kinetic analysis of an in vitro reconstituted yeast translation system demonstrated that Rli1 induces eRF1 ribosome accommodation in an ATP-independent manner. Ribosome accommodation allows eRF1 to catalyze peptidyl-tRNA hydrolysis through its conserved GGQ (Gly-Gly-Gln) motif, which releases the newly synthesized peptide. ATP hydrolysis driven by Rli1 promotes ribosome subunit dissociation, demonstrating that eukaryotic translation termination and ribosome recycling are combined within the same release factor-mediated process. Such combination contrasts with the separation of the two processes observed in bacteria (Shoemaker and Green, 2011). In this manner, the 60S subunit is disassembled from the 40S subunit, which is then released from the deacylated tRNA and mRNA molecules. During all this process of ribosome recycling, Rli1 remains bound to the 40S subunit and has been suggested to preclude 60S rejoining until a late-stage in the initiation complex (Heuer et al., 2017; Mancera-Martínez et al., 2017). In the case of archaea, it has been proposed that ribosome dissociation is caused by a conformational change following aABCE1-ribosome interaction, and that ATP hydrolysis is required to separate aABCE1 from the 30S subunit following ribosome dissociation (Barthelme et al., 2011; Kiosze-Becker et al., 2016).
Termination of translation can be inefficient. One of the known causes of inefficient translation termination is the continued association of ribosomes with defective mRNA molecules, which impairs translation and produces potentially deleterious peptides. To circumvent this, different mRNA surveillance pathways can degrade defective mRNA molecules and their translation products, and recycle the associated ribosomes. These mRNA surveillance pathways have been extensively reviewed (Franckenberg et al., 2012; Graille and Séraphin, 2012; Shoemaker and Green, 2012; Brandman and Hegde, 2016), so they are only briefly discussed here.
Each of the three primary mRNA surveillance mechanisms targets a different cause of ribosome stalling and Rli1 participates in all three mechanisms. In no-go decay (NGD), a physical obstruction slows down or stops ribosome progression on the mRNA molecule. Physical obstructions can include an inhibitory secondary structure, chemical damage, or a polybasic sequence within the nascent protein (Doma and Parker, 2006; Kuroha et al., 2010). Non-stop decay (NSD) occurs when the mRNA lacks a genuine stop codon, possibly due to truncation or premature polyadenylation of the mRNA molecule. In NSD, the ribosome continues translation until it encounters an in-frame stop codon on the 3′ UTR or comes to the poly(A) mRNA sequence (tail). Translation of the poly(A) tail generates a positively charged poly-lysine region that disturbs ribosome movement by interacting with its negatively charged translation tunnel (Frischmeyer et al., 2002; Ito-Harashima et al., 2007; Lu and Deutsch, 2008; Guydosh and Green, 2014). Lastly, non-functional 18S rRNA decay (18S-NRD) repairs errors in translation that are caused by dysfunctional ribosomes carrying an inactive or immature 40S subunit. 18S-NRD rapidly removes these faulty ribosomes that have initiated translation, but cannot produce an elongating peptide (LaRiviere et al., 2006; Soudet et al., 2010).
Each of these surveillance systems uses the same basic ribosome rescue machinery components. Ribosome rescue starts with recognition of the stalled ribosome by a ternary complex formed by Dom34 and Hsp70 subfamily B Suppressor 1 (Hbs1·GTP), which are paralogs of eRF1 and eRF3, respectively (Cole et al., 2009; Shoemaker et al., 2010; Tsuboi et al., 2012). Following this, Rli1 dissociates the ribosome into the 40S and 60S subunits via a similar mechanism as during the normal termination of translation. Dom34 lacks the conserved NIKS motif that is involved in stop codon recognition and the GGQ motif that catalyzes peptide release, which are characteristic features of eRF1 (Graille et al., 2008; Shoemaker et al., 2010).
The mRNA surveillance pathways appear to be conserved among all eukaryotes and archaea. For example, Pelota, the Dom34 paralog in Drosophila melanogaster, can restore NGD in Dom34-depleted yeast cells (Passos et al., 2009). Also, the human and fly Pelota-Hbs1 complex, together with ABCE1/Pixie, participates in NSD (Pisareva et al., 2011; Saito et al., 2013; Kashima et al., 2014). In archaea, the elongation factor aEF1α, an ortholog of eRF3, interacts with aRF1 during the normal termination of translation and aPelota during mRNA surveillance, resulting in ribosome dissociation via aABCE1 action (Saito et al., 2010; Barthelme et al., 2011; Becker et al., 2012).
Rli1 is required for ribosome biogenesis and reactivation
Yeast ribosome biogenesis begins in the nucleolus where the 35S and 5S rDNAs are transcribed. Following this, pre-35S and pre-5S rRNAs are cotranscriptionally assembled with ribosomal proteins, ribosome biogenesis factors (RBFs), and small nucleolar ribonucleoproteins (snoRNPs) to form the 90S or small subunit (SSU) processome, which is the earliest ribosome precursor. Cleavage of the 35S pre-rRNA creates the pre-60S and pre-40S particles, and the maturation of these continues in the nucleoplasm and the cytoplasm (Gerhardy et al., 2014). Once in the cytoplasm, RBFs prevent premature translation initiation on immature pre-60S and pre-40S particles (Gartmann et al., 2010; Strunk et al., 2011; Greber et al., 2012).
Maturation of the pre-60S particle is complete when the pre-6S rRNA is processed to form 5.8S rRNA, all ribosomal proteins are assembled, and the last RBFs are released (Lo et al., 2010). The last step in 40S subunit maturation is performed in a translation-like cycle, whereby the initiation factor eIF5B links a pre-40S particle to a mature 60S subunit to form an empty 80S-like ribosome, which is necessary for 20S pre-rRNA cleavage into mature 18S rRNA. This process also serves as an additional checkpoint that, together with those performed during subunit maturation, ensures functionality of the ribosome (Strunk et al., 2012; Karbstein, 2013). Finally, Rli1, together with Dom34 and possibly Hbs1, dissociates the ribosomal subunits, which are then ready to enter the translation cycle (Shoemaker and Green, 2011; Strunk et al., 2012). Likewise, human Pelota and ABCE1 can dissociate empty ribosomes in vitro (Pisareva et al., 2011).
Protein synthesis is a cyclic process in which ribosomal subunits dissociate once they have processed an mRNA molecule and subsequently can reinitiate translation. However, in yeast cells subjected to stress conditions, some ribosomal subunits are complexed into inactive 80S ribosomes to reduce translation rates and increase the probability of surviving (Ashe et al., 2000; Uesono and Toh, 2002). In yeast, these inactive ribosomes are stabilized by the Suppressor of ToM1 (Stm1) factor (Balagopal and Parker, 2011; Ben-Shem et al., 2011; Van Dyke et al., 2013). Ribosomal inactivation is reversed once stress conditions are relieved and the ribosomal subunits can reenter the translation cycle. In glucose-starved yeast cells, dissociation of Stm1-bound ribosome requires the combined action of the Dom34-Hbs1 complex and Rli1. When yeast cells are grown in glucose-deficient media, translation rates rapidly decrease, associated with a decline in polysome levels and the accumulation of Stm1-inactivated 80S ribosomes. With the addition of glucose, translation rapidly resumes; however, Dom34 or Hbs1 loss-of-function prevents the recovery of translation, which causes a cessation in growth (Ashe et al., 2000; van den Elzen et al., 2014).
The role of ABCE proteins in development
Yeast Rli1 is the best-characterized ABCE protein. In yeast and the unicellular protist Trypanosoma brucei, ABCE1 loss-of-function arrests growth (Dong et al., 2004; Estévez et al., 2004). Similar observations have been made in multicellular eukaryotes, showing that in some species, ABCE1 loss-of-function and overexpression both result in impaired growth (Table 1; Amsterdam et al., 2004; Estévez et al., 2004; Zhao et al., 2004; Coelho et al., 2005a; Chen et al., 2006; Kougioumoutzi et al., 2013). Consistent with the fundamental role of ABCE1 in ribosome biogenesis and recycling, ABCE1 expression is detected in most tissues and developmental stages in all studied organisms (Du et al., 2003; Zhao et al., 2004; Maeda et al., 2005; Sarmiento et al., 2006; Kougioumoutzi et al., 2013).
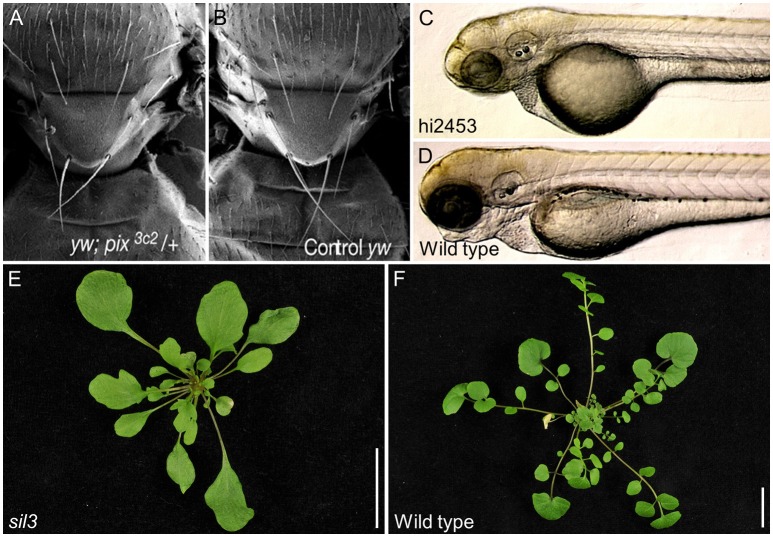
Many of the effects caused by ABCE1 loss-of-function in eukaryotes have been revealed through genetic screens. For example, the pixie alleles were identified in a screen for ethyl methanesulfonate-induced dominant modifiers of a small-wing phenotype in Drosophila melanogaster (Coelho et al., 2005b). Null pixie alleles are recessive lethal, whereas hypomorphic alleles produce a severe delay in growth (Table 1). Nevertheless, the final body size of pixie mutants is comparable to that of the wild type. The pixie mutant phenotype resembles the Minute phenotype, which is associated with mutant alleles of genes encoding ribosomal proteins. Adult pixie flies display short thoracic bristles, occasional eye roughening, and an increased wing size relative to body size (Figures 3A,B). The increased wing size relative to body size is proposed to be due to extra cell divisions that act as a compensation mechanism triggered by high-level apoptosis observed in the wing imaginal discs during the development of pixie larvae (Coelho et al., 2005a).
Figure 3.
Developmental effects of ABCE gene dysfunction in different species. (A) Drosophila melanogaster pixie mutants display thoracic bristles that are slenderer and shorter than those of (B) the wild type. (C) An insertional allele of Danio rerio ABCE1 reduces head size compared with (D) wild type and produces pericardial edema and lethality at 5 days post-fertilization. (E) A hypomorphic allele of Cardamine hirsuta SIL3 triggers loss of the leaflets that characterize (F) wild type leaves. Pictures of (A,B), as well as (C,D), were taken at the same scale. (E,F) Scale bars indicate 2 cm. Adapted with permission of authors and journals from (A,B) Coelho et al. (2005a), (C,D) Amsterdam et al. (2004) Copyright 2004 National Academy of Sciences, and (E,F) Kougioumoutzi et al. (2013).
ABCE proteins also function in vertebrate development. In a large screen for essential genes in embryo and early larval development in Danio rerio, mutations in ABCE1 were found to cause lethality 5 days after fertilization. These abce1 mutants exhibited underdeveloped liver/gut, pericardial edema, and small heads. Like the hypomorphic pixie alleles, the zebrafish ABCE1 alleles also limit eye development (Table 1 and Figures 3C,D; Amsterdam et al., 2004). The abce1 gene is also essential in Caenorhabditis elegans and Xenopus laevis as its suppression by RNAi or antisense morpholino oligonucleotides, respectively, arrests growth (Table 1; Kamath et al., 2003; Zhao et al., 2004; Chen et al., 2006). The ability of the ABCE1-suppressed zebrafish and Xenopus laevis embryos to develop up to a certain point has been suggested to be due to the ABCE1 maternal supply to the egg. The growth arrest would therefore occur after depletion of the maternal supply (Amsterdam et al., 2004; Chen et al., 2006).
ABCE1 genes also have essential functions in plants. In Cardamine hirsuta, SIMPLE LEAF3 (SIL3, also named ChRLI2) plays a role in leaf complexity. While wild-type Cardamine hirsuta plants display compound leaves, which are divided into leaflets, homozygous plants carrying the putatively hypomorphic sil3 allele show a substantial decrease in leaflet but not leaf number (Table 1 and Figures 3E,F; Kougioumoutzi et al., 2013). The sil3 mutant has a reduced growth and its phenotype suggests alterations in auxin homeostasis at the whole-organism level. Further proof of perturbed auxin homeostasis was found by analyzing sil3 leaves, which are small and exhibit aberrant venation patterns, as usually observed in mutants affected in auxin signaling. In addition, the lack of leaflets in sil3 plants was explained by reduced cell proliferation on the regions where leaflets were expected to emerge. Nevertheless, leaf and leaflet initiation are controlled by the same general mechanisms that consist in the accumulation of auxin by its polarized flow through the PIN-FORMED1 (PIN1) transporters in the regions where leaf initiation occurs. Auxin accumulation then triggers leaf or leaflet initiation (Scarpella et al., 2010). Although the study of the sil3 mutant is consistent with auxin homeostasis and signaling being sensitive to perturbation of ribosomal activity, the sil3 mutant is interesting because leaflet number, but not leaf number, is reduced. To explain this observation, it has been suggested that the high energy demand from cells proliferating during leaflet development could not be satisfied in sil3 plants due to suboptimal ribosome function (Kougioumoutzi et al., 2013). The two Arabidopsis ABCE genes have not been studied at a developmental level.
In addition, virus-induced gene silencing (VIGS) has been used to suppress expression of RLIh gene(s) in Nicotiana benthamiana, in which the number of ABCE paralogs remains to be established. For VIGS, 4-week-old plants were infected with potato virus X (PVX) or pea early browning virus (PEBV) vectors carrying partial RLIh cDNAs. RLIh silencing caused vein whitening, leaf distortion, and delayed growth, with silenced plants reaching only half of the height of controls, due to a reduction in cell size and number in shoot internodes (Table 1; Petersen et al., 2004). Again, these observations corroborate the important role of ABCE proteins in whole-organism development. Whether these developmental defects are due to the disruption of the ABCE function as a ribosome-dissociating factor (Franckenberg et al., 2012), an endogenous suppressor of RNA silencing (Sarmiento et al., 2006; Kärblane et al., 2015) or both, remains to be clarified.
Concluding remarks
In this review, we have discussed the essential ribosome-dissociation activity of ABCE proteins, which is required for ribosome biogenesis and recycling. Furthermore, we have described the general growth defects associated with compromised ABCE protein function in all studied organisms. Most of these growth defects can be attributed to defective mRNA translation, which reduces protein levels and in turn prevents cells from generating the energy required for normal growth and/or proliferation. However, not all abce mutant phenotypes can be explained by a general depletion of cellular energy. As is the case for mutants affected in translation in several species, some phenotypes appear to be associated with compromised regulatory networks that remain uncharacterized. For instance, it is striking the fact that the Cardamine hirsuta sil3 mutant shows a reduction in leaflet but not leaf number, given that both processes share common pathways. Future work is needed to clarify these and other questions and to determine whether there are specific factors responsible for the differential requirements for ABCEs during development. Additional studies will also determine how ABCE dysfunction causes specific developmental aberrations in all studied organisms, as has been previously described for other proteins involved in ribosome biogenesis or function. In addition, the presence of more than one ABCE subfamily member in some genomes remains to be explained. Some plants and insects have two ABCE genes while other plants or Drosophila melanogaster have a single ABCE gene. The presence of two ABCE genes might be interpreted as an in-progress pseudogenization or a developing functional redundancy.
The ribosome has been proposed to represent a layer of post-transcriptional regulation of gene expression. Under the so-called filter hypothesis, the ribosome is viewed as a machine able to selectively influence or filter the translation of various mRNAs (Mauro and Edelman, 2002). There is increasing evidence of the existence of specialized ribosomes in different cell types, which are heterogeneous in either ribosomal protein composition, in their interactions with ribosome-associated factors, or both (Xue and Barna, 2012; Shi and Barna, 2015). These specialized ribosomes would differentially translate different mRNAs or mRNA groups. It follows from these assumptions that mutation of genes encoding discrete ribosomal proteins or ribosome-associated factors would render tissue- or organ-specific phenotypes. The ABCE proteins of Drosophila melanogaster, Caenorhabditis elegans, and Cardamine hirsuta might represent one such factors.
Author contributions
JLM designed the review. JLM, EM-B and CN-Q analyzed all literature data, prepared Figures 1–3, and Table 1 and wrote the manuscript. All authors have accepted the final version of the manuscript and agreed to be accountable for all aspects of the work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Enrique Lopez-Juez for his useful comments on the manuscript. Research in the laboratory of JLM is supported by grants from the Ministerio de Economía y Competitividad of Spain (BIO2014-53063-P) and the Generalitat Valenciana (PROMETEOII/2014/006). CN-Q and EM-B hold predoctoral fellowships from the Universidad Miguel Hernández and Ministerio de Educación, Cultura y Deporte of Spain (FPU13/00371), respectively.
References
- Alhebshi A., Sideri T. C., Holland S. L., Avery S. V. (2012). The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Mol. Biol. Cell 23, 3582–3590. 10.1091/mbc.e12-05-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A., Nissen R. M., Sun Z., Swindell E. C., Farrington S., Hopkins N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. U.S.A. 101, 12792–11297. 10.1073/pnas.0403929101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen C. B., Becker T., Blau M., Anand M., Halic M., Balar B., et al. (2006). Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443, 663–668. 10.1038/nature05126 [DOI] [PubMed] [Google Scholar]
- Andersen D. S., Leevers S. J. (2007). The essential Drosophila ATP-binding cassette domain protein, Pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J. Biol. Chem. 282, 14752–14760. 10.1074/jbc.M701361200 [DOI] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B. (2000). Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11, 833–848. 10.1091/mbc.11.3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V., Parker R. (2011). Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17, 835–842. 10.1261/rna.2677311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D., Dinkelaker S., Albers S. V., Londei P., Ermler U., Tampé R. (2011). Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci. U.S.A. 108, 3228–3233. 10.1073/pnas.1015953108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D., Scheele U., Dinkelaker S., Janoschka A., Macmillan F., Albers S. V., et al. (2007). Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 282, 14598–14607. 10.1074/jbc.M700825200 [DOI] [PubMed] [Google Scholar]
- Becker T., Franckenberg S., Wickles S., Shoemaker C. J., Anger A. M., Armache J. P., et al. (2012). Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506. 10.1038/nature10829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau De Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011). The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Bisbal C., Martinand C., Silhol M., Lebleu B., Salehzada T. (1995). Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270, 13308–13317. 10.1074/jbc.270.22.13308 [DOI] [PubMed] [Google Scholar]
- Brandman O., Hegde R. S. (2016). Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 23, 7–15. 10.1038/nsmb.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broehan G., Kroeger T., Lorenzen M., Merzendorfer H. (2013). Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics 14, 1–18. 10.1186/1471-2164-14-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Q., Dong J., Ishimura A., Daar I., Hinnebusch A. G., Dean M. (2006). The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J. Biol. Chem. 281, 7452–7457. 10.1074/jbc.M510603200 [DOI] [PubMed] [Google Scholar]
- Coelho C. M., Kolevski B., Bunn C., Walker C., Dahanukar A., Leevers S. J. (2005a). Growth and cell survival are unevenly impaired in pixie mutant wing discs. Development 132, 5411–5424. 10.1242/dev.02148 [DOI] [PubMed] [Google Scholar]
- Coelho C. M., Kolevski B., Walker C. D., Lavagi I., Shaw T., Ebert A., et al. (2005b). A genetic screen for dominant modifiers of a small-wing phenotype in Drosophila melanogaster identifies proteins involved in splicing and translation. Genetics 171, 597–614. 10.1534/genetics.105.045021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. E., LaRiviere F. J., Merrikh C. N., Moore M. J. (2009). A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell 34, 440–450. 10.1016/j.molcel.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Annilo T. (2005). Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genom. Hum. Genet. 6, 123–142. 10.1146/annurev.genom.6.080604.162122 [DOI] [PubMed] [Google Scholar]
- Dean M., Rzhetsky A., Allikmets R. (2001). The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166. 10.1101/gr.GR-1649R [DOI] [PubMed] [Google Scholar]
- Dermauw W., Van Leeuwen T. (2014). The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110. 10.1016/j.ibmb.2013.11.001 [DOI] [PubMed] [Google Scholar]
- des Georges A., Hashem Y., Unbehaun A., Grassucci R. A., Taylor D., Hellen C. U., et al. (2014). Structure of the mammalian ribosomal pre-termination complex associated with eRF1·eRF3·GDPNP. Nucleic Acids Res. 42, 3409–3418. 10.1093/nar/gkt1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R. (2006). Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564. 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Lai R., Nielsen K., Fekete C. A., Qiu H., Hinnebusch A. G. (2004). The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 279, 42157–42168. 10.1074/jbc.M404502200 [DOI] [PubMed] [Google Scholar]
- Dooher J. E., Lingappa J. R. (2004). Conservation of a stepwise, energy-sensitive pathway involving HP68 for assembly of primate lentivirus capsids in cells. J. Virol. 78, 1645–1656. 10.1128/JVI.78.4.1645-1656.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X. L., Wang D., Qian X. Y., Jiang L. Z., Chun W., Li K. G., et al. (2003). cDNA cloning and expression analysis of the rice (Oryza sativa L.) RNase L inhibitor. DNA Seq. 14, 295–301. 10.1080/1085566031000141162 [DOI] [PubMed] [Google Scholar]
- Estévez A. M., Haile S., Steinbüchel M., Quijada L., Clayton C. (2004). Effects of depletion and overexpression of the Trypanosoma brucei ribonuclease L inhibitor homologue. Mol. Biochem. Parasitol. 133, 137–141. 10.1016/j.molbiopara.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Franckenberg S., Becker T., Beckmann R. (2012). Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr. Opin. Struct. Biol. 22, 786–796. 10.1016/j.sbi.2012.08.002 [DOI] [PubMed] [Google Scholar]
- Frischmeyer P. A., Van Hoof A., O'donnell K., Guerrerio A. L., Parker R., Dietz H. C. (2002). An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–22561. 10.1126/science.1067338 [DOI] [PubMed] [Google Scholar]
- Gartmann M., Blau M., Armache J. P., Mielke T., Topf M., Beckmann R. (2010). Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. Biol. Chem. 285, 14848–14851. 10.1074/jbc.C109.096057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardy S., Menet A. M., Peña C., Petkowski J. J., Panse V. G. (2014). Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 123, 327–344. 10.1007/s00412-014-0463-z [DOI] [PubMed] [Google Scholar]
- Graille M., Chaillet M., Van Tilbeurgh H. (2008). Structure of yeast Dom34: a protein related to translation termination factor eRF1 and involved in no-go decay. J. Biol. Chem. 283, 7145–7154. 10.1074/jbc.M708224200 [DOI] [PubMed] [Google Scholar]
- Graille M., Séraphin B. (2012). Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol. 13, 727–735. 10.1038/nrm3457 [DOI] [PubMed] [Google Scholar]
- Greber B. J., Boehringer D., Montellese C., Ban N. (2012). Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1228–1233. 10.1038/nsmb.2425 [DOI] [PubMed] [Google Scholar]
- Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J. J., Seedorf M., et al. (2007). The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315, 646–649. 10.1126/science.1134641 [DOI] [PubMed] [Google Scholar]
- Guydosh N. R., Green R. (2014). Dom34 rescues ribosomes in 3' untranslated regions. Cell 156, 950–962. 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A., Gerovac M., Schmidt C., Trowitzsch S., Preis A., Kötter P., et al. (2017). Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat. Struct. Mol. Biol. 24, 453–460. 10.1038/nsmb.3396 [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., et al. (1986). A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323, 448–450. 10.1038/323448a0 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2002). The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399–414. 10.1101/gad.955102 [DOI] [PubMed] [Google Scholar]
- Holland I. B., Blight M. A. (1999). ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293, 381–399. 10.1006/jmbi.1999.2993 [DOI] [PubMed] [Google Scholar]
- Hopfner K. P. (2016). Architectures and mechanisms of ATP binding cassette proteins. Biopolymers 105, 492–504. 10.1002/bip.22843 [DOI] [PubMed] [Google Scholar]
- Hwang J. U., Song W. Y., Hong D., Ko D., Yamaoka Y., Jang S., et al. (2016). Plant ABC transporters enable many unique aspects of a terrestrial plant's lfestyle. Mol. Plant 9, 338–355. 10.1016/j.molp.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Hyde S. C., Emsley P., Hartshorn M. J., Mimmack M. M., Gileadi U., Pearce S. R., et al. (1990). Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346, 362–365. 10.1038/346362a0 [DOI] [PubMed] [Google Scholar]
- Inagaki Y., Blouin C., Susko E., Roger A. J. (2003). Assessing functional divergence in EF-1α and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 31, 4227–4237. 10.1093/nar/gkg440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S., Kuroha K., Tatematsu T., Inada T. (2007). Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524. 10.1101/gad.1490207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kärblane K., Gerassimenko J., Nigul L., Piirsoo A., Smialowska A., Vinkel K., et al. (2015). ABCE1 is a highly conserved RNA silencing suppressor. PLoS ONE 10:e0116702. 10.1371/journal.pone.0116702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K. (2013). Quality control mechanisms during ribosome maturation. Trends Cell Biol. 23, 242–250. 10.1016/j.tcb.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher A., Büttner K., Märtens B., Jansen R. P., Hopfner K. P. (2005). X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure 13, 649–659. 10.1016/j.str.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Karcher A., Schele A., Hopfner K. P. (2008). X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J. Biol. Chem. 283, 7962–7971. 10.1074/jbc.M707347200 [DOI] [PubMed] [Google Scholar]
- Kashima I., Takahashi M., Hashimoto Y., Sakota E., Nakamura Y., Inada T. (2014). A functional involvement of ABCE1, eukaryotic ribosome recycling factor, in nonstop mRNA decay in Drosophila melanogaster cells. Biochimie 106, 10–16. 10.1016/j.biochi.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Kerr I. D. (2004). Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem. Biophys. Res. Commun. 315, 166–173. 10.1016/j.bbrc.2004.01.044 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S., Gross T., Rotte C., Baierlein C., Ficner R., Krebber H. (2010). The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 11, 214–219. 10.1038/embor.2009.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosze-Becker K., Ori A., Gerovac M., Heuer A., Nürenberg-Goloub E., Rashid U. J., et al. (2016). Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry. Nat. Commun. 7, 1–19. 10.1038/ncomms13248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G., Sipos K., Lange H., Fekete Z., Bedekovics T., Janáky T., et al. (2005). Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24, 589–598. 10.1038/sj.emboj.7600541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kougioumoutzi E., Cartolano M., Canales C., Dupré M., Bramsiepe J., Vlad D., et al. (2013). SIMPLE LEAF3 encodes a ribosome-associated protein required for leaflet development in Cardamine hirsuta. Plant J. 73, 533–545. 10.1111/tpj.12072 [DOI] [PubMed] [Google Scholar]
- Kuroha K., Akamatsu M., Dimitrova L., Ito T., Kato Y., Shirahige K., et al. (2010). Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11, 956–961. 10.1038/embor.2010.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere F. J., Cole S. E., Ferullo D. J., Moore M. J. (2006). A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell 24, 619–626. 10.1016/j.molcel.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Liu S., Li Q., Liu Z. (2013). Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS ONE 8:e63895. 10.1371/journal.pone.0063895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K. Y., Li Z., Bussiere C., Bresson S., Marcotte E. M., Johnson A. W. (2010). Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39, 196–208. 10.1016/j.molcel.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Xu Y., Cui F. (2016). Phylogenetic analysis of the ATP-binding cassette transporter family in three mosquito species. Pestic. Biochem. Physiol. 132, 118–124. 10.1016/j.pestbp.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Lu J., Deutsch C. (2008). Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86. 10.1016/j.jmb.2008.08.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Lee J. M., Miyagawa Y., Koga K., Kawaguchi Y., Kusakabe T. (2005). Cloning and characterization of a ribonuclease L inhibitor from the silkworm, Bombyx mori. DNA Seq. 16, 21–27. 10.1080/10425170400028871 [DOI] [PubMed] [Google Scholar]
- Mancera-Martínez E., Brito Querido J., Valasek L. S., Simonetti A., Hashem Y. (2017). ABCE1: a special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol. 14, 1279–1285. 10.1080/15476286.2016.1269993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V. P., Edelman G. M. (2002). The ribosome filter hypothesis. Proc. Natl. Acad. Sci. U.S A. 99, 12031–12036. 10.1073/pnas.192442499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos D. O., Doma M. K., Shoemaker C. J., Muhlrad D., Green R., Weissman J., et al. (2009). Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell 20, 3025–3032. 10.1091/mbc.e09-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumi C. M., Chuk M., Snider J., Stagljar I., Michaelis S. (2009). ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593. 10.1128/MMBR.00020-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen B. O., Jørgensen B., Albrechtsen M. (2004). Isolation and RNA silencing of homologues of the RNase L inhibitor in Nicotiana species. Plant Sci. 167, 1283–1289. 10.1016/j.plantsci.2004.06.030 [DOI] [Google Scholar]
- Pisareva V. P., Skabkin M. A., Hellen C. U., Pestova T. V., Pisarev A. V. (2011). Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817. 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D. E., Berninghausen O., et al. (2014). Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 8, 59–65. 10.1016/j.celrep.2014.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Kobayashi K., Wada M., Kikuno I., Takusagawa A., Mochizuki M., et al. (2010). Omnipotent role of archaeal elongation factor 1 alpha (EF1α) in translational elongation and termination, and quality control of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 19242–19247. 10.1073/pnas.1009599107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Hosoda N., Hoshino S. (2013). The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 288, 17832–17843. 10.1074/jbc.M112.448977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J., Bedwell D. M. (2004). GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24, 7769–7778. 10.1128/MCB.24.17.7769-7778.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento C., Nigul L., Kazantseva J., Buschmann M., Truve E. (2006). AtRLI2 is an endogenous suppressor of RNA silencing. Plant Mol. Biol. 61, 153–163. 10.1007/s11103-005-0001-8 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Barkoulas M., Tsiantis M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2:a001511. 10.1101/cshperspect.a001511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Barna M. (2015). Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Ann. Rev. Cell. Dev. Biol. 31, 31–54. 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- Shoemaker C. J., Eyler D. E., Green R. (2010). Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372. 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Green R. (2011). Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, E1392–E1398. 10.1073/pnas.1113956108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Green R. (2012). Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601. 10.1038/nsmb.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Mugnier P., Das A. K., Webb H. M., Evans D. R., Tuite M. F., et al. (2000). The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321. 10.1016/S0092-8674(00)80667-4 [DOI] [PubMed] [Google Scholar]
- Soudet J., Gélugne J. P., Belhabich-Baumas K., Caizergues-Ferrer M., Mougin A. (2010). Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 29, 80–92. 10.1038/emboj.2009.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B. S., Loucks C. R., Su M., Vashisth H., Cheng S., Schilling J., Skiniotis G., et al. (2011). Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333, 1449–1453. 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B. S., Novak M. N., Young C. L., Karbstein K. (2012). A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150, 111–121. 10.1016/j.cell.2012.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Beek J., Guskov A., Slotboom D. J. (2014). Structural diversity of ABC transporters. J. Gen. Physiol. 143, 419–435. 10.1085/jgp.201411164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomii K., Kanehisa M. (1998). A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 8, 1048–1059. 10.1101/gr.8.10.1048 [DOI] [PubMed] [Google Scholar]
- Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., et al. (2012). Dom34:Hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3' end of aberrant mRNA. Mol. Cell 46, 518–529. 10.1016/j.molcel.2012.03.013 [DOI] [PubMed] [Google Scholar]
- Uesono Y., Toh E. A. (2002). Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277, 13848–13855. 10.1074/jbc.M108848200 [DOI] [PubMed] [Google Scholar]
- van den Elzen A. M., Schuller A., Green R., Séraphin B. (2014). Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 33, 265–276. 10.1002/embj.201386123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke N., Chanchorn E., Van Dyke M. W. (2013). The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochem. Biophys. Res. Commun. 430, 745–750. 10.1016/j.bbrc.2012.11.078 [DOI] [PubMed] [Google Scholar]
- Vasiliou V., Vasiliou K., Nebert D. W. (2009). Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290. 10.1186/1479-7364-3-3-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier P. J., Bird D., Burla B., Dassa E., Forestier C., Geisler M., et al. (2008). Plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. 10.1016/j.tplants.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Wang B., Dukarevich M., Sun E. I., Yen M. R., Saier M. H., Jr. (2009). Membrane porters of ATP-binding cassette transport systems are polyphyletic. J. Membr. Biol. 231, 1–10. 10.1007/s00232-009-9200-6 [DOI] [PubMed] [Google Scholar]
- Xie X., Cheng T., Wang G., Duan J., Niu W., Xia Q. (2012). Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Mol. Biol. Rep. 39, 7281–7291. 10.1007/s11033-012-1558-3 [DOI] [PubMed] [Google Scholar]
- Xue S., Barna M. (2012). Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369. 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A., Panse V. G., Petfalski E., Dez C., Tollervey D., Hurt E. C. (2005). Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24, 580–588. 10.1038/sj.emboj.7600540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Fang L. L., Johnsen R., Baillie D. L. (2004). ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 323, 104–111. 10.1016/j.bbrc.2004.08.068 [DOI] [PubMed] [Google Scholar]
- Zheng W. H., Västermark A., Shlykov M. A., Reddy V., Sun E. I., Saier M. H., et al. (2013). Evolutionary relationships of ATP-Binding Cassette (ABC) uptake porters. BMC Microbiol. 13, 1–20. 10.1186/1471-2180-13-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman C., Klein K. C., Kiser P. K., Singh A. R., Firestein B. L., Riba S. C., et al. (2002). Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415, 88–92. 10.1038/415088a [DOI] [PubMed] [Google Scholar]