Abstract
Bicoid interacting 3 domain containing RNA methyltransferase (BCDIN3D) is a member of the Bin3 methyltransferase family and is evolutionary conserved from worm to human. BCDIN3D is overexpressed in breast cancer, which is associated with poor prognosis of breast cancers. However, the biological functions and properties of BCDIN3D have been enigmatic. Recent studies have revealed that human BCDIN3D monomethylates 5′-monophsosphate of cytoplasmic tRNAHis in vivo and in vitro. BCDIN3D recognizes the unique and exceptional structural features of cytoplasmic tRNAHis and discriminates tRNAHis from other cytoplasmic tRNA species. Thus, BCDIN3D is a tRNAHis-specific 5′-monophosphate methyltransferase. Methylation of the 5′-phosphate group of tRNAHis does not significantly affect tRNAHis aminoacylation by histidyl-tRNA synthetase in vitro nor the steady state level or stability of tRNAHis in vivo. Hence, methylation of the 5′-phosphate group of tRNAHis by BCDIN3D or tRNAHis itself may be involved in certain unknown biological processes, beyond protein synthesis. This review discusses recent reports on BCDIN3D and the possible association between 5′-phosphate monomethylation of tRNAHis and the tumorigenic phenotype of breast cancer.
Keywords: Bicoid interacting 3 domain containing RNA methyltransferase, methylation, tRNA, breast cancer, protein synthesis
Introduction
Bicoid interacting 3 domain containing RNA methyltransferase (BCDIN3D) contains an S-(5′-adenosyl)-L-methionine (AdoMet) binding motif, and is homologous to a conserved family of eukaryotic protein methyltransferases acting on RNA-binding proteins (Zhu and Hanes, 2000). The BCDIN3D is evolutionary conserved and has been identified in various animals from worms to human (Xhemalce et al., 2012), however, its biological properties and functions are unclear. BCDIN3D mRNA overexpression has been reported in human breast cancer cells, which is associated with cellular invasion and poor prognosis in triple-negative breast cancer (Liu et al., 2007; Yao et al., 2016). The molecular basis of involvement of BCDIN3D in the tumorigenic phenotype of breast cancer has remained elusive. This review discusses recent studies on human BCDIN3D. We describe herein that a specific tRNA for histidine (tRNAHis) is now identified as a primary target of BCDIN3D and discuss the association between the tumorigenic phenotype of breast cancer and the methylation of tRNAHis by BCDIN3D.
How Does Human Bcdin3D Recognize Specific Rna?
Xhemalce et al. (2012) reported that BCDIN3D catalyzes dimethylation of 5′-monophosphate of specific precursor microRNAs (pre-miRNAs) (Xhemalce et al., 2012), such as tumor suppressor miR145 and miR23b (He et al., 2007; Shi et al., 2007; Sachdeva et al., 2009; Spizzo et al., 2010), using AdoMet as a methyl-group donor. Dimethylation of the 5′-monophosphate of pre-miRNA nullifies the negative charge at the 5′-terminal of pre-miRNA. Since Dicer recognizes the negative charge at the 5′-terminal of pre-miRNAs for efficient and accurate cleavage (Park et al., 2011), the dimethylation of 5′-phosphate of pre-miRNA inhibits subsequent processing. Consequently, mature miRNAs are down-regulated. They also reported that the depletion of BCDIN3D mRNA by specific shRNAs suppressed the tumorigenic phenotype of MDA-MB231 breast cancer cells (Xhemalce et al., 2012). Therefore, it was proposed that BCDIN3D promotes the cellular invasion of breast cancer cells by downregulating tumor suppressor miRNAs through dimethylation of the 5′-phosphate group of the corresponding pre-miRNAs. However, there are no apparent common features including primary or secondary structures among the corresponding pre-miRNAs of downregulated miRNAs in breast cancer cells. Thus, the mechanisms by which BCDIN3D recognizes only a specific group of pre-miRNAs and downregulates mature miRNAs in breast cancer cells are unclear.
Cytoplasmic tRnaHis is Co-Purified With Bcdin3D and Contains a 5′-Monomethylmonophosphate Group
To identify other potential RNA substrates of BCDIN3D in vivo and to elucidate the mechanism underlying the recognition and regulation of specific RNAs by BCDIN3D, recently, BCDIN3D-binding RNAs in human HEK293T cells were analyzed (Martinez et al., 2017). When BCDIN3D, expressed in HEK293T cells, was purified from the cell extracts, a distinct 70–80-nucleotie-long RNA molecule was co-purified with BCDIN3D protein.
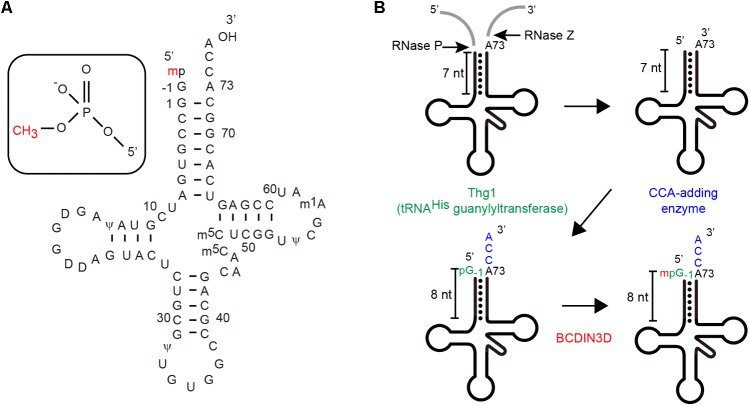
It was assumed that this co-purified RNA might be cytoplasmic tRNAHis (Figure 1A), since the nucleotide sequences of cytoplasmic tRNAHis from human and fruit fly reportedly contained a 5′-monomehtylphosphate group (Cooley et al., 1982; Rosa et al., 1983). Analysis of the RNA co-purified with BCDIN3D via RT-PCR and sequencing confirmed that cytoplasmic tRNAHis is co-purified with BCDIN3D from the cell extracts, but not other tRNAs, such as tRNAPhe. Subsequent direct analysis of the RNA via liquid chromatography and mass spectrometry (LC-MS) revealed that this RNA is cytoplasmic tRNAHis. Moreover, the 5′-monophosphate of cytoplasmic tRNAHis was fully monomethylated, but not dimethylated at all. Furthermore, 5′-monophsophate of tRNAHis is reportedly fully monomethylated even under normal physiological conditions in HEK293T cells, as observed previously in cytoplasmic tRNAHis from HeLa cells (Rosa et al., 1983).
FIGURE 1.
(A) The nucleotide sequence of human cytoplasmic tRNAHis including modified nucleosides. Human cytoplasmic tRNAHis in a clover-leaf structure, wherein the 5′-phosphate group is monomethylated (Rosa et al., 1983). (B) Maturation of human cytoplasmic tRNAHis (Gu et al., 2003; Betat et al., 2014) Cytoplasmic tRNAHis has an additional guanosine residue at position-1 (G-1) and an 8-nucleotide-long acceptor helix with G-1:A72 mis-pairing at the top of the acceptor helix (Rosa et al., 1983).
Cytoplasmic tRnaHis is Methylated by Bcdin3D in Vitro
The enzymatic activity of recombinant human BCDIN3D expressed in E. coli was examined using human cytoplasmic tRNAHis transcript as a substrate and S-(5′-adenosyl)-L- methionine (SAM) as a methyl-group donor in vitro (Martinez et al., 2017). Cytoplasmic tRNAHis transcript was reportedly efficiently methylated by BCDIN3D in vitro; however, unexpectedly, human pre-miR-145, which was previously reportedly dimethylated by BCDIN3D (Xhemalce et al., 2012), is hardly methylated under the same conditions assessed. The reaction products were further analyzed via LC-MS and it was confirmed that BCDIN3D monomethylates 5′-monophosphate of cytoplasmic tRNAHis. Almost 100% of the tRNAHis reaction product comprised 5′-monomethylphosphate. Moreover, BCDIN3D does not dimethylate 5′-monophosphate of tRNAHis or pre-miR145 in vitro. Steady-state kinetics of methylation of these RNA substrates revealed that cytoplasmic tRNAHis is a greater than 2–3 orders of magnitude better substrate than pre-miR145.
BCDIN3D reportedly dimethylates pre-miR145 (Xhemalce et al., 2012). However, only a small fraction (less than 1%) of pre-miR145 substrates was methylated at the reaction end points (Xhemalce et al., 2012). The lower methylation of pre-miR145 by BCDIN3D is consistent with that in the recent study (Martinez et al., 2017). Furthermore, BCDIN3D reportedly transfers two methyl-groups from SAM to 5′-monophosphate of pre-miR145 (Xhemalce et al., 2012). This is inconsistent with the recent findings of Martinez et al. (2017), wherein neither tRNAHis nor pre-miR145 are dimethylated by BCDIN3D in vitro. Perhaps, the efficiency of dimethylation of 5′-phosphate of pre-miR145 by BCDIN3D would be much lower than that of monomethylation of 5′-phosphate of pre-miR145 in vitro. The previously observed methylation of pre-miR145 (Xhemalce et al., 2012) is at baseline levels, as compared with that in cytoplasmic tRNAHis, and is not significant.
Cytoplasmic tRnaHis is Methylated by Bcdin3D in Vivo
BCDIN3D-knockout HEK293T cells, established via CRISPR/Cas9 editing, are viable, although they exhibit a slightly reduced growth rate than the parental cells (Martinez et al., 2017). Cytoplasmic tRNAHis isolated from BCDIN3D-knockout cells completely lost their methyl moiety at the 5′-monophosphate group, as evident from LC-MS. Exogenous expression of BCDIN3D in the BCDIN3D-knockout cell restored the 5′-monomethylphosphate modification of cytoplasmic tRNAHis. Thus, the BCDIN3D is responsible for the monomethylation of 5′-monophosphate of cytoplasmic tRNAHis in HEK293T cells under normal physiological conditions. In BCDIN3D-knockout cells, no other RNAs except for cytoplasmic tRNAHis, are significantly methylated by recombinant BCDIN3D in vitro (Martinez et al., 2017).
A recent study using HEK293T cells reported that BCDIN3D-knockout or BCDIN3D overexpression do not alter mature miR145 expression levels (Martinez et al., 2017), concurrent with the recent in vitro results showing that BCDIN3D does not dimethylate the 5′-monophosphate group of either tRNAHis or pre-miR145. Only pre-miRNA with 5′-dimethylated phosphate, but not pre-miRNA with 5′-monomethylated phosphate, is processed at lower levels by Dicer (Xhemalce et al., 2012). These observations also suggest that BCDIN3D does not dimethylate the 5′-phosphate group of pre-miR145.
Together with the recent results of in vitro methylation assays using recombinant BCDIN3D and tRNAHis transcript and the pre-miR145 transcript (Martinez et al., 2017), the primary target of BCDIN3D is cytoplasmic tRNAHis rather than pre-miRNAs. BCDIN3D displays monomethylation activity on the 5′-phosphate group of RNA. Considering the significantly lower activity of BCDIN3D toward pre-miR145 and that miR145 expression is not regulated by BCDIN3D in vivo, methylation of pre-miR145 probably does not occur in HEK293T cells. Under certain biological process or specific conditions in breast cancer cells, BCDIN3D might recognize specific pre-miRNAs, such as pre-miR145, through the regulatory factors which assist BCDIN3D in recognizing specific RNA species. Elucidation of the regulatory mechanism of specific pre-miRNA (di)methylation process by BCDIN3D in breast cancer cells awaits further study.
tRnaHis Recognition by Bcdin3D
Human cytoplasmic tRNAHis is matured through unique processes and has unique structural features among cytoplasmic tRNA species (Jackman et al., 2012; Betat et al., 2014; Figure 1B). After transcription by RNA polymerase-III, the 5′-leader and 3′-tailer sequences of precursor tRNAHis are cleaved. Thereafter, a single guanosine residue (G) is attached to the 5′-end (at position -1) in the 3′–5′ direction by tRNAHis-specific guanylyltransferase (Thg1) (Gu et al., 2003; Jackman and Phizicky, 2006; Jackman et al., 2012) and the CCA is added at the 3′-end (positions 74–76) (Tomita and Yamashita, 2014). Consequently, the mature form of cytoplasmic tRNAHis has an 8-nucleotide-long acceptor helix with G-1:A73 mis-paring at the top the helix, while other cytoplasmic tRNAs have 7-nucleotide-long acceptor helices.
In vitro steady-state kinetics of methylation of mutant cytoplasmic tRNAHis transcripts by recombinant BCDIN3D revealed that BCIDN3D recognizes G-1, G-1:A73 mis-pairing at the top of the acceptor stem and 8-nucleotide-long extended acceptor helix. The minihelix of tRNAHis is also methylated efficiently by BCIN3D. Thus, BCDIN3D recognizes the unique structural features of cytoplasmic tRNAHis, especially in the top-half region of tRNAHis, and discriminates cytoplasmic tRHAHis from other tRNA species (Martinez et al., 2017).
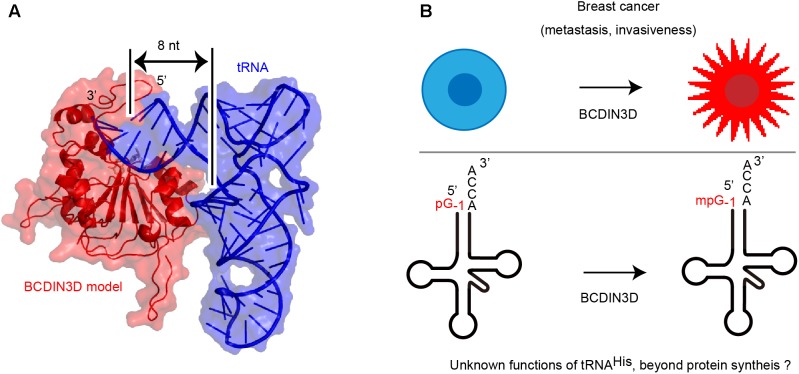
The structure of human BCDIN3D is still unclear. The amino acid sequence of BCDIN3D is homologous to that of the catalytic domain of methylphosphate capping enzyme (MePCE), which uses SAM to transfer a methyl group onto the γ-phosphate of the 5′-guanosine of 7SK RNA (Jeronimo et al., 2007; Shuman, 2007). Structural modeling of human BCDIN3D using the catalytic domain of MePCE and possible tRNA-binding modeling suggest that BCDIN3D recognizes the acceptor stem of tRNAHis and measures the length of acceptor helix of tRNAHis (Figure 2A). Only the 5′-end of tRNA with an 8-nucleotide-long acceptor helix and G-1:A72 mis-pairing at the top the acceptor helix could enter the catalytic pocket of BCDIN3D, and the 5′-phosphate would be monomethylated. The mechanism underlying the recognition of tRNA by BCDIN3D would differ from those for tRNA recognition by the CCA-adding enzymes (Tomita et al., 2004; Yamashita et al., 2014, 2015; Yamashita and Tomita, 2016),which recognize the TΨC loop of tRNA.
FIGURE 2.
(A) A model of human Bicoid interacting 3 domain containing RNA methyltransferase (BCDIN3D)-tRNAHis complex structure. Human BCDIN3D structure was modeled using SWISS-MODEL (Biasini et al., 2014), based on the structure of the catalytic domain of methylphosphate capping enzyme (MePCE) (Jeronimo et al., 2007) as a template model. tRNA was manually docked onto the surface of the modeled BCDIN3D and the 5′-terminal of tRNA can enter the catalytic SAM binding site. BCDIN3D measures the length of the acceptor helix of tRNA. (B) A potential association between methylation of tRNAHis and breast cancer. tRNAHis might have unknown functions, beyond its established function in protein synthesis.
The kinetics of methylation of tRNAHis mutants and the structural model of BCDIN3D and tRNAHis complex also suggest that human BCDIN3D is tRNAHis-specific 5′-monophosphate methyltransferase, and cytoplasmic tRNAHis is a primary target of human BCDIN3D.
The Biological Role of 5′-Methylation of Cytoplasmic tRnaHis
The biological role of 5′-monomethylphosphate of cytoplasmic tRNAHis remains unclear. While the 5′-monomethylphosphate of cytoplasmic tRNAHis decreases the affinity of tRNAHis toward histidyl-tRNA synthetase (Martinez et al., 2017), as expected from the complex structure of bacterial histidyl-tRNA synthetase with tRNAHis (Tian et al., 2015) and biochemical evaluation (Fromant et al., 2000), the overall aminoacylation efficiency is not affected by the modification. The steady-state level of cytoplasmic tRNAHis in BCDIN3D-knockout cells and its parental HEK293T cells are not significantly different. Furthermore, the stabilities of cytoplasmic tRNAHis from HEK293T cells and BCDIN3D-knockout cells after treatment with actinomycin-D do not show significant differences (Martinez et al., 2017). However, 5′-monomethylmonophosphate protects cytoplasmic tRNAHis from degradation in vitro in cytoplasmic cell extracts. Thus, methylation of 5′-monophosphate of cytoplasmic tRNAHis might be involved in its stability under specific conditions or in certain biological processes.
Perspective
The correlation between methylation of the 5′-monophosphate group of cytoplasmic tRNAHis and tumorigenic phenotype of breast cancer remains unknown (Figure 2B). tRNAs are involved in various biological process in cells (Sobala and Hutvagner, 2011; Raina and Ibba, 2014; Megel et al., 2015; Kumar et al., 2016; Park and Kim, 2018; Schimmel, 2018) beyond their established functions, as adaptors in protein synthesis.
In breast cancer cells, initiator tRNAMet is reportedly upregulated (Pavon-Eternod et al., 2013). Furthermore, in highly metastatic breast cancer cells, the upregulation of specific tRNAs, such as tRNAGluUUC and tRNAArgCCG, stabilizes mRNAs containing the corresponding codons and enhances translation (Goodarzi et al., 2016). However, knockout of BCDIN3D in HEK293T does not affect the steady-state level of cytoplasmic tRNAHis (Martinez et al., 2017). Thus, 5′-monophosphate methylation of cytoplasmic tRNAHis would not enhance the translation of specific mRNAs, although this warrants further investigation. Small RNA fragments have reportedly been derived from tRNAs, i.e., tRNA fragments (tRFs), and participate in various cellular functions (Sobala and Hutvagner, 2011; Raina and Ibba, 2014; Kumar et al., 2016). Under various cellular stress conditions, tRFs are often produced (Ivanov et al., 2011, 2014; Durdevic et al., 2013; Durdevic and Schaefer, 2013; Gebetsberger and Polacek, 2013). In breast and prostate cancer, specific tRNAs, such as cytoplasmic tRNALys and tRNAHis, are cleaved by angiogenin, and the tRNA half fragments are abundantly expressed in a sex hormone-dependent manner (Honda et al., 2015). These tRNA half fragments also promote proliferation of breast and prostate cancer cells by a yet unknown mechanism. In human and mouse cells, 3′- or 5′- terminal tRFs (3′-tRF or 5′-tRF) are produced and accumulate in an asymmetric manner. These tRFs associate with Ago2 and the tRFs probably serve as typical miRNAs. The 3′-tRF, but not the 5′-tRF, derived from cytoplasmic tRNAHis is complementary to human endogenous retroviral sequences in the genome (Li et al., 2012).
It would be noteworthy to assume that 5′-monomethylation of 5′-phosphate of tRNAHis regulates the expression of the tRNA half fragments and/or tRFs derived from tRNAHis in breast cancer cells or under specific biological or stress conditions. The production of tRNA half fragments and/or tRFs, in turn, might regulate the genes involved in tumorigenesis in breast cancers. Future studies are required to understand whether methylation of the 5′-phosphate group of tRNAHis by BCDIN3D is involved in the tumorigenic phenotype of breast cancer and other cancers and to potentially elucidate the unknown functions of tRNAs, beyond their established functions.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The study in our laboratory was supported in part by grants to KT from the Funding Program for Next Generation World-Leading Researchers of JSPS, by Grants-in-Aid for Scientific Research (A), and Grant-in-Aid for Scientific Research on Innovative Areas from JSPS, Takeda Science Foundation, Japan Foundation for Applied Enzymology, Terumo Foundation for Life Science and Art, and Princess Takamatsu Cancer Research Foundation.
References
- Betat H., Long Y., Jackman J. E., Morl M. (2014). From end to end: tRNA editing at 5′- and 3′-terminal positions. Int. J. Mol. Sci. 15 23975–23998. 10.3390/ijms151223975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42 W252–W258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Appel B., Soll D. (1982). Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc. Natl. Acad. Sci. U.S.A. 79 6475–6479. 10.1073/pnas.79.21.6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdevic Z., Mobin M. B., Hanna K., Lyko F., Schaefer M. (2013). The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 4 931–937. 10.1016/j.celrep.2013.07.046 [DOI] [PubMed] [Google Scholar]
- Durdevic Z., Schaefer M. (2013). tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? Bioessays 35 323–327. 10.1002/bies.201200158 [DOI] [PubMed] [Google Scholar]
- Fromant M., Plateau P., Blanquet S. (2000). Function of the extra 5′-phosphate carried by histidine tRNA. Biochemistry 39 4062–4067. 10.1021/bi9923297 [DOI] [PubMed] [Google Scholar]
- Gebetsberger J., Polacek N. (2013). Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 10 1798–1806. 10.4161/rna.27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H., Nguyen H. C. B., Zhang S., Dill B. D., Molina H., Tavazoie S. F. (2016). Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165 1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Jackman J. E., Lohan A. J., Gray M. W., Phizicky E. M. (2003). tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 17 2889–2901. 10.1101/gad.1148603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lowe S. W., Hannon G. J. (2007). microRNAs join the p53 network - another piece in the tumour-suppression puzzle. Nat. Rev. 7 819–822. 10.1038/nrc2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Loher P., Shigematsu M., Palazzo J. P., Suzuki R., Imoto I., et al. (2015). Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 112 E3816–E3825. 10.1073/pnas.1510077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Emara M. M., Villen J., Gygi S. P., Anderson P. (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., O’Day E., Emara M. M., Wagner G., Lieberman J., Anderson P. (2014). G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 111 18201–18206. 10.1073/pnas.1407361111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. E., Gott J. M., Gray M. W. (2012). Doing it in reverse: 3′-to-5′ polymerization by the Thg1 superfamily. RNA 18 886–899. 10.1261/rna.032300.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. E., Phizicky E. M. (2006). tRNAHis guanylyltransferase adds G-1 to the 5′ end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA 12 1007–1014. 10.1261/rna.54706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., et al. (2007). Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27 262–274. 10.1016/j.molcel.2007.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Kuscu C., Dutta A. (2016). Biogenesis and function of transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 41 679–689. 10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ender C., Meister G., Moore P. S., Chang Y., John B. (2012). Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40 6787–6799. 10.1093/nar/gks307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Wang X., Chen G. Y., Dalerba P., Gurney A., Hoey T., et al. (2007). The prognostic role of a gene signature from tumorigenic breast-cancer cells. N. Engl. J. Med. 356 217–226. 10.1056/NEJMoa063994 [DOI] [PubMed] [Google Scholar]
- Martinez A., Yamashita S., Nagaike T., Sakaguchi Y., Suzuki T., Tomita K. (2017). Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 45 5423–5436. 10.1093/nar/gkx051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megel C., Morelle G., Lalande S., Duchene A. M., Small I., Marechal-Drouard L. (2015). Surveillance and cleavage of eukaryotic tRNAs. Int. J. Mol. Sci. 16 1873–1893. 10.3390/ijms16011873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. J., Kim T. H. (2018). Fine-tuning of gene expression by tRNA-derived fragments during abiotic stress signal transduction. Int. J. Mol. Sci. 19:E518. 10.3390/ijms19020518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. E., Heo I., Tian Y., Simanshu D. K., Chang H., Jee D., et al. (2011). Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475 201–205. 10.1038/nature10198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M., Gomes S., Rosner M. R., Pan T. (2013). Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19 461–466. 10.1261/rna.037507.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M., Ibba M. (2014). tRNAs as regulators of biological processes. Front. Genet. 5:171 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M. D., Hendrick J. P., Jr., Lerner M. R., Steitz J. A., Reichlin M. (1983). A mammalian tRNAHis-containing antigen is recognized by the polymyositis-specific antibody anti-Jo-1. Nucleic Acids Res. 11 853–870. 10.1093/nar/11.3.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., et al. (2009). p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 106 3207–3212. 10.1073/pnas.0808042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 19 45–58. 10.1038/nrm.2017.77 [DOI] [PubMed] [Google Scholar]
- Shi B., Sepp-Lorenzino L., Prisco M., Linsley P., deAngelis T., Baserga R. (2007). Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 282 32582–32590. 10.1074/jbc.M702806200 [DOI] [PubMed] [Google Scholar]
- Shuman S. (2007). Transcriptional networking cap-tures the 7SK RNA 5′-γγ-Methyltransferase. Mol. Cell 27 517–519. 10.1016/j.molcel.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Sobala A., Hutvagner G. (2011). Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2 853–862. 10.1002/wrna.96 [DOI] [PubMed] [Google Scholar]
- Spizzo R., Nicoloso M. S., Lupini L., Lu Y., Fogarty J., Rossi S., et al. (2010). miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 17 246–254. 10.1038/cdd.2009.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Wang C., Liu Y., Xie W. (2015). Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase. Nucleic Acids Res. 43 2980–2990. 10.1093/nar/gkv129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Fukai S., Ishitani R., Ueda T., Takeuchi N., Vassylyev D. G., et al. (2004). Structural basis for template-independent RNA polymerization. Nature 430 700–704. 10.1038/nature02712 [DOI] [PubMed] [Google Scholar]
- Tomita K., Yamashita S. (2014). Molecular mechanisms of template-independent RNA polymerization by tRNA nucleotidyltransferases. Front. Genet. 5:36 10.3389/fgene.2014.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B., Robson S. C., Kouzarides T. (2012). Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell 151 278–288. 10.1016/j.cell.2012.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Martinez A., Tomita K. (2015). Measurement of acceptor-TPsiC helix length of tRNA for terminal A76-addition by A-adding enzyme. Structure 23 830–842. 10.1016/j.str.2015.03.013 [DOI] [PubMed] [Google Scholar]
- Yamashita S., Takeshita D., Tomita K. (2014). Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase. Structure 22 315–325. 10.1016/j.str.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Yamashita S., Tomita K. (2016). Mechanism of 3′-matured tRNA discrimination from 3′-immature tRNA by class-II CCA-adding enzyme. Structure 24 918–925. 10.1016/j.str.2016.03.022 [DOI] [PubMed] [Google Scholar]
- Yao L., Chi Y., Hu X., Li S., Qiao F., Wu J., et al. (2016). Elevated expression of RNA methyltransferase BCDIN3D predicts poor prognosis in breast cancer. Oncotarget 7 53895. 10.18632/oncotarget.9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Hanes S. D. (2000). Identification of drosophila bicoid-interacting proteins using a custom two-hybrid selection. Gene 245 329–339. 10.1016/S0378-1119(00)00048-2 [DOI] [PubMed] [Google Scholar]