Abstract
Circular RNAs (circRNAs) are a large class of noncoding RNAs characterized with closed loop structures without 3′ and 5′ polar ends. They can roughly be divided into exonic circRNAs, exon–intron circRNAs and circular intronic RNAs. CircRNAs are characterized with stability, prevalence, specificity and conservation, which arouse great interest in circRNAs as disease biomarkers. Their abilities to sponge to miRNAs, cis-regulate parent genes, bind to proteins and encode proteins endow circRNAs a critical role of regulation in eukaryotic cells. This concise review focuses on circRNAs as functional biomarkers and therapeutic targets in both tumor and nontumorous diseases.
Keywords: : biomarkers, characterization, circRNAs, function, human disease, regulators
Lay abstract
Although they were discovered in 1970s, circular RNAs (circRNAs) have attracted great interest only relatively recently. Instead of genome ‘junk matters’, circRNAs are now considered as promising biomarkers and treatment targets. CircRNAs are involved in numerous cancer-related and noncancer diseases, such as lung cancer, gastric cancer, cardiovascular diseases, diabetes mellitus and so on. This review outlines the classification, characterization and function of circRNAs, with a specific focus on recent studies concerning the role of circRNAs in human diseases.
Circular RNAs (circRNAs) are a class of endogenous noncoding RNAs characterized with closed ring structure without 3′ and 5′ ends. Although circRNA transcripts were firstly described decades ago [1], they have long been considered as low-level ‘transcriptional noise’ with little or no regulatory potential [2]. This phenomenon may result from traditional RNA detection methods depending on poly-A ends, which unavoidably filter out back-splicing sequences without free ends. In recent years, with the development of deep sequencing and bioinformatic approaches, a large number of circRNAs have been uncovered with stable expression in eukaryotic cells [3,4].
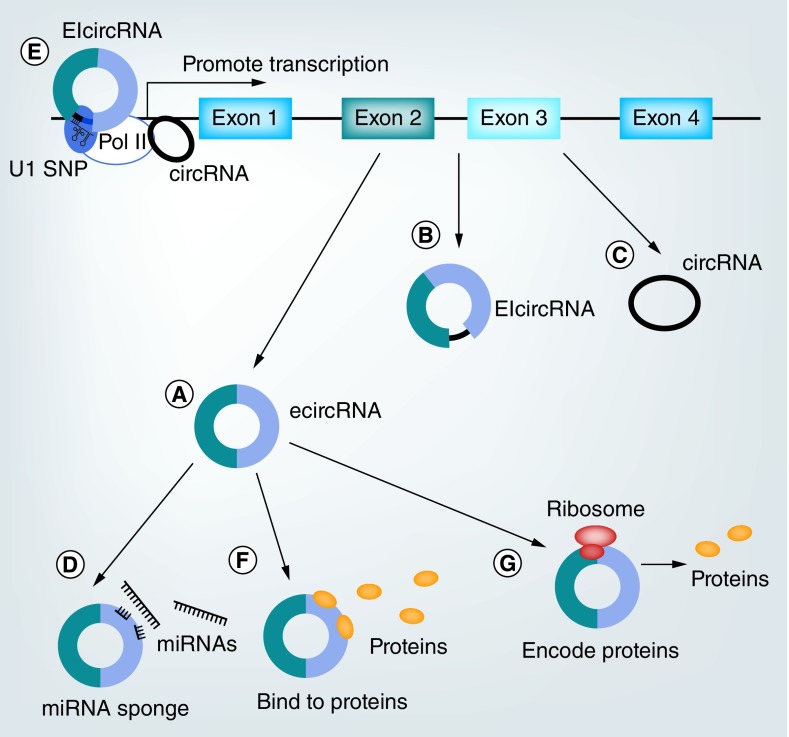
CircRNAs can mainly be divided into three subtypes (Figure 1), namely exonic circRNAs (ecircRNAs) [4], exon–intron circRNAs (EIciRNAs) [5] and circular intronic RNAs (ciRNAs) [6]. EcircRNAs are predominantly localized in the cytoplasm, such as Cdr1as, Sry and circ-HIPK3 [4]. Most ecircRNAs are generated from back-splicing of pre-mRNAs, in which downstream donor-exons splice to upstream acceptor-exons [7]. EIciRNAs are abundant in the nucleus. These circRNAs are formed with introns ‘retained’ between exons during the back-splicing process [5]. CiRNAs, containing two or more connected introns, localize mainly in the nucleus. Their processing depends on consensus motifs containing 7 nt GU-rich elements close to the 5′ splice site and 11 nt C-rich elements proximal to the branch point site [6].
Figure 1. . Regulatory functions of different types of circular RNAs.
(A–C) Three types of circular RNAs (circRNAs). (A) EcircRNAs are generated from exons. (B) EIcircRNAs contain both exons and introns. (C) CircRNAs are formed by introns. (D–G) Functions of circRNAs. (D) CircRNAs can act as miRNA sponges. (E) CircRNAs are able to promote transcription. (F) CircRNAs can bind to proteins. (G) CircRNAs encode proteins.
CircRNA: Circular intronic RNA; EcircRNA: Exonic RNA; EIcircRNA: exon–intron RNA.
The expression of circRNAs is stable, prevalent, specific and conserved. Compared with their linear counterparts, circRNAs are more resistant to RNase R-mediated degradation and have longer half-lives in some cases [4]. CircRNAs are abundant in various eukaryotic cells and human tissues, with parts of the circRNAs highly conserved among different species [3,8]. The expression of circRNAs shows spatio-temporal specificity, and changes during the disease process, indicating an interesting function of circRNAs in physiological and pathological conditions [9]. In 2013, the ‘sponging’ function of circRNA was first demonstrated, and it was found that circRNAs can suppress miRNA function by targeting seed sequences [10,11]. Later on, the role of circRNAs as promising noninvasive biomarkers with regulatory potency was investigated in various diseases such as cardiovascular [12], endocrine [13], autoimmune [14] and CNS diseases [15], and cancer [16]. Other functions of circRNAs, such as cis-regulation and protein coding, also suggest the potential role of circRNAs in human disease. This review summarizes the characterization and function of circRNAs and investigates the association between circRNAs and numerous diseases including cancer and noncancer diseases.
Characteristics
CircRNAs are shown to demonstrate stable, specific and conservative expression patterns.
Stability & prevalence
CircRNA transcriptions have no free 3′ and 5′ ends, thus are more resistant to RNase R and more stable than corresponding linear RNAs in most cases. In cells, although the circRNAs exhibit average half-lives of more than 48 h, the half-lives of associated linear RNAs are less than 20 h [4]. The stability may lead to a more abundant expression of circRNAs. For example, the abundance of circular transcripts of HIPK3 is roughly fivefold more than their linear counterparts. In some cases, the circular transcripts are estimated to be tenfold more abundant than cognate linear mRNA [4,9,17].
The prevalence of circRNAs is not unique in mammal cells, but is common in the eukaryotic world, from mammals to insects, plants and fungi [4,10]. In human beings, a large number of circRNAs are detected in diverse clinical specimens, such as whole blood [18], plasma [19] and saliva [20]. It is reported that in human whole blood samples, while circRNA isoforms are already detectable, in hundreds of cases the corresponding linear transcripts are absent [18].
Specificity
Oftentimes, the expression of circRNAs alters in different cell lines. For instance, the circular ratios of gene AFAP1, ASPH and SH3PXD2A are significantly different in BJ-T, HEK293 and HeLa cells [7].
CircRNAs also have specific expression in different tissues. In human beings, some circRNAs accumulate more in the CNS, especially synapses, compared with other tissues such as thyroid gland, liver and muscle [9]. Similar expression patterns can be observed in mouse and Drosophila. For example, circRNA generated from Rims2 is highly expressed in adult mouse brain while it can rarely be detected in other mouse tissues [9]. In Drosophila, more than 90% of circRNAs identified in any fly tissues can also be detected in the head while half of the circles observed in the head cannot be detected in other tissues [21]. Tissue-specific expression is also observable in plants. For instance, in polyploidy Gossypium species, the overall expression level of circRNAs is higher in ovule samples than leaf samples. In addition, more than 80% circRNAs express only in ovule tissues [22].
Moreover, circRNAs are expressed in a developmental-stage specific manner. In mammalian and Drosophila brains, the global expression of circRNAs varies in different stages. For instance, mouse circRNAs derived from the Staufen2 gene exhibit reciprocal expression during neuronal differentiation [9]. In Drosophila, circRNA accumulation is associated with the aging process of the brain. The level of brain circRNAs is elevated from embryo to larva and pupa, and is even higher in adult heads [21].
Conservation
A small proportion of circRNAs is highly conserved across different species. In a research performed by Dong et al., the authors found about 15,000 circRNAs in both human and mouse genomes, indicating that 15% of total human circRNAs and 40% of total mouse circRNAs are conserved [23]. In another study concerning circRNA expression in heart, the authors discovered that about 10% cardiac circRNAs are conserved across human, mouse and rat, and about 30% cardiac circRNAs can be found simultaneously in mouse and rat [24]. In a recent study conducted by Stoll et al., among the 3441 explored human pancreatic islet circRNAs, 497 orthologous circRNAs can be determined in parallel mouse islet samples [25]. The conserved expression of circRNAs is associated with complementary intronic sequences flanking back-spliced exons [23]. Orthologous circRNAs across species often exhibit longer flanking introns than species-specific ones [22].
Nomenclature
With the advancement of next generation sequencing and bioinformatic technology, a large number of circRNAs are detected for the first time. Some newly discovered circRNAs are nominated by their founders concerning the type, location or function of circRNAs (Figure 1). For example, ci-ankrd52, a ciRNA generated from parent gene ANKRD52, is named after its genome location. The name of a well-known ecircRNA, ciRS-7, contains the meaning of circRNA sponge for miR-7. However, such nomenclature can be confusing because one gene can generate several circular transcripts and one miRNA can become targets for different circRNAs. Luckily, existing databases, such as circBase (http://circbase.org/), allow one to assess the variety of names, forms and functions of circRNAs.
Functions
Although the exact regulatory mechanism of circRNAs is still unclear, current research concentrates mainly on the following four aspects, namely sponging of miRNAs, cis-regulating of parent genes, binding to proteins and encoding proteins (Figure 1). What is more, the categorization and localization of circRNAs are important to analyze their functions (Table 1).
Table 1. . Circular RNAs classification and function.
| Type | Derivation | Location | Function | Examples | Ref. |
|---|---|---|---|---|---|
| EcircRNA | Exons | Mainly cytoplasm | MiRNA sponge; cis-regulation at post-transcriptional level; protein binding; encode functional proteins | Cdr1as; circ-ITCH; hsa_circ_0031288; circ-FBXW7 | [10,26–28] |
| EIcircRNA | Exons and introns | Mainly nucleus | Cis-regulation of parent gene | circEIF3J; circPAIP2 | [5] |
| CircRNA | Introns | Mainly nucleus | Cis-regulation of parent gene | circ-ankrd52; circ-mcm5; circ-sirt7 | [6] |
CircRNA: Circular intronic RNA; EcircRNA: Exonic RNA; EIcircRNA: exon–intron RNA.
CircRNAs as miRNA sponges
Lines of evidence indicate that natural circRNAs serve as effective miRNA sponges (Figure 1D). MiRNAs are small noncoding RNAs (19–22 nt) which negatively modulate mRNA expression in post-transcriptional stage via binding to 3′-untranslated regions [29]. CircRNAs localized in cytoplasm can interact with both miRNA and AGO, and sequentially eliminate the suppression of miRNAs on mRNA. For example, the first seriously studied circRNA, Cdr1as, harbors more than 70 conserved miR-7 binding site. The effect of Cdr1as expression simulates that of miR-7 silencing, indicating a role of miRNA binding [10,11]. Another well-known circRNA, Sry, can serve as competitive inhibitor for miR-138 by binding to target sites [11]. Although one paper published in 2014 indicated that few circRNAs could act as efficient miRNA sponges [30], more and more circRNAs have been demonstrated in recent years to shape gene expression via inhibiting miRNAs. For example, circHIPK3 contains 18 potential binding sites for nine different miRNAs [31]. CircHIPK3 plays an important role in hepatocellular carcinoma (HCC) by sponging miR-124, leading to abnormal cancer cell proliferation and migration [32]. CircHIPK3 is also able to regulate endothelial proliferation and vascular dysfunction in diabetic retinopathy (DR) via blocking miR-30a-3p [33].
Cis-regulation of parent genes
CiRNAs are able to regulate the expression of their parent genes in cis (Figure 1E). Nuclear ciRNAs localized near the transcription sites of their parent genes can interact with RNA Pol II elongation machinery and act as positive regulators for transcription. For example, knockdown of ci-ankrd52, ci-mcm5 and ci-sirt7 results in suppressed expression of their parental mRNAs [6]. EIciRNAs are also capable to cis-regulate their parental genes. EIciRNAs are able to promote transcription of RNA Pol II through interacting with U1 snRNP [34]. Silencing of two EIciRNAs, circEIF3J and circPAIP2, reduces the expression level of their parental mRNAs [5].
The majority of ecircRNAs acts as miRNA sponges and does not regulate the expression of their cognate genes. However, circ-ITCH shares the same miRNA response elements with the 3′-untranslated region of ITCH mRNA. By binding to miRNAs, circ-ITCH relieves the suppressive effect of miRNA on its own parent mRNA. The enhanced expression of ITCH at post-transcriptional level by circ-ITCH finally results in the suppressed activity of Wnt/β-catenin pathway. Dysregulation of circ-ITCH and Wnt/β-catenin pathway are involved in the progression of lung cancer, esophageal squamous cell carcinoma and colorectal cancer (CRC) [26,35,36].
Protein binding
Except miRNAs, circRNAs are also able to interact with other entities, such as proteins (Figure 1F). In a study conducted by Du et al., circ-Foxo3 was demonstrated to influence cell cycle progression by binding to two cell cycle proteins (CDK2 and p21) [37]. PES1 is an important protein essential for ribosome biogenesis. CircANRIL can attach to PES1 and increase apoptosis of cells [38].
RNA binding proteins are a specific class of proteins which can regulate the formation and function of mRNAs through binding to ACUAA motifs in 3′UTR region [39]. HuR is a well-studied RNA binding protein which can adhere to PABPN1 mRNA and promote its expression. However, hsa_circ_0031288 (CircPABPN1) is able to reduce PABPN1 expression via sequestering and suppressing HuR [27].
Protein coding
It was reported in 1995 that synthetic circRNAs with continuous open reading frames were capable to be translated into long-repeating polypeptide chains [40]. Several recent researches indicate that endogenous circRNAs containing open reading frames are also able to encode functional proteins (Figure 1G) [41]. For example, FBXW7–185aa translated from circ-FBXW7 and SHPRH-146aa translated from circ-SHPRH are tumor suppressors in glioblastoma [28,42].
Nontumor diseases
In accordance with the disclosure of circRNA functions, the pivotal roles of circRNA in diseases as biomarker and regulator are catching more attention (Table 2).
Table 2. . Circular RNAs in nontumor diseases.
| Disease | CircRNA | Target | Method involved | Expression | Function | Ref. |
|---|---|---|---|---|---|---|
| Atherosclerosis | circANRIL | PES1 protein | PCR and RT-qPCR | Decrease | Atheroprotective | [38] |
| hsa_circ_0003575 | – | Microarray analysis; RT-qPCR | Increase | Repress endothelial cell proliferation and angiogenesis | [43] | |
| CAD | circ11783–2 | – | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for CAD combined with T2DM | [44] |
| MI and heart failure | circMFACR | miR-652–3p/MTP18 | PCR and RT-qPCR | Increase | Promote apoptosis and MI | [45] |
| MICRA | – | RT-qPCR | Decrease | Potential biomarker for risk stratification | [46] | |
| Hypertension | hsa_circ_0005870 | – | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for hypertension diagnosis | [47] |
| Myocardial fibrosis | circ_000203 | miR-26b-5p/Col1a2, CTGF | Microarray analysis; PCR; RT-qPCR | Increase | Promote fibrosis | [48] |
| circ_010567 | miR-141/TGF-β1 | Microarray analysis; RT-qPCR | Increase | Promote fibrosis | [49] | |
| Cardiac hypertrophy | circHRCR | miR-223/ARC | RT-qPCR | Decrease | Repress cardiac hypertrophy | [50] |
| Diabetes mellitus and prediabetes | hsa_circ_0054633 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for T2DM and prediabetes diagnosis | [13] |
| DR | circ_0005015 | miR-519d-3p | Microarray analysis; RT-qPCR | Increase | Potential biomarker for DR diagnosis; promote proliferation | [51] |
| circHIPK3 | miR-30a-3p/VEGFC, WNT2, FZD4 | RT-qPCR | Increase | Promote proliferation and vascular dysfunction | [33] | |
| RA | circRNA_104871 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for RA diagnosis | [52] |
| LN | circHLA-C | miR-150 | RNA-seq; RT-qPCR | Increase | Potential biomarker for LN diagnosis | [53] |
CAD: Coronary artery disease; circANRIL: Circular ANRIL; CircRNA: Circular RNA; DR: Diabetic retinopathy; LN: Lupus nephritis; MI: Myocardial infarction; RA: Rheumatoid arthritis; RT-qPCR: Reverse transcription-quantitative polymerase chain reaction; T2DM: Type 2 diabetes mellitus.
Cardiovascular diseases
Atherosclerosis
Atherosclerosis is a disease in which the inside of an artery narrows due to building up of plaques, and may lead to coronary artery disease (CAD), stroke and peripheral artery disease. In a 2016 research conducted by Holdt et al., the atheroprotective function of circANRIL was investigated. Forced expression of circANRIL could increase cell apoptosis and decrease proliferation in vitro [38]. Instead of sponging miRNAs [11] or cis-regulating parental genes [6], circANRIL implemented its protective function through binding to PES1 protein, which finally resulted in the impaired ribosome maturation as well as apoptosis in smooth muscle cells and macrophages [38].
In a more recent study, Li et al. constructed an oxLDL-treated endothelial cell injury model in order to find potential diagnostic biomarker and elucidate molecular mechanism for atherosclerosis. They found that hsa_circ_0003575 was significantly upregulated in oxLDL-treated endothelial cells. Hsa_circ_0003575 silencing could promote cell proliferation and angiogenesis in ox-LDL-treated endothelial cells [43].
Coronary artery disease
CAD, also named coronary atherosclerotic heart disease, is caused by coronary artery atherosclerotic stenosis or occlusion and the sequential myocardial ischemia and hypoxia.
In a current study, Pan et al. investigated circRNAs in three pairs of plasma samples from CAD patients and control subjects. 24 different circRNAs were identified, and in the network constructed by bioinformatic technology, nine circRNAs could together promote TRPM3 expression by inhibiting hsa-miR-130a-3p [19]. Although this research provides a new insight into pathological regulation of CAD, the interactions in this network are mainly based on bioinformatics analysis, and demand further investigation.
CAD is closely related with Type 2 diabetes mellitus (T2DM), and the metabolism abnormalities in T2DM directly increase CAD risk [54]. By using microarray analysis, Li et al. explored circRNA expression profile in CAD patients with hyperglycemia. Logistic regression analysis of two independent cohorts showed that hsa-circ11783–2 was more correlated with both CAD and T2DM than other selected circRNAs [44]. This is the first study to examine the association of circRNAs with both CAD and T2DM, and the function of hsa-circRNA11783–2 requires further exploration.
Myocardial infarction & heart failure
Most myocardial infarction (MI) occurs on the bases of CAD, when blood supply suddenly decreases or stops to a part of the heart. Severe MI causes cardiomyocyte death and may provoke heart failure.
In a study concerning the pathological mechanisms of MI, the authors discovered that circRNA MFACR regulated cardiomyocyte death via circMFACR/miR-652–3p/MTP18 axis [45]. MTP18 was able to induce mitochondrial fission [55] and promote cardiomyocyte apoptosis in MI [45]. MiR-652–3p was capable of inhibiting MTP18 expression and was negatively regulated by circMFACR. CircRNA MFACR elevated apoptotic cell death in MI through eliminating miR-652–3p suppression on MTP18. This research sheds new light on understanding molecular mechanism of MI [45].
A large percentage of acute MI (AMI) patients develop into heart failure due to maladaptive left ventricular remodeling. Salgado-Somoza et al. identified circRNA MICRA as a prognostic biomarker to improve the risk stratification after AMI. Patients with decreased MICRA level were more likely to be classified into reduced ejection fraction group. Both ordinal regression analysis and bootstrap internal validation were utilized to demonstrate the value of MICRA in prognostic stratification of AMI induced heart failure [46].
Hypertension
In a research published in 2016, Wu et al. identified circRNA expression profiles in hypertension patients’ peripheral blood. Hsa_circ_0005870 showed a significant down expression in hypertension group. Both GO and KEGG pathway analysis indicated the involvement of hsa_circ_0005870 in hypertension [47]. Hsa_circ_0005870 may represent a novel biomarker for the diagnosis of hypertension. However, the exact regulation mechanism of hsa_circ_0005870 needs further investigation.
In another study, Cheng et al. investigated characteristic profile of circRNA in kidney samples from four kinds of hypertension rat models. Aberrant circRNAs were identified and verified with RT-qPCR. Bioinformatics technologies were used to predict the circRNA/miRNA/mRNA network [56]. This study serves as a primary foundation for further researches concerning hypertension combined with kidney diseases.
Cardiomyopathy
As a kind of cardiomyopathy, the pathological progression of myocardial fibrosis is characterized with the activation of cardiac fibroblasts (CFs), in which fibroblasts transform into myofibroblasts, resulting in collagen deposition in extracellular matrix [57]. Myocardial fibrosis can be caused by deformity of cardiomyocytes resulting from metabolism disorders in diabetes mellitus patients. In two recent researches concerning circRNAs in myocardial fibrosis, both diabetic mouse and mice CFs were used as research models. CircRNA circRNA_000203 [48] and circRNA_010567 [49] were found to be remarkably upregulated in diabetic mouse myocardium and Ang-II-treated CFs. Elevated level of circRNA_000203 and circRNA_010567 accelerated fibrosis-associated protein expression. For mechanism, circRNA_000203 was demonstrated to inhibit miR-26b-5p and eliminate miRNA suppressive effect on Col1a2 and CTGF [48]. Another circRNA, circRNA_010567, was proved to sponge miR-141 and increase the expression level of TGF-β1. [49]. These two studies shed new light on the pro-fibrosis effect of circRNA_000203 and circRNA_010567, and the regulatory function of circRNA/miRNA/mRNA axis in myocardial fibrosis.
Cardiac hypertrophy is characterized by maladaptive thickening of the myocardium. Wang et al. revealed the circHRCR/miR-223/ARC regulatory axis in cardiac hypertrophy, which might finally develop into abnormal cardiac remodeling and heart failure. MiR-223 was capable to induce heart failure in vivo and cardiomyocyte hypertrophy in vitro [50]. ARC, a reported protein involved in pathological hypertrophy inhibition [58], was demonstrated to be the downstream target of miR-223. In order to find the antihypertrophy molecule, the authors selected 100 published circRNAs from online databases. Among them, circHRCR was significantly downregulated in pathological conditions. Further investigation demonstrated that circHRCR could repress abnormal cardiac hypertrophy and heart failure through the circHRCR/miR-223/ARC axis [50].
Diabetes mellitus
Diabetes mellitus
Diabetes mellitus is a kind of metabolic disorder in which patients are affected by hyperglycemia due to inadequate insulin or insulin resistance. Diabetes mellitus can be divided into Type 1 diabetes mellitus and T2DM.
In 2016, Zhao et al. delineated the expression profile of circRNAs in T2DM and prediabetes patients’ peripheral blood for the first time. They selected five circRNAs as candidate biomarkers and verified in two independent cohorts. The results showed that hsa_circ_0054633 presented the highest diagnostic ability among the chosen circRNAs [13]. This study provides an insight into novel biomarker for prediabetes and T2DM.
Long-term diabetes mellitus often leads to vascular complications, including microvascular and macrovascular diseases, which are the major causes for morbidity and mortality in diabetes mellitus. CircRNAs are involved in diabetes mellitus correlated vasculopathy. For example, circWDR77 is upregulated in high glucose treated vascular smooth muscle cells. CircWDR77 regulates vascular smooth muscle cells proliferation and migration via directly binding to miR-124 and alleviating suppression for target FGF-2 [59].
Diabetic retinopathy
DR is one of the common complications caused by diabetes mellitus. In 2017, Gu et al. analyzed the altered circRNA profiles in DR patients’ serum. This is the first circRNA study concerning DR and lays the first stone for later biomarker detection and mechanism elucidation [60]. In a following study, Zhang et al. revealed the distinctive expression profile of circRNAs in diabetic retinas. Circ_0005015 expression was significantly upregulated in retina samples, vitreous samples, peripheral plasma samples and fibrovascular membranes of DR patients. Circ_0005015 silencing reduced human retinal vascular endothelial cells (HRVECs) proliferation, migration and tube formation. Luciferase activity assays found miR-519d-3p as the direct target for circ_0005015 [51]. Circ_0005015 is manifested as a regulatory biomarker for the DR diagnosis and treatment. In another research, Shan et al. demonstrated that circHIPK3 was elevated in hyperglycemia treated retinal endothelial cells and diabetic mouse retinas. CircHIPK3 served as a miRNA sponge to block miR-30a-3p activity and thus induced increase in levels of VEGFC, WNT2 and FZD4 [33]. Increment of VEGFC, WNT2 and FZD4 was reported in various retina disorders [61,62]. CircHIPK3 silencing could alleviate diabetes-induced endothelial proliferation and retina microvascular dysfunction [33].
Regulate insulin secretion
Several reports manifest that circRNAs are involved in the regulation of islet cells vitality. For example, Cdr1as, perhaps the best-identified endogenous mammalian circRNA, can be increased in islet cells by long-term forskolin and PMA stimulation [63]. As an miR-7 sponge [30], Cdr1as is able to improve insulin secretion and transcription through inhibiting miR-7 and accelerating Myrip and Pax6 expression [63]. In another study, the authors analyzed circRNAs in human islets and cognate ones in mouse islets. They revealed that Cdr1as and circHIPK3 were abundant in normal islets, but declined in diabetic mouse. Cdr1as and circHIPK3 silencing in wild-type animal models caused defective insulin secretion and diminished islet cell proliferation. While Cdr1as performed such regulatory function by blocking miR-7, circHIPK3 regulated islet cell function by sequestering miR-124–3p and miR-338–3p and elevating Slc2a2, Akt1 and Mtpn [64].
Autoimmune diseases
Rheumatoid arthritis
Zheng et al. [65] and Ouyang et al. [52] screened the expression profile of circRNAs in rheumatoid arthritis (RA) patients’ peripheral mononuclear cells by microarray analysis. The assay results were verified by RT-qPCR method. Zheng et al. predicted the circRNA/miRNA interaction utilizing bioinformatic software [65]. Ouyang et al. analyzed the correlation between differential circRNAs and clinicopathological factors, finding that circRNA_104871 exhibited the largest diagnostic ability [52].
System lupus erythematosus
Li et al. screened circRNA profiles in system lupus erythematosus patients’ peripheral blood plasma. CircRNA candidates were selected and validated. Potential circRNA/miRNA interaction networks were constructed [66]. In another study, Luan et al. determined circRNA profiles in renal samples from lupus nephritis patients and health controls [53]. In their preceding investigation, the authors noticed that miR-150 was positively correlated with renal chronicity scores [67]. In the current study, they spotted circHLA-C as a probable regulator for miR-150. CircHLA-C and miR-150 exhibited a negative correlation. As a potential biomarker, circHLA-C was positively correlated with clinical factors, such as serum creatinine, renal activity index, proteinuria and crescentic glomeruli [53].
Cancers
As it has been revealed by numerous studies, circRNAs are involved in the initiation and progression of various human cancers and may become potential diagnostic biomarkers, as it is shown in Table 3.
Table 3. . Circular RNAs in cancers.
| Disease | CircRNA | Target | Method involved | Expression | Function | Ref. |
|---|---|---|---|---|---|---|
| Lung cancer | hsa_circ_000064 | – | RT-qPCR | Increase | Potential biomarker for lung cancer diagnosis; promote proliferation and invasion | [68] |
| circRNA_100876 | – | RT-qPCR | Increase | Potential biomarker for NSCLC diagnosis | [69] | |
| hsa_circ_0014130 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for NSCLC diagnosis | [70] | |
| hsa_circ_0013958 | miR-134/CCND1 | Microarray analysis; RT-qPCR | Increase | Potential biomarker for LAC diagnosis; promote proliferation and metastasis | [71] | |
| hsa_circ_0012673 | miR-22/ErbB3 | RT-qPCR | Increase | Potential biomarker for LAC diagnosis; promote proliferation | [72] | |
| Breast cancer | circRNA-000911 | miR-449a/Notch1 | Microarray analysis; RT-qPCR | Decrease | Promote apoptosis | [73] |
| circ-ABCB10 | miR-1271 | Microarray analysis; RT-qPCR | Increase | Promote proliferation | [74] | |
| hsa_circ_0001982 | miR-143 | Microarray analysis; RT-qPCR | Increase | Promote proliferation | [75] | |
| circGFRA1 | miR-34a | Microarray analysis; RT-qPCR | Increase | Promote proliferation | [76] | |
| GC | hsa_circ_0074362 | – | RT-qPCR | Decrease | Potential biomarker for GC diagnosis | [77] |
| hsa_circ_002059 | – | RT-qPCR | Decrease | Potential biomarker for GC diagnosis | [78] | |
| hsa_circ_0014717 | – | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for GC diagnosis | [79] | |
| hsa_circ_0000190 | – | RT-qPCR | Decrease | Potential biomarker for GC diagnosis | [80] | |
| hsa_circ_0000520 | – | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for GC diagnosis | [81] | |
| hsa_circ_0001017 and hsa_circ_0061276 | – | Microarray analysis; RT-qPCR; RT-ddPCR | Decrease | Potential biomarker for GC diagnosis | [82] | |
| hsa_circ_100269 | miR-630 | RT-qPCR | Decrease | Potential biomarker for GC diagnosis; repress proliferation | [83] | |
| circLARP4 | miR-424–5p/LATS1 | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for GC prognosis; repress proliferation and invasion | [84] | |
| HCC | hsa_circ_0001649 | – | RT-qPCR | Decrease | Potential biomarker for HCC diagnosis | [85] |
| hsa_circ_0005075 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for HCC diagnosis | [86] | |
| circZKSCAN1 | – | RT-qPCR | Decrease | Potential biomarker for HCC diagnosis; repress proliferation, migration and invasion | [87] | |
| circMTO1 | miR-9/p21 | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for HCC prognosis; repress proliferation and invasion | [88] | |
| cSMARCA5 | miR-17–3p, miR-181b-5p/TIMP3 | RNA-seq; RT-qPCR | Decrease | Potential biomarker for HCC prognosis; repress proliferation and migration | [89] | |
| circ_0067934 | miR-1324/FZD5 | RT-qPCR | Increase | Promote proliferation and migration | [90] | |
| circHIPK3 | miR-124/AQP3 | RT-qPCR | Increase | Promote proliferation and migration | [32] | |
| circRNA_100338 | miR-141–3p | Microarray analysis; RT-qPCR | Increase | Promote migration and invasion | [91] | |
| CRC | circRNA0003906 | – | RT-qPCR | Decrease | Potential biomarker for CRC diagnosis | [92] |
| hsa_circ_001988 | – | RT-qPCR | Decrease | Potential biomarker for CRC diagnosis | [93] | |
| hsa_circ_103809 and hsa_circ_104700 | – | Microarray analysis; RT-qPCR | Decrease | Potential biomarker for CRC diagnosis | [94] | |
| hsa_circ_0000069 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for CRC diagnosis; promote proliferation, migration and invasion | [95] | |
| hsa_circ_0007534 | – | Microarray analysis; RT-qPCR | Increase | Potential biomarker for CRC diagnosis; promote proliferation | [96] | |
| hsa_circ_0020397 | miR-138/TERT, PD-L1 | RT-qPCR | Increase | Potential biomarker for CRC diagnosis; promote invasion | [97] | |
| Bladder cancer | circTCF25 | miR-103a-3p, miR-107/CDK6 | Microarray analysis; RT-qPCR | Increase | Promote proliferation and migration | [98] |
| circRNA-MYLK | miR-29a/VEGFA | Microarray analysis; RT-qPCR | Increase | Promote cancer proliferation | [99] | |
| circBCRC4 | miR-101/EZH2 | RT-qPCR | Decrease | Promote apoptosis | [100] | |
| circ-ITCH | miR-17, miR-224/p21, PTEN | RT-qPCR | Decrease | Repress proliferation and migration | [101] | |
| Glioma | hsa_circ_0046701 | miR-142–3p/ITGB8 | RT-qPCR | Increase | Promote proliferation and invasion | [102] |
| circ-FBXW7 | – | RNA-seq; RT-qPCR | Decrease | Code protein | [28] | |
| circ-SHPRH | – | RNA-seq; RT-qPCR | Decrease | Code protein | [42] | |
CircRNA: Circular RNA; CRC: Colorectal cancer; GC: Gastric cancer; HCC: Hepatocellular carcinoma; LAC: Lung adenocarcinoma; NSCLC: Non-small-cell lung cancer; RT-ddPCR: Reverse transcription-droplet digital polymerase chain reaction; RT-qPCR: Reverse transcription-quantitative polymerase chain reaction.
Lung cancer
Lung cancer, also known as lung carcinoma, is the leading cause of cancer death globally [103]. According to histopathological classification, lung cancer can generally be divided into non-small-cell lung cancer (NSCLC) and small cell lung cancer.
Luo et al. reported that hsa_circ_0000064 exhibited elevated expression in both lung cancer tissues and lung cancer cell lines (A549 and H1229). There was a close correlation between hsa_circ_0000064 augmentation and tumor differentiation, tumor-lymph node-metastasis (TNM) stage and lymphatic metastasis. Hsa_circ_0000064 silencing suppressed the proliferation and migration of cancer cells, and promoted cell apoptosis. For molecular mechanisms, the authors discovered that hsa_circ_0000064 regulated apoptotic-related proteins, cycle-related proteins and invasion-related proteins [68].
NSCLC accounts for nearly 85% of all primary lung cancers. Yao et al. revealed that circRNA_100876 was elevated in NSCLC tissues compared with pair-matched nontumor tissues. The upregulated expression was correlated with TNM stage and lymphatic metastasis. Furthermore, NSCLC patients with higher circRNA_100876 level had an overall shorter survival time than NSCLC patients with lower expression level [69]. In another study, hsa_circ_0014130 was found to be notably upregulated in NSCLC tissues and the expression was also associated with both TNM stage and lymphatic metastasis. The AUC was 0.878. Then, the circRNA/miRNA interaction network was predicted by bioinformatics technologies [70].
Lung adenocarcinoma (LAC) is currently the most common subtype of NSCLC in lifelong nonsmokers [104]. Zhao et al. investigated circRNA profile in early-stage LAC patients’ tumor tissue and adjacent normal tissue. Five dysregulated circRNAs were validated and potential circRNA/miRNA network was predicted [105]. In another study, hsa_circ_0013958 was detected as a LAC biomarker with regulatory potency. Hsa_circ_0013958 was increased in LAC cell lines, cancer tissues and cancer patients’ plasma. The expression level of hsa_circ_0013958 was associated with tumor staging and lymph node metastasis and the AUC was 0.815. Biological function experiments validated that hsa_circ_0013958 was involved in cellular proliferation and metastasis. Finally, the hsa_circ_0013958/miR-134/CCND1 regulatory axis was constructed [71]. Similarly, the expression level of hsa_circ_0012673 was significant overexpressed in LAC tissues, and was associated with tumor size. Hsa_circ_0012673 could accelerate LAC proliferation through sequestering miR-22, resulting in elevated level of targeted ErbB3 [72].
Breast cancer
The most commonly diagnosed cancer among women has been breast cancer in recent years, which is also the leading cause of cancer death in women younger than 45 years [106]. In a present study, a circRNA with antioncogenic role was found in breast cancer. CircRNA-000911 was significantly decreased in breast cancer tissues and cancer cell lines. Overexpression of circRNA-000911 could induce decreased cell proliferation, migration and invision as well as increased apoptosis. CircRNA-000911 implemented its regulatory function via inhibiting miR-449a and promoting Notch1 and NF-κB signaling pathway [73]. In the study performed by Liang et al., the authors revealed that circ-ABCB10 was significantly increased in breast cancer tissues. SiRNA induced circ-ABCB10 silencing contributed to decreased cancer cell proliferation and elevated apoptosis. Function analysis demonstrated that circ-ABCB10 carried out its tumorigenesis role via binding to miR-1271 [74]. Similar tumor-promoting effect was also observed in hsa_circ_0001982 targeting miR-143 [75].
Triple negative breast cancer is a kind of breast cancer not sensitive to hormone therapies targeting ER, PR or Her2/neu. He et al. analyzed circRNA patterns in cancer cell lines and verified one upregulated circRNA circGFRA1. Silencing of circGFRA1 relieved suppression for target miR-34a, which results in inhibited cancer cell proliferation and promoted apoptosis [76]. Yan et al. revealed that circVRK1 was reduced in breast cancer stem cells. CircVRK1 repressed stemness-maintenance ability of breast cancer stem cells [107]. Furthermore, miR-153–5p, the predicted target of circVRK1, was previously reported to be involved in stemness-maintenance of triple negative breast cancer [108].
Gastric cancer
Gastric cancer (GC) is among the leading causes of cancer death in developing countries nowadays [109]. Several researches delineated the global expression patterns of circRNAs in GC tissues or patients’ plasma [79,82,110,111]. Part of the results was validated with RT-qPCR. These investigations lay the foundation for future exploration in GC.
From 2015 to 2017, quite a few circRNAs were detected as potential GC biomarkers, such as hsa_circ_0074362 [77], hsa_circ_002059 [78] and hsa_circ_0014717 [79]. All of these diagnostic biomarkers were dysregulated in cancer tissue compared with paired noncancerous tissue and were correlated with several clinical-pathological factors. For example, Hsa_circ_0074362 was significantly down-expressed in GC tissues, cancer cell lines and gastritis. The level of hsa_circ_0074362 in GC was significantly lower compared with moderate gastritis. The expression level of hsa_circ_0074362 was associated with CA19–9 and lymphatic metastasis. The receiver operator curve was 0.630 [77]. These suggest that hsa_circ_0074362 may have potential values in the screening of GC.
Some circRNAs as biomarkers not only exist in tumor tissues, but also exist in patients’ plasma, such as hsa_circ_0000190 [80], hsa_circ_0000520 [81], hsa_circ_0001017 and hsa_circ_0061276 [82]. For instance, Li et al. demonstrated that hsa_circ_0001017 and hsa_circ_0061276 expression levels were downregulated in both cancer tissues and paired plasma and were associated with tumor size and distal metastasis. Through combining the expression levels of hsa_circ_0001017 and hsa_circ_0061276 in tissue and plasma, the AUC could ascend to 0.966 [82].
Among these disclosed circular biomarkers, a few are manifested as functional miRNA sponges. In their previous study, Zhang et al. noticed the remarkable decline of hsa_circRNA_100269 in recurrent GC tissue [112]. In their follow-up examination, they manifested that hsa_circRNA_100269 overexpression could prohibit tumor cell proliferation via absorbing miR-630 [83]. Another example of regulatory circRNAs in GC is circLARP4. Based on their preceding research concerning mRNA LATS1 as a tumor suppressor in GC [84], Zhang et al. explored the upstream regulatory network for LATS1. They revealed that circLARP4 could impede GC cell proliferation and invasion by targeting miR-424–5p, which led to raised LATS1. However, only early stage patients with a higher circLARP4 expression had a better outcome than patients with circLARP4 low expression. Similar trends could not be observed in late stage patients [113].
Hepatocellular carcinoma
HCC is the most common type of primary liver cancer and the third leading cause of cancer mortality in many countries [114]. Early researches demonstrated diagnostic potency of circRNAs in HCC. For instance, both hsa_circ_0001649 and hsa_circ_0005075 were identified as novel biomarkers for HCC [85,86]. Later researches concern not only about diagnostic ability but also regulatory potency of circRNAs in HCC. Some circRNAs display antioncogenic effects. Yao discovered that, in HCC tissue samples, both circZKSCAN1 and ZKSCAN1 mRNA were significantly lower compared with noncancer tissues. Blocking circZKSCAN1 and/or ZKSCAN1 mRNA would promote cancer cell proliferation, migration and invasion. Furthermore, circZKSCAN1 was associated with tumor numbers, cirrhosis, vascular invasion and tumor grade [87]. Another two circRNAs, circMTO1 and cSMARCA5, were also downregulated in HCC tissue. Both of them could inhibit HCC cell proliferation and migration. CircMTO1 exerted tumor suppressive role via circMTO1/miR-9/p21 axis [88], and cSMARCA5 via cSMARCA5/miR-17–3p, miR-181b-5p/TIMP3 axis [89]. Some circRNAs act as tumor-promoting molecules in HCC. Circ_0067934 and circHIPK3 were all highly expressed in HCC tissues. Knockdown of these circRNAs resulted in suppressed HCC cell proliferation and migration [32,90]. Circ_0067934 achieved its regulatory function through circ_0067934/miR-1324/FZD5 pathway [90], and circHIPK3 through circHIPK3/miR-124/AQP3 axis [32].
A majority of HCC in Asian area arises from chronic hepatocellular B virus (HBV) infection and subsequent cirrhosis [115]. Cui et al. clarified circRNA expression profiles in HBV-related HCC by microarray analysis [116]. Huang et al. discovered that circRNA_100338 was upregulated in HBV-related HCC, and correlated with both low survival rate of patients and invasive process of cancer cells. CircRNA_100338 could facilitate cancer cell invasion and migration by sponging and inhibiting miR-141–3p [91].
Colorectal cancer
In recent years, several circRNAs are detected as candidate biomarkers for CRC. For instance, the expression level of circRNA0003906 was significantly decreased in CRC tissues and CRC cell lines, and was correlated with clinicopathological factors, such as lymphatic metastasis and poor differentiation. The AUC was 0.818, which demonstrated the diagnostic ability of circRNA0003906 [92]. Similarly, hsa_circ_001988 [93], hsa_circRNA_103809 and hsa_circRNA_104700 [94] were also demonstrated as potential diagnostic biomarkers. Their expression levels were dysregulated in CRC and correlated with clinicopathological features. The AUC for hsa_circ_001988, hsa_circRNA_103809 and hsa_circ_104700 was 0.788, 0.699 and 0.616, respectively [93,94].
Several ectopic circRNAs were verified to have regulatory potential in CRC. Gua et al. revealed that hsa_circ_0000069 silencing notably attenuated tumor cell proliferation, migration and invasion [95]. Zhang et al. testified that hsa_circ_0007534 silencing led to suppressed proliferation and induced apoptosis of CRC cells [96]. In another research, the authors delineated that hsa_circ_0020397 performed its tumor-generating effects via hsa_circ_0020397/miR-138/TERT, PD-L1 axis [97].
Metastasis is among the major causes of tumor death and often occurs in late stages of tumor development [117]. Two recent studies investigated the metastasis in CRC by utilizing circRNA profiling. Jiang et al. detected differential circRNAs in CRC metastasis cells [118], and Zeng et al. compared circRNA expression in CRC patients with and without lung metastasis [119]. These findings provided candidate circRNAs for later investigations concerning CRC metastasis.
Bladder cancer
Recent studies reveal certain circRNAs with regulatory function in bladder cancer. Zhong et al. discovered that forced expression of circTCF25 would sequester miR-103a-3p and miR-107, which led to the increased CDK6 expression and promoted cancer cell proliferation and migration [98]. In another study, the authors manifested that circRNA-MYLK and its downstream target miR-29a/VEGFA, VEGFR2 signaling pathway were related with bladder cancer cell proliferation and epithelial–mesenchymal transition process [99]. Li et al. discovered that increased expression of circRNA BCRC4 could induce bladder cancer cell apoptosis by inhibiting miR-101 ability, which relieved suppression for EZH2 [100].
Circ-ITCH is generated from itchy E3 ubiquitin protein ligase (ITCH) coding region and is involved in bladder cancer. For instance, Yang et al. found that circ-ITCH was downregulated in bladder cancer tissues and cell lines. Decreased expression of circ-ITCH was correlated with shortened survival in bladder cancer patients. Expression of circ-ITCH represses cancer cell proliferation, migration and metastasis. This consequence of circ-ITCH was achieved by inhibiting miR-17 and miR-224 and promoting p21 and PTEN [101].
Glioma
In 2016, Song et al. constructed a computational filter named UROBORUS and disclosed circRNA expression in gliomas for the first time [120]. Then Li et al. revealed that hsa_circ_0046701 was increased in glioma tissue and cell lines. Hsa_circ_0046701 was able to regulate tumor cell proliferation and invasion by sponging miR-142–3p and increasing ITGB8 [102]. Interestingly, the open reading frame equipped in circ-FBXW7 and circ-SHPRH allowed them to translate functional proteins, FBXW7–185aa and SHPRH-146aa [28,42]. Both FBXW7–185aa and SHPRH-146aa displayed antioncogenic potency in glioma. These two researches indicated a potential new role for circRNAs.
Conclusion & future perspective
Unlike linear RNAs, continuous circRNAs have no free ends and are more resistant to RNase. The properties of circRNAs include stability, prevalence, specificity and conservation. CircRNAs have been demonstrated to regulate cellular function through sponging other factors (miRNAs or proteins), promoting transcription or coding proteins. From cancers to noncancerous disorders, large quantities of studies have revealed the involvement of circRNAs in human diseases as biomarkers and/or regulators. These potential circular biomarkers/regulators, combined with currently widely used diagnostic and treating methods, may improve future clinical activities. However, in order to contribute to later diagnosis and treatment, the precise role of circRNA in both physiological and pathological conditions needs further investigation. Besides the reported mechanism, new hypothesis may be proposed and demonstrated. What is more, the curative effects and the side effects of circRNAs as treatment targets in vivo should be evaluated and examined.
Executive summary.
Circular RNAs (circRNAs) are generated without 3′ and 5′ free ends.
CircRNAs include exonic RNAs, exon–intron RNAs and circular intronic RNAs.
CircRNAs are stably, prevalently, specifically and conservatively expressed.
CircRNAs regulate gene expression and transcription through binding to miRNAs, Pol II and proteins. Some endogenous circRNAs code proteins themselves.
CircRNAs are related with diverse human diseases.
Footnotes
Authors’ contributions
Y Fang collected information and wrote the manuscript.
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Creative Commons Attribution 4.0 License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Discovery of circular RNAs (circRNAs).
- 2.Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 3.Wang PL, Bao Y, Yee MC, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9(6):e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Features of circRNAs.
- 4.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Basic characteristics of circRNAs.
- 5.Li Z, Huang C, Bao C, et al. exon–intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]; • Formation of circRNAs.
- 8.Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl. 6):680. doi: 10.1186/s12864-017-4029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58(5):870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]; •• Demonstration of circRNA function as miRNA sponges.
- 11.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]; •• The first study concerning well-known circRNA Cdr1as.
- 12.Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. Biomed. Res. Int. 2017:5135781. doi: 10.1155/2017/5135781. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li LJ, Huang Q, Pan HF, Ye DQ. Circular RNAs and systemic lupus erythematosus. Exp. Cell Res. 2016;346(2):248–254. doi: 10.1016/j.yexcr.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 15.Idda ML, Munk R, Abdelmohsen K, Gorospe M. Noncoding RNAs in Alzheimer's disease. Wiley Interdiscip. Rev. RNA. 2018;9(2) doi: 10.1002/wrna.1463. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: a novel type of biomarker and genetic tools in cancer. Oncotarget. 2017;8(38):64551–64563. doi: 10.18632/oncotarget.18350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs As a new class of putative biomarkers in human blood. PLoS ONE. 2015;10(10):e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The potential of circRNAs as biomarkers in human blood.
- 19.Pan RY, Liu P, Zhou HT, et al. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget. 2017;8(36):60280–60290. doi: 10.18632/oncotarget.19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61(1):221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westholm JO, Miura P, Olson S, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao T, Wang L, Li S, Xu M, Guan X, Zhou B. Characterization of conserved circular RNA in polyploid Gossypium species and their ancestors. FEBS Lett. 2017;591(21):3660–3669. doi: 10.1002/1873-3468.12868. [DOI] [PubMed] [Google Scholar]
- 23.Dong R, Ma XK. Increased complexity of circRNA expression during species evolution. 2017;14(8):1064–1074. doi: 10.1080/15476286.2016.1269999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. BioMed Res. Int. 2016:1579490. doi: 10.1155/2016/1579490. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl Cancer Inst. 2018;110(3) doi: 10.1093/jnci/djx166. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A relatively new function of circRNAs.
- 29.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 34.Huang C, Shan G. What happens at or after transcription: insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS ONE. 2015;10(6):e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 41.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66(1):22.e29–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 43.Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed. Pharmacother. 2017;95:1514–1519. doi: 10.1016/j.biopha.2017.09.064. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Zhao Z, Jian D, Li W, Tang H, Li M. Hsa-circRNA11783–2 in peripheral blood is correlated with coronary artery disease and Type 2 diabetes mellitus. Diab. Vasc. Dis. Res. 2017;14(6):510–515. doi: 10.1177/1479164117722714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Gan TY, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salgado-Somoza A, Zhang L, Vausort M, Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu N, Jin L, Cai J. Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin. Exp. Hypertens. 2017;39(5):454–459. doi: 10.1080/10641963.2016.1273944. [DOI] [PubMed] [Google Scholar]
- 48.Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487(4):769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 51.Zhang SJ, Chen X, Li CP, et al. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest. Ophthalmol. Vis. Sci. 2017;58(14):6500–6509. doi: 10.1167/iovs.17-22698. [DOI] [PubMed] [Google Scholar]
- 52.Ouyang Q, Wu J, Jiang Z, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell. Physiol. Biochem. 2017;42(2):651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 53.Luan J, Jiao C, Kong W, et al. circHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol. Ther. Nucleic Acids. 2018;10:245–253. doi: 10.1016/j.omtn.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu DY, Pan CY, Yu JM. The relationship between coronary artery disease and abnormal glucose regulation in China: the China Heart Survey. Eur. Heart J. 2006;27(21):2573–2579. doi: 10.1093/eurheartj/ehl207. [DOI] [PubMed] [Google Scholar]
- 55.Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 2005;118(Pt 14):3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 56.Cheng X, Joe B. Circular RNAs in rat models of cardiovascular and renal diseases. Physiol. Genomics. 2017;49(9):484–490. doi: 10.1152/physiolgenomics.00064.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czubryt MP. Common threads in cardiac fibrosis, infarct scar formation, and wound healing. Fibrogenesis & Tissue Repair. 2012;5(1):19. doi: 10.1186/1755-1536-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murtaza I, Wang HX, Feng X, et al. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J. Biol. Chem. 2008;283(10):5996–6004. doi: 10.1074/jbc.M706466200. [DOI] [PubMed] [Google Scholar]
- 59.Chen J, Cui L, Yuan J, Zhang Y, Sang H. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017;494(1–2):126–132. doi: 10.1016/j.bbrc.2017.10.068. [DOI] [PubMed] [Google Scholar]
- 60.Gu Y, Ke G, Wang L, Zhou E, Zhu K, Wei Y. Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res. 2017;58(3):176–184. doi: 10.1159/000479156. [DOI] [PubMed] [Google Scholar]
- 61.Korn C, Augustin HG. Mechanisms of vessel pruning and regression. Dev. Cell. 2015;34(1):5–17. doi: 10.1016/j.devcel.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Campochiaro PA. Ocular neovascularization. J. Mol. Med. (Berl.) 2013;91(3):311–321. doi: 10.1007/s00109-013-0993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metabol. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng F, Yu X, Huang J, Dai Y. Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology. Mol. Med. Rep. 2017;16(6):8029–8036. doi: 10.3892/mmr.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Li K, Lai W, et al. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin. Chim. Acta. 2018;480:17–25. doi: 10.1016/j.cca.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 67.Zhou H, Hasni SA, Perez P, et al. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J. Am. Soc. Nephrol. 2013;24(7):1073–1087. doi: 10.1681/ASN.2012080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed. Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 69.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol. Res. Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 70.Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci. Rep. 2018;8(1):2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Zhu X, Zhang H, et al. Increased circular RNA hsa_circ_0012673 acts as a sponge of miR-22 to promote lung adenocarcinoma proliferation. Biochem. Biophys. Res. Commun. 2018;496(4):1069–1075. doi: 10.1016/j.bbrc.2018.01.126. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int. J. Oncol. 2018;52(3):743–754. doi: 10.3892/ijo.2018.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am. J. Cancer Res. 2017;7(7):1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 75.Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36(11):901–908. doi: 10.1089/dna.2017.3862. [DOI] [PubMed] [Google Scholar]
- 76.He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36(1):145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomarkers Med. 2018;12(1):11–20. doi: 10.2217/bmm-2017-0114. [DOI] [PubMed] [Google Scholar]
- 78.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 79.Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 81.Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 82.Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J. Mol. Med. (Berl.) 2018;96(1):85–96. doi: 10.1007/s00109-017-1600-y. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging. 2017;9(6):1585–1594. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, Wang G, Chu SJ, et al. Loss of large tumor suppressor 1 promotes growth and metastasis of gastric cancer cells through upregulation of the YAP signaling. Oncotarget. 2016;7(13):16180–16193. doi: 10.18632/oncotarget.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 86.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol. Oncol. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 89.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018 doi: 10.1016/j.jhep.2018.01.012. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 90.Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem. Biophys. Res. Commun. 2018;497(2):626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 91.Huang XY, Huang ZL, Xu YH, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141–3p pathway in hepatitis B-related hepatocellular carcinoma. Sci. Rep. 2017;7(1):5428. doi: 10.1038/s41598-017-05432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int. J. Clin. Exp. Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang P, Zuo Z, Shang W, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39(3) doi: 10.1177/1010428317694546. 1010428317694546. [DOI] [PubMed] [Google Scholar]
- 95.Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. OncoTargets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur. Rev. Med. Pharm. Sci. 2018;22(1):118–126. doi: 10.26355/eurrev_201801_14108. [DOI] [PubMed] [Google Scholar]
- 97.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 98.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 100.Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ, Xiao XY. Overexpression of CircRNA BCRC4 regulates cell apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2017;37(6):886–890. doi: 10.1007/s11596-017-1822-9. [DOI] [PubMed] [Google Scholar]
- 101.Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol. Cancer. 2018;17(1):19. doi: 10.1186/s12943-018-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li G, Yang H, Han K, Zhu D, Lun P, Zhao Y. A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis by increasing the expression of miR-142–3p target ITGB8 in glioma. Biochem. Biophys. Res. Commun. 2018;498(1):254–261. doi: 10.1016/j.bbrc.2018.01.076. [DOI] [PubMed] [Google Scholar]
- 103.Aggarwal A, Lewison G, Idir S, et al. The state of lung cancer research: a global analysis. J. Thorac. Oncol. 2016;11(7):1040–1050. doi: 10.1016/j.jtho.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 104.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J. Clin. Oncol. 2007;25(5):561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 105.Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell. Physiol. Biochem. 2017;44(6):2138–2146. doi: 10.1159/000485953. [DOI] [PubMed] [Google Scholar]
- 106.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 107.Yan N, Xu H, Zhang J, et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8(56):95704–95718. doi: 10.18632/oncotarget.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu R, Shi P, Nie Z, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6(4):533–544. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 110.Sui W, Shi Z, Xue W, et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol. Rep. 2017;37(3):1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 111.Dang Y, Ouyang X, Zhang F, et al. Circular RNAs expression profiles in human gastric cancer. Sci. Rep. 2017;7(1):9060. doi: 10.1038/s41598-017-09076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Y, Li J, Yu J, et al. Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget. 2017;8(14):22936–22943. doi: 10.18632/oncotarget.15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424–5p and regulating LATS1 expression. Mol. Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu RX, Seto WK, Lai CL, Yuen MF. Epidemiology of hepatocellular carcinoma in the Asia-Pacific region. Gut Liver. 2016;10(3):332–339. doi: 10.5009/gnl15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pineau P, Tiollais P. [Hepatitis B vaccination: a major player in the control of primary liver cancer] Pathol. Biol. (Paris) 2010;58(6):444–453. doi: 10.1016/j.patbio.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 116.Cui S, Qian Z, Chen Y, Li L, Li P, Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol. Lett. 2018;15(1):423–432. doi: 10.3892/ol.2017.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 118.Jiang W, Zhang X, Chu Q, et al. The circular RNA profiles of colorectal tumor metastatic cells. Front. Genet. 2018;9:34. doi: 10.3389/fgene.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zeng Y, Xu Y, Shu R, et al. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. Int. J. Mol. Med. 2017;40(6):1818–1828. doi: 10.3892/ijmm.2017.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song X, Zhang N, Han P, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9):e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]