Abstract
Aim:
Imprinted genes exhibit expression in a parent-of-origin-dependent manner and are critical for child development. Recent limited evidence suggests that prenatal exposure to phthalates, ubiquitous endocrine disruptors, can affect their epigenetic dysregulation.
Materials & methods:
We quantified DNA methylation of nine imprinted gene differentially methylated regions by pyrosequencing in 296 cord blood DNA samples in a Mexican–American cohort. Fetal exposure was estimated by phthalate metabolite concentrations in maternal urine samples during pregnancy.
Results:
Several differentially methylated regions of imprinted genes were associated with high molecular weight phthalates. The most consistent, positive, and false discovery rate significant associations were observed for MEG3.
Conclusion:
Phthalate exposure in utero may affect methylation status of imprinted genes in newborn children.
Keywords: : cord blood, differentially methylated region, DNA methylation, endocrine disruptors, imprinted genes, in utero exposure, Mexican–Americans, newborns, phthalates, pyrosequencing
Epigenetic influences on gene expression have been implicated as a mediator of the relationship between environmental exposures and health status. Early-life adverse environments can cause epigenetic shifts, establishing disease trajectories into adulthood, an idea described by the Developmental Origins of Health and Disease hypothesis [1,2]. Epigenetic modifications can alter gene expression without changing the underlying nucleotide sequence. DNA methylation is the epigenetic mechanism most often studied [1,3,4]. It is simultaneously heritable and susceptible to environmental insults, and is able to reveal cumulative effects of environmental exposures throughout the life course [5,6].
Genomic imprinting involves the expression of only one allele of the gene and is dependent on the parental origin of the expressed allele [7]. The determination of active versus inactive allele is regulated by DNA methylation that is established during the process of epigenetic reprogramming that occurs in the developing gametes [8]. The majority of the known imprinted genes within the human genome have differentially methylated regions (DMRs) whose methylation is also parent-of-origin-dependent [9,10]. Methylation patterns in the DMRs are remodeled and established prior to germ layer specification, and are maintained in somatic tissues throughout life [8]. A number of human studies have demonstrated the effects of environmental exposures on imprinted gene DNA methylation, including the influence of diet [11–14], cigarette smoke [15], maternal antibiotic use [16], metal exposure [17–22] and maternal stress [23,24]. Differentially methylated regions of imprinted genes represent a unique opportunity to assess the effects of in utero exposure since they influence development and growth in early life [18].
Limited but growing evidence indicates that exposure to phthalates is associated with DNA methylation changes of imprinted genes [25–27]. Phthalates, diesters of phthalic acid, are a family of chemicals often found in consumer products, resulting in common exposure in the USA [28,29]. Some phthalates, such as di-(2-ethylhexyl) phthalate (DEHP), are added to plastics to increase flexibility and can be found in toys, plastic containers and medical supplies. Other phthalates, such as diethyl phthalate, are used as solvents in personal care products such as perfumes and lotions [30]. Common routes of exposure to phthalates, which are not chemically bound to their substrates and leach into the environment, include ingestion, dermal absorption and inhalation. Previous studies in humans have demonstrated associations between phthalate exposure during or prior to pregnancy and adverse impacts on health outcomes in children, including altered birth weight [31–33], gestational age [34,35], preterm birth [36,37], child growth and development [38,39], child behavior [40] and asthma [41].
Research in animals [42–45] has shown associations between phthalates and global and site-specific methylation; however, there is a paucity of information on imprinted gene DMR methylation in relation to phthalate exposure in utero. Only two human studies are available at this time that have examined the relationship between phthalate exposure during pregnancy and placental imprinted gene methylation (H19 and IGF2) [25,27]. Currently there are no data available on the relationship between cord blood imprinted gene methylation profiles and prenatal exposure to phthalates.
Previously, we reported on the prenatal phthalate metabolite concentrations in participants of the Center for the Health Assessment of Mothers and Children of Salinas (CHAMACOS) study and observed inverse associations of MEP concentrations with DNA methylation of repetitive elements in newborns and children [46]. We have also assessed the relationship between prenatal phthalate exposure in CHAMACOS mothers and both genome-wide site-specific and region DNA methylation as assessed by the 450K array in newborns, identifying 27 different regions within the human genome associated primarily with DEHP metabolites [47]. The objective of the present study is to assess the impact of prenatal phthalate exposure on DNA methylation of imprinted genes in CHAMACOS newborns.
Materials & methods
Study subjects
CHAMACOS is a longitudinal birth cohort study assessing neurological, developmental and respiratory health effects of environmental exposure in pregnant Mexican–American women and their children residing in the agricultural region of Salinas Valley, CA, USA [48]. From 1999 to 2000, a total of 601 pregnant women were enrolled in the study and 527 women delivered live, singleton newborns. At the time of enrollment, CHAMACOS mothers were at least 18 years of age, Spanish or English speaking, eligible for low-income health insurance, and were receiving prenatal care at one of several participating clinics. During pregnancy, CHAMACOS mothers were interviewed twice (13.2 ± 5.1 and 26.0 ± 2.7 weeks gestation) by trained bilingual, bicultural staff regarding reproductive and medical history, sociodemographic factors and pregnancy-specific lifestyle and environmental exposures [48,49]. The pregnancy visits will subsequently be referred to as either the early or late pregnancy visits.
Immediately following delivery, whole cord blood was collected from the umbilical vein in BD vacutainers (Becton, Dickinson and Company, NJ, USA) without anticoagulant. Blood specimens were allowed to clot for at least 30 min, centrifuged at 1200 rpm for 10 min, divided into serum and blood clot aliquots, and stored at -80°C. The QIAamp Blood DNA Maxi kit (Qiagen, Inc., CA, USA) was used to isolate genomic DNA from clots as previously described [50]. Methylation of nine imprinted gene DMRs was measured in 296 newborn children (148 girls and 148 boys) that had sufficient DNA available for analysis. The subset was not significantly different from the main CHAMACOS cohort in many demographic and exposure characteristics (child sex, poverty index, education, gestational age and parity). However, mothers in this sample tended to be slightly younger and more obese, lived fewer years in the USA and gave birth to children that were less likely to have a low birth weight. The number of CHAMACOS participants with pyrosequencing data is approximately double the subjects included in existing literature assessing the relationship between phthalate exposure and imprinted genes [25,27] and we anticipate that we will have sufficient power to detect a minimum of a 0.05 linear correlation between phthalates and imprinted genes.
CHAMACOS study protocols were approved by the University of California (UC), Berkeley Committee for Protection of Human Subjects. All mothers provided written informed consent at the time of enrollment.
Phthalate metabolite measurements
Maternal urine samples were collected during the early and late pregnancy visits. The urine samples were aliquoted, barcoded, and stored at -80°C in the UC Berkeley School of Public Health Biorepository. Eleven phthalate metabolites were quantified using online solid phase extraction coupled with isotope dilution high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry, as previously described [46,49,51,52]. The metabolites measured included three low molecular weight (LMW) metabolites (monoethyl phthalate [MEP], mono-n-butyl phthalate [MBP], mono-isobutyl phthalate [MiBP]), four DEHP metabolites (mono[2-ethylhexyl] phthalate [MEHP], mono[2-ethyl-5-hydroxyhexyl] phthalate [MEHHP], mono[2-ethyl-5-oxohexyl] phthalate [MEOHP], mono[2-ethyl-5-carboxypentyl] phthalate [MECPP]), and four high molecular weight (HMW) metabolites (monobenzyl phthalate [MBzP], mono[3-carboxypropyl] phthalate [MCPP], monocarboxyoctyl phthalate [MCOP], monocarboxynonyl phthalate [MCNP]). Quality control procedures comprised incorporation of laboratory and field blanks, calibration standards and spiked controls with low and high concentrations into the experimental runs.
The limits of detection (LOD) for all the phthalate metabolites have been previously described [46,51,53]. The instrumental reading values were used when the measured phthalate metabolite concentrations fell below the LOD. In instances, where the measured concentrations were below the LOD and instrumental signals were unavailable, the ‘fill-in’ method, described in Lubin et al. [54] was used to impute phthalate metabolite concentrations from a log-normal distribution. Summary measurements of the DEHP metabolites in units of micrograms per liter were generated as described previously [55]. Specifically, molar concentrations of each DEHP metabolite were calculated by dividing the concentration of the metabolite by its molecular weight. The molar concentrations for each DEHP phthalate metabolite were summed and then multiplied by the average molecular weight of the metabolites. All phthalate metabolites, with the exception of the DEHP metabolites, were analyzed individually and HMW and LMW sum variables were not generated since values are largely driven by DEHP and diethyl phthalate metabolites, respectively. Phthalate metabolite concentrations (micrograms per liter) were divided by creatinine concentrations (grams per liter), measured using a commercially available diagnostic enzyme method (Vitros CREA slides; Ortho Clinical Diagnostics, NJ, USA) concurrently with the phthalate metabolites, to generate values (micrograms per gram creatinine) adjusted for urinary dilution as previously described [49].
Since, we anticipate that average phthalate metabolite concentrations during pregnancy will be a more stable indicator of pregnancy exposure, we assessed in utero phthalate exposure as the average of the log10 transformed concentrations of creatinine-corrected phthalate metabolites from the two prenatal visits [52]. Participants with measurements greater or less than three-times the interquartile range for a particular phthalate were removed from analyses. All descriptive analyses, figures and regression models included creatinine adjusted phthalate metabolite concentrations averaged across pregnancy. Additionally, we performed a sensitivity analysis that included adjusting for urinary dilution using specific gravity instead of creatinine, and we observed that it did not affect the findings. Specific gravity was quantified with a refractometer (National Instrument Company, Inc., MD, USA), and urinary phthalate metabolites were adjusted for specific gravity as previously described [46]. The results of the principal component and mean methylation analyses of the imprinted gene and average phthalate exposure during pregnancy when adjusting for specific gravity were similar to the creatinine adjusted models.
DNA methylation analyses
The EZ DNA Methylation kit (Zymo Research, CA, USA) was used for bisulfite conversion of 800 ng of DNA, following the manufacturer's instructions. Treating DNA with sodium bisulfite converts unmethylated cytosines to uracils and leaves methylated cytosines unchanged.
The PyroMark PCR Kit (Qiagen, CA, USA) was used to prepare bisulfite-treated DNA for PCR amplification, using published primer sequences for nine imprinted gene DMRs and assays previously developed and validated at Duke University [8,56]. These imprinted genes were selected based on their biological significance and availability of reliable assays [25,57–61]. Pyrosequencing of the amplified and bisulfite converted DNA was achieved using the Pyromark Q96 MD system (Qiagen), yielding percent methylation estimates for the CpG sites within the sequence analyzed for each DMR. Methylation was measured for the following imprinted genes: H19, IGF2, MEG3 and MEG3-IG, MEST, NNAT, PLAGL1, PEG3 and SGCE/PEG10. Specifically, methylation was measured at the DMRs upstream of IGF2 exon 3 (chr 11p15.5; three CpG sites), upstream of the H19 gene (chr 11p15.5; four CpG sites), two DMRs involved in regulating the DLK1/MEG3 imprinted domain (chr 14q32.2; MEG3-IG: four CpG sites; MEG3: seven CpG sites), the MEST promoter (chr 7q32.2; four CpG sites), the NNAT locus (chr 20q11.23; three CpG sites), the PEG3 promoter (chr 19q13.43; ten CpG sites), the PLAGL1 locus (chr 6q24.2; six CpG sites) and the SGCE/PEG10 promoter (chr 7q21.3; six CpG sites). The paternal allele is expressed for the genes IGF2, DLK1, MEST, NNAT, PEG3, PLAGL1 and SGCE; whereas the maternal allele is expressed in MEG3 and H19. The selected imprinted genes are critical in the regulation of early growth [61], and alterations of DNA methylation by environmental insults, such as phthalate exposure, could have implications for imprinted gene expression and downstream effects on growth trajectories. In the CHAMACOS cohort, we have observed that in utero exposure to MEP is associated with increased odds of being overweight or obese that persists from age 5 to 12 [52]. Quality assurance measures involved inclusion of technical repeats and positive and negative controls. Samples whose methylation values exceeded two standard deviations from the mean for the plate were reanalyzed. The CV of intraplate repeat measures ranged from 1–3%. Using mixtures of DNA with fully methylated and unmethylated sequences [8,61], we have previously shown that pyrosequencing allows for the detection of 0.5–5% methylation differences.
Three approaches were used to account for cell composition in the analysis based on cytological differential cell counts (DCC) [62] and 450K-array estimates [47]. The methods included adjusting for cytological DCC in a subset of cord blood samples [46,62] and estimating cell-type proportions from CHAMACOS 450K cord blood DNA methylation data [47] using adult [63] and cord blood [64] flow sorted reference panels. The cord blood reference panel includes estimates of nucleated red blood cells (nRBCs).
Gene expression analysis
Validation of significant hits of the relationship between average phthalate metabolites during pregnancy and imprinted gene methylation in newborns was performed using a two-step RT-PCR in a subset of the CHAMACOS participants with imprinted gene data and available isolated RNA. Specifically, we assessed the overall expression of MEG3 in 119 CHAMACOS children with imprinted gene data, since methylation changes due to environment exposure could lead to alteration of gene expression. As a negative control, we also examined overall expression patterns in DLK1 and its relationship with MEG3-IG methylation, an imprinted gene DMR where we did not expect to see changes in gene expression since the mean methylation across the DMR was not significantly associated with phthalate exposure. Expression of the reference gene GAPDH was used as a control. SuperScript™ IV VILO™ Master Mix was used to convert RNA to cDNA. Predesigned TaqMan Gene Primer Assays for MEG3, MEG3-IG and GAPDH, as well as, TaqMan Fast Advanced Master Mix were used in the RT-PCR analysis according to the manufacturer's protocols. Reactions were performed on a Rotor-Gene 6000 (Qiagen, formerly Corbett Life Science). Amplicon specificity was verified by reviewing melt curves. Ct values were calculated for each gene, and ΔCt values, calculated as the difference between the imprinted genes (MEG3 or MEG3-IG) and GAPDH Ct values, were used in statistical analysis. Interplate replicates and negative and positive controls were included in the expression analysis for quality control.
Statistical analyses
Covariates included in the regression models were selected from factors related to phthalate metabolite levels in the CHAMACOS cohort, after performing bivariate analyses (i.e., linear regressions, Student's t-test, ANOVA) and important covariates identified in previous studies examining the relationship between imprinted genes and prenatal phthalate exposure. The relevant covariates include prepregnancy body mass index (BMI), years in the USA, parity and child sex. Prepregnancy BMI and years in the USA were coded as continuous variables, while parity and sex were coded as shown in Table 1. Additionally, a plate (batch) variable was included in the regression models to adjust for technical effects.
Table 1. . Demographic characteristics of Center for the Health Assessment of Mothers and Children of Salinas mothers and children (1999–2000).
| Characteristic | CHAMACOS mothers and their children with imprinted gene DMR methylation data (n = 296) n (%) |
|---|---|
| Child sex: | |
| – Boy | 148 (50.0) |
| – Girl | 148 (50.0) |
| Child gestational age at birth: | |
| – 34–36 weeks | 20 (6.8) |
| – ≥37 weeks | 276 (93.2) |
| Child birth weight: | |
| – Low birth weight (<2500 g) | 10 (3.4) |
| – Normal birth weight (≥2500 g) | 286 (96.6) |
| Maternal age at pregnancy: | |
| – 18–24 | 137 (46.3) |
| – 25–29 | 101 (34.1) |
| – 30–34 | 46 (15.5) |
| – 35–45 | 12 (4.1) |
| Number of years mother lived in USA at pregnancy: | |
| – Less than 1 year | 55 (18.6) |
| – 1–5 years | 102 (34.5) |
| – 6–10 years | 70 (23.6) |
| – 11 or more years | 69 (23.3) |
| Maternal prepregnancy BMI:† | |
| – Underweight (<18.5 kg/m2) | 2 (0.7) |
| – Normal (18.5–24.9 kg/m2) | 118 (40.7) |
| – Overweight (25–29.9 kg/m2) | 115 (39.7) |
| – Obese (≥30 kg/m2) | 55 (19.0) |
| Parity: | |
| – 0 | 106 (35.8) |
| – ≥1 | 190 (64.2) |
| Education: | |
| – ≤6th grade | 125 (42.2) |
| – 7–12th grade | 114 (38.5) |
| – ≥High school graduate | 57 (19.3) |
| Poverty status: | |
| – ≤Poverty level | 184 (62.2) |
| – Poverty level | 112 (37.8) |
†Total number of observations for maternal prepregnancy BMI varies due to missing data.
BMI: Body mass index; CHAMACOS: Center for the Health Assessment of Mothers and Children of Salinas; DMR: Differentially methylated region.
Prior to performing regression analyses, we logit-transformed the methylation fractions to ‘M-values’ to reduce dependence of their variance on their mean levels. Methylation fractions have more intuitive biological interpretation, but the M-value is more statistically valid for the differential analysis of methylation levels [65,66]. Methylation outliers, specifically M-values that were below the 25th percentile minus three-times the interquartile range and values greater than the 75th percentile plus three-times the interquartile range, were designated as not available in the dataset, resulting in one to four substitutions for all imprinted genes, with the exception of the MEST and PLAGL1 DMRs. As a result of the correlation between CpG sites within a DMR, we decided to conduct DMR-level principal components (PCs) analyses of the data to determine the independent M-value methylation signals, represented as linear combinations of CpGs that explain at least 95% of variability at the locus. DMR-level PC analyses yielded a total of 26 PCs across the nine investigated imprinted gene DMRs (number of PCs: H19 = 2, IGF2 = 2, MEG3 = 1, MEG3-IG = 3, MEST = 2, NNAT = 2, PLAGL1 = 4, PEG3 = 6 and SGCE/PEG10 = 4). The first PCs for all imprinted gene DMRs demonstrated similar contributions for each CpG to the variance explained by PC1. We fit separate regression models for each combination of the 26 PCs and 12 phthalate metabolite variables (11 phthalate metabolites and the DEHP summary variable) averaged across pregnancy, with the PCs as the response and the average phthalate metabolite concentrations and confounder variables (mentioned above) as covariates. Since the analysis with the PCs does not provide information regarding direction of association, we also performed separate regression analyses for each imprinted gene DMR using the average M-value methylation across the CpGs within the DMR as the outcome. We confirmed regression findings of the relationship between average phthalate metabolites and mean imprinted gene methylation by implementing a bootstrap analysis, sampling individuals with replacement for 1000 iterations.
Since many imprinted genes impact early growth patterns and MEG3 has been associated previously with growth outcomes at birth, we performed additional analyses to examine the relationship between average percent methylation of the MEG3 DMR and birth weight. A dichotomous birth weight variable was generated at a cut-off of 2500 g to distinguish between low and normal/high birth weight newborns. We performed a two-sample t-test to compare mean methylation values in the MEG3 DMR in the two birth weight groups. We also ran regressions to test the association between MEG3 mean methylation and birth weight, coded as a continuous or binary variable, adjusting for infant sex, maternal prepregnancy BMI, methylation plate, route of delivery and gestational age.
We controlled for the false discovery rate (FDR). The FDR can be defined as the average number of false rejections of the null hypothesis divided by the total number of rejections [67]. All statistical analyses were performed in R Version 3.2.3 (R Foundation for Statistical Computing, Vienna, Austria, 2013). p-values <0.05 were considered significant, and two-sided statistical tests were used.
Results
CHAMACOS participants
Demographic characteristics of CHAMACOS mothers and children are presented in Table 1. Most mothers were young, living below the poverty level, had previously delivered children, had low levels of education, and were overweight or obese (BMI ≥25 kg/m2) prior to pregnancy. Approximately, half of the women resided in the USA for over 5 years at the time of delivery (mean = 6.9 years, SD = 6.9 years). The majority of the children, equally represented by boys and girls, were born term (93.2%) and with a normal birth weight (96.6%).
Phthalate exposure
Table 2 shows the distributions of average phthalate metabolite concentrations across pregnancy for the CHAMACOS mothers included in this study. Detection frequencies of phthalate metabolites during pregnancy were 90–100%. Compared with ten other phthalate metabolites, MEP had the highest urinary concentrations during pregnancy (interquartile range [IQR]: 90.7–444.7 μg/g creatinine) in the CHAMACOS cohort [49,52], consistent with trends observed in the general US population of adults [28]. Levels of urinary MEHHP (IQR: 9.5–29.2), MEOHP (IQR: 7.0–20.7) and MECPP (IQR: 17.7–45.3) contributed most to the DEHP sum concentration.
Table 2. . Distribution of phthalate metabolite concentrations averaged across pregnancy.
| Phthalate metabolite | Pregnancy average (n = 265) | |||
|---|---|---|---|---|
| Median | IQR | Minimum | Maximum | |
| MEP | 214.2 | (90.7–444.7) | 7.2 | 6607.7 |
| MBP | 24.4 | (14.2–45.3) | 3.2 | 228.6 |
| MiBP | 2.7 | (1.6–5.3) | 0.1 | 192.0 |
| MEHP | 3.9 | (2.2–7.0) | 0.1 | 101.6 |
| MEHHP | 16.1 | (9.5–29.2) | 1.4 | 478.6 |
| MEOHP | 12.1 | (7.0–20.7) | 0.9 | 353.4 |
| MECPP | 26.7 | (17.7–45.3) | 5.3 | 665.2 |
| ΣDEHP | 60.9 | (37.1–98.5) | 8.0 | 1524.5 |
| MBzP | 8.2 | (4.7–13.8) | 0.5 | 97.4 |
| MCPP | 2.0 | (1.3–2.9) | 0.1 | 28.3 |
| MCOP | 3.4 | (2.1–4.7) | 0.5 | 82.7 |
| MCNP | 1.9 | (1.3–2.6) | 0.3 | 17.1 |
All units are in μg/g creatinine.
DEHP: Di-2-ethylhexyl phthalate; IQR: Interquartile range; MBP: Mono-n-butyl phthalate; MBzP: Monobenzyl phthalate; MCNP: Monocarboxynonyl phthalate; MCOP: Monocarboxyoctyl phthalate; MCPP: Mono(3-carboxypropyl) phthalate; MECPP: Mono(2-ethyl-5-carboxypentyl) phthalate; MEHHP: Mono(2-ethyl-5-hydroxyhexyl) phthalate; MEHP: Mono(2-ethylhexyl) phthalate; MEOHP: Mono(2-ethyl-5-oxohexyl) phthalate; MEP: Monoethyl phthalate; MiBP: Mono-isobutyl phthalate.
DNA methylation of imprinted genes
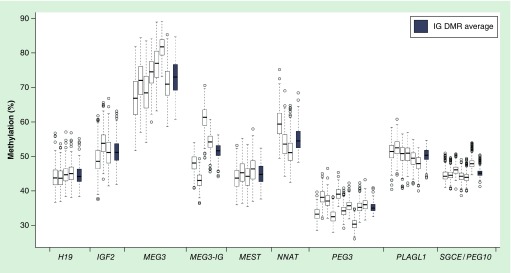
Distributions of DNA methylation values at individual CpG sites within each DMR and average methylation for each of the nine imprinted gene DMRs are shown in Figure 1. The percentage of DNA methylation varies between imprinted genes and within the individual CpG sites within DMRs. MEG3 exhibited the highest (73 ± 5%) and PEG3 had the lowest (35 ± 1%) average methylation compared with the other seven imprinted genes DMRs, which were closer to the expected methylation level of 50% and ranged from 44 to 55%. Moreover, whereas some of the imprinted genes (H19, MEST, PLAGL1, SGCE) have relatively similar CpG methylation values across each DMR, the MEG3-IG and NNAT DMRs demonstrate a wide range of methylation in CpGs from different individuals (MEG3-IG: 36–71%; NNAT: 42–75%).
Figure 1. . Distribution of average and CpG specific imprinted gene differentially methylated region percent methylation in Center for the Health Assessment of Mothers and Children of Salinas newborns.
The white box plots represent distributions of the % methylation of individual CpGs in the nine imprinted gene DMRs in 296 newborns. The leftmost box plot for each imprinted gene DMR represents the first CpG assessed in the DMR. The gray box plots to the right of each group of white boxes represent the average % methylation across all CpG sites within an IG DMR. The total number of observations for each imprinted gene varies slightly due to missing data.
DMR: Differentially methylated region.
Phthalates & DMR methylation principal component analyses
Significant results of the regression analyses of associations between PCs of imprinted gene DMR methylation in cord blood DNA and phthalate metabolite concentrations during pregnancy are shown in Table 3. After adjusting for years in the USA at the time of pregnancy, parity, prepregnancy BMI, child sex and batch, HMW phthalate metabolites were related to PCs of the MEG3 DMRs and PC3 of PEG3. All DEHP metabolites assessed were significantly associated with PC1 of the MEG3 DMR (MEHP: p = 0.03; MEHHP: p = 0.002; MEOHP: p = 0.0003; MECPP: p = 0.0001). MBzP was also related to PC1 of the MEG3 DMR (p = 0.01). MCPP was associated with PC2 of the MEG3-IG DMR (p = 0.01). Average pregnancy levels of MCOP (p = 0.02) and MCNP (p = 0.03) were associated with the third principal component of PEG3. Out of the observed associations between the average phthalate metabolites during pregnancy and the MEG3 DMRs, only the associations between MEG3 and certain DEHP metabolites (MEOHP, MECPP, Σ DEHP) remained significant after FDR adjustment (all p < 0.04).
Table 3. . Regression models of average maternal phthalate metabolite concentrations during pregnancy with principal components of child imprinted gene differentially methylated region M-value methylation at delivery.
| Imprinted gene DMR | PC # | Significant exposure | β† | 95% CI | p-value |
|---|---|---|---|---|---|
| MEG3 | PC 1 | MEHP | -0.321 | (-0.602 to -0.039) | 0.03 |
| PC 1 | MEHHP | -0.544 | (-0.885 to -0.203) | 2E-03 | |
| PC 1 | MEOHP | -0.615 | (-0.944 to -0.286) | 3E-04* | |
| PC 1 | MECPP | -0.812 | (-1.211 to -0.412) | 9E-05* | |
| PC 1 | Σ DEHP | -0.719 | (-1.099 to -0.338) | 3E-04* | |
| PC 1 | MBzP | -0.448 | (-0.765 to -0.132) | 0.01 | |
| MEG3-IG | PC 2 | MCPP | 0.050 | (0.013–0.087) | 0.01 |
| PEG3 | PC 3 | MCOP | -0.051 | (-0.095 to -0.007) | 0.02 |
| PC 3 | MCNP | -0.055 | (-0.104 to -0.006) | 0.03 | |
Covariates in each regression model included batch, years in the USA, parity, prepregnancy BMI and child sex. Models included log10 transformed concentrations of creatinine-corrected phthalate metabolites.
†β for PCs are not interpretable; for direction of association, refer to mean methylation analysis
*Significant after FDR adjustment (all p < 0.04). p < 0.05 statistically significant.
BMI: Body mass index; DEHP: Di-2-ethylhexyl phthalate; DMR: Differentially methylated region; MBzP: Monobenzyl phthalate; MCNP: Monocarboxynonyl phthalate; MCOP: Monocarboxyoctyl phthalate; MCPP: Mono(3-carboxypropyl) phthalate; MECPP: Mono(2-ethyl-5-carboxypentyl) phthalate; MEHHP: Mono(2-ethyl-5-hydroxyhexyl) phthalate; MEHP: Mono(2-ethylhexyl) phthalate; MEOHP: Mono(2-ethyl-5-oxohexyl) phthalate; PC: Principal component.
Phthalates & imprinted gene average DMR methylation
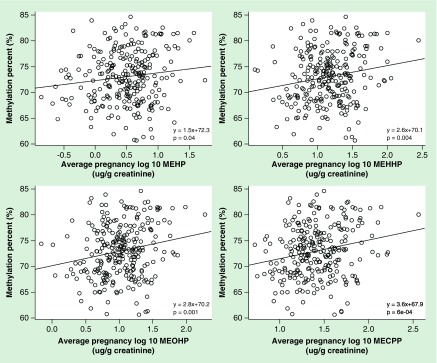
In order to determine directions of association, we analyzed the relationship between phthalate exposure and DNA methylation averaged across all CpG sites within each imprinted gene DMR (Figure 2, Table 4). In crude models without adjustment for confounders, we found a positive association between average pregnancy DEHP metabolites and mean methylation percent at the MEG3 DMR (Figure 2, all p < 0.05). In models adjusting for covariates, MEG3-IG and PEG3, which were significantly associated with some of the phthalate metabolites in the PC analysis, were no longer significant in the analysis with mean M-value methylation as the outcome (MEG3-IG and MCPP; PEG3 and MCOP; PEG3 and MCNP; all p-values >0.05). MBzP (β = 0.17; p = 0.01) and DEHP metabolites (MEHP: β = 0.12, p = 0.03; MEHHP: β = 0.21, p = 0.002; MEOHP: β = 0.23, p = 0.0003; MECPP: β = 0.31, p = 0.00007) were positively associated with mean methylation across the MEG3 DMR. All of the observed relationships remained significant after FDR adjustment (all p < 0.05), with the exception of the associations with MEHP and MBzP. Bootstrap analysis confirmed the findings; specifically, the significant and positive association between HMW phthalates and mean methylation across CpG sites within the MEG3 DMR.
Figure 2. . Scatter plots of the crude relationships between prenatal di-2-ethylhexyl phthalate metabolites and mean percent methylation of the MEG3 differentially methylated region.
MECPP: Mono(2-ethyl-5-carboxypentyl) phthalate; MEHHP: Mono(2-ethyl-5-hydroxyhexyl) phthalate; MEHP: Mono(2-ethylhexyl) phthalate; MEOHP: Mono(2-ethyl-5-oxohexyl) phthalate.
Table 4. . Regression models of average maternal phthalate metabolite concentrations during pregnancy with mean child imprinted gene differentially methylated region M-value methylation at delivery.
| Imprinted gene DMR | Significant exposure | β | 95% CI | p-value |
|---|---|---|---|---|
| MEG3 | MEHP | 0.120 | (0.015–0.225) | 0.03 |
| MEHHP | 0.206 | (0.078–0.333) | 2E-03* | |
| MEOHP | 0.232 | (0.110–0.335) | 3E-04* | |
| MECPP | 0.306 | (0.157–0.455) | 7E-05* | |
| Σ DEHP | 0.271 | (0.130–0.413) | 2E-04* | |
| MBzP | 0.168 | (0.050–0.286) | 0.01 | |
Covariates in each regression model included batch, years in the USA, parity, prepregnancy BMI and child sex. Models included log10 transformed concentrations of creatinine-corrected phthalate metabolites.
*Significant after FDR adjustment (all p < 0.05). p < 0.05 statistically significant.
BMI: Body mass index; DEHP: Di-2-ethylhexyl phthalate; FDR: False discovery rate; MBzP: Monobenzyl phthalate; MECPP: Mono(2-ethyl-5-carboxypentyl) phthalate; MEHHP: Mono(2-ethyl-5-hydroxyhexyl) phthalate; MEHP: Mono(2-ethylhexyl) phthalate; MEOHP: Mono(2-ethyl-5-oxohexyl) phthalate.
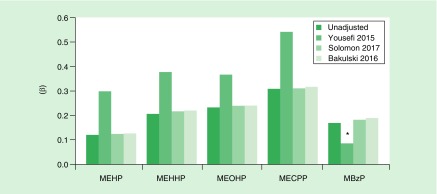
Adjustment for cell composition
Figure 3 presents the regression estimates for the significant relationships of the mean MEG3 methylation and prenatal phthalate metabolite concentrations adjusting for cell-type composition using three different methodologies as described in the methods section. The array-based estimates of cell composition [47,64] produced estimates of the regression coefficients similar to the model without cell-type adjustment (unadjusted). Coefficients from models adjusting for cytological DCC [62] showed even stronger relationships between phthalate exposure and MEG3 methylation in the same direction. Adjusting for white blood cell composition estimates using CHAMACOS 450K array data and the cord blood flow sorted reference panel replicated the crude models of the imprinted gene mean methylation and average pregnancy phthalate metabolites, suggesting limited bias due to cell-type composition.
Figure 3. . Regression coefficients of the relationships between prenatal di-2-ethylhexyl phthalate metabolites and mean methylation of the MEG3 DMR with and without cell-type adjustment.
The methods included adjusting for cytological differential cell counts in a subset of cord blood samples (Yousefi 2015 [62]) and estimating cell-type proportions from CHAMACOS 450K cord blood DNA methylation data using adult (Solomon 2017 [47]) and cord blood (Bakulski 2016 [64]) flow sorted reference panels. The cord blood reference panel includes estimates of nucleated red blood cells (nRBCs).
*Not statistically significant at p < 0.05; all other presented relationships were significant at p < 0.05.
MBzP: Monobenzyl phthalate; MECPP: Mono(2-ethyl-5-carboxypentyl) phthalate; MEHP: Mono(2-ethylhexyl) phthalate; MEHHP: Mono(2-ethyl-5-hydroxyhexyl) phthalate; MEOHP: Mono(2-ethyl-5-oxohexyl) phthalate.
Gene expression validation
Following identification of significant hits for the association between DEHP and BzBP phthalate metabolites and MEG3 methylation, we assessed gene expression of the MEG3 gene in a subset of 119 CHAMACOS participants. Ct values for MEG3 ranged from 22 to 36, with a mean of 27.6. As expected [6], MEG3 M-value methylation was inversely correlated with MEG3 expression relative to the housekeeping gene GAPDH (Pearson r = -0.10; p = 0.29). As a control, we also assessed the relationship between DLK1 expression and MEG3-IG mean methylation, an imprinted gene DMR that was not significantly associated with phthalate exposure. As expected, we did not observe a significant relationship (Pearson r = -0.05; p = 0.58). Additionally, we tested the relationships between phthalate metabolite concentrations and MEG3 expression, adjusting for the covariates identified in the methylation analysis. We observed consistent negative associations between DEHP metabolites and MEG3 expression (MEHP: β = -0.44, MEHHP: β = -1.14, MEOHP: β = -0.63, MECPP: β = -1.21); however, none of the associations reached statistical significance.
Imprinted gene methylation & birth weight
We found a significant difference in mean MEG3 percent methylation levels comparing low birth weight newborns and newborns with normal to high birth weights (t = -2.35; p = 0.04), with lower average methylation values in the low birth weight group. However, the regression analyses did not show a significant relationship between MEG3 methylation and continuous and binary birth weight in the CHAMACOS newborns.
Discussion
We examined the association of prenatal phthalate exposure and imprinted gene methylation profiles in newborn children using DNA isolated from cord blood. Previous analysis in the CHAMACOS cohort of repetitive element methylation and array-based DMRs uncovered significant associations of DNA methylation in cord blood with MEP and DEHP metabolites, respectively [46,47]. In the current study of nine imprinted gene DMRs, the most consistent findings were the associations between DEHP and BzBP phthalate metabolites and DNA methylation of the MEG3 DMR, located within an intron, downstream of the MEG3 promoter [68]. DNA methylation profiles in the CHAMACOS cohort were similar to values reported for imprinted genes in other cohorts with DNA isolated from cord blood samples [68].
To the best of our knowledge, our study is the first to demonstrate the relationship between MEG3 DNA methylation and prenatal phthalate exposure. In CHAMACOS newborns, prenatal exposure to several DEHP metabolites was positively and significantly associated with DNA methylation at the MEG3 DMR. Validation analysis demonstrated an inverse relationship between MEG3 methylation and expression, which is consistent with expected effects of hypermethylation. However, this result did not quite reach statistical significance. Relatively weak relationships between differential DNA methylation and expression of the same genes is common, as regulatory mechanisms may also include noncoding RNAs and chromatin modifications [1].
Hypomethylation or increased expression of MEG3 in humans have previously been associated with higher birth weight and large for gestational age status, respectively [69,70]. Conversely, decreased expression of MEG3 has been observed in human Intrauterine Growth Restriction placenta samples [71]. In the current study, we did not find a significant relationship between MEG3 methylation and continuous birth weight, which corroborates a previous report by Vidal et al. (2013) [16]. However, we did observe a significant difference when we contrasted MEG3 methylation in low birth weight newborns and normal weight ones. In addition to its relationship with birth outcomes, hypermethylation of the MEG3 gene in human tumor samples and cell lines has previously been associated with tumorigenesis [57,58,72–76]. Additionally, epigenetic and genetic studies have also uncovered links between MEG3 variants and methylation patterns and diabetes [77]. Specifically, Kameswaram et al. [78] found decreased expression of MEG3 in human islets from T2D organ donors, which was highly correlated with hypermethylation of MEG3. Studies in mice have demonstrated that decreased expression of the Meg3 gene can impact glucose tolerance and reduce insulin secretion [79]. Modification of epigenetic profiles in MEG3 following environmental exposure could lead to shifts in hormonal and metabolic markers, as well as early growth.
Only two studies have assessed the relationship between pregnancy phthalate exposure and imprinted gene methylation, both in placenta samples. Zhao et al. [27] measured urinary phthalate concentrations of five metabolites (two LMW and three HMW) and DNA methylation of IGF2 (two CpGs) in 181 placenta samples collected in China. The researchers observed that DEHP metabolites were significantly and negatively associated with IGF2 DNA methylation. Additionally, LaRocca et al. [25] examined the association between phthalate metabolite concentrations and DNA methylation of the H19 and IGF2 imprinted genes in 179 placenta samples from two cohorts, the Harvard Epigenetic Birth Cohort and the Predictors of Preeclampsia Study. The authors found a significant inverse relationship between H19 methylation in placenta and the sum of the LMW phthalates metabolite concentrations and the sum of all the 11 phthalate metabolites measured. A similar pattern was observed for the IGF2 DMR0. They also observed an inverse association between DEHP metabolites and the IGF2 DMR0, similar to the results seen in the study by Zhao et al. However, both of these studies did not analyze MEG3 methylation in respect to phthalates.
The number of CpG sites in the studies assessing placenta DNA methylation and the cord blood analysis in CHAMACOS differ between the studies. Additionally, only two imprinted genes were assessed in the placenta studies, while our study involved eight imprinted genes with nine DMRs. We are also mindful that the difference between the tissues used (placenta vs cord blood) may also be the reason for observed differences in methylation results with phthalate exposure.
A recent study by Goodrich et al. [26] examined the relationship between prenatal and childhood exposure to heavy metals and endocrine disrupting chemicals, including phthalates and DNA methylation of DMRs of two imprinted genes (H19: 4 CpG sites, IGF2: 7 CpG sites) in periadolescents. They found that phthalate metabolite levels (MBzP and MIBP) in maternal urine samples were associated with H19 hypermethylation in blood from 247 8- to 14-year-old children, but this finding was attenuated to nonsignificance after adjustment for multiple testing. Overall, the CHAMACOS results as well as the findings by LaRocca et al. [25], Zhao et al. [27] and Goodrich et al. [26] suggest potential epigenetic modifications in imprinted genes involved in fetal growth, in association with prenatal phthalate exposure from the time of birth that may persist into adolescence.
A hypothesized mechanism by which exposure to environmental chemicals may induce epigenetic shifts involves altered transcription factor occupancy. Martin and Fry [80] performed an analysis across 11 studies that included altered cord blood and placental methylation patterns of CpG sites within 341 genes related to five environmental contaminants (lead, arsenic, cadmium, tobacco smoke and mercury). They found that 56 transcription factor binding sites were enriched in the promoter regions of the genes, which could impact the access of DNA methyltransferase to CpG sites, inhibiting the process of DNA methylation and leading to hypomethylation of the gene. Additionally, the authors provided suggestive evidence that glucocorticoid-receptor dependent signaling may mediate the relationship between environmental chemical exposure and transcription factor activity, since the promoter regions of 38 of the 56 genes encoding the transcription factors contained glucocorticoid-responsive-related sequence elements. It is possible that prenatal exposure to phthalates could initiate a similar response of altered transcription factor activity in the current study in the MEG3 DMR, located downstream of the promoter region, resulting in methylation shifts. Future studies should further explore the mechanism proposed by Martin and Fry in additional environmental contaminants, including phthalates.
One of the notable strengths of our study involves the novel investigation of DNA methylation profiles of multiple imprinted genes in newborns in relation to prenatal phthalates, a ubiquitous endocrine disruptor. Two previous studies [25,27] assessed the role of pregnancy phthalate levels on DNA methylation in imprinted genes; however, they used placenta samples and only for three or fewer imprinted regions. In addition to the advantages of assessing a greater number of imprinted genes in our study, our analyses also benefited from a larger sample size.
There were also several limitations in our study. In the umbilical cord, nRBCs and leukocytes mixed in the clots may contribute to DNA methylation levels [64]. However, we were able to account for variable leukocyte composition typical in umbilical cord blood in sensitivity analyses using cytological differential cell count [62], as well as a cord blood reference panel including nRBCs [47,64]. The results for the unadjusted and adjusted models were very similar. Another potential limitation involves the applicability of the findings to other populations, given that the CHAMACOS participants are Mexican–Americans from families with low socioeconomic status. Since observed exposure levels to phthalates in CHAMACOS women were similar to the general population of US adults, the data are likely relevant to many other populations. In this study, we have focused on nine imprinted gene DMRs that have been previously established and validated, and we intend to explore other regions with parental-origin-dependent differential methylation in the future.
Conclusion
Several significant associations were observed in CHAMACOS newborns between prenatal phthalate exposure, primarily DEHP metabolites, and DNA methylation in DMRs of MEG3. MEG3 is known for its involvement in early growth, tumorigenesis and metabolic processes. This new data complement our earlier reports on significant effects of prenatal phthalates on Alu and LINE-1 repeats, as well as differential methylation of several genes revealed by 450K analysis. Our findings would need to be replicated in other cohorts in the future, but they do provide further insight into the complex relationship between environmental exposure to EDCs and biological pathways leading to altered health status in children, including modifications of DNA methylation of imprinted genes.
Future perspective
Understanding the molecular mechanisms that mediate the effects of chemical exposure in the prenatal environment on health is critical for disease prevention. DNA methylation, an epigenetic mechanism that has been shown to relate to both exposures and disease status, may be one pathway through which endocrine disrupting chemicals, including phthalates, exert their effect. There is limited, yet suggestive evidence that prenatal phthalate exposure can impact DNA methylation profiles of imprinted genes, as well as other genes. Future research should focus on understanding the role of imprinted gene methylation on mediating the effects of environmental exposure on health outcomes through adulthood.
Summary points.
Epigenetic mechanisms are possible mediators of the relationship between environmental exposure and disease status.
DNA methylation is the most widely examined epigenetic mechanism.
DNA methylation is involved in the regulation of imprinted genes, which are characterized by the expression of one of the two parental alleles.
In 296 newborns from the Center for the Health Assessment of Mothers and Children of Salinas study, we assessed the relationship between prenatal phthalate exposure and methylation of nine imprinted gene differentially methylated regions.
Prenatal phthalate exposure averaged across pregnancy was associated with multiple imprinted gene differentially methylation regions.
Average phthalate metabolites concentrations across pregnancy of several di-2-ethylhexyl phthalate metabolites were significantly and positively associated with methylation at the MEG3 DMR, a gene associated with early growth, tumorigenesis and diabetes. Many of these relationships remained significant after false discovery rate adjustment.
Sensitivity analysis, including adjusting for specific gravity instead of creatinine to account for urinary dilution and accounting for cord blood cell composition by multiple methods, yielded similar results to crude models.
Early-life environmental exposure to endocrine disrupting chemicals, specifically phthalates, may contribute to dysregulation of DNA methylation of imprinted genes.
Acknowledgements
The authors would like to thank all CHAMACOS participants and field staff for contributing to this study. We are also grateful to A Calafat and X Ye for their help with measuring phthalate metabolite concentrations.
Footnotes
Author's contributions
G Tindula, N Holland, C Hoyo, B Eskenazi and SK Murphy conceived and designed the study; A Bradman, K Huen and Z Huang provided expert guidance in study design and manuscript review; G Tindula, C Grenier and ME Fung analyzed the data; and G Tindula and N Holland wrote the manuscript. All authors read, edited and approved the manuscript prior to submission.
Disclaimer
The interpretations of the study findings pertain to the authors alone, and do not reflect the official views of NIEHS, the US EPA, Duke University, NC State University or the University of California, Berkeley.
Financial & competing interests disclosure
This publication was made possible by grants from the National Institute of Environmental Health Sciences (NIEHS) [P01 ES009605, R01 ES021369, R01 ES023067, R24 ES028529, F31 ES027751], the NIH [UG3OD023356], and from the US Environmental Protection Agency (EPA) [R82670901, and RD83451301] (G Tindula, K Huen, ME Fung, A Bradman, B Eskenazi, N Holland). Contributions by SK Murphy, C Hoyo, Z Huang and C Grenier were supported by the National Institute of Environmental Health Sciences [P01 ES022831] and by the US Environmental Protection Agency [RD83543701]. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical disclosure
The authors state that they have obtained appropriate institutional review board approval for all human experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Breton CV, Marsit CJ, Faustman E, et al. Small-magnitude effect sizes in epigenetic end points are important in children's environmental health studies: the Children's Environmental Health and Disease Prevention Research Center's Epigenetics Working Group. Environ. Health Perspect. 2017;125(4):511–526. doi: 10.1289/EHP595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ. The origins of the developmental origins theory. J. Intern. Med. 2007;261(5):412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 3.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009;21(2):243–251. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feil R, Fraga M. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 5.Foley DL, Craig JM, Morley R, et al. Prospects for epigenetic epidemiology. Am. J. Epidemiol. 2009;169(4):389–400. doi: 10.1093/aje/kwn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014;6(2):1–20. doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy SK, Huang Z, Hoyo C. Differentially methylated regions of imprinted genes in prenatal, perinatal and postnatal human tissues. PLoS ONE. 2012;7(7):e40924. doi: 10.1371/journal.pone.0040924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkins JF, Ubeda F, Van Cleve J. The evolving landscape of imprinted genes in humans and mice: conflict among alleles, genes, tissues and kin. Bioessays. 2016;38:482–489. doi: 10.1002/bies.201500198. [DOI] [PubMed] [Google Scholar]
- 10.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 11.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. USA. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6(7):928–936. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HS, Barraza-Villarreal A, Biessy C, et al. Dietary supplementation with polyunsaturated fatty acid during pregnancy modulates DNA methylation at IGF2/H19 imprinted genes and growth of infants. Physiol. Genomics. 2014;46(23):851–857. doi: 10.1152/physiolgenomics.00061.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian YY, Huang XL, Liang H, et al. Effects of maternal folic acid supplementation on gene methylation and being small for gestational age. J. Hum. Nutr. Diet. 2016;29(5):643–651. doi: 10.1111/jhn.12369. [DOI] [PubMed] [Google Scholar]
- 15.Murphy SK, Adigun A, Huang Z, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012;494(1):36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal AC, Murphy SK, Murtha AP, et al. Associations between antibiotic exposure during pregnancy, birth weight and aberrant methylation at imprinted genes among offspring. Int. J. Obes. (Lond.) 2013;37(7):907–913. doi: 10.1038/ijo.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidal AC, Semenova V, Darrah T, et al. Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol. Toxicol. 2015;16 doi: 10.1186/s40360-015-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nye MD, Hoyo C, Murphy SK. In vitro lead exposure changes DNA methylation and expression of IGF2 and PEG1/MEST. Toxicol. In Vitro. 2015;29(3):544–550. doi: 10.1016/j.tiv.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Xie C, Murphy SK, et al. Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ. Health Perspect. 2016;124(5):666–673. doi: 10.1289/ehp.1408577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nye MD, King KE, Darrah TH, et al. Maternal blood lead concentrations, DNA methylation of MEG3 DMR regulating the DLK1/MEG3 imprinted domain and early growth in a multiethnic cohort. Environ. Epigenet. 2016;2(1):1–8. doi: 10.1093/eep/dvv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrich JM, Sanchez BN, Dolinoy DC, et al. Quality control and statistical modeling for environmental epigenetics: a study on in utero lead exposure and DNA methylation at birth. Epigenetics. 2015;10(1):19–30. doi: 10.4161/15592294.2014.989077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas D, Rager JE, Smeester L, et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol. Sci. 2015;143(1):97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansell T, Novakovic B, Meyer B, et al. The effects of maternal anxiety during pregnancy on IGF2/H19 methylation in cord blood. Transl. Psychiatry. 2016;6:e765. doi: 10.1038/tp.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal AC, Benjamin Neelon SE, Liu Y, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet. Epigenet. 2014;6:37–44. doi: 10.4137/GEG.S18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaRocca J, Binder AM, McElrath TF, Michels KB. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ. Res. 2014;133:396–406. doi: 10.1016/j.envres.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Inverse associations between IGF2 and H19 differentially methylated regions and high molecular weight prenatal phthalate metabolites were observed.
- 26.Goodrich JM, Dolinoy DG, Sanchez BN, et al. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolscence. Environ. Epigenet. 2016;2(3):1–11. doi: 10.1093/eep/dvw018. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The effects of prenatal and childhood exposure to heavy metals and phthalates on DNA methylation of H19 and IGF2 were analyzed in 8–14 year old children. Maternal phthalate metabolite levels of monobenzyl phthalate and mono-isobutyl phthalate were positively associated with H19 methylation.
- 27.Zhao Y, Chen J, Wang X, Song Q, Xu HH, Zhang YH. Third trimester phthalate exposure is associated with DNA methylation of growth-related genes in human placenta. Sci. Rep. 2016;6:33449. doi: 10.1038/srep33449. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The relationship of urinary phthalate concentrations of two low molecular weight and three high molecular weight metabolites and DNA methylation of IGF2 in placenta was characterized. They found that di-2-ethylhexyl phthalate metabolites were significantly and negatively associated with IGF2 DNA methylation.
- 28.Silva MJ, Barr DB, Reidy JA, et al. Urinary levels of seven phthalate metabolites in the US population from the National Health and Nutrition Examination Survey (NHANES) 1999–2000. Environ. Health Perspect. 2004;112(3):331–338. doi: 10.1289/ehp.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodruff TJ, Zota AR, Schwartz JM. Environmental chemicals in pregnant women in the United States: NHANES 2003–2004. Environ. Health Perspect. 2011;119(6):878–885. doi: 10.1289/ehp.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CDC, Fourth national report on human exposure to environmental chemicals. 2009. www.cdc.gov/exposurereport/pdf/FourthReport.pdf [PubMed]
- 31.Smarr MM, Grantz KL, Sundaram R, Maisog JM, Kannan K, Louis GM. Parental urinary biomarkers of preconception exposure to bisphenol A and phthalates in relation to birth outcomes. Environ. Health. 2015;14:1–11. doi: 10.1186/s12940-015-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Lin L, Cao Y, Chen B, Zheng L, Ge RS. Phthalate levels and low birth weight: a nested case–control study of Chinese newborns. J. Pediatr. 2009;155(4):500–504. doi: 10.1016/j.jpeds.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 33.de Cock M, De Boer MR, Lamoree M, Legler J, Van De Bor M. Prenatal exposure to endocrine disrupting chemicals and birth weight – a prospective cohort study. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 2016;51(2):178–185. doi: 10.1080/10934529.2015.1087753. [DOI] [PubMed] [Google Scholar]
- 34.Whyatt RM, Adibi JJ, Calafat AM, et al. Prenatal di(2-ethylhexyl)phthalate exposure and length of gestation among an inner-city cohort. Pediatrics. 2009;124(6):e1213–e1220. doi: 10.1542/peds.2009-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberger B, Vetrano AM, Archer FE, et al. Effects of maternal exposure to phthalates and bisphenol A during pregnancy on gestational age. J. Matern. Fetal Neonatal Med. 2014;27(4):323–327. doi: 10.3109/14767058.2013.815718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferguson KK, McElrath TF, Meeker JD. Environmental phthalate exposure and preterm birth. JAMA Pediatr. 2014;168(1):61–67. doi: 10.1001/jamapediatrics.2013.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meeker JD, Hu H, Cantonwine DE, et al. Urinary phthalate metabolites in relation to preterm birth in Mexico city. Environ. Health Perspect. 2009;117(10):1587–1592. doi: 10.1289/ehp.0800522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valvi D, Casas M, Romaguera D, et al. Prenatal phthalate exposure and childhood growth and blood pressure: evidence from the Spanish INMA-Sabadell birth cohort study. Environ. Health Perspect. 2015;123(10):1022–1029. doi: 10.1289/ehp.1408887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y, Ha EH, Kim EJ, et al. Prenatal exposure to phthalates and infant development at 6 months: prospective Mothers and Children's Environmental Health (MOCEH) study. Environ. Health Perspect. 2011;119(10):1495–1500. doi: 10.1289/ehp.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel SM, Miodovnik A, Canfield RL, et al. Prenatal phthalate exposure is associated with childhood behavior and executive functioning. Environ. Health Perspect. 2010;118(4):565–571. doi: 10.1289/ehp.0901470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whyatt RM, Perzanowski MS, Just AC, et al. Asthma in inner-city children at 5–11 years of age and prenatal exposure to phthalates: the Columbia Center for Children's Environmental Health Cohort. Environ. Health Perspect. 2014;122(10):1141–1146. doi: 10.1289/ehp.1307670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostka G, Urbanek-Olejnik K, Wiadrowska B. Di-butyl phthalate-induced hypomethylation of the c-myc gene in rat liver. Toxicol. Ind. Health. 2010;26(7):407–416. doi: 10.1177/0748233710369124. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Arguelles DB, Papadopoulos V. Identification of hot spots of DNA methylation in the adult male adrenal in response to in utero exposure to the ubiquitous endocrine disruptor plasticizer di-(2-ethylhexyl) phthalate. Endocrinology. 2015;156(1):124–133. doi: 10.1210/en.2014-1436. [DOI] [PubMed] [Google Scholar]
- 44.Pogribny IP, Tryndyak VP, Boureiko A, et al. Mechanisms of peroxisome proliferator-induced DNA hypomethylation in rat liver. Mutat. Res. 2008;644(1–2):17–23. doi: 10.1016/j.mrfmmm.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S, Zhu J, Li Y, et al. Dynamic epigenetic changes involved in testicular toxicity induced by di-2-(ethylhexyl) phthalate in mice. Basic Clin. Pharmacol. Toxicol. 2010;106(2):118–123. doi: 10.1111/j.1742-7843.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- 46.Huen K, Calafat AM, Bradman A, Yousefi P, Eskenazi B, Holland N. Maternal phthalate exposure during pregnancy is associated with DNA methylation of LINE-1 and Alu repetitive elements in Mexican–American children. Environ. Res. 2016;148:55–62. doi: 10.1016/j.envres.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Inverse associations of prenatal MEP concentrations with DNA methylation of repetitive elements was observed in Center for the Health Assessment of Mothers and Children of Salinas newborns and 9-year-old children.
- 47.Solomon O, Yousefi P, Huen K, et al. Prenatal phthalate exposure and altered patterns of DNA methylation in cord blood. Environ. Mol. Mutagen. 2017;58(6):398–410. doi: 10.1002/em.22095. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The study, using 450K array data from the Center for the Health Assessment of Mothers and Children of Salinas cohort, identified 27 differentially methylated regions associated primarily with di-2-ethylhexyl phthalate metabolites.
- 48.Eskenazi B, Kogut K, Huen K, et al. Organophosphate pesticide exposure, PON1, and neurodevelopment in school-age children from the CHAMACOS study. Environ. Res. 2014;134:149–157. doi: 10.1016/j.envres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holland N, Huen K, Tran V, et al. Urinary phthalate metabolites and biomarkers of oxidative stress in a Mexican–American cohort: variability in early and late pregnancy. Toxics. 2016;4(1) doi: 10.3390/toxics4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holland N, Furlong C, Bastaki M, et al. Paraoxonase polymorphisms, haplotypes, and enzyme activity in Latino mothers and newborns. Environ. Health Perspect. 2006;114(7):985–991. doi: 10.1289/ehp.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva MJ, Samandar E, Preau JL, Jr, Reidy JA, Needham LL, Calafat AM. Quantification of 22 phthalate metabolites in human urine. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2007;860(1):106–112. doi: 10.1016/j.jchromb.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 52.Harley KG, Berger K, Rauch S, et al. Association of prenatal urinary phthalate metabolite concentrations and childhood BMI and obesity. Pediatr. Res. 2017;82(3):405–415. doi: 10.1038/pr.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parlett LE, Calafat AM, Swan SH. Women's exposure to phthalates in relation to use of personal care products. J. Expo. Sci. Environ. Epidemiol. 2013;23(2):197–206. doi: 10.1038/jes.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lubin JH, Colt JS, Camann D, et al. Epidemiologic evaluation of measurement data in the presence of detection limits. Environ. Health Perspect. 2004;112(17):1691–1696. doi: 10.1289/ehp.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zota AR, Calafat AM, Woodruff TJ. Temporal trends in phthalate exposures: findings from the National Health and Nutrition Examination Survey, 2001–2010. Environ. Health Perspect. 2014;122(3):235–241. doi: 10.1289/ehp.1306681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nye MD, Hoyo C, Huang Z, et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS ONE. 2013;8(2):e56325. doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 2005;90(4):2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 58.Gejman R, Batista DL, Zhong Y, et al. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab. 2008;93(10):4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Otsuka S, Maegawa S, Takamura A, et al. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 2009;85(4):157–165. doi: 10.2183/pjab.85.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rezvani G, Lui JC, Barnes KM, Baron J. A set of imprinted genes required for normal body growth also promotes growth of rhabdomyosarcoma cells. Pediatr. Res. 2012;71(1):32–38. doi: 10.1038/pr.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soubry A, Guo L, Huang Z, et al. Obesity-related DNA methylation at imprinted genes in human sperm: results from the TIEGER study. Clin. Epigenetics. 2016;8:1–11. doi: 10.1186/s13148-016-0217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yousefi P, Huen K, Quach H, et al. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ. Mol. Mutagen. 2015;56(9):751–758. doi: 10.1002/em.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:1–16. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yousefi P, Huen K, Aguilar Schall R, et al. Considerations for normalization of DNA methylation data by Illumina 450K BeadChip assay in population studies. Epigenetics. 2013;8(11):1141–1152. doi: 10.4161/epi.26037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Du P, Zhang X, Huang CC, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:1–9. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wasserman L. All of Statistics: A Concise Course in Statistical Inference. Springer; NY, USA: 2004. Multiple testing; pp. 165–168. [Google Scholar]
- 68.Soubry A, Murphy SK, Wang F, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int. J. Obes. (Lond.) 2015;39(4):650–657. doi: 10.1038/ijo.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoyo C, Daltveit AK, Iversen E, et al. Erythrocyte folate concentrations, CpG methylation at genomically imprinted domains, and birth weight in a multiethnic newborn cohort. Epigenetics. 2014;9(8):1120–1130. doi: 10.4161/epi.29332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kappil MA, Green BB, Armstrong DA, et al. Placental expression profile of imprinted genes impacts birth weight. Epigenetics. 2015;10(9):842–849. doi: 10.1080/15592294.2015.1073881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMinn J, Wei M, Schupf N, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27(6–7):540–549. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X, Gejman R, Mahta A, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70(6):2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawakami T, Chano T, Minami K, Okabe H, Okada Y, Okamoto K. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum. Mol. Genet. 2006;15(6):821–830. doi: 10.1093/hmg/ddl001. [DOI] [PubMed] [Google Scholar]
- 74.Benetatos L, Dasoula A, Hatzimichael E, Georgiou I, Syrrou M, Bourantas KL. Promoter hypermethylation of the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin. Lymphoma Myeloma. 2008;8(3):171–175. doi: 10.3816/CLM.2008.n.021. [DOI] [PubMed] [Google Scholar]
- 75.Benetatos L, Hatzimichael E, Dasoula A, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 2010;34(2):148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 76.Astuti D, Latif F, Wagner K, et al. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br. J. Cancer. 2005;92(8):1574–1580. doi: 10.1038/sj.bjc.6602478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kameswaran V, Kaestner KH. The missing lnc(RNA) between the pancreatic beta-cell and diabetes. Front. Genet. 2014;5:200. doi: 10.3389/fgene.2014.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kameswaran V, Bramswig NC, McKenna LB, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell. Metab. 2014;19(1):135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You L, Wang N, Yin D, et al. Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J. Cell. Physiol. 2016;231(4):852–862. doi: 10.1002/jcp.25175. [DOI] [PubMed] [Google Scholar]
- 80.Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ. Epigenet. 2016;2(1):1–9. doi: 10.1093/eep/dvv011. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Martin and Fry presents a novel biological mechanism, transcription factor activity, which can mediate the effect of environmental chemicals on CpG methylation profiles.