Abstract
Biochemical assay interference is becoming increasingly recognized as a significant waste of resource in drug discovery, both in industry and academia. A seminal publication from Baell and Holloway raised the awareness of this issue, and they published a set of alerts to identify what they described as PAINS (pan-assay interference compounds). These alerts have been taken up by drug discovery groups, even though the original paper had a somewhat limited data set. Here, we have taken Lilly’s far larger internal data set to assess the PAINS alerts on four criteria: promiscuity (over six assay formats including AlphaScreen), compound stability, cytotoxicity, and presence of a high Hill slope as a surrogate for non-1:1 protein–ligand binding. It was found that only three of the alerts show pan-assay promiscuity, and the alerts appear to encode primarily AlphaScreen promiscuous molecules. Although not enriching for pan-assay promiscuity, many of the alerts do encode molecules that are unstable, show cytotoxicity, and increase the prevalence of high Hill slopes.
Keywords: Assay interference, promiscuity, HTS, PAINS, cytotoxicity, data analysis
The issue of biochemical assay interference resulting in false positives has been well documented in the literature1−6 and can result in significant waste of resources for the original investigators, or worse, reporting of incorrect information, potentially polluting the scientific literature, and wasting the resources of others. Understanding the causes of assay interference is of benefit to both academic and industrial drug discovery teams to enable them to reduce waste through focusing on hits most likely to be genuine actives. A complete list of mechanisms causing this behavior is not fully understood, but reported mechanisms include protein alkylation by electrophilic moieties,7,8 redox cycling,9,10 compound aggregation,11−13 sample impurities,14 or interference in the assay technology itself (e.g., metal chelation15 or compound fluorescence16,17).
Working toward identifying mechanisms and substructures prone to assay interference, Baell and Holloway3 analyzed six AlphaScreen assays and described a set of alerts they suggested captured the majority of their assay false positives (referred to as BH2010). These alerts were proposed to be PAINS (pan-assay interference compounds), despite being derived from a series of AlphaScreen assays with some validation using two fluorescence polarization assays. These alerts have become commonly used in drug discovery, despite the data set limitations in the original analysis, although it appears this use has gone beyond what was initially intended in some cases.18,19
Some of the PAINS alerts have been independently assessed in the literature since the original publication. Nissink and Blackburn describe their method for assessing “frequent-hitters” and suggest that 10 out of the 15 PAINS alerts considered did show an increased propensity for promiscuity.20 More recently, assessments from Capuzzi et al. and Jasial et al. describe the assessment of PAINS alerts using the PubChem data set.21,22 Both reports come to similar conclusions and suggest that the PAINS alerts do not encode pan-assay promiscuity, with matching compounds frequently found inactive, and urge caution when applying them to novel hits, although a subsequent criticism of this analysis has also been published.19
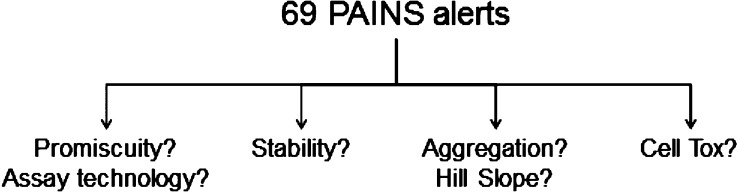
Herein, we describe the assessment of 69 of the alerts (those that matched 15 or more molecules in the BH2010 paper) over a large, high quality data set, with assays separated by the technology used, to determine if any of the alerts do show pan-assay behavior. In addition to this promiscuity assessment, we have looked at three other end points (compound stability, Hill slope, and cytotoxicity) to try to further understand these alerts and possible mechanisms of assay interference.
Our first step in the process was to translate the original PAINS alerts from the format described in that publication: SLN, into more robust SMARTS queries.23 This mostly revolved around aromaticity perception and is a challenge within cheminformatics. This is discussed further in the Supporting Information (SI). The tools and queries used can be found as part of a larger open source release of internally developed cheminformatics methods: https://github.com/EliLillyCo/LillyMol.
Having curated a set of the PAINS alerts that we felt maximally identified what was initially intended from the original alerts, we gathered the necessary data to assess them. We started by looking at promiscuity data, and 14,424,191 data points from 3,048 assays covering six different commonly used biochemical assay formats were collected. For each compound tested in one of these assays, an active was defined as a compound reporting XC50 (compound concentration that produces a 50% effect in the assay whether that be an antagonist or agonist effect) lower than the top screening concentration; an inactive was defined as a compound reporting an XC50 greater than the top screening concentration (typically 10–20 μM) or reporting ≤30% inhibition if tested at a single point (SP) concentration. Compounds reporting >30% inhibition in a SP assay were excluded from the analysis as the majority of these would be forwarded to a dose response. This is a difference between our analysis and that of the BH2010 paper, whereby they used SP data only. For each assay, the target and the gene family for that target were also considered in the analysis. Compounds for which any sample had been measured to be less than 80% pure (see SI for method) or matched substructures used to filter the screening set in the BH2010 paper were excluded from the analysis (assessment of the PAINS alerts for increased instability can be found later in the Letter). Molecules included in the analysis are broadly similar for each assay format regarding both timing of testing and heavy atom count. A summary of the activity data included in the analysis can be seen in Table 1. Having gathered the data, the next step was to assess the compounds that match each of the PAINS alerts for promiscuity. Here, a degree of normalization was required. Overall, for each assay format, and for each rule, we calculated a “PAINS Activity Enrichment”, which is the active rate of the PAINS compounds divided by the active rate of the entire data set.24 A random selection of compounds from the total set would be expected to yield an activity enrichment of 1.0, whereas a subset of the whole that showed an increased active rate over random we would suggest is promiscuous. In addition to the enrichment values, which are based on the raw data and can be biased, we have calculated odds ratios and false discovery rates (FDR) where appropriate on a subset normalized by target and corrected for multiple hypothesis testing. The approaches here are described in the SI.
Table 1. Summary of All Activity Data Included in Analysis of PAINS Alerts.
| assay format | actives | data points | active rate (%) | unique compounds | unique assays | unique gene families |
|---|---|---|---|---|---|---|
| AS | 81704 | 1,688652 | 4.8 | 498,977 | 214 | 7 |
| ELISA | 82,514 | 416,841 | 19.8 | 304,478 | 228 | 7 |
| FB | 317,865 | 4,008,981 | 7.9 | 459,068 | 982 | 8 |
| FP | 138,393 | 1,791,893 | 7.7 | 492,831 | 225 | 6 |
| FRET | 486,748 | 4,565,466 | 10.7 | 537,795 | 932 | 11 |
| SPA | 277,196 | 1,952,358 | 14.2 | 534,701 | 467 | 11 |
| overall | 1,384,420 | 14,424,191 | 9.6 | 921,054 | 3,048 | 16 |
Table 2 shows the data included, overall active rates, and enrichment over random for the compounds that have been tested in each of the assay formats that match the PAINS alerts. Across all of the compounds tested, the compounds hitting the PAINS alerts show 1.4-fold over the random active rate, suggesting there is some enrichment for promiscuity; however, this is not the case for every assay format. Enrichment >1.5-fold for promiscuity is seen for the AlphaScreen (AS), fluorescence polarization (FP), and fluorescence resonance energy transfer (FRET) assay formats with similar odds ratios (FDR < 0.0001), but not for the enzyme-linked immunosorbent assay (ELISA), filter binding (FB), and scintillation proximity assay (SPA) assay formats, suggesting that the alerts may not be pan-assay from a promiscuity perspective. Also, even though there is some enrichment over random, many compounds containing these structural motifs have been tested and found to be inactive at a greater rate (similar to the findings of Capuzzi et al. and Jasial et al.), suggesting that, although somewhat promiscuous, there is some specificity to their inhibition (although not necessarily via the intended mechanism).
Table 2. Summary of Data for Compounds Matching the PAINS Alerts.
| format | actives | data points | active rate (%) | unique compounds | unique assays | unique gene families | PAINS activity enrichment (odds ratio) |
|---|---|---|---|---|---|---|---|
| AS | 3,490 | 32,471 | 10.8 | 9,813 | 173 | 7 | 2.2 (2.5) |
| ELISA | 1,769 | 7,945 | 22.2 | 5,749 | 134 | 6 | 1.1 (1.1) |
| FB | 8,523 | 102,141 | 8.3 | 11,134 | 721 | 8 | 1.1 (1.1) |
| FP | 5,032 | 39,755 | 12.7 | 11,645 | 201 | 5 | 1.7 (1.5) |
| FRET | 20,794 | 103,973 | 20.0 | 11,101 | 702 | 10 | 1.9 (2.5) |
| SPA | 5,223 | 37,833 | 13.8 | 11,274 | 309 | 10 | 1.0 (1.6) |
| overall | 44,831 | 324,118 | 13.8 | 20,492 | 2,240 | 16 | 1.4 (1.7) |
Following on from the overall promiscuity statistics, we looked at each rule contained within the PAINS rule set to see if any particular alerts showed increased promiscuity. For compounds matching each rule, we calculated the active rate, the PAINS activity enrichment and associated FDRs (SI Tables S1 and S7). Interestingly, only one rule shows significant overall enrichment independent of assay format here: anil_di_alk_A (1.9-fold), suggesting that most rules do not enrich for pan-assay promiscuity. We compared the results for each assay format by looking at the number of alerts that achieved ≥1.5-fold (50% increase) over the active rate expected at random and those showing a statistically significant increase (FDR < 0.1) for the normalized subset. When looking at each assay format, we observed that, for the AS format, more alerts showed some promiscuity compared to the other formats (Table 3). For the other two assay formats that overall showed an increase in promiscuity (FP and FRET), this was coming from a smaller number of alerts (anil_di_alk_A, ene_rhod_A, and anil_alk_ene, in particular). Together, this suggests that the original PAINS alerts code primarily for AS interferers and may not necessarily be pan-assay as originally proposed. For ELISA and FB assays, there appears to be very little enrichment for promiscuity.
Table 3. Numbers of PAINS Alerts Showing >1.5-Fold Activity Enrichment for Each Assays Format.
| assay format | AS | ELISA | FB | FP | FRET | SPA |
|---|---|---|---|---|---|---|
| alerts hit | 33 | 10 | 2 | 12 | 11 | 13 |
| alerts hit signficantlya | 11 | 5 | 2 | 8 | 6 | 9 |
| alerts tested | 57 | 55 | 61 | 61 | 60 | 61 |
FDR < 0.1.
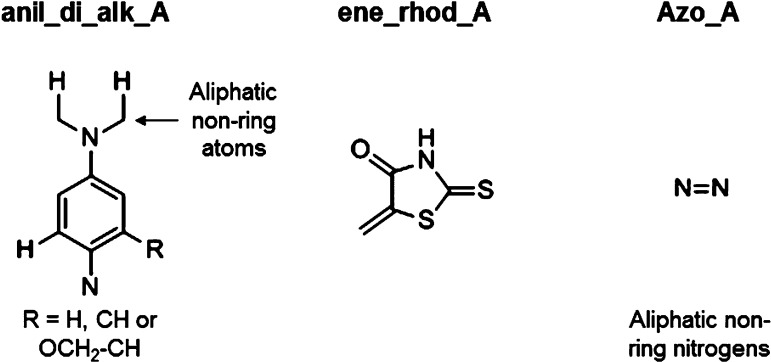
We looked over each of the alerts to identify which of them was truly pan-assay in their promiscuous behavior. We defined this as showing ≥1.5-fold activity enrichment on the raw data and statistical significance for the normalized subset for at least four of the six assay formats tested (61 of the alerts matched molecules that had been tested against at least four assay formats). Out of the 63 alerts to which we matched compounds, only two of the alerts met this criteria. These were anil_di_alk_A and ene_rhod_A, which hit five and four of the assay formats, respectively (Figure 1). Alert azo_A shows enrichment in five of the assay formats in the raw data, but only one of these reaches significance in the normalized subset. All three of these were also assessed by Nissink and Blackburn, and consistent with our analysis, these alerts showed an increase in frequent hitter behavior relative to what would be expected at random.18
Figure 1.
PAINS alerts showing pan-assay promiscuous behavior.
A difference between our analysis and the BH2010 paper is that we have filtered out compounds with measured instability. One mechanism of assay interference could be compound degradation yielding an electrophilic or other interfering motif. We speculated that some of the PAINS alerts may encode inherently unstable motifs rather than be interferers themselves. To investigate this, 172,613 compounds with measured purity were analyzed. A compound was considered pure if all measured samples were ≥80% of the parent structure, those with any sample <80% were considered impure (assuming that they would be >90% at submission). Samples that identified a single molecule, but with a correspondingly incorrect m/z, were discarded through concerns said molecule may have been incorrectly registered and not necessarily unstable. Of the total number of compounds considered, 5.3% were classified as impure. For each of the compounds matching the PAINS alerts, an instability enrichment was calculated similarly to the activity enrichment. Compounds matching the PAINS alerts that show instability at a higher rate than the overall set may have achieved their assay interference by degradation products in the BH2010 paper but may be missed by our analysis, which filtered known impure compounds.
Interestingly, 25 of the 46 alerts for which we had stability data showed ≥1.5-fold enrichment for instability, 13 of which reach statistical significance. Three of these showed greater than 10-fold enrichment with statistical significance, suggesting these motifs are frequently unstable (pyrrole_B, keto_keto_beta_A, and anil_alk_ene). Of the two alerts that showed pan-assay promiscuity, ene_rhod_A showed instability, suggesting this as a possible mechanism, but molecules matching the anil_di_alk_A rule showed no enrichment in instability. This stability data is summarized in SI Table S2.
In addition to instability and promiscuity, we also looked at cytotoxicity. Frequently, primary actives showing a dose response in a biochemical assay are evaluated in cellular assays to show target or pathway modulation in a cellular context, or to establish a cellular phenotype. For some of these assays, say in oncology, cytotoxicity may be the primary end point, or cytotoxicity may confound an assay result and provide an additional mechanism of assay interference. Molecules that interfere in a primary biochemical assay, but also in a cellular assay through a toxicity mechanism, may get reported in the literature as a true positive, despite interfering in both assays. To assess the cytotoxicity of molecules matching the PAINS alerts, 1,099,163 data points from 1,261 cytotoxicity assays were considered and treated in the same manner as the promiscuity data, looking for enrichment over what would be expected at random. Of the 56 PAINS alerts for which we had data, 15 of these showed an enrichment of ≥1.5-fold (nine of which are statistically significant) suggesting they show some enrichment for cytotoxicity, with quinone_A being the worst offender here with 4-fold enrichment. Of the two alerts that showed pan-assay promiscuity, anil_di_alk_A also shows statistically significant enrichment for cytotoxicity. The data for molecules matching this rule identified by a biochemical assay and “confirmed” in a cellular assay should be used with caution. Something to consider when interpreting this data is the time frame of the assay; typically a cytotoxicity assay will run for a period of days, but for assay readouts that occur in a shorter time frame, this data may not be relevant. The cytotoxicity data is summarized in SI Table S3.
Although many of the alerts do not appear to code for promiscuity, we sought to try to gather some information as to whether, when found active, the compound may be acting via an MOA that may not be 1:1 protein/ligand binding. We have used the Hill slope of the dose–response curve as a surrogate for this. A high Hill slope can be an indicator of an undesirable MOA such as compound aggregation, which is generally regarded as an interference mechanism and almost impossible to optimize to a molecule that would be effective in the clinic.25,26 To assess this, the Hill slopes of 1,447,886 active compounds were imported from 1,845 biochemical assays. A High hill slope was defined as >2.0, a normal Hill slope between 0.5 and 1.5, and slopes <0.5 or 1.5–2.0 were discarded. Overall, the PAINS alerts did not show enrichment in the presence of a high Hill slope or for any particular assay format. However, when we looked at each of the alerts, 41 out of the 61 alerts for which molecules with dose–response data was available showed >1.5-fold enrichment for the presence of a high Hill slope, 27 of which reach statistical significance. Although many of the alerts did not show enrichment for promiscuity, it appears that when found as hits, many of the PAINS alerts code for molecules that deliver a high Hill slope, with the inference being that the mechanism of action (MOA) may not be 1:1 protein/ligand binding. The rule anil_di_alk_A, which showed both pan-assay promiscuity and cytotoxicity, did not show overall enrichment in Hill slope, and given that there were many matches to this rule, this resulted in overall no enrichment for the PAINS alerts despite many of the alerts enriching for a high Hill slope. The worst offending rules regarding Hill slope are the amino_acridine_A and thiophene_amino_Aa rules, which both overall show a 3.5-fold enrichment for the presence of a high Hill slope overall (FDR < 0.1) and show enrichment of >1.5-fold for molecules tested in at least five of the assay formats considered here. Given the large, flat, and aromatic nature of these substructures, there is a high probability these represent aggregation prone molecules causing the high Hill slopes.
If we look at the Hill slope data by assay format, it is clear that FP and FRET assays are the most prone to PAINS alert containing molecules showing a high hill slope. For FP and FRET assays, 10 and 15 of the alerts, respectively, show statistically significant enrichment of high Hill slopes with a further 17 and 15, respectively, showing enrichment but not reaching significance. By comparison, the other formats have <6 alerts each showing this enrichment for high Hill slopes, suggesting that FP and FRET assays may be most susceptible to colloidal aggregation interference. This Hill slope data is summarized in SI Tables S4 to S7.
In summary, assay interference continues to be an issue that reduces productivity in drug discovery, and publications such BH2010 have helped the field recognize and understand the issue. Here, we hoped to build on this knowledge through analysis of the performance of those published alerts on a larger pharmaceutical company database with respect to promiscuity, but also looking at the behavior of the alerts in compound stability, cytotoxicity, and Hill slope data types. For promiscuity, only two of the alerts were found to show pan-assay promiscuity, and the alerts appear to mostly enrich for AlphaScreen promiscuity, the assay format used in the original publication. Despite not showing significant enrichment for promiscuity, a much larger fraction showed enrichment for compound instability and presence of a high Hill slope (although most prevalent in FP and FRET assays) suggesting that the alerts may code for other undesirable behaviors resulting in assay interference. In addition to this biochemical assay interference, we find that 15 of the alerts enrich for cytotoxicity. This may mean that cellular assays alone could be inappropriate confirmatory assays for molecules matching these alerts. We would always recommend that direct binding to a target is established possibly through a biophysical assay and that mechanistic studies are performed to understand the function.
Overall, we hope that this analysis will help scientists understand further when a compound hits one of the PAINS alerts, what the issue might be, and whether it is relevant to their assay format. Some assays do not show any interference behavior for certain substructures, and inaccurate accusations of the compound being a false positive can be reduced.
Acknowledgments
The authors thank the Quantitative Biology and Analytical Technology groups at Lilly for generation of the data used here, James Lumley and his team for providing infrastructure to access the data included in this manuscript, and David Evans and John Strelow for proofreading the manuscript.
Glossary
ABBREVIATIONS
- ELISA
enzyme-linked immunosorbent assay
- FB
filter binding
- FDR
false discovery rate
- FP
fluorescence polarization
- FRET
fluorescence resonance energy transfer
- MOA
mechanism of action
- PAINS
pan-assay interference compounds
- SLN
SYBYL line notation
- SMARTS
smiles arbitrary target specification
- SP
single point
- SPA
scintillation proximity assay
Biography
Dr. Lewis Vidler is a senior research scientist at Eli Lilly and Company in the U.K. He achieved a first class chemistry degree at the University of Oxford in 2009, followed by a Ph.D. in computational medicinal chemistry at the Institute of Cancer research in London, publishing a number of highly cited papers on bromodomains. Lewis then joined Lilly in 2013 as a computational medicinal chemist. His role at Lilly is primarily computational drug discovery project support, but he is interested in all aspects of drug discovery, especially medicinal chemistry and compound design.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00097.
Data tables, a discussion around the SLN to SMARTS translation, statistical methods used, and experimental method for purity determination (PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): L.R.V., I.A.W., B.J.M., and M. B. are employees and stockholders of Eli Lilly and Company.
Supplementary Material
References
- Walters W. P.; Stahl M. T.; Murcko M. A. Virtual screening—an overview. Drug Discovery Today 1998, 3 (4), 160–178. 10.1016/S1359-6446(97)01163-X. [DOI] [Google Scholar]
- Walters W. P.; Namchuk M. A guide to drug discovery: Designing screens: how to make your hits a hit. Nat. Rev. Drug Discovery 2003, 2, 259–266. 10.1038/nrd1063. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53 (7), 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Ferrins L.; Falk H.; Nikolakopoulos G. PAINS: Relevance to Tool Compound Discovery and Fragment-Based Screening. Aust. J. Chem. 2013, 66, 1483–1494. 10.1071/CH13551. [DOI] [Google Scholar]
- Dahlin J. L.; Walters M. A. The essential roles of chemistry in high-throughput screening triage. Future Med. Chem. 2014, 6, 1265–1290. 10.4155/fmc.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B.; Walters M. A. Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- Rishton G. Reactive compounds and in vitro false positives in HTS. Drug Discovery Today 1997, 2 (9), 382–384. 10.1016/S1359-6446(97)01083-0. [DOI] [Google Scholar]
- Dahlin J. L.; Nissink J. W. M.; Strasser J. M.; Francis S.; Higgins L.; Zhou H.; Zhang Z.; Walters M. A. PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 2015, 58 (5), 2091–2113. 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares K.; Blackmon N.; Shun T. Y.; Shinde S. N.; Takyi H. K.; Wipf P.; Lazo J. S.; Johnston P. A. Profiling the NIH small molecule repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev. Technol. 2010, 8, 152–174. 10.1089/adt.2009.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. A. Redox cycling compounds generate H2O2 in HTS buffers containing strong reducing reagents—real hits or promiscuou.s artifacts?. Curr. Opin. Chem. Biol. 2011, 1, 174–182. 10.1016/j.cbpa.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Simeonov A.; Jadhav A.; Babaoglu K.; Inglese J.; Shoichet B. K.; Austin C. P. A High-Throughput Screen for Aggregation-Based Inhibition in a Large Compound Library. J. Med. Chem. 2007, 50, 2385–2390. 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Helfand B. T.; Feng B.; Shoichet B. K. A Specific Mechanism of Nonspecific Inhibition. J. Med. Chem. 2003, 46 (20), 4265–4272. 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- McGovern S. L.; Caselli E.; Grigorieff N.; Shoichet B. K. A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem. 2002, 45 (8), 1712–1722. 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Hermann J. C.; Chen Y.; Wartchow C.; Menke J.; Gao L.; Gleason S. K.; Haynes N.-E.; Scott N.; Petersen A.; Gabriel S.; Vu B.; George K. M.; Narayanan A.; Li S. H.; Qian H.; Beatini N.; Niu L.; Gan Q.-F. Metal impurities cause false positives in high-throughput screening campaigns. ACS Med. Chem. Lett. 2013, 4, 197–200. 10.1021/ml3003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorpp K.; Rothenaigner I.; Salmina E.; Reinshagen J.; Low T.; Brenke J. K.; Gopalakrishnan J.; Tetko I. V.; Gul S.; Hadian K. Identification of small-molecule frequent hitters from AlphaScreen high-throughput screens. J. Biomol. Screening 2014, 19, 715–726. 10.1177/1087057113516861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov A.; Jadhav A.; Thomas C. J.; Wang Y.; Huang R.; Southall N. T.; Shinn P.; Smith J.; Austin C. P.; Auld D. S.; Inglese J. Fluorescence spectroscopic profiling of compound libraries. J. Med. Chem. 2008, 51, 2363–2371. 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- Gul S.; Gribbon P. Exemplification of the challenges associated with utilising fluorescence intensity based assays in discovery. Expert Opin. Drug Discovery 2010, 5, 681–690. 10.1517/17460441.2010.495748. [DOI] [PubMed] [Google Scholar]
- Kenny P. W. Comment on The Ecstasy and Agony of Assay Interference Compounds. J. Chem. Inf. Model. 2017, 57 (11), 2640–2645. 10.1021/acs.jcim.7b00313. [DOI] [PubMed] [Google Scholar]
- Baell J. B.; Nissink J. W. M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol. 2018, 13, 36. 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissink J. W. M.; Blackburn S. Quantification of frequent-hitter behavior based on historical high-throughput screening data. Future Med. Chem. 2014, 06 (10), 1113–1126. 10.4155/fmc.14.72. [DOI] [PubMed] [Google Scholar]
- Capuzzi S. J.; Muratov E. N.; Tropsha A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J. Chem. Inf. Model. 2017, 57 (3), 417–427. 10.1021/acs.jcim.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasial S.; Hu Y.; Bajorath J. How Frequently Are Pan-Assay Interference Compounds Active? Large-Scale Analysis of Screening Data Reveals Diverse Activity Profiles, Low Global Hit Frequency, and Many Consistently Inactive Compounds. J. Med. Chem. 2017, 60 (9), 3879–3886. 10.1021/acs.jmedchem.7b00154. [DOI] [PubMed] [Google Scholar]
- Bruns R. F.; Watson I. A. Rules for Identifying Potentially Reactive or Promiscuous Compounds. J. Med. Chem. 2012, 55 (22), 9763–9772. 10.1021/jm301008n. [DOI] [PubMed] [Google Scholar]
- Gao C.; Margolis B. J.; Strelow J. M.; Vidler L. R.; Mader M. M. Beyond PAINs: Chemotype Sensitivity of Protein Methyltransferases in Screens. ACS Med. Chem. Lett. 2016, 7 (2), 156–161. 10.1021/acsmedchemlett.5b00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet B. K. Interpreting Steep Dose-Response Curves in Early Inhibitor Discovery. J. Med. Chem. 2006, 49, 7274–7277. 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- Prinz H. Hill coefficients, dose–response curves and allosteric mechanisms. J. Chem. Biol. 2010, 3 (1), 37–44. 10.1007/s12154-009-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.