Abstract
Recent studies of hydrogen sulfide (H2S) signaling implicate low molecular weight (LMW) thiol persulfides and other reactive sulfur species (RSS) as signaling effectors. Here, we show that a CstR protein from the human pathogen Enterococcus faecalis (E. faecalis), previously identified in Staphylococcus aureus (S. aureus), is an RSS-sensing repressor that transcriptionally regulates a cst-like operon in response to both exogenous sulfide stress and Angeli’s salt, a precursor of nitroxyl (HNO). E. faecalis CstR reacts with coenzyme A persulfide (CoASSH) to form interprotomer disulfide and trisulfide bridges between C32 and C61′, which negatively regulate DNA binding to a consensus CstR DNA operator. A ΔcstR strain exhibits deficiency in catheter colonization in a catheter-associated urinary tract infection (CAUTI) mouse model, suggesting sulfide regulation and homeostasis is critical for pathogenicity. Cellular polysulfide metabolite profiling of sodium sulfide-stressed E. faecalis confirms an increase in both inorganic polysulfides and LMW thiols and persulfides sensed by CstR. The cst-like operon encodes two authentic thiosulfate sulfurtransferases and an enzyme we characterize here as an NADH and FAD-dependent coenzyme A (CoA) persulfide reductase (CoAPR) that harbors an N-terminal CoA disulfide reductase (CDR) domain and a C-terminal rhodanese homology domain (RHD). Both cysteines in the CDR (C42) and RHD (C508) domains are required for CoAPR activity and complementation of a sulfide-induced growth phenotype of a S. aureus strain lacking cstB, encoding a nonheme FeII persulfide dioxygenase. We propose that S. aureus CstB and E. faecalis CoAPR employ orthogonal chemistries to lower CoASSH that accumulates under conditions of cellular sulfide toxicity and signaling.
Graphical Abstract

Emerging studies suggest that hydrogen sulfide (H2S) and/or H2S-derived reactive sulfur species (RSS) play beneficial roles as a signaling molecule in mammalian systems,1 and contribute to the cellular response to reactive nitrogen species (RNS), reactive oxygen species (ROS),2–7 and antibiotics stress.8,9 These growth-promoting functions of RSS are balanced by a potent inhibition by H2S of heme-containing enzymes and cellular respiration through poisoning of cytochrome c oxidase10 and the ability to precipitate transition metals into insoluble complexes.11 H2S can freely pass through biological membranes, existing mainly as hydrosulfide anion (HS−) once inside the cell at physiological pH.12 H2S can also be endogenously produced by enzymes of reverse trans-sulfuration and cysteine degradation pathways, extensively studied in mammalian systems, and include cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), 3-mercaptopyruvate sulfurtransferase (3-MST), and D-amino acid oxidase (DAO).13,14 Considering both endogenous and exogenous production of H2S and H2S-derived RSS in living systems as well as myriad beneficial and deleterious roles, efficient regulatory strategies that control cellular H2S concentrations and speciation are required. In certain organisms, excess H2S is effluxed via a specific hydrosulfide ion channel,15 whereas other organisms chemically assimilate H2S using a sulfide oxidation system.13
We previously characterized the cst operon from the major human pathogen Staphylococcus aureus (S. aureus).16 The cst operon is transcriptionally regulated by the repressor, CstR, which responds to cellular sulfide stress through sensing low molecular weight (LMW) persulfides and inorganic polysulfides (Sn2−) collectively termed RSS.16 LMW persulfides can be produced enzymatically in a number of ways, including the oxidation of S2− by sulfide:quinone oxidoreductase (SQR) and a LMW thiol,17,18 via 3-MST,19 which produces reactive inorganic polysulfides,20,21 and via cysteinyl tRNA synthetase, which catalyzes the formation of cysteine persulfides in a PLP-dependent fashion that can also be incorporated into proteins translationally.22 Nonenzymatic LMW persulfide production, on the other hand, can result from the reaction of sulfide with LMW disulfides, attack of inorganic dihydropolysulfides (HSnH, n ≥ 2) or organic polysulfides (RSnH, n ≥ 2) on sulfenylated (S-hydroxylated) thiols,6 or reaction of inorganic polysulfides with organic thiols.19 Recent work establishes that H2S and nitric oxide (NO) also react to form a number of bioactive species, including highly reactive polysulfides that activate TRPA1 channels,23 LMW persulfides,7 and the one-electron reduced form of the radical NO•, nitroxyl (HNO), which is a potent, thiophilic electrophile.3–5,7,24,25
These processes collectively create a pool of labile sulfane (sulfur-bonded) sulfur found in LMW persulfides, which can accumulate to significant levels in both mammalian and bacterial cells,26 with CBS and CSE as significant sources of endogenous H2S.2,27 LMW persulfides, as well as significant proteome sulfuration (also referred to as S-sulfhydration or persulfuration28) documented to occur in both mammalian and bacterial cells,26,27,29,30 are consistent with emerging evidence to suggest that RSS may be protective against irreversible and inactivating oxidative and electrophilic modifications31 and may also impact the extent of proteome nitrosation.32 Indeed, coupling of endogenous H2S and NO is reported to protect diverse bacterial pathogens, including S. aureus against general microbial stress induced by antibiotics, the mechanism of which remains under investigation.8,9,33,34 How mammalian and bacterial cells reductively control RSS accumulation and proteome sulfuration is not fully understood, but recent studies implicate thioredoxins and thioredoxin-like proteins and glutaredoxins in this process.26,30
In this work, we present a functional characterization of CstR from a second major human pathogen, Enterococcus faecalis (E. faecalis or Ef), that is also conserved in highly pathogenic strains of Bacillus anthracis (B. anthracis; Figure 1). The genomic region adjacent to cstR suggests a divergently transcribed cst-like operon, encoding CstR, two putative sulfurtransferases, RhdA and RhdB, and what we hypothesize is a coenzyme A (CoA) persulfide reductase, CoAPR, encoded by a gene we designate coaP (Figure 1). A closely related CoA disulfide reductase-rhodanese fusion protein (CDR-RHD) has been structurally characterized in B. anthracis str. Ames alongside the authentic CoA disulfide reductase (CDR)35,36 prior to the recognition that this operon might be regulated by RSS.37 Here, we confirm various predictions of this regulatory model and further show that unregulated expression of the cst-like operon in a ΔcstR E. facaelis strain inhibits catheter colonization in a catheter-associated urinary tract infection (CAUTI) mouse model. We provide evidence that cst-encoded gene products in both S. aureus and E. facaelis underpin a common physiological strategy to limit the accumulation of CoASSH and perhaps other RNS-derived CoA adducts38,39 under conditions of sulfide stress and/or H2S/NO cross-talk, which may be particularly important in the urinary tract of infected animals.26
Figure 1.
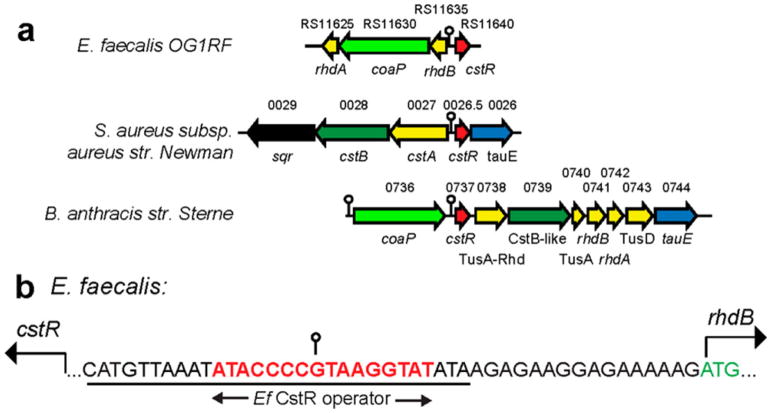
Identification of cst-like operons. (a) Genomic organization of the E. faecalis strain OG1RF cst-like operon region (top) compared to the previously characterized genomic region of S. aureus (middle)16 and an analogous region of B. anthracis str. Sterne (bottom) derived from a Synteny analysis using either cstR or Ef coaP genes as query.75 B. anthracis str. Ames (not shown) is organized as in the Sterne strain. Genes are shaded according to function: red, sulfurtransferase repressor CstR;43 yellow, sulfurtransferase or sulfur trafficking function analogous to that found in the multidomain sulfurtransferase S. aureus CstA (Rhd-TusA-TusD);42 green, nonheme FeII persulfide dioxygenase–rhodanese fusion CstB;37 light green, persulfide reductase–rhodanese fusion;36 blue, candidate sulfite efflux transporter TauE. In E. faecalis, the cst-like operon is composed of a cstR (OG1RF_RS11640, 86 residues), rhdB (OG1RF_RS11635, 104 residues), a candidate CoA persulfide reductase coaP (OG1RF_RS11630, 549 residues), and rhdA (OG1RF_RS11625, 99 aa). The structurally characterized B. anthracis CDR-RHD is encoded by locus tag BAS0774, which is analogous to BA0736 shown here.36 (b) Nucleotide sequence upstream of the Ef rhdB ORF to show the characteristic CstR operator.43 The consensus 16-bp operator site for Ef CstR43 is highlighted in red bold with the 29-bp Ef CstR duplex DNA used for the DNA binding assays underlined.
RESULTS
Identification of cst-like Operons in E. faecalis and B. anthracis
In an effort to further understand bacterial sulfide homeostasis and detoxification beyond S. aureus, we identified a candidate CstR in E. faecalis strain OG1RF, denoted Ef CstR, which is conserved in all sequenced E. faecalis strains, as well as in pathogenic B. anthracis species (Figure 1a). Although distinct from the sulfide oxidation system in S. aureus, this genomic region is hypothesized to encode proteins involved in sulfur trafficking, including two candidate single-domain sulfurtransferases (rhodaneses),40 denoted RhdA (OG1RF_RS11625) and RhdB (OG1RF_RS11635), and an enzyme annotated as a CDR-RHD,36 which we propose here is a CoA persulfide reductase (denoted coaP, encoding CoAPR; OG1RF_RS11630; Figure 1a).41 In the single B. anthracis strain shown, this core cst operon region appears to be significantly expanded to include other known cellular persulfide carrier proteins involved in sulfur shuttling and H2S resistance previously characterized in S. aureus (Figure 1a).16,26,37,42 Given the consensus GC-rich CstR operator, 5′-ATA|C4G/CxxxG2|TAT,43 found upstream of rhdB (Figure 1b), we hypothesized that this cst-like operon is inducible by H2S with encoded proteins comprising a novel bacterial strategy for the clearance of cellular RSS.
The Ef cst-like Operon Is Transcriptionally Induced by Na2S and Angeli’s Salt
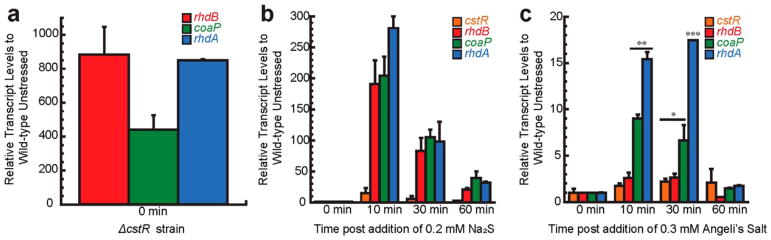
Using quantitative real-time PCR (qRT-PCR), we first showed CstR functions as a repressor since the coaP, rhdA, and rhdB genes are strongly expressed in ΔcstR mutant strain relative to an uninduced wild-type strain (Figure 2a and Table S1). As anticipated from earlier work in S. aureus,16 the addition of 0.2 mM Na2S to early mid log liquid cultures grown microaerophilically results in significant induction of all three genes 10 min postaddition of Na2S, with comparatively little induction of cstR (Figure 2b). The mRNA levels of all three induced genes is decreased by 30 min and further by 60 min, kinetics similar to that observed for sulfide-induction of the S. aureus cst operon.16 This suggests an initial bolus of intracellular sulfide or other downstream RSS is cleared by CstR-regulated gene products and/or via one of a number of nonenzymatic mechanisms described above.
Figure 2.
E. faecalis cst -like operon induced by both sulfide and Angeli’s salt in vivo. Quantitative RT-PCR was performed on (a) the ΔcstR mutant and wild-type E. faecalis strain OG1RF with the addition of (b) 0.2 mM Na2S and (c) 0.3 mM Angeli’s salt for early mid log cultures. The fold changes of induction for Ef cstR (orange), rhdB (red), coaP (green), and rhdA (blue) were normalized relative to the level of gyrase. Values represent relative transcript levels to untreated wild-type cells and are shown as mean ± SD from replicate cultures with statistical significance established using a paired t test relative to unstressed wild-type strain under the same conditions (***p<0.001, **p<0.01, *p<0.05). Primers for the analysis are listed in Table S1.
Previous studies have shown that endogenously produced H2S and nitric oxide protect major bacterial species from the effects of antibiotic stress8 through a mechanism we have suggested involves the intermediacy of HNO.3,5,7,16,24 To investigate the impact of HNO on expression of CstR-regulated genes, we added 0.3 mM Angeli’s salt (Na2N2O3), a commonly used precursor of HNO and nitrite (NO2−)44 to early mid log cultures grown microaerophilically and monitored cst transcription by qRT-PCR.44,45 A weaker but significant, acute-phase induction of ~4–15-fold of coaP, rhdA, and rhdB was observed 10 min post-addition of Angeli’s salt, with comparatively little induction of cstR (Figure 2c), superimposed on a modest growth phenotype (Figure S1c). We observe an analogous induction of the S. aureus cst genes by Angeli’s salt under aerobic culture conditions, which occurs with a concomitant increase of cellular RSS.7 These observations appear to be attributable to HNO since neither sodium nitrite nor an NO• donor, diethylamine-NONOate (DEA NONOate), induce the cst-like operon nor give a growth phenotype relative to unstressed cells (Figure S1).16
Profiling of LMW Thiols, Organic, and Inorganic RSS in E. faecalis under Sulfide Stress
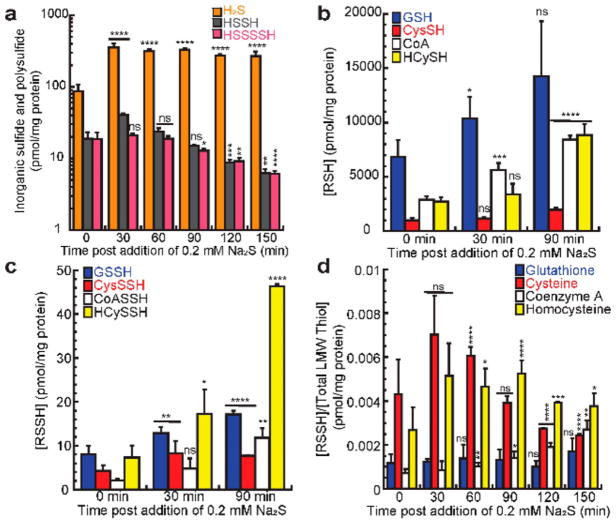
We next carried out polysulfide metabolite profiling experiments to further elucidate the physiological role of RSS in this process utilizing a recently developed UPLC-ESI-MS/MS method (Figure 3).26 These profiling experiments reveal detectable endogenous H2S and inorganic polysulfides (Figure 3a), with a ~4–5-fold increase in inorganic sulfide upon the addition of 0.2 mM Na2S to these microaerophilically grown cultures. These concentrations of H2S are some ~2-fold (endogenous) to 200-fold (postsulfide addition) lower than in S. aureus (Figure 3a).26 H2S remains consistently elevated 30 min postsulfide addition, while inorganic polysulfides significantly decrease after 90 min over the course of the experiment as the cells enter the late-log phase (Figure S2a). This is likely due to the high reactivity of these inorganic RSS, which are capable of transferring sulfur to LMW thiols present in these cells.19
Figure 3.
LMW inorganic species and organic thiol and persulfide concentrations for wild-type E. faecalis strain OG1RF following induction of the cst-like operon at t = 0 by 0.2 mM Na2S. (a) Level changes of inorganic species H2S (orange), HSSH (gray), and HSSSSH (pink). (b) Level changes of reduced glutathione (GSH, blue), cysteine (CysSH, red), CoA (white), and homocysteine (HCySH, yellow). (c) Level changes of glutathione (GSSH, blue), cysteine (CysSSH, red), CoA (CoASSH, white), and homocysteine (HCySSH, yellow) persulfides. (d) Ratio of persulfide to total cellular LMW thiol of glutathione (blue), cysteine (red), CoA (white), and homocysteine (yellow). Values represent mean ± SD of the mean derived from results of triplicate experiments with statistical significance established using a paired t test relative to wild-type strain under the same conditions (****p ≤ 0.0001, ***p ≤ 0.001, **p ≤ 0.01, *p ≤ 0.05; ns, no significant difference). For endogenous levels (t = 0), values represent means ± SD from n = 12 measurements, and values outside one standard deviation of the mean were excluded in final mean and standard deviation calculations. Additional time points for these conditions and cellular concentrations of wild-type E. faecalis strain OG1RF stressed with 0.4 mM Na2S are found in Figures S2 and S4.
Organic RSS profiling experiments (Figure 3b–d) establish that the metabolically important thiol, CoA, is present at ~3000 pmol per milligram of protein in unstressed cells, or ~4 fold higher than cysteine and on the same order of magnitude as glutathione (GSH) and homocysteine (HCySH; Figure 3b). The addition of Na2S causes a modest increase in LMW thiols of ~2–3 fold by 90 min, increasing further by 150 min (Figures 3b and S2b). The increase in LMW thiols is likely attributed to the fact that cysteine is a metabolic precursor to CoA, HCySH, and GSH as evident by the larger accumulation of these thiols relative to cysteine (Figures 3b and S2b). Cysteine does, however, accumulate ~2-fold by 150 min, which may be due to the fact that H2S is a substrate for cysteine synthase but is kinetically limited by cellular O-acetylserine concentrations.46 Strikingly, CoA persulfide (CoASSH) is endogenously present at just ~2 pmol per milligram of protein, which is ~2–5-fold lower than other LMW persulfides (Figure 3c) and ~30-fold lower than in S. aureus.26 The addition of Na2S, however, causes LMW persulfides to increase ~2–3 fold within 30 min. After this time, cysteine persulfide (CysSSH) levels return to approximately endogenous concentrations while all other LMW persulfides are further increased ~3–5 fold (Figures 3c and S2c). Interestingly, CoASSH is the only LMW persulfide to continuously accumulate over the course of the experiment, whereas other LMW persulfides remain relatively constant or decrease after 90 min postsulfide addition (Figures S2b and S3).
These data establish that LMW persulfides are present endogenously at less than 1% total LMW thiol under these microaerophilic culture conditions, which is ~10-fold lower than observed in S. aureus26 under aerobic culture conditions and with thiosulfate (TS) as the sole sulfur source (Figure 3d). In addition, CoASSH is the only LMW persulfide whose ratio to total thiol increases in a statistically significant manner over time. Similar results for cellular profiling of LMW inorganic and organic species were observed when wild-type E. faecalis was stressed with 0.4 mM Na2S (Figure S4).
Negative Regulation of DNA Binding by Ef CstR upon Reaction with CoASSH
The experiments described above make the prediction that CstR cysteine thiols would react with more electrophilic RSS to form more oxidized thiol species.16 LC-ESI-MS analysis reveals that unreacted and reduced Ef CstR exists mainly as a non-cross-linked monomer (12 802 Da, Figure S5a) and that anaerobic incubation with CoA, CoA disulfide, or Na2S does not detectably change the extent of dimer formation (Figures S5b–d). In contrast, incubation of the reduced Ef CstR with CoASSH gives rise largely to a covalently cross-linked dimer (25 602 Da, Figure S5e), confirmed by high-resolution ESI-MS/MS in both +2 (Figure S6a) and +4 (Figure S6b) charge states. Further analysis reveals that a small amount of trisulfide-cross-linked C32- and C61-containing peptide in the +5-charge state (Figures S6c) is also present, to ~1% of the disulfide. These anaerobic derivatization experiments are consistent with initial attack of one of two Cys (C32 or C61) on CoASSH, with the release of HS− or CoAS− followed by the attack of the resolving Cys of the mixed (hydro)disulfide to generate a disulfide bond.
Fluorescent anisotropy-based DNA binding titrations were next carried out to assess the DNA binding affinities of Ef CstR vs CoASSH-derivatized Ef CstR to a 29-bp fluorescein-labeled DNA harboring the cstR operator (see Figure 1b). The binding isotherm obtained can be fit to a two-CstR tetramer binding model and reveal a significant attenuation of binding affinity for CoASSH-derivatized Ef CstR (Figure S7a and Table 1), as previously observed for S. aureus CstR.16,43 While only CoASSH was investigated as a CstR oxidant in this way, any persulfide identified in our profiling experiments (Figure 3) could potentially react with CstR to inhibit DNA binding as found previously in S. aureus.16,43 In control experiments, Ef CstR exposed to air for an extended period of time confirms no cysteine oxidation or formation of covalently cross-linked dimeric Ef CstR (Figure S8).
Table 1.
Equilibrium Binding Parameters for Reduced Ef CstR and Ef CstRs Derivatized with CoASSH and Angeli’s Salta
| reactant | r0 | rcomplex | K1b (× 107 M−1) | K2b (× 107 M−1) | Ktetb (× 107 M−1) |
|---|---|---|---|---|---|
| 0.073 | 0.121 | 1.7 ± 0.9 | 42 ± 7 | 8.4 ± 2.3 | |
| CoASSH | 0.079 | 0.126c | 2.8 ± 0.4 | 0.2 ± 0.1 | 0.7 ± 0.1 |
| Angeli’s salt | 0.077 | 0.124c | 0.7 ± 0.2 | 0.4 ± 0.1 | 0.5 ± 0.1 |
Conditions: 10 nM fluorescein–DNA duplex, 2–240 nM Ef CstR (protomer), 25 mM HEPES, 200 mM NaCl, at pH 7.0, 2 mM EDTA, at 25 °C (2 mM TCEP added for the reduced Ef CstR).
Determined from a model that assumes two tetramers bind to each operator DNA with stepwise association constants of K1 and K2, and Ktet is the average macroscopic tetramer association constant (Ktet = sqrt(K1· K2)).16
Normalized to a change in anisotropy for unreacted Ef CstR.43
Negative Regulation of DNA Binding by Ef CstR upon Reaction with Angeli’s Salt
Given that the cst-like operon in E. faecalis is inducible by Angeli’s salt (Figure 2c), we next determined the DNA binding affinity of Angeli’s salt-derivatized Ef CstR. We find an attenuation of DNA binding affinity of Ef CstR comparable to that of CoASSH-reacted Ef CstR (Figure S7b and Table 1). Although the chemical modification on Ef CstR was not investigated here, Angeli’s salt is expected to induce the formation of interprotomer disulfide bridges as previously observed in S. aureus.7 These data suggest that HNO reacts directly with Ef CstR thiols to inhibit DNA binding, thus providing a potential second mechanism beyond HNO-induced perturbation of sulfide and persulfide levels to explain the induction of the cst-like operon by HNO.7 Given similar degrees of inhibition of DNA binding by oxidation of CstR with Angeli’s salt vs CoASSH, the relatively weaker induction by Angeli’s salt (Figure 2) suggests that a smaller fraction of CstR thiols may be derivatized by HNO in cells.
Thiosulfate Sulfurtransferase Activities of RhdA, RhdB, and CDR-RHD
Both rhdA and rhdB are predicted to encode canonical rhodaneses,40 which are thought to transfer sulfane sulfur from a persulfide donor to an appropriate cellular acceptor(s); however, their persulfide target specificity, if any, remains undefined. Interestingly, homology models created for RhdA and RhdB reveal largely opposite electrostatic surface potentials and have been designated as such. RhdA is largely acidic, and RhdB is basic (Figure S9). These electrostatic properties may impact their as yet undefined physiological roles in maintaining cellular sulfide and RSS homeostasis of both small molecules and the proteome,18 but this was not investigated further here. As anticipated, RhdA and RhdB possess significant thiosulfate sulfurtransferase (TST) activity, with steady-state kinetic parameters comparable to those of S. aureus CstA rhodanese previously characterized (Figure S9a,b).42
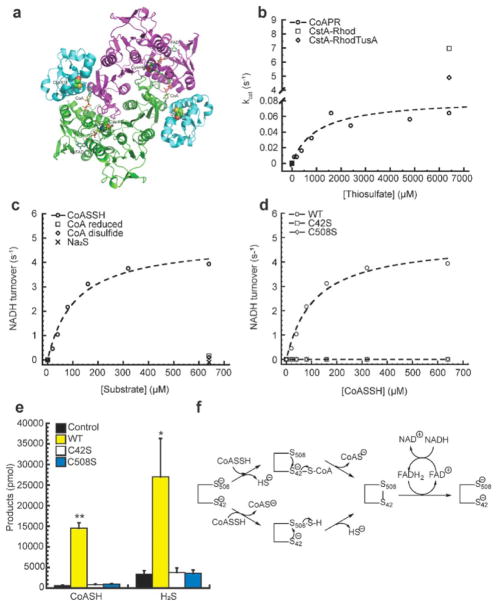
In S. aureus CstA, the N-terminal rhodanese possesses significant TST activity; the same is true of the C-terminal rhodanese domain of the persulfide dioxygenase, CstB.37,42 As described below, the CDR-RHD encoded by coaP (see Figure 1a) also harbors a C-terminal rhodanese homology domain with a putative active site cysteine, C508 (Figure 4a). We find only minimal TST activity (Figure 4b); kcat is some 90-fold lower than the CstARhod and authentic rhodaneses RhdA and RhdB.42 This suggests that the TST activity of this domain is not functionally relevant in cells, thus implicating this domain in performing some other function (vide infra).
Figure 4.
Structural model and function of Ef CDR-RHD. (a) B. anthracis CDR-RHD adopts a homodimeric assembly state, shown in ribbon representation (PDB: 3ICT).36 The N-terminal CDR domains (residues 1–450) from two protomers are shaded in purple and green, respectively, with the RHD (residues 451–554) colored in cyan. Each CDR-RHD protomer binds to one CoA and one FAD molecule. Active site cysteine residues from CDR (C44; C42 in Ef CDR-RHD) and RHD (C514; C508 in Ef CDR-RHD) are highlighted as spheres. (b) The initial thiosulfate turnover rate as a function of thiosulfate concentration catalyzed by Ef CDR-RHD, S. aureus CstARhod, and CstARhodTusA, with the continuous fit to the Michaelis–Menten equation shows Ef CDR-RHD has minimal TST activity. Ef CDR-RHD Km determined as 1.0 ± 0.6 mM and Vmax determined as 0.08 ± 0.1 μmol·min−1·mg−1, and kcat is some 90-fold lower than CstARhod and CstARhodTusA.42 (c) The initial NADH turnover rate plotted as a function of the concentration of CoASSH, CoA, CoA disulfide, and Na2S, with the continuous line a fit to the Michaelis–Menten eq (Table 2) and demonstrating Ef CDR-RHD has authentic NADH and FAD-dependent persulfide reductase activity only toward CoASSH. (d) The initial NADH turnover rate plotted as a function of the concentration of wild-type (WT), C42S, and C508S CDR-RHDs, with the continuous lines fit to the Michaelis–Menten eq (Table 2) shows no detectable persulfide reductase activity for either cysteine mutant compared to wild-type. (e) Product analysis for persulfide activity of WT, C42S, and C508S CDR-RHDs where only WT shows an increase in both CoA and H2S products compared to C42S and C508S CDR-RHDs and control experiments without the addition of enzyme. Values represent mean ± SD derived from replicate experiments with statistical significance established using a paired t-test relative to control under the same conditions (**p ≤ 0.01, *p ≤ 0.05). (f) Possible models for Ef CDR-RHD as a coenzyme A persulfide reductase, CoAPR acting upon a single CoASSH molecule. Top route: C42 attacks the CoASSH to form a C42–S–S–CoA mixed disulfide with the release of HS−, and C508 then reduces the mixed disulfide with the release of CoA and formation of C42–S–S–C508 disulfide. Bottom route: C508 attacks the CoASSH to form an enzyme-bound persulfide with CoA, and then C42 attacks the C508 persulfide with the release of HS−, forming C42–S–S–C508 disulfide. In both cases, FADH2 eventually reduces the disulfide bond between C42 and C508, which is rereduced by NADH, and the enzyme turns over. An alternative model that incorporates features of the bottom route that invokes a CoA thiol as a cofactor has also been proposed.47
The coaP-Encoded CDR-RHD Is a Bona Fide CoA Persulfide Reductase
The structures of previously characterized CDR-RHD fusions from B. anthracis (Figure 1a) and Shewanella loihica47 reveal a homodimeric assembly state where each protomer contains an N-terminal CoA disulfide reductase (CDR) domain and a C-terminal rhodanese homology domain (RHD; Figure 4a).36 As expected from these studies, fully reduced Ef CDR-RHD adopts a dimeric assembly state (Figure S10a).35,48 In contrast to a typical CDR49 or a CDR-RHD, Ef CDR-RHD as purified does not show a characteristic FAD absorption (Figure S10b). As a result, stoichiometric FAD was added to the purified enzyme in order to detect any reductase activity. Although CoASSH, CysSSH, HCySSH, and GSSH are all found in cells (Figure 3), we find that only CoASSH is used by Ef CDR-RHD as a substrate, with a Km of 111 (±23) μM and kcat of 4.9 (±0.4) s−1, obtained by monitoring NADH oxidation (Table 2). These steady-state parameters compare favorably with those measured previously for the polysulfide reductase from P. f uriosus,41 yielding similar catalytic efficiencies, kcat/Km, of 4.4 (±1.0) × 104 M−1 s−1 and 5.4 × 104 M−1 s−1 for the Ef CDR-RHD and P. furiosus enzymes, respectively.41 Ef CDR-RHD exhibits no detectable FAD-dependent reductase activity toward other sulfur-containing species in the CoASSH substrate mixture (see Methods), which includes CoA, CoA disulfide, or Na2S (Figure 4c), and the observed persulfide reductase activity is specific for NADH over NADPH47 (Table 2). A lack of activity toward CoA disulfide is consistent with the structure, which reveals a binding site for a single pantothenate arm that bridges the two domains within a pseudoprotomer (Figure 4a).
Table 2.
Persulfide Reductase Activity of CoAPR towards LMW Persulfide Substrates in the Presence of FAD and Either NADHa or NADPHb and in the Presence of Excess Product or Other Persulfides
| LMW persulfide substrate | reaction additives | Km (μ M) | Vmax (μ mol min−1 mg−1) | kcat (s−1) | kcat/Km (× 104 M−1 s−1) | kcat ratiof |
|---|---|---|---|---|---|---|
| CoASSHa | none | 111 ± 23 | 4.9 ± 0.4 | 4.9 ± 0.4 | 4.4 (±1.0) | 1.0 ± 0.12 |
| CoASSHb | NADPH | N.D.c | 0.37 ± 0.12 | 0.31 ± 0.07 | N.D. | 0.06 ± 0.24 |
| CoASSHa | 200 μM CoASH | 111e | 4.0 ± 0.3 | 4.0 ± 0.3 | N.D. | 0.82 ± 0.11 |
| CoASSHa | 1 mM CoASH | 111e | 2.2 ± 0.2 | 2.2 ± 0.2 | N.D. | 0.45 ± 0.12 |
| CoASSHa | 200 μM CysSSH | 111e | 5.3 ± 0.4 | 5.2 ± 0.4 | N.D. | 1.1 ± 0.11 |
| CoASSHa | 1 mM CysSSH | 111e | 3.4 ± 0.4 | 3.4 ± 0.4 | N.D. | 0.69 ± 0.14 |
| CysSSHa,d | none | 270 ± 568 | 0.11 ± 0.11 | 0.11 ± 0.11 | N.D. | 0.022 ± 1.0 |
| GSSHa,d | none | 150 ± 293 | 0.08 ± 0.06 | 0.08 ± 0.06 | N.D. | 0.016 ± 0.75 |
Conditions: 100 nM CoAPR (protomer), 100 nM FAD, 100 μM NADH, 25 mM Tris-HCl, 200 mM NaCl, pH 8.0, 25 °C.
Conditions: 100 nM CoAPR (protomer), 100 nM FAD, 100 μM NADPH, 25 mM Tris-HCl, 200 mM NaCl, pH 8.0, 25 °C.
N.D., not determined.
Very low activity, making it difficult to measure Km and Vmax accurately.
Km fixed to 111 μM during data fitting.
Relative kcat determined relative to kcat for CoASSH substrate and NADH cofactor, first row of table.
Product analysis of these CDR-RHD-catalyzed persulfide reductions reveals the expected products of a two-electron reduction of CoASSH, namely, the free thiol CoASH and HS(Figure 4e). We have therefore renamed Ef CDR-RHD CoA persulfide reductase “CoAPR.” Additional steady-state kinetics experiments reveal that Ef CDR-RHD is moderately inhibited by the product thiol CoA and exhibits some inhibition when the poor substrate CysSSH is added to these reactions (Table 2). The weak inhibition by CysSSH can be explained by the fact that cysteine, in both the thiol and persulfide forms, defines the business end of CoA and may well bind to CoAPR with some affinity. The extent to which this would occur in vivo is not known, but cysteine thiol and persulfide are present at lower concentrations in cells relative to those derived from CoA (Figures 3 and S2).
C42S and C508S CoAPR mutants are devoid of CoA persulfide reductase activity as they exhibit no NADH oxidation (Figure 4d), nor do they generate either of the expected products (Figure 4e). C42 is conserved in all CDRs48,49 and in CDR-RHDs,36,41,47 and forms an obligatory mixed disulfide with CoA as the redox center in the resting state of this group 3 flavoprotein disulfide reductase that is required for both disulfide and persulfide reductase activities.50 In contrast, the role of C508 is less clear but may play a role in persulfide sulfur transfer.36,47 Although the two active sites of CoAPR must cooperate to generate the products of NADH oxidation (Figures 4e,f), the existing structures of CDR-RHDs (Figure 4a) provides little insight as to how this might occur, since the two active sites are far apart from one another.36,47 Indeed, either mechanism shown (Figure 4f) would seem to require substantial changes in the relative orientations of the CDR and RHD domains within or between protomers of the dimer, and/ or a “swinging pantothenate arm” as suggested previously.36,47
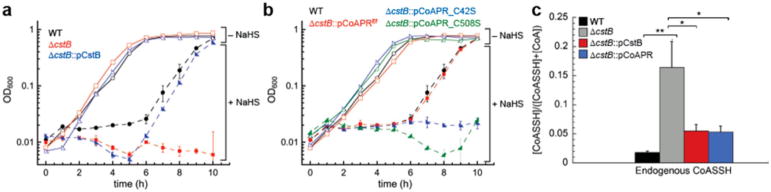
CoaP Complements a Sulfide-Induced Growth Phenotype in a S. aureus ΔcstB Strain
We next investigated a potential biological role of CoAPR motivated by our characterization of CstB as a nonheme FeII persulfide dioxygenase (PDO) that catalyzes the oxidation of RSSH to thiosulfate and reduced thiols, and our finding that a ΔcstB strain of S. aureus strain Newman exhibits a sulfide-induced growth phenotype.37 This growth phenotype can be complemented by expression of the wild-type allele from an extrachromosomal plasmid (Figure 5a).37 Remarkably, a wild-type-like growth phenotype is obtained in 0.2 mM NaHS stress when the ΔcstB strain is complemented with the E. faecalis coaP gene encoded on the same plasmid (Figure 5b). In contrast, a plasmid encoding C42S or C508S alleles of coaP fails to complement the ΔcstB growth phenotype, which is consistent with the essentiality of both cysteine residues in the enzymatic activity of CoAPR (Figure 4d). In addition, a near wild-type CoASSH/CoA ratio is achieved when a ΔcstB strain is complemented by expression of either the wild-type cstB allele or the E. faecalis coaP gene encoded on a plasmid (Figure 5c).
Figure 5.
Complementation of sulfide-induced growth phenotype and increased CoASSH levels in a ΔcstB S. aureus Newman strain by E. faecalis CoAPR. Representative growth curves for wild-type (WT), ΔcstB, and plasmid-complemented ΔcstB allelic S. aureus Newman strains on HHWm media in the presence of 0 mM and 0.2 mM NaHS added at t = 0 h. (a) WT (black squares), ΔcstB (blue squares), and ΔcstB:pCstB (red squares) strains in the presence of 0 mM NaHS; WT (black circles), ΔcstB (blue circles), and ΔcstB:pCstB (red circles) strains in the presence of 0.2 mM NaHS. (b) ΔcstB:pCoAPR (violet squares), ΔcstB:pCoAPRC42S (green squares), and ΔcstB:pCoAPRC508S (pink squares) strains in the presence of 0 mM NaHS; ΔcstB:pCoAPR (violet circles), ΔcstB:pCoAPRC42S (green circles), and ΔcstB:pCoAPRC508S (pink circles) strains in the presence of 0.2 mM NaHS. (c) Ratio of CoASSH to total CoA thiol for WT (black), ΔcstB (gray), ΔcstB:pCstB (red), and ΔcstB:pCoAPR (blue). Values represent mean ± SD of the mean derived from replicate experiments with statistical significance established using a paired t-test relative to wild-type strain under the same conditions (**p ≤ 0.01, *p ≤ 0.05).
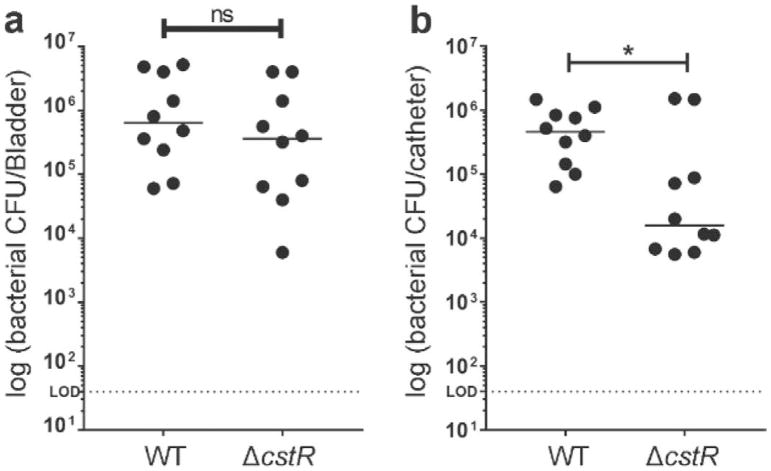
A ΔcstR E. faecalis Mutant Exhibits Reduced Virulence in a Murine Mouse CAUTI Model
We next investigated the effects of RSS on the pathophysiology of an E. faecalis-mediated catheter-associated urinary tract infection (CAUTI) model by introducing 4- to 5-mm-long pieces of silicone tubing into the bladders of C57BL/6Ncr female mice. Following catheter implantation, mice were immediately infected with ~2 × 107 CFU of OG1RF wild-type (WT) or ΔcstR mutant (Table S1) in phosphate-buffered saline (PBS) introduced transurethrally into the lumen of the bladder. Twenty-four hours after infection, bacteria were recovered from the bladder at a median value of 6.0 × 105 CFU for animals infected with WT OG1RF and 4.0 × 105 CFU for animals infected with the ΔcstR mutant, exhibiting no statistical difference in colonization (Figure 6a). In contrast, bacteria recovered from the implants at a median value of 4.2 × 105 CFU for animals infected with WT OG1RF, and a significant decrease was observed for animals infected with ΔcstR mutant, 10.7 × 104 CFU (Figure 6b). These data suggest that projected hyper-clearance of cytoplasmic RSS26 in E. faecalis results in decreased virulence in the CAUTI model, thus implicating RSS regulation as important in the pathophysiology of this opportunistic pathogen.
Figure 6.
A ΔcstR mutant strain deficient in colonization of silicone implants in a murine model of CAUTI. Graphs represent bacterial titers in log scale recovered from the (a) bladders and (b) silicone implants from female C57BL/6Ncr mice infected with OG1RF wild-type or ΔcstR mutant for 24 h. Horizontal dashed lines represent the limits of detection for viable bacteria. Each symbol represents the value for an individual mouse, and each experiment was done three times with four or five mice for each strain. The horizontal bars indicate the median value for each group of mice. Values that are significantly different by the Mann–Whitney U test are indicated as follows: *p < 0.05; ns, difference not statistically significant.
DISCUSSION
Enterococci, normally commensal bacteria of the human gastrointestinal tract (GIT), have become important opportunistic pathogens. They are a leading cause of hospital-acquired urinary tract infections and are frequently associated with other severe diseases such as endocarditis and bacteremia.51,52 While sulfide plays a role in various physiological processes that protect the GIT,53–55 increased H2S has also been implicated in several gut-associated diseases including colonic inflammation, ulcerative colitis,56 colorectal cancer,57 and irritable bowel syndrome.58 A number of reports suggest that the GIT is exposed to millimolar endogenous sulfide from a variety of sources,59,60 thus necessitating homeostatic mechanisms that regulate cellular sulfide and associated RSS in bacteria that inhabit this niche. We propose that CstR is a bona fide RSS sensor16,43 and that cst-like operon-encoded proteins mediate RSS speciation in E. faecalis.
The importance of the expression of the cst-like operon genes for E. faecalis colonization of the GIT and pathogen virulence is not yet known, but elevated levels of sulfide can be detected in this particular niche.9,14,61 In addition, treatment of some bacteria with β-lactam antibiotics upregulates enzymes that biosynthesize pantothenic acid and CoA perhaps in an effort to overcome antibiotics-induced H2S upregulation8 and the resulting increase in CoA persulfides (Figure 3) that occur as a result. Using a murine mouse CAUTI model, we observe a significant decrease in bacterial colonization of silicone catheter implants when mice are infected with a ΔcstR mutant strain (Figure 6b). This suggests that uncontrolled clearance of RSS mediated by unregulated expression of CstR-regulated genes (Figure 2a)26 reduces the virulence of this opportunistic pathogen and implies that regulation of these species in E. faecalis is important to its pathophysiology in this niche. It is interesting to note that mice infected with a S. aureus ΔcstR mutant strain also show a virulence phenotype in the kidney, an organ of the upper urinary tract.26 It is well established that urinary tract infections give rise to a significant burden of the ROS and oxidative damage in this niche,62–66 and the production and proper regulation of RSS may provide the bacteria a growth advantage in this microenvironment. Indeed, early studies in S. aureus showed that nitrite- and NO-induced dispersal of an abiotic biofilm results in upregulation of the cst operon,67 consistent with our findings in the CAUTI model.
Glutathione, cysteine, CoA, and homocysteine are major constituents of the LMW thiol pool in E. faecalis under these growth conditions (Figure 3b), with homocysteine ~15-fold higher than in S. aureus.26 L-Homocysteine can be formed via the trans-sulfuration process that converts L-cysteine, formed by cysteine synthase, to L-cystathionine via cystathionine γ-synthase (CGS), which is then converted to L-homocysteine via cystathionine-β-lyase (CBL).68 Exogenous Na2S clearly drives the biosynthesis of L-homocysteine (Figure 3b), likely via this pathway. Additionally, L-cysteine can be formed from L-homocysteine via the reverse transsulfuration process, which produces L-cysteine through a L-cystathionine intermediate, and L-methionine can be produced by a transmethylation reaction utilizing a methyl donor, e.g., tetrahydrofolate, to methylate L-homocysteine to biosynthesize L-methionine.69,70 The intestinal flora are a major site of L-homocysteine production,69,71 which drives glutathione biosynthesis;72,73 exposure of gut micro-organisms to homocysteine may therefore impact sulfide and RSS homeostasis in this niche, consistent with our profiling experiments (Figure 3). E. faecalis also maintains a significant pool of CoA that is strongly perturbed upon addition of exogenous sulfide to cells (Figure 3).
Our polysulfide metabolite profiling experiments reveal that CoASSH, while readily detected in ambiently grown cells, is maintained at very low endogenous levels relative to other organic persulfides, in stark contrast to what was observed in S. aureus.26 The addition of exogenous Na2S uniquely increases these levels by ~5–10-fold over several cell doublings (Figure 3), giving rise to a measurable growth phenotype (Figure S2c). This suggests that increases in RSS of this magnitude overruns the cst-like operon expression, and the organism is unable to effectively clear CoASSH and other RSS; this, in turn suggests that E. faecalis must protect itself from accumulating CoASSH. Consistent with this, we show here that one of the CstR-regulated genes, coaP, encodes a bona fide CoA persulfide reductase, CoAPR. Although the structure and biochemical characterization is available only for closely related enzymes,36,41,47 we show here that CoAPR requires both domains to reduce the hydrodisulfide of CoA, producing the free thiol CoA and H2S in an FAD-dependent and NADH-requiring fashion (Figure 4e). In contrast to the cst operon of S. aureus, which clearly functions in sulfide detoxification, the E. faecalis cst-like operon-encoded CoAPR, which while regenerating CoA for metabolic use as in S. aureus, also produces H2S, a finding that appears counter to a role in sulfide detoxification. However, the ultimate fate of this CoAPR-generated H2S is not yet known nor is the degree to which H2S versus CoASSH is deleterious to E. faecalis growth.
The finding that a sulfide-induced ΔcstB growth phenotype of S. aureus can be complemented by a wild-type coaP allele, but not C42S or C508S alleles (Figure 5), strongly suggests that in S. aureus, the origin of the growth defect may well be an elevated level of CoASSH. In fact, a single cst-encoded gene (cstB) deletion in S. aureus also gives rise to a striking increase specifically of CoASSH to ≈15% of the endogenous CoA pool (Figure 5c). Although we do not yet know how CoAPR is able to complement a lack of CstB, their orthogonal chemistries suggest to us that these sulfide/RSS homeostasis systems may have evolved in part to protect the integrity of the cellular CoA pool, derivatization of which will negatively impact cellular metabolism through disruption of the TCA cycle and other acyl-transfer requiring processes.74 For example, if CoASSH is a substrate for pyruvate dehydrogenase, the product of that reaction would be the mixed disulfide between thioacetate and CoA, which would simply regenerate CoA upon cellular reduction, negatively impacting levels of short chain thioesters, including acetyl-CoA. We also show that when E. faecalis is stressed with Angeli’s salt, a smaller but significant induction of the cst-like operon is observed (Figure 2b) and control experiments with NO2− and a representative NO donor suggest that HNO is the actual physiological effector of Angeli’s salt, as previously established for S. aureus.7,16 In addition, since we observe a similar attenuation of DNA operator binding affinity by both Angeli’s salt- and CoASSH-oxidized Ef CstR, the cst-like operon may be capable of responding to multiple cellular stressors via similar or distinct modifications of Ef CstR. Since HNO is an inducer of both operons, it will be very interesting to determine if CoAPR and CstB possess catalytic activity toward CoA sulfinamides (CoA-SONH2) as the anticipated reaction product upon derivatization by HNO,39 and as recently observed in yeast cells subjected to nitric oxide stress as the product of CoA-SNO reductase.38 Experiments are underway to further evaluate these ideas.
METHODS
All experimental methods can be found in the Supporting Information (SI).
Supplementary Material
Acknowledgments
We thank A. Lee for performing the DNA binding experiments presented in this paper. The authors gratefully acknowledge support by the U.S. National Institutes of Health (R01 GM097225 and R35 GM118157 to D.P.G.). B.J.C.W. gratefully acknowledges receipt of a predoctoral fellowship from the Graduate Training Program in Quantitative and Chemical Biology (T32 GM109825). This work was supported by grants R01 DK051406, R01 AI108749, and P50 DK064540 from the National Institutes of Health to A.L.F.-M. and S.J.H.
ABBREVIATIONS
- Cst
copper-sensing operon repressor (CsoR)-like sulfurtransferase
- RSS
reactive sulfur species
- HNO
nitroxyl
- H2S
hydrogen sulfide
- NO
nitric oxide
- CoA
coenzyme A
- CDR
CoA disulfide reductase
- CoASSH
CoA persulfide
- CysSH
cysteine
- CysSSH
cysteine persulfide
- GSH
glutathione
- GSSH
glutathione persulfide
- HCySH
homocysteine
- HCySSH
homocysteine persulfide
- Rhd
rhodanese
- RHD
rhodanese homology domain
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschem-bio.8b00230.
Supporting Tables S1–S3, supporting Figures S1–S10, and a detailed description of the experimental materials and methods used in this study (PDF)
References
- 1.Kolluru GK, Shen X, Bir SC, Kevil CG. Hydrogen sulfide chemical biology: pathophysiological roles and detection. Nitric Oxide. 2013;35:5–20. doi: 10.1016/j.niox.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, Yamamoto M, Ono K, Devarie-Baez NO, Xian M, Fukuto JM, Akaike T. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipovic MR, Eberhardt M, Prokopovic V, Mijuskovic A, Orescanin-Dusic Z, Reeh P, Ivanovic-Burmazovic I. Beyond H2S and NO interplay: hydrogen sulfide and nitroprusside react directly to give nitroxyl (HNO). A new pharmacological source of HNO. J Med Chem. 2013;56:1499–1508. doi: 10.1021/jm3012036. [DOI] [PubMed] [Google Scholar]
- 4.Cortese-Krott MM, Fernandez BO, Kelm M, Butler AR, Feelisch M. On the chemical biology of the nitrite/sulfide interaction. Nitric Oxide. 2015;46:14–24. doi: 10.1016/j.niox.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Cortese-Krott MM, Kuhnle GG, Dyson A, Fernandez BO, Grman M, DuMond JF, Barrow MP, McLeod G, Nakagawa H, Ondrias K, Nagy P, King SB, Saavedra JE, Keefer LK, Singer M, Kelm M, Butler AR, Feelisch M. Key bioactive reaction products of the NO/H2S interaction are S/N-hybrid species, polysulfides, and nitroxyl. Proc Natl Acad Sci U S A. 2015;112:E4651–4660. doi: 10.1073/pnas.1509277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuevasanta E, Lange M, Bonanata J, Coitino EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Reaction of Hydrogen Sulfide with Disulfide and Sulfenic Acid to Form the Strongly Nucleophilic Persulfide. J Biol Chem. 2015;290:26866–26880. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng H, Shen J, Edmonds KA, Luebke JL, Hickey AK, Palmer LD, Chang FJ, Bruce KA, Kehl-Fie TE, Skaar EP, Giedroc DP. Sulfide homeostasis and nitroxyl intersect via formation of reactive sulfur species in Staphylococcus aureus. mSphere. 2017;2:e00082–17. doi: 10.1128/mSphere.00082-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 9.Shukla P, Khodade VS, SharathChandra M, Chauhan P, Mishra S, Siddaramappa S, Pradeep BE, Singh A, Chakrapani H. On demand redox buffering by H2S contributes to antibiotic resistance revealed by a bacteria-specific H2S donor. Chem Sci. 2017;8:4967–4972. doi: 10.1039/c7sc00873b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- 11.Luther GW, Rickard DT, Theberge S, Olroyd A. Determination of metal (Bi)Sulfide stability constants of Mn2+, Fe2+, Co2+, Ni2+, Cu2+, and Zn2+ by voltammetric methods. Environ Sci Technol. 1996;30:671–679. [Google Scholar]
- 12.Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel ML, Lee JK, Pohl P. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci U S A. 2009;106:16633–16638. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibuya N, Koike S, Tanaka M, Ishigami-Yuasa M, Kimura Y, Ogasawara Y, Fukui K, Nagahara N, Kimura H. A novel pathway for the production of hydrogen sulfide from D-cysteine in mammalian cells. Nat Commun. 2013;4:1366. doi: 10.1038/ncomms2371. [DOI] [PubMed] [Google Scholar]
- 15.Czyzewski BK, Wang DN. Identification and characterization of a bacterial hydrosulphide ion channel. Nature. 2012;483:494–497. doi: 10.1038/nature10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol. 2014;94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishanina TV, Yadav PK, Ballou DP, Banerjee R. Transient Kinetic Analysis of Hydrogen Sulfide Oxidation Catalyzed by Human Sulfide Quinone Oxidoreductase. J Biol Chem. 2015;290:25072–25080. doi: 10.1074/jbc.M115.682369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen J, Peng H, Zhang Y, Trinidad JC, Giedroc DP. Staphylococcus aureus sqr Encodes a Type II Sulfide:Quinone Oxidoreductase and Impacts Reactive Sulfur Speciation in Cells. Biochemistry. 2016;55:6524–6534. doi: 10.1021/acs.biochem.6b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura Y, Koike S, Shibuya N, Lefer D, Ogasawara Y, Kimura H. 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci Rep. 2017;7:10459. doi: 10.1038/s41598-017-11004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura Y, Mikami Y, Osumi K, Tsugane M, Oka J, Kimura H. Polysulfides are possible H2S-derived signaling molecules in rat brain. FASEB J. 2013;27:2451–2457. doi: 10.1096/fj.12-226415. [DOI] [PubMed] [Google Scholar]
- 21.Kimura Y, Toyofuku Y, Koike S, Shibuya N, Nagahara N, Lefer D, Ogasawara Y, Kimura H. Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci Rep. 2015;5:14774. doi: 10.1038/srep14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akaike T, Ida T, Wei FY, Nishida M, Kumagai Y, Alam MM, Ihara H, Sawa T, Matsunaga T, Kasamatsu S, Nishimura A, Morita M, Tomizawa K, Nishimura A, Watanabe S, Inaba K, Shima H, Tanuma N, Jung M, Fujii S, Watanabe Y, Ohmuraya M, Nagy P, Feelisch M, Fukuto JM, Motohashi H. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat Commun. 2017;8:1177. doi: 10.1038/s41467-017-01311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto R, Koike S, Takano Y, Shibuya N, Kimura Y, Hanaoka K, Urano Y, Ogasawara Y, Kimura H. Polysulfides (H2Sn) produced from the interaction of hydrogen sulfide (H2S) and nitric oxide (NO) activate TRPA1 channels. Sci Rep. 2017;7:45995. doi: 10.1038/srep45995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberhardt M, Dux M, Namer B, Miljkovic J, Cordasic N, Will C, Kichko TI, de la Roche J, Fischer M, Suarez SA, Bikiel D, Dorsch K, Leffler A, Babes A, Lampert A, Lennerz JK, Jacobi J, Marti MA, Doctorovich F, Hogestatt ED, Zygmunt PM, Ivanovic-Burmazovic I, Messlinger K, Reeh P, Filipovic MR. H2S and NO cooperatively regulate vascular tone by activating a neuroendocrine HNO-TRPA1-CGRP signalling pathway. Nat Commun. 2014;5:4381. doi: 10.1038/ncomms5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianco CL, Toscano JP, Bartberger MD, Fukuto JM. The chemical biology of HNO signaling. Arch Biochem Biophys. 2017;617:129–136. doi: 10.1016/j.abb.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng H, Zhang Y, Palmer LD, Kehl-Fie TE, Skaar EP, Trinidad JC, Giedroc DP. Hydrogen Sulfide and Reactive Sulfur Species Impact Proteome S-Sulfhydration and Global Virulence Regulation in Staphylococcus aureus. ACS Infect Dis. 2017;3:744–755. doi: 10.1021/acsinfecdis.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedmann R, Onderka C, Wei S, Szijarto IA, Miljkovic JL, Mitrovic A, Lange M, Savitsky S, Yadav PK, Torregrossa R, Harrer EG, Harrer T, Ishii I, Gollasch M, Wood ME, Galardon E, Xian M, Whiteman M, Banerjee R, Filipovic MR. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci. 2016;7:3414–3426. doi: 10.1039/c5sc04818d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppenol WH, Bounds PL. Signaling by sulfur-containing molecules. Quantitative aspects. Arch Biochem Biophys. 2017;617:3–8. doi: 10.1016/j.abb.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, Wang B, Bebek G, Evans CR, Fox PL, Gerson SL, Hoppel CL, Liu M, Arvan P, Hatzoglou M. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife. 2015;4:e10067. doi: 10.7554/eLife.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doka E, Pader I, Biro A, Johansson K, Cheng Q, Ballago K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, Arner ES, Nagy P. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci Adv. 2016;2:e1500968. doi: 10.1126/sciadv.1500968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millikin R, Bianco CL, White C, Saund SS, Henriquez S, Sosa V, Akaike T, Kumagai Y, Soeda S, Toscano JP, Lin J, Fukuto JM. The chemical biology of protein hydro-persulfides: Studies of a possible protective function of biological hydropersulfide generation. Free Radical Biol Med. 2016;97:136–147. doi: 10.1016/j.freeradbiomed.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, Carroll KS. Persulfide reactivity in the detection of protein s-sulfhydration. ACS Chem Biol. 2013;8:1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mironov A, Seregina T, Nagornykh M, Luhachack LG, Korolkova N, Lopes LE, Kotova V, Zavilgelsky G, Shakulov R, Shatalin K, Nudler E. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc Natl Acad Sci U S A. 2017;114:6022–6027. doi: 10.1073/pnas.1703576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korshunov S, Imlay KR, Imlay JA. The cytochrome bd oxidase of Escherichia coli prevents respiratory inhibition by endogenous and exogenous hydrogen sulfide. Mol Microbiol. 2016;101:62–77. doi: 10.1111/mmi.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallen JR, Paige C, Mallett TC, Karplus PA, Claiborne A. Pyridine nucleotide complexes with Bacillus anthracis coenzyme A-disulfide reductase: a structural analysis of dual NAD(P)H specificity. Biochemistry. 2008;47:5182–5193. doi: 10.1021/bi8002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallen JR, Mallett TC, Boles W, Parsonage D, Furdui CM, Karplus PA, Claiborne A. Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking. Biochemistry. 2009;48:9650–9667. doi: 10.1021/bi900887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen J, Keithly ME, Armstrong RN, Higgins KA, Edmonds KA, Giedroc DP. Staphylococcus aureus CstB Is a Novel Multidomain Persulfide Dioxygenase-Sulfurtransferase Involved in Hydrogen Sulfide Detoxification. Biochemistry. 2015;54:4542–4554. doi: 10.1021/acs.biochem.5b00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anand P, Hausladen A, Wang YJ, Zhang GF, Stomberski C, Brunengraber H, Hess DT, Stamler JS. Identification of S-nitroso-CoA reductases that regulate protein S-nitrosylation. Proc Natl Acad Sci U S A. 2014;111:18572–18577. doi: 10.1073/pnas.1417816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keceli G, Toscano JP. Reactivity of nitroxyl-derived sulfinamides. Biochemistry. 2012;51:4206–4216. doi: 10.1021/bi300015u. [DOI] [PubMed] [Google Scholar]
- 40.Cipollone R, Ascenzi P, Visca P. Common themes and variations in the rhodanese superfamily. IUBMB Life. 2007;59:51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- 41.Herwald S, Liu AY, Zhu BE, Sea KW, Lopez KM, Sazinsky MH, Crane EJ., 3rd Structure and substrate specificity of the pyrococcal coenzyme A disulfide reductases/ polysulfide reductases (CoADR/Psr): implications for S(0)-based respiration and a sulfur-dependent antioxidant system in Pyrococcus. Biochemistry. 2013;52:2764–2773. doi: 10.1021/bi3014399. [DOI] [PubMed] [Google Scholar]
- 42.Higgins KA, Peng H, Luebke JL, Chang FM, Giedroc DP. Conformational analysis and chemical reactivity of the multidomain sulfurtransferase. Biochemistry. 2015;54:2385–2398. doi: 10.1021/acs.biochem.5b00056. [DOI] [PubMed] [Google Scholar]
- 43.Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, Skaar EP, Giedroc DP. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011;286:13522–13531. doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amatore C, Arbault S, Ducrocq C, Hu S, Tapsoba I. Angeli’s salt (Na2N2O3) is a precursor of HNO and NO: a voltammetric study of the reactive intermediates released by Angeli’s salt decomposition. ChemMedChem. 2007;2:898–903. doi: 10.1002/cmdc.200700016. [DOI] [PubMed] [Google Scholar]
- 45.Dutton AS, Fukuto JM, Houk KN. Mechanisms of HNO and NO production from Angeli’s salt: density functional and CBS-QB3 theory predictions. J Am Chem Soc. 2004;126:3795–3800. doi: 10.1021/ja0391614. [DOI] [PubMed] [Google Scholar]
- 46.Cook PF, Wedding RT. Cysteine synthetase from Salmonella typhimurium LT-2. Aggregation, kinetic behavior, and effect of modifiers. J Biol Chem. 1978;253:7874–7879. [PubMed] [Google Scholar]
- 47.Warner MD, Lukose V, Lee KH, Lopez K, Sazinsky HM, Crane EJ. Characterization of an NADH-dependent persulfide reductase from Shewanella loihica PV-4: implications for the mechanism of sulfur respiration via FAD-dependent enzymes. Biochemistry. 2011;50:194–206. doi: 10.1021/bi101232y. [DOI] [PubMed] [Google Scholar]
- 48.delCardayre SB, Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus Purification and characterization of the native enzyme. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 49.Mallett TC, Wallen JR, Karplus PA, Sakai H, Tsukihara T, Claiborne A. Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution. Biochemistry. 2006;45:11278–11289. doi: 10.1021/bi061139a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: advances in chemistry and function. Prog Nucleic Acid Res Mol Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 51.Agudelo Higuita NI, Huycke MM. Enterococcal Disease, Epidemiology, and Implications for Treatment. In: Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary; Boston: 2014. Internet. [Google Scholar]
- 52.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo C, Liang F, Shah Masood W, Yan X. Hydrogen sulfide protected gastric epithelial cell from ischemia/ reperfusion injury by Keap1 s-sulfhydration, MAPK dependent anti-apoptosis and NF-kappaB dependent anti-inflammation pathway. Eur J Pharmacol. 2014;725:70–78. doi: 10.1016/j.ejphar.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Fiorucci S, Distrutti E, Cirino G, Wallace JL. The emerging roles of hydrogen sulfide in the gastrointestinal tract and liver. Gastroenterology. 2006;131:259–271. doi: 10.1053/j.gastro.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 55.Wallace JL. Physiological and pathophysiological roles of hydrogen sulfide in the gastrointestinal tract. Antioxid Redox Signaling. 2010;12:1125–1133. doi: 10.1089/ars.2009.2900. [DOI] [PubMed] [Google Scholar]
- 56.Pitcher MC, Cummings JH. Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut. 1996;39:1–4. doi: 10.1136/gut.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 58.Xu GY, Winston JH, Shenoy M, Zhou S, Chen JD, Pasricha PJ. The endogenous hydrogen sulfide producing enzyme cystathionine-beta synthase contributes to visceral hypersensitivity in a rat model of irritable bowel syndrome. Mol Pain. 2009;5 doi: 10.1186/1744-8069-5-44. 1744-8069-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macfarlane GT, Gibson GR, Cummings JH. Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol. 1992;72:57–64. doi: 10.1111/j.1365-2672.1992.tb04882.x. [DOI] [PubMed] [Google Scholar]
- 60.Barton LL, Ritz NL, Fauque GD, Lin HC. Sulfur Cycling and the Intestinal Microbiome. Dig Dis Sci. 2017;62:2241–2257. doi: 10.1007/s10620-017-4689-5. [DOI] [PubMed] [Google Scholar]
- 61.Bouillaud F, Blachier F. Mitochondria and sulfide: a very old story of poisoning, feeding, and signaling? Antioxid Redox Signaling. 2011;15:379–391. doi: 10.1089/ars.2010.3678. [DOI] [PubMed] [Google Scholar]
- 62.Zhao C, Hartke A, La Sorda M, Posteraro B, Laplace JM, Auffray Y, Sanguinetti M. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect Immun. 2010;78:3889–3897. doi: 10.1128/IAI.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mundi H, Bjorksten B, Svanborg C, Ohman L, Dahlgren C. Extracellular release of reactive oxygen species from human neutrophils upon interaction with Escherichia coli strains causing renal scarring. Infect Immun. 1991;59:4168–4172. doi: 10.1128/iai.59.11.4168-4172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lundberg JO, Carlsson S, Engstrand L, Morcos E, Wiklund NP, Weitzberg E. Urinary nitrite: more than a marker of infection. Urology. 1997;50:189–191. doi: 10.1016/S0090-4295(97)00257-4. [DOI] [PubMed] [Google Scholar]
- 65.Carlsson S, Govoni M, Wiklund NP, Weitzberg E, Lundberg JO. In Vitro Evaluation of a New Treatment for Urinary Tract Infections Caused by Nitrate-Reducing Bacteria. Antimicrob Agents Chemother. 2003;47:3713–3718. doi: 10.1128/AAC.47.12.3713-3718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlsson S, Wiklund NP, Engstrand L, Weitzberg E, Lundberg JO. Effects of pH, nitrite, and ascorbic acid on nonenzymatic nitric oxide generation and bacterial growth in urine. Nitric Oxide. 2001;5:580–586. doi: 10.1006/niox.2001.0371. [DOI] [PubMed] [Google Scholar]
- 67.Schlag S, Nerz C, Birkenstock TA, Altenberend F, Gotz F. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol. 2007;189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aitken SM, Kirsch JF. The enzymology of cystathionine biosynthesis: strategies for the control of substrate and reaction specificity. Arch Biochem Biophys. 2005;433:166–175. doi: 10.1016/j.abb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 69.Bauchart-Thevret C, Stoll B, Burrin DG. Intestinal metabolism of sulfur amino acids. Nutr Res Rev. 2009;22:175–187. doi: 10.1017/S0954422409990138. [DOI] [PubMed] [Google Scholar]
- 70.Brosnan JT, Brosnan ME. The sulfur-containing amino acids: an overview. J Nutr. 2006;136:1636S–1640S. doi: 10.1093/jn/136.6.1636S. [DOI] [PubMed] [Google Scholar]
- 71.Riedijk MA, Stoll B, Chacko S, Schierbeek H, Sunehag AL, van Goudoever JB, Burrin DG. Methionine transmethylation and transsulfuration in the piglet gastrointestinal tract. Proc Natl Acad Sci U S A. 2007;104:3408–3413. doi: 10.1073/pnas.0607965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beatty PW, Reed DJ. Involvement of the cystathionine pathway in the biosynthesis of glutathione by isolated rat hepatocytes. Arch Biochem Biophys. 1980;204:80–87. doi: 10.1016/0003-9861(80)90009-0. [DOI] [PubMed] [Google Scholar]
- 73.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39:13005–13011. doi: 10.1021/bi001088w. [DOI] [PubMed] [Google Scholar]
- 74.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 75.Oberto J. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinf. 2013;14:4. doi: 10.1186/1471-2105-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.