Abstract
Genetic influences play a significant role in risk for psychiatric disorders, prompting numerous endeavors to further understand their underlying genetic architecture. In this paper, we summarize and review evidence from traditional twin studies and more recent genome-wide molecular genetic analyses regarding two important issues that have proven particularly informative for psychiatric genetic research. First, emerging results are beginning to suggest that genetic risk factors for some (but not all) clinically diagnosed psychiatric disorders or extreme manifestations of psychiatric traits in the population share genetic risks with quantitative variation in milder traits of the same disorder throughout the general population. Second, there is now evidence for substantial sharing of genetic risks across different psychiatric disorders. This extends to the level of characteristic traits throughout the population, with which some clinical disorders also share genetic risks. In this review, we summarize and evaluate the evidence for these two issues, for a range of psychiatric disorders. We then critically appraise putative interpretations regarding the potential meaning of genetic correlation across psychiatric phenotypes. We highlight several new methods and studies which are already using these insights into the genetic architecture of psychiatric disorders to gain additional understanding regarding the underlying biology of these disorders. We conclude by outlining opportunities for future research in this area.
Key words: genetic correlation, twin studies, genetics, pleiotropy, GWAS
Introduction
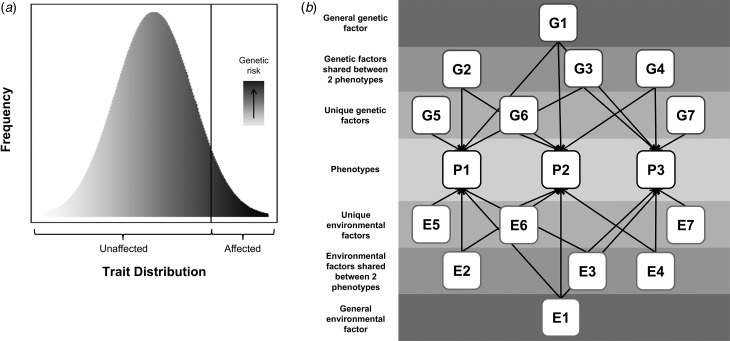
Psychiatric disorders are relatively common in terms of lifetime prevalence and are associated with considerable distress and functional impairment (Whiteford et al. 2013). Understanding the etiology of these disorders is of critical importance to developing effective treatments and reducing suffering. There is strong evidence that these disorders are complex and partly genetic in origin, with twin study heritability estimates of 40–80% (Polderman et al. 2015). Environmental factors also contribute and possibly moderate genetic risk. This review will consider two important related hypotheses: that psychiatric disorders share genetic risks with variation in relevant population traits (illustrated in Fig. 1a) and that there are shared genetic contributions across different psychiatric phenotypes (illustrated in Fig. 1b).
Fig. 1.
Hypothesized models of: (a) shared genetic risks across disorder and population trait variation, where the extreme end of a continuous distribution of a trait is associated with a continuous underlying genetic liability, and (b) shared genetic risks across different disorders, where squares labeled ‘P’ represent phenotypes, and squares labeled ‘G’ and ‘E’, represent genetic or environmental contributions, respectively, that can be shared or unique (indicated by the number of arrows pointing to phenotypes). All G factors are uncorrelated with one another and thus the entire genetic contribution to a phenotype can be modelled as the sum of the genetic factors contributing to it (e.g. for P1 this would be G1 + G2 + G3 + G5). The same is true for environmental factors (i.e. environmental contribution to P1 is E1 + E2 + E3 + E5). As an illustrative example, if P1 were ADHD, P2 were ASD, and P3 were MDD, then G1 represents any genetic variants that are shared between ADHD, ASD, and MDD; G2–G4 represents genetic variants shared between only two of these disorders (e.g. G2 would be genetic risk for ADHD and ASD but not MDD); and G5–G7 represent unique genetic risks (e.g. G5 is genetic risk that is unique to ADHD and not shared with either ASD or MDD). N.B. The shapes are not indicative of whether a variable is latent or measured.
The hypothesis that psychiatric disorders are extreme manifestations of continuously distributed population traits is not new [e.g. for a theoretical review see (Plomin et al. 2009)]. However, studies specifically testing whether categorical, clinical disorders share genetic risks with continuous variation in related sub-diagnostic traits in the population have been sparse until recently. A pressing matter that needs to be evaluated for specific psychiatric phenotypes, is the extent to which the current evidence supports this hypothesis. Recent years have also seen a dramatic increase in studies examining the related issue of shared genetic risks across different psychiatric disorders. Given the fast-growing body of research on this subject, the time is ripe to assess the strength of the evidence of shared risks for specific pairs of psychiatric phenotypes. In this review, we summarize and evaluate the evidence for the two hypotheses illustrated in Fig. 1, for a range of psychiatric phenotypes that have been extensively studied using both traditional twin and molecular genetic methods. We also discuss possible interpretations and implications for genetic research and clinical practice. Based on a non-exhaustive literature search, studies were included if they formally tested for shared genetic risks across a psychiatric disorder and traits related to the same disorder or across different psychiatric phenotypes (either defined as disorder or traits). Twin studies using DeFries-Fulker analysis were also included, although these studies do not directly test for genetic correlation; see additional discussion below.
Overview of twin and molecular genetic methods
Most evidence concerning shared genetic risks within and across phenotypic constructs comes from twin studies and common variant genome-wide analyses. The twin design, which relies on comparison of identical (monozygotic) and non-identical (dizygotic) individuals, is commonly used to estimate the heritability of individual traits. Of particular relevance is the DeFries-Fulker analytic method, which estimates group heritability. Group heritability indicates the degree to which the mean difference between a proband group and the rest of a given sample is influenced by genetic factors. Significant group heritability indicates similar etiology for milder variation in continuous traits and more severe manifestations. An extension of the twin design, the bivariate twin model, allows one to estimate the degree of genetic correlation (rg) between two phenotypes. Complementarily, molecular genetic methods directly test for shared genetic risks across phenotypes. One method is the estimation of genetic correlation [e.g. using LDSC or GREML-GCTA (Yang et al. 2011; Bulik-Sullivan et al. 2015a)] from millions of common variants (single nucleotide polymorphisms; SNPs), for example using a case-control sample of one psychiatric disorder and another sample assessed for a relevant continuous trait or a different disorder. Such methods provide correlation estimates of the degree to which genetic risks are shared. However, practical limitations include a need for very large sample sizes and for some methods (e.g. GREML-GCTA), access to raw genotypes, limiting the application of these tools. A second approach uses a genome-wide association study (GWAS) ‘discovery’ sample to calculate polygenic risk scores (PRS) (Wray et al. 2014) for individuals in an independent ‘target’ sample. PRS for a phenotype of interest can be tested for association with another phenotype (e.g. another psychiatric disorder or trait variation) in the target sample, to establish whether there are shared genetic risks across phenotypes. Although studies using PRS methods can show direct evidence for shared genetic risks, typically modest effect sizes are observed (Wray et al. 2014), whereas molecular genetic studies that estimate genetic correlation provide a more precise assessment of the degree of shared genetic risks across phenotypes using different definitions.
It is important to note several differences in the meaning of results obtained from twin and molecular genetic analyses. For a more thorough review of different methods for estimation of univariate heritability and genetic correlation, please see Yang et al. (2017). In brief, the correlation estimates from twin studies capture all inherited genetic variants shared by monozygotic twins. These estimates are likely to be different and higher than those from SNP-based studies as the latter is only based on additive common variant effects tagged by genotyping arrays. However, the source of any shared genetic effects cannot be discerned from twin studies; effects may be driven by or limited to specific types of variants (e.g. rare mutations) but not to other classes of variants (e.g. SNPs). Genetic studies assessing multiple classes of variants are needed to determine the source of genetic correlations estimated using twin studies. It is also worth noting that evidence from common and rare variant studies regarding shared genetic risks between two phenotypes might not be consistent.
Shared genetic risks across categorical disorders and population trait variation
Genetic studies have consistently demonstrated that thousands of common variants of small effect, as well as rare variants of larger effect, increase the risk for psychiatric disorders (Sullivan et al. 2012; Cross-Disorder Group of the PGC, 2013a; Davis et al. 2013; Robinson et al. 2015). This complex polygenic architecture supports a model where a quantitatively distributed liability (influenced by numerous genes) is associated with one or more continuous phenotypes that underlie the diagnostic distinction between cases and controls. According to such a model (Fig. 1a), genetic risks that contribute to clinical diagnoses will also influence variation in related quantitative traits in the general population. See Table 1 for a summary of studies that have addressed this hypothesis for specific psychiatric disorders.
Table 1.
Summary of studies investigating shared genetic risks across disorders and trait variation
| Disorder | Evidence from studies |
|---|---|
| ASD |
Twin studies: persistently high group heritability across varying cut-offs; Genetic correlation of 0.70 between clinical diagnoses of ASD and autistic traits (Robinson et al.
2011; Lundström et al.
2012; Colvert et al.
2015) LDSC: ASD and population social-communication traits: rg = 0.27; replicated in independent clinical sample: rg = 0.30; genetic correlation is highest at age 8 years (rg = 0.34) and drops and is no longer significant for traits measured at ages 11 (0.16), 14 (0.21), and 17 (0.01) years (Robinson et al. 2016; St Pourcain et al. 2017) PRS: PRS derived using a clinical ASD discovery GWAS showed significant association with social-communication problems at age 8 years but not ages 11, 14, and 17 years in the general population (St Pourcain et al. 2017) and self-reported autistic traits in adults (in particular symptoms related to attention to detail, but also rigidity and childhood behaviors) (Bralten et al. 2017), although 1 smaller study does not find an association with autistic traits in children (Krapohl et al. 2016) Other: rare de novo loss-of-function and missense mutations associated with Vineland composite scores in ASD probands and unaffected siblings (Robinson et al. 2016) |
| ADHD |
Twin studies: high group heritability from DeFries-Fulker analysis (Levy et al.
1997; Larsson et al.
2011; Greven et al.
2016) LDSC: ADHD and population traits of ADHD: rg = 0.96 (Middeldorp et al. 2016); replicated in a larger study: rg = 0.94 (Demontis et al. 2017). ADHD and traits of extraversion in the population: rg = 0.30 (Lo et al. 2016) PRS: multiple analyses of independent target samples find associations between clinically-defined ADHD PRS and population traits of ADHD and vice versa, although 1 smaller study does not find an association (Groen-Blokhuis et al. 2014; Martin et al. 2014a; Stergiakouli et al. 2015, 2017; Krapohl et al. 2016; Riglin et al. 2016; Brikell et al. 2017; Jansen et al. 2017) |
| ID |
Twin studies: Cognitive abilities in the general population seem to share genetic risks with milder forms of ID (Spinath et al.
2004; Reichenberg et al.
2016) and extremely high IQ (Shakeshaft et al.
2015). There is some evidence, however, of discontinuity between cognitive abilities and severe forms of ID (Reichenberg et al.
2016) Other: Rare, likely pathogenic copy number variants (which are associated with developmental delay) are also associated with lower cognitive ability in the population (Männik et al. 2015; Kendall et al. 2016) |
| Anxiety disorders | Twin studies: extreme over-anxiety and specific fears, such as of animals, seem to share genetic risks with milder trait anxiety (Stevenson et al. 1992; Goldsmith & Lemery, 2000). Notably, the evidence is lacking on links between anxiety disorders and anxiety traits in samples that are closer in age to the typical age of onset for anxiety disorders |
| OCD |
Twin studies: although heritable OCD traits are present throughout the general population (van Grootheest et al.
2005), no study has focused on the extreme presentation of these traits PRS: OCD PRS were associated with OCD traits in the general population (den Braber et al. 2016) |
| MDD |
Twin studies: smaller studies yielded inconsistent findings; one study did not find significant group heritability, although a subsequent study of children and adolescents did (Rende et al.
1993; Eley, 1997) PRS: MDD PRS associated with depressive symptoms in elderly (Demirkan et al. 2011) but not with internalizing traits at ages 3–10 years (Jansen et al. 2017) LDSC: MDD and depressive symptoms in the population: rg = 0.91–1.00; MDD and traits of neuroticism in the population: rg = 0.56–0.70 (Direk et al. 2016; Lo et al. 2016; Major Depressive Disorder Working Group of the PGC et al. 2017) |
| SCZ & psychosis |
Twin studies: Significant group heritability for severe and milder manifestations of adolescent psychotic experiences, suggesting genetic links between mild and severe psychotic experiences; however, the association with psychotic disorders such as SCZ is unclear from these studies (Ronald et al.
2014b; Zavos et al.
2014) PRS: no association of SCZ PRS and psychotic experiences in adolescents; evidence of association of SCZ PRS and adolescent negative schizophrenia-like symptoms as well as ‘thought problems’ at age 10 years; increased PRS in unaffected relatives and with increasing severity in probands (Zammit et al. 2013; Bigdeli et al. 2014; Sieradzka et al. 2014; Jones et al. 2016; Krapohl et al. 2016; Meier et al. 2016; Jansen et al. 2017) |
ASD, autism spectrum disorder; ADHD, attention-deficit hyperactivity disorder; ID, intellectual disability; OCD, obsessive-compulsive disorder; MDD, major depressive disorder; SCZ, schizophrenia
Group heritability (implemented in DeFries-Fulker analysis) (DeFries & Fulker, 1985) refers to the degree to which genetic factors influence the mean difference between extreme groups and the rest of a sample; significant group heritability implies a genetic link between milder and more severe manifestations of a trait
Linkage disequilibrium score correlation (LDSC) (Bulik-Sullivan et al. 2015a, b) estimates the contribution of all SNPs from genome-wide data and indexes this as an estimate of SNP-heritability; which is different to twin heritability (Wray et al. 2014). This method can be applied to examine shared genetic risks between disorders and population traits to give an estimate of genetic correlation. Genome-wide association studies (GWAS) directly assess the independent association of many millions of common genetic variants (single nucleotide polymorphisms; SNPs) with a phenotype. Polygenic risk score (PRS) analysis, uses a GWAS ‘discovery’ sample to calculate genetic risk scores for individuals in an independent ‘target’ sample with genetic data; scores are derived by calculating the number of risk alleles weighted by the discovery effect size for each SNP and then summing these values for the set of SNPs, for each target individual (The International Schizophrenia Consortium, 2009). Regression analyses are used to test whether PRS for the discovery phenotype (e.g. clinical disorder) are associated with phenotypes of interest in the independent target sample (e.g. symptom variation in the population)
Disorders with early onset
Twin studies have reported significant group heritability using several different definitions of ASD (Robinson et al. 2011; Lundström et al. 2012). One study employed a novel twin model to estimate the genetic correlation between ASD diagnoses and traits (rg = 0.70) (Colvert et al. 2015). PRS studies show mixed results, with association between clinical ASD PRS with social-communication problems at age 8 but not later ages (St Pourcain et al. 2017), with self-assessed autistic traits in adults (Bralten et al. 2017) and null results in a third study (Krapohl et al. 2016). Modest, genetic correlation (rg = 0.27–0.34) was estimated between clinical ASD and social-communication traits at age 8, with non-significant estimates at ages 11–17 years (Robinson et al. 2016; St Pourcain et al. 2017). The rate of rare de novo mutations was associated with autism-related behaviors not only in children with ASD but also in unaffected siblings (Robinson et al. 2016).
Twin studies of attention-deficit hyperactivity disorder (ADHD) traits have also revealed substantial group heritability for extreme scores on ADHD traits (Levy et al. 1997; Larsson et al. 2011), albeit extremely low ADHD scores are a potential exception (Greven et al. 2016). Multiple PRS analyses have demonstrated that genetic risk for clinically-diagnosed ADHD is shared with ADHD traits assessed between ages 3 and 17 years (Groen-Blokhuis et al. 2014; Martin et al. 2014a; Stergiakouli et al. 2015, 2017; Riglin et al. 2016; Brikell et al. 2017; Jansen et al. 2017). Estimates of genetic correlation between ADHD diagnosis and traits are very high (rg = 0.94–0.96) (Middeldorp et al. 2016; Demontis et al. 2017), with a moderate genetic correlation (rg = 0.30) between ADHD diagnosis and extraversion traits in the population (Lo et al. 2016).
Cognitive abilities display a similar pattern of significant group heritability in studies of mild intellectual disability (ID) (Spinath et al. 2004), different quantiles of reading assessments (Logan et al. 2012), and high levels of intelligence (Shakeshaft et al. 2015). However, severe ID appears to be an exception to this pattern (Reichenberg et al. 2016). Molecular genetic studies of ID have focused on very rare mutations (Girirajan et al. 2011; The Deciphering Developmental Disorders Study, 2014) and there is some evidence that rare, likely pathogenic copy number variants (CNVs) are associated with poor performance on cognitive tasks in the population (Männik et al. 2015; Kendall et al. 2016). Studies assessing the degree of shared common variants between ID and cognition in the population are lacking.
Converging evidence from twin and molecular genetic methods so far shows reasonably strong support for certain child-onset neurodevelopmental disorders (i.e. ADHD, ASD, and mild ID) as the extreme ends of continuous distributions of population traits.
Disorders with onset in adolescence and adulthood
There is a lack of studies testing for shared genetic risks across disorder and traits for anxiety disorders and obsessive-compulsive disorder (OCD). Although twin studies have established the heritability of anxiety traits, only two studies reported significant group heritability for anxiety disorders (Stevenson et al. 1992; Goldsmith & Lemery, 2000). Twin studies of OCD indicate that traits characteristic of OCD are heritable and present throughout the population (van Grootheest et al. 2005), although no twin studies have tested whether extreme OCD traits share genetic risks with milder traits. One recent study found associations between OCD PRS and continuously-distributed obsessive-compulsive traits in the population (den Braber et al. 2016).
Twin studies of group heritability for depressive traits have found mixed results (Rende et al. 1993; Eley, 1997). Shared genetic influences across major depressive disorder (MDD) and depressive traits have been reported in an elderly population using PRS analysis (Demirkan et al. 2011) but not in a childhood sample assessing internalizing traits at ages 3–10 years (Jansen et al. 2017). Recent common variant analyses showed very high genetic correlation (rg = 0.91–1.00) between MDD and depressive symptoms (Direk et al. 2016; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) and moderate correlation between MDD and personality measures, notably neuroticism (rg = 0.56–0.74), in the general population (Lo et al. 2016; Major Depressive Disorder Working Group of the PGC et al. 2017).
The genetic evidence for a continuous spectrum of psychosis in the population is more complex. Psychotic experiences (e.g. paranoia and hallucinations) show low-to-moderate heritability (15–59%), with significant group heritability implying a genetic link between mild and severe psychotic experiences (Zavos et al. 2014). However, it is unclear from twin studies whether psychotic experiences are related to schizophrenia. Findings from PRS studies are mixed, with several studies finding no association of schizophrenia or bipolar disorder (BD) PRS with adolescent psychotic experiences (Sieradzka et al. 2014; Krapohl et al. 2016), others reporting an association in the opposite direction to that expected (Zammit et al. 2013) and others finding associations between schizophrenia PRS and adolescent negative symptoms (e.g. apathy or lack of energy) related to schizophrenia (Jones et al. 2016) and ‘thought problems’ at age 10 (Jansen et al. 2017). Schizophrenia PRS are higher in unaffected relatives of schizophrenia probands compared with controls (Bigdeli et al. 2014) and in individuals with more strictly defined schizophrenia, in terms of chronicity or severity of disorder (Meier et al. 2016).
Evidence for shared genetic risks across disorders and traits is limited for adolescent- and adult-onset psychiatric disorders. Preliminary supporting evidence is seen for OCD and MDD. The picture is quite complex for schizophrenia and there is insufficient evidence to conclude whether anxiety disorders share genetic risks with related population traits.
Limitations and interpretation
There are several limitations of existing studies and important issues that have not been sufficiently addressed. First, many twin studies use percentile-based cut-offs to identify probands, rather than using clinical diagnoses. Second, twin studies have largely employed DeFries-Fulker analysis, which does not directly estimate genetic correlation between psychiatric disorders and related traits; rather, significant group heritability suggests a link between extreme values of a trait and variation in the trait. Direct estimation of the genetic correlation, as done for ASD (Colvert et al. 2015), would likely be informative in future twin research.
Although analyses of population traits do not include many individuals who have psychiatric diagnoses, it is important to determine whether associations persist when such individuals are excluded. If not, this might suggest that any association signal is driven by extreme cases and not continuous variation in the trait of interest. Another important issue is the strength of any observed genetic correlations. It is entirely likely that even if there is some degree of shared genetic risk between a disorder and related traits, this will be partial and unique genetic effects will also contribute [e.g. as may be the case with ASD and social-communication traits, given somewhat modest genetic correlations (Robinson et al. 2016)].
Given that most psychiatric disorders consist of multiple domains, another challenge is identifying whether relevant population traits show different degrees of shared genetic risk with a given psychiatric disorder, as seems to be the case for schizophrenia genetic risk in relation to psychotic experiences and negative symptoms in the population (Jones et al. 2016). Another difficulty with analyzing continuously distributed psychiatric traits is capturing the full spectrum of a relevant behavior, as most measurement instruments are optimized for detecting difficulties not abilities, thereby resulting in highly zero-inflated and skewed distributions that often violate modeling assumptions. It is unknown whether normalizing such scores through transformations or by regressing out covariates and rank-transforming a variable is an optimal solution and such methods may introduce technical artifacts (Pain et al. 2017). Skewed variables need to be analysed using models that appropriately account for non-normal distributions of data. Ideally, measures that better capture the full variability of behavioral phenotypes are also needed.
We suggest that the assessment of the degree to which a heritable disorder can be considered as an extreme manifestation of population traits should include the following investigations: estimation of the heritability of relevant population traits, estimation of genetic correlation between the disorder and traits, and sensitivity analyses to determine whether any correlation is explained entirely by inclusion of individuals scoring at the extreme end of the trait distribution.
Shared genetic risks across different psychiatric phenotypes
Whilst the degree to which many specific psychiatric disorders share genetic risk with related population traits is yet to be determined, there is much more evidence regarding shared genetic risks across different disorders. Below we consider the strength of the evidence examining this hypothesis, as illustrated in Fig. 1b. See Table 2 for a summary. It is important to note that many studies have examined shared genetic risk between one psychiatric disorder and population traits related to another phenotype, thereby providing additional, albeit indirect, evidence for sharing of genetic risks across psychiatric disorders and continuous traits.
Table 2.
Summary of studies investigating shared genetic risks across disorders
| Disorder | ASD | ADHD | ID | SCZ | BD | MDD | AXD | AN&ED | OCD |
|---|---|---|---|---|---|---|---|---|---|
| ADHD |
Twin rg: 0.54–0.87 (Reiersen et al.
2008; Ronald et al.
2008; Lichtenstein et al.
2010) SNP rg: ns (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017) PRS: mixed evidence (Cross-Disorder Group of the PGC, 2013b; Martin et al. 2014a; Krapohl et al. 2016; Brikell et al. 2017; Jansen et al. 2017) Other: Overlap of CNV loci (Lionel et al. 2011; Williams et al. 2012) |
||||||||
| ID |
Twin rg: 0.04–0.71 (disorder) & −0.27 (traits) (Hoekstra et al.
2009, 2010; Lichtenstein et al.
2010) PRS: positive association (Clarke et al. 2016) SNP rg: 0.21–0.38 between ASD and general cognition (Anttila et al. 2017; Sniekers et al. 2017) Other: Overlap of CNV loci & genes hit by rare loss-of-function mutations (Guilmatre et al. 2009; Pescosolido & Gamsiz, 2013; De Rubeis et al. 2014; Iossifov et al. 2014; Samocha et al. 2014) |
Twin rg: −0.16 to −0.41 (Greven et al.
2011, 2014) PRS: negative association with IQ, positive association with learning difficulties (Martin et al. 2014b; Clarke et al. 2016; Brikell et al. 2017) SNP rg: −0.27 to −0.41 between ADHD and general cognition (Demontis et al. 2017; Sniekers et al. 2017) Other: Overlap of CNV loci (Lionel et al. 2011; Williams et al. 2012) |
|||||||
| SCZ |
SNP rg: 0.16–0.23 (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al.
2015a; Anttila et al.
2017; The ASD Working Group of The PGC, 2017) PRS: association with disorder & ASD traits (Cross-Disorder Group of the PGC, 2013b; Krapohl et al. 2016; St Pourcain et al. 2017) Other: Overlap of CNV loci (Guilmatre et al. 2009) |
SNP rg: 0.22–0.23 (Bulik-Sullivan et al.
2015a; Anttila et al.
2017) PRS: mixed evidence for ADHD disorder (Cross-Disorder Group of the PGC, 2013b; Hamshere et al. 2013); mixed for population traits of ADHD (Krapohl et al. 2016; Jansen et al. 2017; Nivard et al. 2017) Other: Overlap of CNV loci (Lionel et al. 2011; Williams et al. 2012) |
Twin rg: 0.15–0.22 between communication impairment and adolescent psychotic-like experiences (Cederlöf et al.
2014a) SNP rg: −0.38 (performance IQ); −0.07 (verbal IQ); −0.20 (general cognition) (Hubbard et al. 2016; Anttila et al. 2017; Sniekers et al. 2017) PRS: negative association with cognition (McIntosh et al. 2013; Lencz et al. 2014; Hagenaars et al. 2016; Hubbard et al. 2016) Other: Overlap of CNV loci (Guilmatre et al. 2009) |
||||||
| BD |
Twin rg: 0.24 (Song et al.
2015) SNP rg: ns (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017) PRS: positive association with disorder; mixed for ASD traits (Cross-Disorder Group of the PGC, 2013b; Krapohl et al. 2016) |
Twin rg: 0.33 (Song et al.
2015) SNP rg: 0.26–0.71 (Bulik-Sullivan et al. 2015a; van Hulzen et al. 2016; Anttila et al. 2017) PRS: ns (Cross-Disorder Group of the PGC, 2013b; Hamshere et al. 2013; Krapohl et al. 2016; Jansen et al. 2017) |
Twin rg: 0.30 between communication impairment and juvenile mania symptoms (Cederlöf et al.
2014b) SNP rg: ns (general cognition) (Anttila et al. 2017; Sniekers et al. 2017) |
Twin rg: 0.28–0.60; 49–68% shared liability (Cardno et al.
2002; Lichtenstein et al.
2009; Song et al.
2015) SNP rg: 0.68–0.79 (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017) PRS: consistent positive association (disorder) (The International Schizophrenia Consortium, 2009; Cross-Disorder Group of the PGC, 2013b) |
|||||
| MDD |
Twin rg: 0.17–0.19 (Hallett et al.
2010) SNP rg: 0.44 between disorders; ns between ASD and depressive symptoms (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) PRS: ns (Cross-Disorder Group of the PGC, 2013b; Krapohl et al. 2016; Jansen et al. 2017) |
Twin rg: 0.34–0.77 (Cole et al.
2009; Chen et al.
2016; Rydell et al. 2017) SNP rg: 0.32–0.52 between disorders; 0.40–0.45 between ADHD and depressive symptoms (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Demontis et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) PRS: ns (Cross-Disorder Group of the PGC, 2013b; Krapohl et al. 2016; Jansen et al. 2017) |
SNP rg: − 0.27 between depressive symptoms and general cognition; ns for MDD (Anttila et al. 2017; Sniekers et al. 2017) |
Twin rg: 0.72–0.78 (traits) (Zavos et al.
2016) SNP rg: 0.34–0.51 between disorders; 0.30 between depressive symptoms and SCZ (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) PRS: positive association (disorder) (Cross-Disorder Group of the PGC, 2013b); mixed evidence (depressive or internalizing traits) (Jones et al. 2016; Nivard et al. 2017) |
Twin rg: 0.35 (Song et al.
2015) SNP rg: 0.32–0.48 between disorders; 0.28 between BD and depressive symptoms (Cross-Disorder Group of the PGC, 2013a; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) PRS: positive association (disorder) (Cross-Disorder Group of the PGC, 2013b); ns (internalizing traits) (Jansen et al. 2017) |
||||
| AXD |
Twin rg: 0.17–0.19 (Hallett et al.
2010) SNP rg: ns (Anttila et al. 2017) PRS: ns (Krapohl et al. 2016; Jansen et al. 2017) |
Twin rg: 0.45–0.58 (Michelini et al.
2015; Chen et al.
2016) SNP rg: ns (Anttila et al. 2017) PRS: ns (Krapohl et al. 2016) |
SNP rg: ns (general cognition) (Anttila et al. 2017; Sniekers et al. 2017) |
SNP rg: ns (Otowa et al.
2016; Anttila et al.
2017) PRS: ns (across disorders); replicated evidence for anxiety symptoms, though 1 study found no effect (Jones et al. 2016; Krapohl et al. 2016; Nivard et al. 2017) |
Twin rg: 0.23 (Song et al.
2015) SNP rg: ns (Otowa et al. 2016; Anttila et al. 2017) PRS: Mixed (disorder) (Otowa et al. 2016); ns (traits) (Krapohl et al. 2016; Jansen et al. 2017) |
Twin rg: 0.70–1.00 (Roy et al.
1995; Thapar & McGuffin, 1997; Kendler et al.
2007; Mosing et al.
2009; Demirkan et al.
2011) SNP rg: 0.68–0.80 between disorders; 0.82 between AXD and depressive symptoms (Otowa et al. 2016; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017) PRS: positive association (disorder) (Otowa et al. 2016); mixed (traits) (Krapohl et al. 2016; Jansen et al. 2017) |
|||
| AN&ED | SNP rg: ns (Bulik-Sullivan et al. 2015a; Anttila et al. 2017) | SNP rg: ns (Bulik-Sullivan et al. 2015a; Anttila et al. 2017) | SNP rg: ns (general cognition) (Anttila et al. 2017; Sniekers et al. 2017) | SNP rg: 0.19–0.22 (Bulik-Sullivan et al. 2015a; Anttila et al. 2017) | SNP rg: ns (Bulik-Sullivan et al. 2015a; Anttila et al. 2017) |
Twin rg: 0.70 (Slane et al.
2011) SNP rg: 0.13 between disorders; ns between AN & depressive symptoms (Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC, et al. 2017) |
SNP rg: ns (Anttila et al. 2017) | ||
| OCD | SNP rg: ns (Anttila et al. 2017) |
SNP rg: ns (Anttila et al.
2017) Other: Heritable latent factor underlying ADHD, OCD, and tics (Pinto et al. 2016) |
SNP rg: ns (general cognition) (Anttila et al. 2017) | SNP rg: 0.33 (Anttila et al. 2017) | SNP rg: 0.31 (Anttila et al. 2017) |
Twin rg: 0.71–0.86 (traits) (Bolhuis et al.
2014) SNP rg: 0.23 between disorders; ns between OCD & depressive symptoms (Anttila et al. 2017) |
SNP rg: ns (Anttila et al.
2017) Other: Common genetic factor underlying OCD and anxiety symptoms (López-Solà et al. 2016) |
Twin rg: 0.52 (Cederlöf et al.
2015) SNP rg: 0.52 (Anttila et al. 2017) |
|
| TS |
Twin rg: 0.60 (Lichtenstein et al.
2010) SNP rg: ns (Anttila et al. 2017) |
Twin rg: 1.00 (Lichtenstein et al.
2010) SNP rg: ns (Anttila et al. 2017) Other: Heritable latent factor underlying ADHD, OCD, and tics (Pinto et al. 2016) |
SNP rg: ns (general cognition)(Anttila et al. 2017) | SNP rg: ns (Anttila et al. 2017) | SNP rg: ns (Anttila et al. 2017) | SNP rg: 0.21 between disorders; ns between TS & depressive symptoms (Anttila et al. 2017) | SNP rg: ns (Anttila et al. 2017) | SNP rg: ns (Anttila et al. 2017) |
SNP rg: 0.41–0.43 (Davis et al.
2013; Anttila et al.
2017) Other: Heritable latent factor underlying ADHD, OCD, and tics (Pinto et al. 2016) |
ADHD, attention-deficit hyperactivity disorder; AN&ED, anorexia nervosa and other eating disorders; ASD, autism spectrum disorder; AXD, anxiety disorders; BD, bipolar disorder; ID, intellectual disability; MDD, major depressive disorder; OCD, obsessive-compulsive disorder; SCZ, schizophrenia; TS, Tourette's syndrome and other tic disorders; SNP, single nucleotide polymorphism; CNV, copy number variant; PRS, polygenic risk score analysis; ns, non-significant estimates based on published studies.
Twin rg is the correlation between the additive genetic variance components from twin studies. Note that the ‘twin rg’ in Lichtenstein et al. (2009) & Song et al. (2015) are estimated from family studies but with a similar approach as in twin studies. SNP rg: is the estimated genetic correlation from genome-wide association studies using LDSC (linkage disequilibrium score correlation) or GCTA (genome-wide complex trait analysis). Only results estimated to be nominally significantly different from zero (p < 0.05) are presented. For a more detailed explanation of the methods, please refer to the caption of Table 1. The GREML-GCTA method (genetic relatedness estimation through maximum likelihood using the GCTA software) (Yang et al. 2011; Lee et al. 2012) is conceptually similar to LDSC; it is used to estimate the contribution of all SNPs from genome-wide data (SNP-heritability) and can be applied to examine shared genetic risks between disorders and population traits to give an estimate of genetic correlation.
Disorders with early onset
Twin studies of neuropsychiatric diagnoses and childhood traits consistently show significant genetic correlations. Associations have been seen between ADHD inattentive symptoms and difficulties in reading and mathematics (Greven et al. 2011, 2014; Wadsworth et al. 2015), categorically and continuously defined ADHD and ASD (Reiersen et al. 2008; Ronald et al. 2008, 2014a; Lichtenstein et al. 2010; Taylor et al. 2012), and ASD with learning difficulties and tics (which are associated with Tourette's syndrome) (Lichtenstein et al. 2010). However, two other twin studies of ASD and intellectual ability have reported low genetic correlations, although this might have been related to measurement differences (Hoekstra et al. 2009, 2010).
Analyses of common genetic variants so far have not confirmed the genetic correlation between ADHD and ASD observed in twin studies (Cross-Disorder Group of the PGC, 2013a, b; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Jansen et al. 2017). Clinical ADHD shares some genetic risk with social-communication traits (Martin et al. 2014a) and other neurodevelopmental and externalizing traits that make up a general factor of childhood psychopathology (Brikell et al. 2017). Clinical ADHD shares genetic risk with lower cognitive abilities in children and adults in the general population (Martin et al. 2014b; Clarke et al. 2016; Stergiakouli et al. 2016; Anttila et al. 2017; Demontis et al. 2017; Riglin et al. 2017; Sniekers et al. 2017). In ASD, there is a positive genetic correlation with common variants associated with cognitive ability, suggesting that these variants operate differently to common risk variants for other psychiatric phenotypes and to rare variants in the context of ASD (Clarke et al. 2016; Robinson et al. 2016; Anttila et al. 2017; Sniekers et al. 2017; Weiner et al. 2017). With regard to rare variants, studies of CNVs have implicated the same genomic regions in multiple disorders, including ASD, ID, and ADHD (Guilmatre et al. 2009; Sebat et al. 2009; Pinto et al. 2010; Williams et al. 2010, 2012; Cooper et al. 2011; Lionel et al. 2011; Sanders et al. 2011; Pescosolido & Gamsiz, 2013). Recent large exome sequencing studies have identified the first robust rare de novo protein-truncating mutations (variants which disrupt protein formation and are likely highly deleterious) associated with ASD, with many of the same genes found to harbor de novo mutations linked to ID (De Rubeis et al. 2014; Iossifov et al. 2014; Samocha et al. 2014; The Deciphering Developmental Disorders Study, 2014).
Twin and molecular studies have yielded some consistent findings, but larger genetic studies are needed to further understand the degree and source of shared genetic risks in these early-onset disorders. The association between ASD and ID is particularly complex, with shared risk for these phenotypes seen at the level of rare risk variants but a positive association seen for common variants; indeed these mixed genetic results may partly explain the low genetic correlations between these phenotypes in twin studies (Hoekstra et al. 2009, 2010).
Disorders with onset in adolescence and adulthood
Twin studies have found substantial evidence of genetic correlations across schizophrenia and BD (Cardno et al. 2002; Lichtenstein et al. 2009), BD and MDD (Song et al. 2015), anxiety disorder subtypes (Mosing et al. 2009), specific anxiety disorders and MDD (Roy et al. 1995; Kendler et al. 2007; Mosing et al. 2009), traits of anxiety and depressive symptoms (Thapar & McGuffin, 1997), MDD and psychotic experiences in adolescence (Zavos et al. 2016), depressive symptoms and disordered eating scores (Slane et al. 2011), OCD and MDD (Bolhuis et al. 2014), and OCD with anxiety-related behaviors and anorexia nervosa (AN) (Cederlöf et al. 2015; López-Solà et al. 2016).
GWAS of adult psychiatric disorders have confirmed that common genetic variants associated with one disorder also play an important role in other disorders. Recent analyses using multiple genome-wide methods report shared genetic risks across schizophrenia, BD, MDD, and OCD, across schizophrenia, AN and OCD, and between MDD with anxiety disorders and AN (Cross-Disorder Group of the PGC, 2013a, b; Bulik-Sullivan et al. 2015a; Anttila et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017). Shared genetic risks are seen across different anxiety disorders (generalized anxiety disorder, panic disorder and phobias) and with MDD, though not with BD or schizophrenia (Otowa et al. 2016). General population studies of schizophrenia PRS report associations with anxiety symptoms, with mixed evidence for association with depressive symptoms between ages 7 and 15 (Jones et al. 2016; Jansen et al. 2017; Nivard et al. 2017). MDD PRS were also associated with anxiety symptoms in an elderly population sample (Demirkan et al. 2011). Thus, there is evidence that a considerable degree of genetic influences are shared across multiple phenotypes, assessed categorically or continuously.
Shared genetic risks across child- and adult-onset disorders
Childhood-onset disorders and disorders with an onset typically in adolescence or adulthood also share genetic risks. For example, twin studies find that early-onset-neurodevelopmental disorders share genetic risk with anxiety (Hallett et al. 2010; Michelini et al. 2015; Chen et al. 2016), MDD (Cole et al. 2009; Lundström et al. 2011), affective problems (Rydell et al. 2017), and OCD (Pinto et al. 2016). In a study of specific intellectual domains, problems with communication shared a modest degree of genetic risk with adolescent hallucinations and mania (Cederlöf et al. 2014b). Molecular genetic studies have reported genetic correlations between both ADHD and ASD with MDD, schizophrenia and BD (Cross-Disorder Group of the PGC, 2013a, b; Bulik-Sullivan et al. 2015a; van Hulzen et al. 2016; Anttila et al. 2017; Demontis et al. 2017; Major Depressive Disorder Working Group of the PGC et al. 2017; The ASD Working Group of The PGC, 2017). Tourette's syndrome shares genetic risks with OCD and MDD (Davis et al. 2013; Anttila et al. 2017). Genetic risk for schizophrenia is associated with numerous traits assessed across ages 3–15 years, including ADHD, aggression, irritability, language, and social abilities (Jansen et al. 2017; Nivard et al. 2017; Riglin et al. 2017). BD and MDD PRS were not found to be associated with early life (age 3–10 years) internalizing and externalizing problems (Jansen et al. 2017).
CNV loci implicated in children with ADHD, ASD, and ID have also been associated with schizophrenia (The International Schizophrenia Consortium, 2008; Guilmatre et al. 2009; Sebat et al. 2009; Williams et al. 2010, 2012; Lionel et al. 2011; Pescosolido & Gamsiz, 2013). Schizophrenia shares genetic risks with cognitive measures throughout the lifespan (McIntosh et al. 2013; Lencz et al. 2014; Hagenaars et al. 2016; Hubbard et al. 2016; Krapohl et al. 2016). General cognitive ability shows negative genetic correlations with schizophrenia and depressive symptoms, though not with BD, anxiety disorder, MDD, OCD or AN (Anttila et al. 2017; Sniekers et al. 2017). Genetic correlations across several psychiatric disorders and personality measures have also been reported (Lo et al. 2016; Anttila et al. 2017). Psychiatric phenotypes also more broadly share genetic contributions with other human complex traits, for example genetic risk for ADHD is shared with behavioral traits (e.g. smoking), brain- (e.g. migraine) and non-brain-based diseases (e.g. type-2-diabetes) and traits (e.g. body mass index) (Anttila et al. 2017; Demontis et al. 2017). A wider review is beyond the scope of this paper.
In summary, studies indicate that a considerable degree of genetic influences on particular disorders are shared with at least one other disorder, regardless of whether one focuses on childhood- or adulthood-onset conditions. It has been hypothesized that a single ‘general genetic factor’ underlies multiple psychiatric phenotypes (Lahey et al. 2012; Caspi et al. 2014). Two twin studies supported this model, with a latent genetic factor accounting for 31% of variance in neurodevelopmental symptoms in a population-based sample (Pettersson et al. 2013) and 10–36% of disorder liability across multiple clinical psychiatric diagnoses (Pettersson et al. 2015). A recent study further confirmed that common genetic risk variants contribute to this general factor, with an estimated SNP-heritability of approximately 0.38 (Neumann et al. 2016). As illustrated in Fig. 1b, the situation is likely to be even more complex, with not only a general genetic factor predisposing to multiple phenotypes but also disorder-specific genetic factors as well as genetic factors relevant only to specific pairs of disorders. Similarly, environmental factors could also be shared or unique and more complex effects, such as gene-environment interactions, could also exist.
Although several of the pairs of psychiatric disorders assessed using GWAS data do not show significant genetic correlations, some of the studies were relatively small and are likely to be underpowered. Notably, genetic correlations are present regardless of whether psychiatric phenotypes are conceptualized continuously or dichotomously, thus providing additional, albeit indirect, support for shared genetic risk across these disorders and related traits.
Interpreting the meaning of genetic correlations
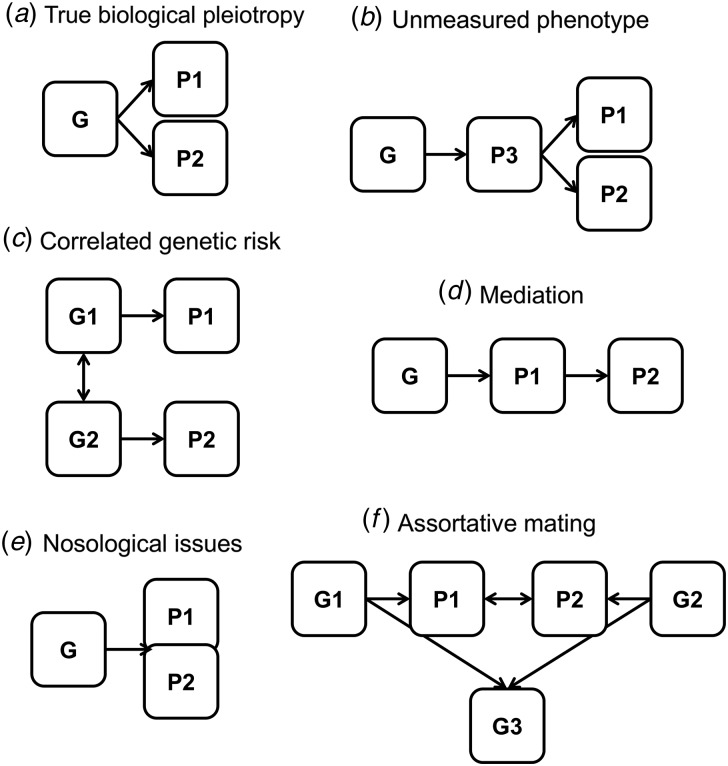
The interpretation of what genetic correlations mean is complex, with a number of possibilities, some of which are not mutually exclusive. One possibility (Fig. 2a) is that of true biological pleiotropy, where the same risk variants (or variants within the same gene) are directly, causally impacting on multiple phenotypes, albeit possibly through separate biological pathways. Alternatively, the same genetic risk variants could be causally affecting a third, unmeasured phenotype which lies on the pathway between risk variants and measured phenotypes (Fig. 2b). A third possibility (Fig. 2c) is that observed genetic correlations are actually capturing different risk variants that are highly correlated but are acting through different mechanisms. For example, even though the same CNV loci have been implicated in multiple disorders (Guilmatre et al. 2009; Lionel et al. 2011; Williams et al. 2012; Pescosolido & Gamsiz, 2013), different variants within these large loci might be associated with different phenotypes. Given that such large, rare variants are also shared by monozygotic twins, this could also influence estimates of genetic correlations based on twin studies. A fourth possibility (Fig. 2d) is that one phenotype mediates the association between genetic risk and a second phenotype and there is no direct causal relationship between the risk variant and this second phenotype. For example, it has been proposed that the genetic correlation between MDD and depressive symptoms in the population could be accounted for by shared genetic risk with low levels of subjective well-being (Direk et al. 2016).
Fig. 2.
Potential interpretations of genetic correlation across phenotypes: (a) true biological pleiotropy, where the same genetic risk variant is causally associated with two phenotypes; (b) unmeasured phenotype, where a third phenotype is on the causal pathway between genetic risk and the outcome phenotypes of interest; (c) correlated genetic risk, where different genetic risk variants that are highly correlated are causally associated with each phenotype; (d) mediation, where a genetic risk variant only acts on one of the phenotypes, which in turn influences a second phenotype; (e) Nosological issues, which blur the distinction between phenotypes, for example comorbidity, ascertainment bias, heterogeneity or diagnostic misclassification; (f) assortative mating, where individuals with the two phenotypes of interest are more likely to mate than expected at random, thereby leading to clustering of genetic risk for both phenotypes in the offspring. N.B. The shapes are not indicative of whether a variable is latent or measured.
Several nosological issues (Fig. 2e) may also explain genetic correlations to an extent. Comorbidity across disorders (e.g. anxiety and MDD) is frequently observed and certain symptom domains show similarities [e.g. manic (BD) or hyperactive (ADHD) symptoms]. Specific symptoms also overlap directly across disorders (e.g. concentration problems in ADHD, MDD or anxiety) and such overlap may largely account for comorbidity [e.g. anxiety and MDD (Cramer et al. 2010)]. Such phenotypic overlap could inflate genetic correlation estimates. Within-disorder heterogeneity could also induce an overall correlation across two phenotypes, when only a sub-group of individuals with one disorder (who may have a specific clinical profile) show genetic correlation with individuals with another phenotype. Another possibility is that of diagnostic misclassification or changes in meeting diagnostic criteria over time (e.g. individuals who are diagnosed with MDD but later develop manic features, leading to a diagnosis of BD). Given the similar diagnostic features across different disorders, accurate diagnosis is difficult. Fortunately, diagnostic changes over time can be taken into consideration using epidemiological family study designs (Song et al. 2015). Simulations show that a 10% rate in misclassification can inflate estimates of genetic correlation (Wray et al. 2012). However, very high degrees of such misclassification would be required to fully account for the observed genetic correlations across psychiatric phenotypes (Anttila et al. 2017). Such issues related to phenotype definition remain to be resolved as the underlying biology of psychiatric disorders is better understood. For now, careful ascertainment and better measurement of frequently co-occurring disorder-level and sub-threshold phenotypes is required.
Another possibility for interpreting observed genetic correlations between psychiatric disorders is that they arise through assortative mating (Fig. 2f). There are substantial effects of assortative mating both within and across multiple psychiatric disorders (Nordsletten et al. 2016). Such assortative mating across disorders would likely increase genetic correlation estimates (Coop & Pickrell, 2016). Finally, there are technical and methodological artifacts (e.g. overlapping or related individuals) that may induce spurious genetic correlations in molecular genetic studies, which need to be ruled out.
More research is needed to determine the extent to which comorbidity, ascertainment bias, heterogeneity, diagnostic misclassification, and assortative mating inflate genetic correlations across psychiatric disorders and how much of these estimates are due to true pleiotropy. Even so, the possible biological interpretations of genetic correlations described above are hard to distinguish using the methods described in this review, as genetic correlations do not pinpoint the source of shared genetic risks. Some clues might be gained by partitioning heritability based on SNP functional category, position or frequency (Finucane et al. 2015), to try to better identify the source of the genetic correlations. Large-scale GWAS meta-analyses and sequencing studies are needed to find robust risk variants associated with multiple disorders.
After identifying specific genetic risk variants that correlate across disorders and considering the above possibilities, well-phenotyped samples and new methods will be needed to interpret the meaning of genetic correlations. Several newly developed methods have the potential to help with interpretation. The method ‘pairwise-GWAS’ aims to determine whether the effect sizes of variants associated with one trait are correlated with effect sizes of those variants for another trait and vice versa (Pickrell et al. 2016). Another method, BUHMBOX, aims to statistically differentiate between situations where there is sub-group heterogeneity (i.e. phenotype misclassification, different biological subtypes of a disorder, ascertainment bias or mediation) or whether there is true pleiotropy (Han et al. 2016).
Implications for research and clinical practice
Despite moderate to high degrees of genetic correlation between some pairs of phenotypes, unique genetic factors are also likely to be important, as illustrated in Fig. 1b. This unique genetic risk is associated with important clinical distinctions that exist between disorders and also between disorders and continuous traits. For example, certain medications are effective for one disorder (e.g. stimulants for ADHD), but do not impact the symptoms of other disorders (Thapar et al. 2017). Also, in the absence of severe impairment resulting from symptoms, the cost-benefit ratio of treatment needs to be considered. Since most genetic correlations are below 1, more insights into the meaning of these correlations are required before clinical practice can be advanced.
The assumption that there is some true sharing of genetic risks has already led to insights into the genetic architecture and biology of psychiatric disorders through combining phenotypes in joint analyses to boost statistical power. For example, a joint GWAS analysis of five psychiatric disorders led to a more powerful approach for identifying genetic variants associated with psychiatric disorders (Cross-Disorder Group of the PGC, 2013b). Similarly, using the results of a GWAS of multiple psychiatric disorders can substantially increase the accuracy of PRS analyses (Maier et al. 2015). Also, a literature review of genetic sequencing studies of several childhood-onset neurodevelopmental disorders has shown the power of pooling information on multiple phenotypes to identify more robust genes implicated in neurodevelopmental disorders (Gonzalez-Mantilla et al. 2016). Gene discovery studies meta-analyzing GWAS of a clinical disorder with GWAS of population traits can benefit from substantially increased power to detect common variants, as can be seen for example for MDD and ADHD (Direk et al. 2016; Demontis et al. 2017). Understanding the nature and degree of shared genetic risks across psychiatric phenotypes will be essential to most effectively using this observation for future research into the genetic architecture of these disorders.
One important limitation of existing molecular genetic studies is that for many psychiatric disorders, sample sizes are still relatively small and analyses are limited in statistical power. PRS studies, in particular, tend to find low effect sizes. As larger and more reliable genetic samples become available in the future, it will be possible to better determine the degree and source of shared genetic risks across psychiatric phenotypes.
Conclusion
Emerging evidence from twin and molecular genetic studies suggests that some genetic risk is shared between diagnosed disorders and variation in psychiatric traits in the population for certain disorders (e.g. ADHD) and across different psychiatric diagnoses (e.g. schizophrenia and BD). More research is needed to investigate the degree of genetic correlation across disorders and traits for other psychiatric phenotypes (e.g. anxiety or BD) and across pairs of different disorders (e.g. anorexia and OCD). Future research should then aim to identify specific genetic loci that are driving any genetic correlations and determine the nature of such correlations. However, recent insights into the genetic architectures of psychiatric disorders are already pointing towards new avenues for further research into the biology of these complex disorders.
Acknowledgements
This work was supported by the Wellcome Trust (grant 106047). Many thanks to Dr Elise Robinson and Prof Anita Thapar for helpful comments on an early draft of the manuscript. Prof Lichtenstein is funded by the Swedish Research Council for Health, Working Life and Welfare.
Declaration of interest
Drs Martin and Taylor report no biomedical financial interests or potential conflicts of interest. Dr Lichtenstein has served as a speaker for Medice.
References
- Anttila V, Bulik-Sullivan B, Finucane HK, Bras J, Duncan L, Escott-Price V et al. (2017) Analysis of shared heritability in common disorders of the brain. bioRxiv, Cold Spring Harbor Labs Journals. doi: 10.1101/048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli TB, Bacanu S-A, Webb BT, Walsh D, O'Neill FA, Fanous AH et al. (2014) Molecular validation of the schizophrenia spectrum. Schizophrenia Bulletin, Oxford University Press 40, 60–65. doi: 10.1093/schbul/sbt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis K, McAdams TA, Monzani B, Gregory AM, Mataix-Cols D, Stringaris A et al. (2014) Aetiological overlap between obsessive-compulsive and depressive symptoms: a longitudinal twin study in adolescents and adults. Psychological Medicine 44, 1439–1449. doi: 10.1017/S0033291713001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bralten J, van Hulzen KJ, Martens MB, Galesloot TE, Arias Vasquez A, Kiemeney LA et al. (2017) Autism spectrum disorders and autistic traits share genetics and biology. Molecular Psychiatry, Nature Publishing Group. doi: 10.1038/mp.2017.98. [DOI] [PubMed] [Google Scholar]
- Brikell I, Larsson H, Lu Y, Pettersson E, Chen Q, Kuja-Halkola R et al. (2017) The contribution of common genetic risk variants for ADHD to a general factor of childhood psychopathology. bioRxiv, Cold Spring Harbor Laboratory. doi: 10.1101/193573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R et al. (2015a) An atlas of genetic correlations across human diseases and traits. Nature Genetics, Nature Research 47, 1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N et al. (2015b) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics, Nature Research 47, 291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Rijsdijk FV, Sham PC, Murray RM and McGuffin P (2002) A twin study of genetic relationships between psychotic symptoms. American Journal of Psychiatry, American Psychiatric Publishing 159, 539–545. doi: 10.1176/appi.ajp.159.4.539. [DOI] [PubMed] [Google Scholar]
- Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S et al. (2014) The p factor: one general psychopathology factor in the structure of psychiatric disorders? Clinical Psychological Science 2, 119–137. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederlöf M, Ohlsson Gotby A, Larsson H, Serlachius E, Boman M, Långström N et al. (2014a) Klinefelter syndrome and risk of psychosis, autism and ADHD. Journal of Psychiatric Research 48, 128–130. [DOI] [PubMed] [Google Scholar]
- Cederlöf M, Ostberg P, Pettersson E, Anckarsäter H, Gumpert C, Lundström S et al. (2014b) Language and mathematical problems as precursors of psychotic-like experiences and juvenile mania symptoms. Psychological Medicine 44, 1293–1302. doi: 10.1017/S0033291713002018. [DOI] [PubMed] [Google Scholar]
- Cederlöf M, Thornton LM, Baker J, Lichtenstein P, Larsson H, Rück C et al. (2015) Etiological overlap between obsessive-compulsive disorder and anorexia nervosa: a longitudinal cohort, multigenerational family and twin study. World Psychiatry 14, 333–338. doi: 10.1002/wps.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-J, Ji C-Y, Wang S-S, Lichtenstein P, Larsson H and Chang Z (2016) Genetic and environmental influences on the relationship between ADHD symptoms and internalizing problems: a Chinese twin study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 171, 931–937. doi: 10.1002/ajmg.b.32411. [DOI] [PubMed] [Google Scholar]
- Clarke T-K, Lupton MK, Fernandez-Pujals AM, Starr J, Davies G, Cox S et al. (2016) Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Molecular Psychiatry, Nature Publishing Group 21, 419–425. doi: 10.1038/mp.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J, Ball HA, Martin NC, Scourfield J and Mcguffin P (2009) Genetic overlap between measures of hyperactivity/inattention and mood in children and adolescents. Journal of the American Academy of Child & Adolescent Psychiatry 48, 1094–1101. doi: 10.1097/CHI.0b013e3181b7666e. [DOI] [PubMed] [Google Scholar]
- Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E et al. (2015) Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry, American Medical Association 72, 415. doi: 10.1001/jamapsychiatry.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G and Pickrell J (2016) What is genetic correlation? Available at https://joepickrell.wordpress.com/2016/04/19/what-is-genetic-correlation/ (Accessed 19 January 2017).
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C et al. (2011) A copy number variation morbidity map of developmental delay. Nature Genetics, Nature Research 43, 838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer AOJ, Waldorp LJ, van der Maas HLJ and Borsboom D (2010) Comorbidity: a network perspective. Behavioral and Brain Sciences, Cambridge University Press 33, 137–150. doi: 10.1017/S0140525X09991567. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the PGC (2013a) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the PGC (2013b) Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM et al. (2013). Partitioning the heritability of tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genetics, Edited by Keller M. C.. Public Library of Science 9, e1003864. doi: 10.1371/journal.pgen.1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFries JC and Fulker DW (1985) Multiple regression analysis of twin data. Behavior Genetics 15, 467–473. [DOI] [PubMed] [Google Scholar]
- Demirkan A, Penninx BWJH, Hek K, Wray NR, Amin N, Aulchenko YS et al. (2011) Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Molecular Psychiatry, Nature Publishing Group 16, 773–783. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E et al. (2017) Discovery of the first genome-wide significant risk loci for ADHD. bioRxiv. doi: 10.1101/145581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber A, Zilhão NR, Fedko IO, Hottenga J-J, Pool R, Smit DJA et al. (2016) Obsessive–compulsive symptoms in a large population-based twin-family sample are predicted by clinically based polygenic scores and by genome-wide SNPs. Translational Psychiatry, Nature Publishing Group 6, e731. doi: 10.1038/tp.2015.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Ercument Cicek A et al. (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direk N, Williams S, Smith JA, Ripke S, Air T, Amare AT et al. (2016) An analysis of two genome-wide association meta-analyses identifies a new locus for broad depression phenotype. Biological Psychiatry, Princeton University Press, Princeton, NJ 110, 9692–9697. doi: 10.1016/j.biopsych.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley TC (1997) Depressive symptoms in children and adolescents: etiological links between normality and abnormality: a research note. Journal of Child Psychology and Psychiatry 38, 861–865. [DOI] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R et al. (2015) Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature Genetics, Nature Research 47, 1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH et al. (2011) Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genetics 7, e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith H and Lemery KS (2000) Linking temperamental fearfulness and anxiety symptoms: a behavior–genetic perspective. Biological Psychiatry 48, 1199–1209. doi: 10.1016/S0006-3223(00)01003-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH and Martin CL (2016) A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry 73, 275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greven CU, Harlaar N, Dale PS and Plomin R (2011) Genetic overlap between ADHD symptoms and reading is largely driven by inattentiveness rather than hyperactivity-impulsivity. Journal of the Canadian Academy of Child and Adolescent Psychiatry 20, 6. [PMC free article] [PubMed] [Google Scholar]
- Greven CU, Kovas Y, Willcutt EG, Petrill SA and Plomin R (2014) Evidence for shared genetic risk between ADHD symptoms and reduced mathematics ability: a twin study. Journal of Child Psychology and Psychiatry 55, 39–48. doi: 10.1111/jcpp.12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greven CU, Merwood A, van der Meer JMJ, Haworth CMA, Rommelse N and Buitelaar JK (2016) The opposite end of the attention deficit hyperactivity disorder continuum: genetic and environmental aetiologies of extremely low ADHD traits. Journal of Child Psychology and Psychiatry 57, 523–531. doi: 10.1111/jcpp.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen-Blokhuis MM, Middeldorp CM, Kan K-J, Abdellaoui A, van Beijsterveldt CEM, Ehli EA et al. (2014) Attention deficit hyperactivity disorder polygenic risk scores predict attention problems in a population-based sample of children. Journal of the American Academy of Child & Adolescent Psychiatry 53, 1123–1129. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca A-LL, Legallic S, Goldenberg A, Drouin-Garraud V et al. (2009) Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Archives of General Psychiatry 66, 947. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DCM, Ritchie SJ et al. (2016) Shared genetic aetiology between cognitive functions and physical and mental health in UK biobank (N = 112 151) and 24 GWAS consortia. Molecular Psychiatry, Nature Publishing Group 21, 1624–1632. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett V, Ronald A, Rijsdijk F and Happé F (2010) Association of autistic-like and internalizing traits during childhood: a longitudinal twin study. American Journal of Psychiatry, American Psychiatric Association 167, 809–817. doi: 10.1176/appi.ajp.2009.09070990. [DOI] [PubMed] [Google Scholar]
- Hamshere ML, Stergiakouli E, Langley K, Martin J, Holmans P, Kent L et al. (2013) A shared polygenic contribution between childhood ADHD and adult schizophrenia. The British Journal of Psychiatry 203, 107–111. doi: 10.1192/bjp.bp.112.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Pouget JG, Slowikowski K, Stahl E, Lee CH, Diogo D et al. (2016) A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases. Nature Genetics, Nature Research 48, 803–810. doi: 10.1038/ng.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RA, Happé F, Baron-Cohen S and Ronald A (2009) Association between extreme autistic traits and intellectual disability: insights from a general population twin study. The British Journal of Psychiatry 195, 531–536. [DOI] [PubMed] [Google Scholar]
- Hoekstra RA, Happé F, Baron-Cohen S, Ronald A, Baron-Cohen S and Ronald A (2010) Limited genetic covariance between autistic traits and intelligence: findings from a longitudinal twin study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, Wiley Subscription Services, Inc., A Wiley Company 153, 994–1007. doi: 10.1002/ajmg.b.31066. [DOI] [PubMed] [Google Scholar]
- Hubbard L, Tansey KE, Rai D, Jones P, Ripke S, Chambert KD et al. (2016) Evidence of common genetic overlap between schizophrenia and cognition. Schizophrenia Bulletin, Oxford University Press 42, 832–842. doi: 10.1093/schbul/sbv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen PR, Polderman TJC, Bolhuis K, van der Ende J, Jaddoe VWV, Verhulst FC et al. (2017) Polygenic scores for schizophrenia and educational attainment are associated with behavioural problems in early childhood in the general population. Journal of Child Psychology and Psychiatry. doi: 10.1111/jcpp.12759. [DOI] [PubMed] [Google Scholar]
- Jones HJ, Stergiakouli E, Tansey KE, Hubbard L, Heron J, Cannon M et al. (2016) Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry 73, 221–228. doi: 10.1001/jamapsychiatry.2015.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall KM, Rees E, Escott-Price V, Einon M, Thomas R, Hewitt J et al. (2016) Cognitive performance among carriers of pathogenic copy number variants: analysis of 152000 UK Biobank subjects. Biological Psychiatry 82, 103–110. doi: 10.1016/j.biopsych.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Gardner CO, Gatz M and Pedersen NL (2007) The sources of co-morbidity between major depression and generalized anxiety disorder in a Swedish national twin sample. Psychological Medicine, Cambridge University Press 37, 453. doi: 10.1017/S0033291706009135. [DOI] [PubMed] [Google Scholar]
- Krapohl E, Euesden J, Zabaneh D, Pingault J-B, Rimfeld K, von Stumm S et al. (2016) Phenome-wide analysis of genome-wide polygenic scores. Molecular Psychiatry, Nature Publishing Group 21, 1188–1193. doi: 10.1038/mp.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey BB, Applegate B, Hakes JK, Zald DH, Hariri AR and Rathouz PJ (2012) Is there a general factor of prevalent psychopathology during adulthood? Journal of Abnormal Psychology, NIH Public Access 121, 971–977. doi: 10.1037/a0028355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson H, Anckarsater H, Råstam M, Chang Z and Lichtenstein P (2011) Childhood attention-deficit hyperactivity disorder as an extreme of a continuous trait: a quantitative genetic study of 8500 twin pairs. Journal of Child Psychology and Psychiatry 53, 73–80. [DOI] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM and Wray NR (2012) Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics, Oxford University Press 28, 2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Knowles E, Davies G, Guha S, Liewald DC, Starr JM et al. (2014) Molecular genetic evidence for overlap between general cognitive ability and risk for schizophrenia: a report from the cognitive genomics consorTium (COGENT). Molecular Psychiatry, Macmillan Publishers Limited 19, 168–174. doi: 10.1038/mp.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy F, Hay DA, McStephen M, Wood C and Waldman I (1997) Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. Journal of the American Academy of Child and Adolescent Psychiatry 36, 737–744. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Carlström E, Råstam M, Gillberg C and Anckarsäter H (2010) The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. American Journal of Psychiatry 167, 1357–1363. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF et al. (2009) Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet 373, 234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel AC, Crosbie J, Barbosa N, Goodale T, Thiruvahindrapuram B, Rickaby J et al. (2011) Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Science Translational Medicine 3, 95ra75. [DOI] [PubMed] [Google Scholar]
- Lo M-T, Hinds DA, Tung JY, Franz C, Fan C-C, Wang Y et al. (2016) Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nature Genetics, Nature Research 49, 152–156. doi: 10.1038/ng.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan JAR, Petrill SA, Hart SA, Schatschneider C, Thompson LA, Deater-Deckard K et al. (2012) Heritability across the distribution: an application of quantile regression. Behavior Genetics, NIH Public Access 42, 256–267. doi: 10.1007/s10519-011-9497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Solà C, Fontenelle LF, Bui M, Hopper JL, Pantelis C, Yücel M et al. (2016) Aetiological overlap between obsessive-compulsive related and anxiety disorder symptoms: multivariate twin study. The British Journal of Psychiatry 208, 26–33. [DOI] [PubMed] [Google Scholar]
- Lundström S, Chang Z, Kerekes N, Gumpert CH, Råstam M, Gillberg C et al. (2011) Autistic-like traits and their association with mental health problems in two nationwide twin cohorts of children and adults. Psychological Medicine, Cambridge University Press 41, 2423–2433. doi: 10.1017/S0033291711000377. [DOI] [PubMed] [Google Scholar]
- Lundström S, Chang Z, Råstam M, Gillberg C, Larsson H, Anckarsäter H et al. (2012) Autism spectrum disorders and autisticlike traits: similar etiology in the extreme end and the normal variation. Archives of General Psychiatry, American Medical Association 69, 46–52. doi: 10.1001/archgenpsychiatry.2011.144. [DOI] [PubMed] [Google Scholar]
- Maier R, Moser G, Chen G-B, Ripke S, Cross-Disorder Working Group of the Psychiatric Genomics Consortium, Coryell W et al. (2015). Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. American Journal of Human Genetics, Elsevier 96, 283–294. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the PGC, Wray NR and Sullivan PF (2017). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. bioRxiv, Cold Spring Harbor Laboratory. doi: 10.1101/167577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik K, Mägi R, Macé A, Cole B, Guyatt AL, Shihab HA et al. (2015) Copy number variations and cognitive phenotypes in unselected populations. JAMA, American Medical Association 313, 2044. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hamshere ML, Stergiakouli E, O'Donovan MC and Thapar A (2014a) Genetic risk for attention-deficit/hyperactivity disorder contributes to neurodevelopmental traits in the general population. Biological Psychiatry 76, 664–671. doi: 10.1016/j.biopsych.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hamshere ML, Stergiakouli E, O'Donovan MC and Thapar A (2014b) Neurocognitive abilities in the general population and composite genetic risk scores for attention-deficit hyperactivity disorder. Journal of Child Psychology and Psychiatry 56, 648–656. doi: 10.1111/jcpp.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh A, Gow A and Luciano M (2013) Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biological Psychiatry, Elsevier 73, 938–943. doi: 10.1016/j.biopsych.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Meier SM, Agerbo E, Maier R, Pedersen CB, Lang M, Grove J et al. (2016) High loading of polygenic risk in cases with chronic schizophrenia. Molecular Psychiatry, Nature Publishing Group 21, 969–974. doi: 10.1038/mp.2015.130. [DOI] [PubMed] [Google Scholar]
- Michelini G, Eley TC, Gregory AM and McAdams TA (2015) Aetiological overlap between anxiety and attention deficit hyperactivity symptom dimensions in adolescence. Journal of Child Psychology and Psychiatry 56, 423–431. doi: 10.1111/jcpp.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp CM, Hammerschlag AR, Ouwens KG, Groen-Blokhuis MM, St. Pourcain B, Greven CU et al. (2016) A genome-wide association meta-analysis of attention-deficit/hyperactivity disorder symptoms in population-based paediatric cohorts. Journal of the American Academy of Child & Adolescent Psychiatry 55, 896–905. doi: 10.1016/j.jaac.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosing MA, Gordon SD, Medland SE, Statham DJ, Nelson EC, Heath AC et al. (2009) Genetic and environmental influences on the co-morbidity between depression, panic disorder, agoraphobia, and social phobia: a twin study. Depression and Anxiety, Wiley Subscription Services, Inc., A Wiley Company 26, 1004–1011. doi: 10.1002/da.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A, Pappa I, Lahey BB, Verhulst FC, Medina-Gomez C, Jaddoe VW et al. (2016). Single nucleotide polymorphism heritability of a general psychopathology factor in children. Journal of the American Academy of Child & Adolescent Psychiatry 55, 1038–1045. e4. doi: 10.1016/j.jaac.2016.09.498. [DOI] [PubMed] [Google Scholar]
- Nivard MG, Gage SH, Hottenga JJ, van Beijsterveldt CEM, Abdellaoui A, Bartels M et al. (2017) Genetic overlap between schizophrenia and developmental psychopathology: longitudinal and multivariate polygenic risk prediction of common psychiatric traits during development. Schizophrenia Bulletin, American Psychiatric Association, Washington, DC 183, 1149–1158. doi: 10.1093/schbul/sbx031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordsletten AE, Larsson H, Crowley JJ, Almqvist C, Lichtenstein P and Mataix-Cols D (2016) Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry, American Medical Association 73, 354. doi: 10.1001/jamapsychiatry.2015.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG et al. (2016) Meta-analysis of genome-wide association studies of anxiety disorders. Molecular Psychiatry, Nature Publishing Group 21, 1391–1399. doi: 10.1038/mp.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain O, Dudbridge F and Ronald A (2017) Are your covariates under control? How normalization can re-introduce covariate effects. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescosolido M and Gamsiz E (2013) Distribution of disease-associated copy number variants across distinct disorders of cognitive development. Journal of the American Academy of Child & Adolescent Psychiatry, Elsevier 52, 414–430, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson E, Anckarsäter H, Gillberg C and Lichtenstein P (2013) Different neurodevelopmental symptoms have a common genetic etiology. Journal of Child Psychology and Psychiatry, and Allied Disciplines 54, 1356–1365. doi: 10.1111/jcpp.12113. [DOI] [PubMed] [Google Scholar]
- Pettersson E, Larsson H and Lichtenstein P (2015) Common psychiatric disorders share the same genetic origin: a multivariate sibling study of the Swedish population. Molecular Psychiatry, Macmillan Publishers Limited 21, 717–721. doi: 10.1038/mp.2015.116. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY and Hinds DA (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nature Genetics, Nature Research 48, 709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R et al. (2010) Functional impact of global rare copy number variation in autism spectrum disorders. Nature, Nature Publishing Group 466, 368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto R, Monzani B, Leckman JF, Rück C, Serlachius E, Lichtenstein P et al. (2016) Understanding the covariation of tics, attention-deficit/hyperactivity, and obsessive-compulsive symptoms: a population-based adult twin study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 171, 938–947. doi: 10.1002/ajmg.b.32436. [DOI] [PubMed] [Google Scholar]
- Plomin R, Haworth CMA and Davis OSP (2009) Common disorders are quantitative traits. Nature Reviews Genetics, Nature Publishing Group 10, 872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM et al. (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics, Nature Research 47, 702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- Reichenberg A, Cederlöf M, McMillan A, Trzaskowski M, Kapara O, Fruchter E et al. (2016) Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proceedings of the National Academy of Sciences of the United States of America, National Academy of Sciences 113, 1098–1103. doi: 10.1073/pnas.1508093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiersen AM, Constantino JN, Grimmer M, Martin NG and Todd RD (2008) Evidence for shared genetic influences on self-reported ADHD and autistic symptoms in young adult Australian twins. Twin Research and Human Genetics 11, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rende RD, Plomin R, Reiss D and Hetherington EM (1993) Genetic and environmental influences on depressive symptomatology in adolescence: individual differences and extreme scores. Journal of Child Psychology and Psychiatry 34, 1387–1398. [DOI] [PubMed] [Google Scholar]
- Riglin L, Collishaw S, Richards A, Thapar AK, Maughan B, O'Donovan MC et al. (2017) Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: a population-based cohort study. The Lancet Psychiatry 4, 57–62. doi: 10.1016/S2215-0366(16)30406-0. [DOI] [PubMed] [Google Scholar]
- Riglin L, Collishaw S, Thapar AK, Dalsgaard S, Langley K, Smith GD et al. (2016) Association of genetic risk variants with attention-deficit/hyperactivity disorder trajectories in the general population. JAMA Psychiatry, Muthén & Muthén, Los Angeles, CA 73, 1285–1292. doi: 10.1001/jamapsychiatry.2016.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, Koenen KC, McCormick MC, Munir K, Hallett V, Happé F et al. (2011) Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%). Archives of General Psychiatry, American Medical Association 68, 1113–1121. doi: 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, Neale BM and Hyman SE (2015) Genetic research in autism spectrum disorders. Current Opinion in Pediatrics, Wolters Kluwer Health 27, 685–691. doi: 10.1097/MOP.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, St Pourcain B, Anttila V, Kosmicki JA, Bulik-Sullivan B, Grove J et al. (2016) Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nature Genetics 48, 552–555. doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Larsson H, Anckarsäter H and Lichtenstein P (2014a) Symptoms of autism and ADHD: a Swedish twin study examining their overlap. Journal of Abnormal Psychology 123, 440–451. doi: 10.1037/a0036088. [DOI] [PubMed] [Google Scholar]
- Ronald A, Sieradzka D, Cardno AG, Haworth CMA, McGuire P and Freeman D (2014b) Characterization of psychotic experiences in adolescence using the specific psychotic experiences questionnaire: findings from a study of 5000 16-year-old twins. Schizophrenia Bulletin, Oxford University Press 40, 868–877. doi: 10.1093/schbul/sbt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Simonoff E, Kuntsi J, Asherson P and Plomin R (2008) Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. Journal of Child Psychology and Psychiatry, Wiley Online Library 49, 535–542. [DOI] [PubMed] [Google Scholar]
- Roy M-A, Neale MC, Pedersen NL, Mathé AA and Kendler KS (1995) A twin study of generalized anxiety disorder and major depression. Psychological Medicine, Cambridge University Press 25, 1037. doi: 10.1017/S0033291700037533. [DOI] [PubMed] [Google Scholar]
- Rydell M, Taylor MJ and Larsson H (2017) Genetic and environmental contributions to the association between ADHD and affective problems in early childhood-A Swedish population-based twin study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 174, 538–546. doi: 10.1002/ajmg.b.32536. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM et al. (2014) A framework for the interpretation of de novo mutation in human disease. Nature Genetics, Nature Research 46, 944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D et al. (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11. 23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL and McCarthy SE (2009) Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends in Genetics 25, 528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeshaft NG, Trzaskowski M, McMillan A, Krapohl E, Simpson MA, Reichenberg A et al. (2015) Thinking positively: the genetics of high intelligence. Intelligence 48, 123–132. doi: 10.1016/j.intell.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieradzka D, Power RA, Freeman D, Cardno AG, McGuire P, Plomin R et al. (2014) Are genetic risk factors for psychosis also associated with dimension-specific psychotic experiences in adolescence? PLoS ONE, Edited by Potash J. B., Public Library of Science 9, e94398. doi: 10.1371/journal.pone.0094398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slane JD, Burt SA and Klump KL (2011) Genetic and environmental influences on disordered eating and depressive symptoms. International Journal of Eating Disorders, Wiley Subscription Services, Inc., A Wiley Company 44, 605–611. doi: 10.1002/eat.20867. [DOI] [PubMed] [Google Scholar]
- Sniekers S, Stringer S, Watanabe K, Jansen PR, Coleman JRI, Krapohl E et al. (2017) Genome-wide association meta-analysis of 78308 individuals identifies new loci and genes influencing human intelligence. Nature Genetics 49, 1107–1112. doi: 10.1038/ng.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bergen SE, Kuja-Halkola R, Larsson H, Landén M and Lichtenstein P (2015) Bipolar disorder and its relation to major psychiatric disorders: a family-based study in the Swedish population. Bipolar Disorders 17, 184–193. doi: 10.1111/bdi.12242. [DOI] [PubMed] [Google Scholar]
- Spinath FM, Harlaar N, Ronald A and Plomin R (2004) Substantial genetic influence on mild mental impairment in early childhood. American Journal on Mental Retardation 109, 34. doi: 10.1352/0895-8017. [DOI] [PubMed] [Google Scholar]
- Stergiakouli E, Davey Smith G, Martin J, Skuse DH, Viechtbauer W, Ring SM et al. (2017) Shared genetic influences between dimensional ASD and ADHD symptoms during child and adolescent development. Molecular Autism, BioMed Central 8. doi: 10.1186/s13229-017-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiakouli E, Martin J, Hamshere ML, Heron J, St Pourcain B, Timpson NJN et al. (2016) Association between polygenic risk scores for attention-deficit hyperactivity disorder and educational and cognitive outcomes in the general population. International Journal of Epidemiology 46, 421–428. doi: 10.1093/ije/dyw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiakouli E, Martin J, Hamshere ML, Langley K, Evans DM, St Pourcain B et al. (2015) Shared genetic influences between attention-deficit/hyperactivity disorder (ADHD) traits in children and clinical ADHD. Journal of the American Academy of Child & Adolescent Psychiatry, Elsevier 54, 322–327. doi: 10.1016/j.jaac.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J, Batten N and Cherner M (1992) Fears and fearfulness in children and adolescents: a genetic analysis of twin data. Journal of Child Psychology and Psychiatry, Blackwell Publishing Ltd 33, 977–985. doi: 10.1111/j.1469-7610.1992.tb00919.x. [DOI] [PubMed] [Google Scholar]
- St Pourcain B, Robinson EBB, Anttila V, Bulik Sullivan B, Maller J, Golding J et al. (2017) ASD and schizophrenia show distinct developmental profiles in common genetic overlap with population-based social-communication difficulties. Molecular Psychiatry, Nature Publishing Group. doi: 10.1038/mp.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ and O'Donovan M (2012) Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews. Genetics 13, 537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Charman T, Robinson EB, Plomin R, Happé F, Asherson P et al. (2012) Developmental associations between traits of autism spectrum disorder and attention deficit hyperactivity disorder: a genetically informative, longitudinal twin study. Psychological Medicine 43, 1735–1746. [DOI] [PubMed] [Google Scholar]
- Thapar A, Cooper M and Rutter M (2017) Neurodevelopmental disorders. The Lancet Psychiatry 4, 339–346. [DOI] [PubMed] [Google Scholar]
- Thapar A and McGuffin P (1997) Anxiety and depressive symptoms in childhood – a genetic study of comorbidity. Journal of Child Psychology and Psychiatry 38, 651–656. [DOI] [PubMed] [Google Scholar]
- The ASD Working Group of The PGC (2017) Meta-analysis of GWAS of over 16000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Molecular Autism 8, 21. doi: 10.1186/s13229-017-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders Study (2014) Large-scale discovery of novel genetic causes of developmental disorders. Nature, Nature Research 519, 223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature, Nature Publishing Group 455, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grootheest DS, Cath DC, Beekman AT and Boomsma DI (2005) Twin studies on obsessive-compulsive disorder: a review. Twin Research and Human Genetics, Cambridge University Press 8, 450–458. doi: 10.1375/twin.8.5.450. [DOI] [PubMed] [Google Scholar]
- van Hulzen KJE, Scholz CJ, Franke B, Ripke S, Klein M, McQuillin A et al. (2016). Genetic overlap between attention-deficit/hyperactivity disorder and bipolar disorder: evidence from genome-wide association study meta-analysis. Biological Psychiatry, Elsevier 82, 634–641. doi: 10.1016/j.biopsych.2016.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth SJ, DeFries JC, Willcutt EG, Pennington BF and Olson RK (2015) The Colorado longitudinal twin study of reading difficulties and ADHD: etiologies of comorbidity and stability. Twin Research and Human Genetics, Cambridge University Press 18, 755–761. doi: 10.1017/thg.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J et al. (2017). Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nature Genetics 49, 978–985. doi: 10.1038/ng.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE et al. (2013) Global burden of disease attributable to mental and substance use disorders: findings from the global burden of disease study 2010. The Lancet 382, 1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E, Anney RJL, Freitag CM, Gill M et al. (2012) Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13. 3. American Journal of Psychiatry, Am Psychiatric Assoc 169, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NM, Zaharieva I, Martin A, Langley K, Mantripragada K, Fossdal R et al. (2010) Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 376, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH and Kendler KS (2012) Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. European Journal of Human Genetics, Nature Publishing Group 20, 668–674. doi: 10.1038/ejhg.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F and Middeldorp CM (2014) Research review: polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry 55, 1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME and Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zeng J, Goddard ME, Wray NR and Visscher PM (2017) Concepts, estimation and interpretation of SNP-based heritability. Nature Genetics 49, 1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
- Zammit S, Hamshere M, Dwyer S, Georgiva L, Timpson N, Moskvina V et al. (2013) A population-based study of genetic variation and psychotic experiences in adolescents. Schizophrenia Bulletin, Oxford University Press 40, 1254–1262. doi: 10.1093/schbul/sbt146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavos HMS, Eley TC, McGuire P, Plomin R, Cardno AG, Freeman D et al. (2016) Shared etiology of psychotic experiences and depressive symptoms in adolescence: a longitudinal twin study. Schizophrenia Bulletin, Oxford University Press 42, 1197–1206. doi: 10.1093/schbul/sbw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavos HMS, Freeman D, Haworth CMA, McGuire P, Plomin R, Cardno AG et al. (2014) Consistent etiology of severe, frequent psychotic experiences and milder, less frequent manifestations. JAMA Psychiatry, American Medical Association 71, 1049. doi: 10.1001/jamapsychiatry.2014.994. [DOI] [PMC free article] [PubMed] [Google Scholar]