Abstract
During oxygenic photosynthesis in cyanobacteria and chloroplasts of plants and eukaryotic algae, conversion of light energy to biologically useful chemical energy occurs in the specialized thylakoid membranes. Light-induced charge separation at the reaction centers of photosystems I and II, two multisubunit pigment-protein complexes in the thylakoid membranes, energetically drive sequential photosynthetic electron transfer reactions in this membrane system. In general, in the prokaryotic cyanobacterial cells, the thylakoid membrane is distinctly different from the plasma membrane. We have recently developed a two-dimensional separation procedure to purify thylakoid and plasma membranes from the genetically widely studied cyanobacterium Synechocystis sp. PCC 6803. Immunoblotting analysis demonstrated that the purified plasma membrane contained a number of protein components closely associated with the reaction centers of both photosystems. Moreover, these proteins were assembled in the plasma membrane as chlorophyll-containing multiprotein complexes, as evidenced from nondenaturing green gel and low-temperature fluorescence spectroscopy data. Furthermore, electron paramagnetic resonance spectroscopic analysis showed that in the partially assembled photosystem I core complex in the plasma membrane, the P700 reaction center was capable of undergoing light-induced charge separation. Based on these data, we propose that the plasma membrane, and not the thylakoid membrane, is the site for a number of the early steps of biogenesis of the photosynthetic reaction center complexes in these cyanobacterial cells.
During the evolution of the modern biosphere, photosynthetic processes in cyanobacteria have played a central role by elevating the oxygen level in the Earth's atmosphere about 3.5 billion years ago (1). Compared with other eubacteria, a unique feature of cyanobacteria, the largest group of oxygenic photosynthetic prokaryotic organisms, is the presence of a differentiated membrane system. Similar to other Gram-negative bacteria, cyanobacteria have an envelope layer consisting of an outer membrane, a peptidoglycan layer, and a plasma membrane (2, 3). In addition, these organisms have a distinct intracellular membrane system, the thylakoids, which are the sites for both oxygenic photosynthesis and respiration (2, 3). Cyanobacteria are the progenitors of chloroplasts in green plants (4). Similar to plants and algae, the thylakoid membranes in cyanobacteria harbor photosystems I and II (PSI and PSII), two large multisubunit pigment protein complexes. The reaction centers of PSI and PSII are the sites for light-induced charge separation required to drive oxygenic photosynthesis in these organisms (5, 6). The polypeptide and cofactor components of both PSI and PSII are reasonably well documented. In particular, x-ray crystallographic structures of these large membrane proteins from a thermophilic cyanobacterium have been elucidated during recent years (7, 8). In comparison, the various events during the biogenesis and assembly of these photosystems remain poorly understood.
The unicellular, naturally transformable cyanobacterium Synechocystis sp. PCC 6803 has been widely used for genetic and biochemical analysis of the form and function of both PSI and PSII (6). It is the first photosynthetic organism with a completely sequenced genome (9) and therefore offers an excellent experimental system to investigate the process of biogenesis of the photosynthetic apparatus.
One of the fundamental questions for all types of cells is how newly synthesized proteins are directed to their appropriate subcellular locations. However, little is known about the mechanisms of protein targeting and localization in cyanobacterial cells. In Synechocystis 6803, the architectures of the plasma and thylakoid membranes, and their interrelations have not been thoroughly investigated. The unique presence of these two membrane systems with different functions in the same prokaryotic cell raises a number of intriguing questions about the sites of synthesis of various membrane proteins and the mechanisms of their targeting. As a first step toward the analysis of the compositions and functions of these two membranes, we have recently developed a procedure for their complete biochemical purification (10). In this method, two separation techniques, one based on surface properties, and the other on the densities of the membrane vesicles, are combined in a two-dimensional manner to yield pure plasma and thylakoid membranes from Synechocystis 6803 cells. In the present article, we have examined the distribution of various polypeptide components of PSI and PSII in such purified thylakoid and plasma membrane preparations. Unexpectedly, we have detected the presence of significant amounts of some, but not all, PSI and PSII proteins in the plasma membranes. Moreover, our data show that such proteins are assembled into pigment-protein complexes in this membrane system. Evidently, the initial events during the biogenesis of PSI and PSII take place exclusively in the plasma membranes of these cyanobacterial cells.
Materials and Methods
Bacterial Strains and Culture Conditions.
Synechocystis sp. PCC 6803 cells were grown at 30°C under 50 μmol photons·m−2·s−1 of white light in BG11 medium (11). Liquid cultures were grown with vigorous bubbling with room air.
Membrane Isolation, Electrophoresis, and Immunodetection.
Thylakoid and plasma membranes from Synechocystis 6803 were purified as described (10). Proteins were fractionated by SDS/PAGE (12). Protein concentrations were estimated by using a protein determination kit (Pierce). Fractionated proteins were blotted onto nitrocellulose filters and reacted with antisera, and the signals were visualized by using enhanced chemiluminescence reagents (Pierce). Antisera against various PSI and PSII components, the NrtA protein, and the Ycf3 and Ycf4 proteins were kind gifts from D. A. Bryant (Pennsylvania State University), P. R. Chitnis (Iowa State University), M. Ikeuchi (University of Tokyo), T. Omata (Nagoya University), and J.-D. Rochaix (University of Geneva), respectively. Each of the antisera was diluted 200- to 1,000-fold, the chemiluminescence signals were developed for 10 s to 2 min, and the data presented were in the linear ranges of detection.
Analysis of Chlorophyll Proteins (CP).
CP compositions of the membranes were examined by PAGE as described (13–15). Samples containing 1 mg protein were solubilized at 4°C for 30 min in the presence of a detergent mixture containing 0.75% (wt/wt) dodecyl maltoside, 0.5% (wt/wt) decyl maltoside, and 0.5% (wt/wt) lithium dodecyl sulfate and centrifuged at 100,000 × g for 25 min at 4°C, and the supernatant was loaded in each lane. The resultant green CP bands on the gels were photographed. The same gels were then illuminated by UV light on a Spectroline TR-302 transilluminator (Spectronic, Westbury, NY) to examine red fluorescence emission from the chlorophyll-containing bands. Individual green bands were excised from such gels, soaked in SDS-containing buffer, fractionated on a second dimension by denaturing SDS/PAGE, and then analyzed by immunoblotting (see above).
Fluorescence Spectroscopy.
Fluorescence emission spectra at 77 K were recorded on a Fluoromax 2 fluorometer (Instruments SA, Edison, NJ). The samples contained purified membrane fractions at 5 μg chlorophyll⋅ml−1.
Electron Paramagnetic Resonance (EPR) Spectroscopy.
EPR spectra of the P700 reaction center chlorophyll(s) of PSI in plasma membrane fractions from Synechocystis 6803 were recorded on a Bruker ESP380E spectrometer at room temperature (courtesy of F. Mamedov and S. Styring, Lund University, Lund). P700+ radical was induced by continuous illumination at room temperature with an 800-W projector lamp filtered through 5 cm of copper sulfate solution and directed into the EPR cavity. EPR conditions were as follows: microwave frequency, 9.76 GHz; microwave power, 20 mW; modulation amplitude, 3 G.
Results
Presence of PSII Proteins in Plasma Membrane.
It is well established that the site for photosynthetic electron transport in cyanobacteria is the thylakoid membrane. Hence, both PSI and PSII exhibit their photosynthetic functions in the thylakoids, and not in the plasma membranes. The PSII holocomplex has numerous protein components, many of which are not essential for its catalytic activity (6, 16–18). As evidenced from the recent x-ray crystallographic data on the structure of PSII from the cyanobacterium Synechococcus elongatus (7), the majority of the functionally important cofactors are bound to a heterodimer of the D1 and D2 proteins (Fig. 1A). As expected, the D1 protein was found primarily in the purified thylakoid membranes (Fig. 1B). However, a small amount of the D1 protein was persistently detected in the purified plasma membrane fraction. NrtA (Sll1450, Cyanobase, www.kazusa.or.jp/cyano/cyano.html), a nitrate-binding protein, is the marker for the plasma membrane (10). As shown in Fig. 1B, there was no detectable contamination of the thylakoid membrane with the plasma membrane. In Synechocystis 6803 cells, the D1 protein is synthesized in a precursor form (pD1) with a carboxyl-terminal extension of 16 amino acid residues (Fig. 1C; refs. 19 and 20). After membrane insertion of the pD1 protein, this C-terminal extension is cleaved off by CtpA (Slr0008), a carboxyl-terminal processing protease (20, 21). This processing event is critically important for the biogenesis of functional PSII complex, because mutants lacking CtpA function cannot assemble a tetra-manganese cluster (denoted as Mn in Fig. 1A) that is essential for the PSII-mediated oxygen evolution process (22). Surprisingly, the CtpA protein was found exclusively in the plasma membrane fraction, and not in the thylakoid membranes (Fig. 1B). It is also noteworthy that the D1 protein in the plasma membrane fraction was in its mature form, consistent with the presence of CtpA in this membrane preparation. We also found small, but significant, amounts of the D2 protein, the α and the β subunits of cytochrome b559, and the manganese stabilizing 33-kDa extrinsic protein (MSP, Sll0427) in the plasma membrane fraction (Fig. 1D). All of these proteins are known to be closely associated with the PSII reaction center (7, 17, 23). On a protein basis, about 5–8% of the total cellular contents of D1, D2, and MSP were present in the plasma membrane fraction. In contrast, 15–20% of the two subunits of cytochrome b559, a protein that has been postulated to be a nucleation center during the assembly PSII (24, 25), were found in this membrane sample. However, the chlorophyll binding proteins CP47 and CP43, comprising the inner light-harvesting antenna of PSII, were not detected in the plasma membrane preparation and found exclusively in the thylakoids.
Figure 1.
(A) A schematic diagram of the membrane protein complex PSII in Synechocystis 6803. Among the nearly 20 polypeptide components of PSII, only those that have been examined in this study are shown (in bold) in this diagram. The pathway of electron transfer from water to the secondary quinone QB with concomitant evolution of oxygen is shown with arrows. (B) Immunoblot detection of the mature form of the D1 protein of PSII and CtpA, a carboxyl-terminal processing protease that acts on the precursor form of D1 (pD1), in the thylakoid (TM) and plasma (PM) membranes isolated from Synechocystis 6803 (6). Fifty micrograms protein-containing sample was loaded in each lane. NrtA, a component of a nitrate transporter in this cyanobacterium, is a marker protein present only in the plasma membrane. (C) A schematic depiction of the folding of the D1 protein in the membrane. The boxed region denotes the C-terminal extension in the pD1 protein. The CtpA protease cleaves off this extension region to form the mature form of the D1 protein. (D) Immunological detection of various structural proteins of PSII in the thylakoid (TM) and plasma (PM) membranes isolated from Synechocystis 6803. Fifty micrograms protein-containing sample was loaded in each lane.
Protein Components of PSI in the Plasma Membrane Fraction.
The structure and function of the PSI complex from cyanobacteria have been well characterized (8, 26, 27). Among the 12 polypeptide components of this pigment-protein complex, all of the functionally important cofactors are coordinated by two large intrinsic membrane proteins, PsaA and PsaB, and a small extrinsic protein PsaC (Fig. 2A). As shown in Fig. 2B, a number of PSI polypeptides were detected in both the thylakoid and plasma membranes, whereas a few others were found only in the thylakoid fraction. Those found in the plasma membrane are PsaA, PsaB, PsaC, and PsaD, four polypeptides that are closely associated with the PSI reaction center (28). Again, 5–8% of the cellular contents of these proteins were present in the plasma membrane fraction. It is noteworthy that these four are the only essential proteins in PSI as judged by gene-disruption studies in Synechocystis 6803 and other cyanobacteria (6, 28). Fig. 2C shows the distribution of three regulatory proteins that are known to be involved in the biogenesis and stability of PSI. Among these, BtpA (Sll0634) was found only in the thylakoid membrane (29), whereas Ycf3 (Slr0823) and Ycf4 (Sll0226) (30) were localized in the plasma membrane and not in the thylakoid membrane. Both Ycf3 and Ycf4 proteins are involved in the biogenesis of the PSI complex in the green alga Chlamydomonas reinhardii (30) and Synechocystis 6803 (ref. 31; E.Z., K. Sonoike, N. B. Ivleva, Y. Hihara, S. V. Shestakov, and H.B.P., unpublished observations; J. Kruip in Cyanomutants database, www.kazusa.or.jp/cyano/mutants), whereas we have shown that the BtpA protein is involved in the maintenance of this reaction center complex (29). Together, these data suggest that the reaction center core of PSI is formed initially in the plasma membrane of Synechocystis 6803.
Figure 2.
(A) A schematic diagram of the membrane protein complex PSI in Synechocystis 6803 cells. The bold letters denote various polypeptide components (PsaA, PsaB, etc.) of PSI. The vertical arrows show the path of electrons in the core region of the PSI complex. (B) Immunological detection of various structural proteins of PSI in the thylakoid (TM) and plasma (PM) membranes in Synechocystis 6803. Fifty micrograms protein-containing sample was loaded in each lane. (C) Immunoblot analysis of three proteins that regulate biogenesis and stability of the PSI complex. TM: thylakoid membrane; PM: plasma membrane. Fifty micrograms protein-containing sample was loaded in each lane.
PSI and PSII Components Are Assembled into Pigment Proteins in Plasma Membranes.
Next, we investigated whether the various polypeptide components of PSI and PSII in the plasma membrane fraction are present as individual polypeptides or assembled into protein complexes. For this purpose, we used a nondenaturing gel system (13–15) to examine the presence of chlorophyll-binding proteins in the thylakoid and plasma membrane fractions. As shown in Fig. 3A Left, at least four green bands could be visualized in the thylakoid membrane sample. Among them, band 1 represents PSI, whereas band 2 represents PSII (13, 14). In cyanobacterial membranes, the relative abundance of PSI is significantly higher than that of PSII (32). Hence, the intensity of the green band 2 was very faint. In fact, the bottom portion of the gel was overexposed in the left panel to visualize this band. The slower migrating green bands in the thylakoid sample contains higher-order aggregates of the PSI holocomplex (33). Interestingly, a green band migrating at a position similar to band 1 was visualized in the plasma membrane. However, in this lane, we did not observe any green band in the band 2 region. However, UV transillumination, a more sensitive technique to detect fluorescent CPs (15), revealed the presence of both band 1 and band 2 in the plasma membrane fraction (Fig. 3A Right).
Figure 3.
(A) Nondenaturing “green” gel electrophoresis to visualize CP complexes in purified thylakoid membrane (TM) and plasma membrane (PM) samples from Synechocystis 6803. (Left) Photographed with white light. (Right) The gel was briefly soaked in 0.1 M HCl and photographed with UV transillumination. See text for further details. (B) The 77K fluorescence emission spectra from excised polyacrylamide gel fragments containing green bands 1 and 2 (see A) isolated from thylakoid (TM) and plasma (PM) membranes, respectively. Excitation was at 440 ± 5 nm. The spectra were corrected for the wavelength characteristics of the emission monochromator and the response of the signal detector. (C) Immunological detection of various proteins present in bands 1 and 2 from the plasma membrane fraction (A). See text for further details.
Bands 1 and 2 from both thylakoid and plasma membranes were excised from the gel and analyzed in two different ways. First, we determined the fluorescence emission spectra of the excised gel fragments that were frozen in liquid nitrogen. Band 1 from both membrane samples had a strong emission band around 720 nm (722 nm in the thylakoid membrane and 719.5 nm in the plasma membrane), indicating the presence of assembled PSI pigment protein. Band 2 from thylakoid membrane had an emission peak at 685 nm, typical for the presence of assembled PSII centers. The emission peak from band 2 from plasma membrane was blue-shifted to 680 nm. It is noteworthy that emission spectrum of purified D1-D2-cyt b559 reaction center preparations from higher plants often exhibit such a blue-shifted peak (34). Next, the excised band 1 and band 2 material from plasma membranes were fractionated by denaturing SDS/PAGE and analyzed by Western blotting for the presence of various PSI and PSII proteins. Band 1 contained PsaA, PsaB, PsaC, and PsaD proteins (Fig. 3C). However, this sample did not contain any other PSI, or any of the PSII components shown in Fig. 1 B and D (data not shown). On the other hand, band 2 contained the D1 and the D2 proteins, as well as the α and β (data not shown) subunits of cytochrome b559. Significantly, CP47 and CP43, as well as PSI proteins, were not detected in this sample (data not shown). Together, these pigment and protein data demonstrate the presence of assembled PSI as well as PSII subcomplexes in the plasma membrane of Synechocystis 6803.
EPR Analysis of PSI in Plasma Membrane.
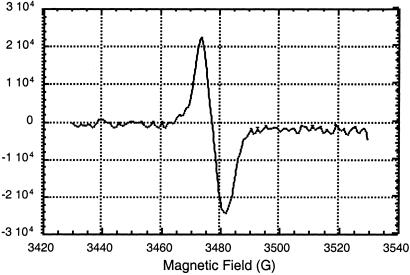
EPR investigations have shown that in the plasma membrane fractions, P700+ can indeed be formed by light (Fig. 4) or by oxidation with potassium ferricyanide (data not shown). Evidently, the PSI complex found in the plasma membranes had assembled reaction center that could undergo light-induced charge separation. A more elaborate functional study of PSI in the plasma membrane fractions will be reported elsewhere.
Figure 4.
Detection of a light-induced EPR signal (g value 2.0030, bandwidth 8 G) from P700+ (light minus dark), indicative of functional charge separation in PSI, in purified plasma membrane preparation from Synechocystis 6803. See text for further details.
Discussion
In this article, we show that a number of subunit proteins that are intimately associated with the reaction center of both photosystems are detected in the plasma membrane fraction of Synechocystis 6803, whereas other subunits were located exclusively in the thylakoid membrane fraction. We also found that the PSI and PSII proteins in the plasma membrane are assembled into CP complexes.
Carboxyl-Terminal Processing of the pD1 Protein in the Plasma Membrane System.
A crucial finding in this work has been the presence of the CtpA protease exclusively in the purified plasma membrane fraction (Fig. 1B). It is noteworthy that although CtpA is hydrophilic in nature, we have found that it is tightly associated with the plasma membrane (E.Z., N. Inagaki, and H.B.P., unpublished observations). As mentioned earlier, the CtpA enzyme is absolutely required for the assembly of the water-oxidizing machinery in PSII. Mutants of Synechocystis 6803 (20), as well as the eukaryotic alga Scenedesmus obliquus (35, 36), without functional CtpA cannot process the carboxyl-terminal extension of the pD1 protein. In these mutants, the PSII complex is assembled, and electron transfer from YZ to QB (Fig. 1A) can occur. However, the Mn cluster cannot assemble in the PSII complex. Thus, the CtpA-mediated processing of pD1 is one of the final steps during the assembly of functionally competent PSII. Hence, the absence of the CtpA protein in the thylakoids (Fig. 1B) indicates that the formation of active PSII cannot occur in these membranes and suggests that the plasma membrane has a significant role in the biogenesis of functional PSII complex.
Assembly of PSI and PSII Subcomplexes in Plasma Membrane.
Another important finding is the presence in the purified plasma membrane fractions of CP complexes that correspond to PSI and PSII subcomplexes, respectively (Fig. 3 A and C). The data presented in Fig. 4 clearly demonstrated that the PSI complex in the plasma membranes has assembled P700 reaction center chlorophylls that can undergo charge separation. Because the overall stoichiometry of PSII/PSI is relatively low in cyanobacteria (32, 37), we have so far been unable to unequivocally determine whether the minute amount of assembled PSII subcomplex in the plasma membrane fraction (band 2 in Fig. 3A) has any functional activity.
A question may be raised about the purity of the plasma membrane preparation used in this study. For example, the presence of small amounts of D1, D2, PsaA, and PsaB proteins may be a consequence of a possible contamination of the plasma membrane preparation by thylakoid membrane. However, the absence of CP47 and CP43 proteins of PSII (Fig. 1D) as well as the PsaF and PsaL proteins of PSI (Fig. 2B), four intrinsic membrane proteins, from the plasma membrane fraction, speaks against such a possibility. In addition, we have recently detected the presence of the MSP protein (Fig. 1A) in a periplasmic preparation from Synechocystis 6803 cells that was isolated by using a procedure that does not rupture the spheroplast (Y. Lewis and H.B.P., unpublished observation).
Biogenesis of Photosystems: A Model.
Based on the data presented here, we propose a model for stepwise biogenesis of the two photosystems in cyanobacteria. Initially, the assembly of the reaction center core (including pigments and other redox components) of both PSI and PSII occurs in the plasma membrane and not in the thylakoid membrane. Subsequently, these reaction center assemblies are translocated to the thylakoid membrane, possibly by membrane vesicle transport, or by lateral movement through connecting membranes. It is noteworthy that Westhoff, Soll, and coworkers (38, 39) have recently provided excellent evidence that a 30-kDa VIPP protein is involved in membrane vesicle transport from the plasma membrane of cyanobacteria, or from the envelope membrane of chloroplasts, for the biogenesis of the thylakoid membranes. At the final step of our biogenesis model, additional regulatory and light-harvesting subunits that are incorporated directly into the thylakoid membrane are assembled into the reaction center core complexes to form complete photosystems. Thus, the photosynthetic reaction centers appear to have maintained a classical prokaryotic route of biogenesis, i.e., they are integrated and assembled in the plasma membrane. In this context, it is noteworthy that Gloeobacter violaceous PCC 7421, one of the very few cyanobacterial genera whose existence can be traced before the endosymbiotic origin of chloroplasts (40), does not have a distinct thylakoid membrane system. PSI and PSII in these cells are localized in the plasma membrane (2, 3, 41). However, G. violaceous cells exhibit relatively low efficiency of photosynthesis and are extremely light sensitive. It is conceivable that the ancestors of the modern-day cyanobacteria had primitive photosystem core complexes assembled in their plasma membrane, whereas the sites of additional pigment-binding and regulatory proteins were determined after the evolution of an efficient photosynthetic apparatus in a distinct thylakoid membrane system.
Acknowledgments
We thank Profs. D. A. Bryant, P. R. Chitnis, M. Ikeuchi, T. Omata, and J.-D. Rochaix for the gifts of various antisera, Dr. F. Mamedov and Prof. S. Styring for EPR analysis of P700, and Ms. Y. Lewis for collegial discussions. This work was supported by the U. S. Department of Energy (to H.B.P.), the National Institutes of Health (to H.B.P.), the National Science Foundation (to H.B.P.), the International Human Frontier Science Program (to H.B.P. and B.A.), the Swedish Natural Science Research Council (to B.A.), and the Carl Trygger Foundation (to B.N.).
Abbreviations
- CP
chlorophyll protein
- EPR
electron paramagnetic resonance
- PSI
photosystem I
- PSII
photosystem II
References
- 1.Nisbet E G, Sleep N H. Nature (London) 2001;409:1083–1091. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]
- 2.Stanier R Y, Cohen-Bazire G. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- 3.Gantt E. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 119–138. [Google Scholar]
- 4.Gray M W. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 5.Bryant D A. Can Bull Fisheries Aquatic Sci. 1986;214:423–500. [Google Scholar]
- 6.Pakrasi H B. Annu Rev Genet. 1995;29:755–776. doi: 10.1146/annurev.ge.29.120195.003543. [DOI] [PubMed] [Google Scholar]
- 7.Zouni A, Witt H T, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Nature (London) 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 8.Jordan P, Fromme P, Witt H T, Klukas O, Saenger W, Krauss N. Nature (London) 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 10.Norling B, Zak E, Andersson B, Pakrasi H. FEBS Lett. 1998;436:189–192. doi: 10.1016/s0014-5793(98)01123-5. [DOI] [PubMed] [Google Scholar]
- 11.Allen M M. J Phycol. 1968;4:1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Guikema J A, Sherman L A. Arch Biochem Biophys. 1983;220:155–166. doi: 10.1016/0003-9861(83)90396-x. [DOI] [PubMed] [Google Scholar]
- 14.Pakrasi H B, Riethman H C, Sherman L A. Proc Natl Acad Sci USA. 1985;82:6903–6907. doi: 10.1073/pnas.82.20.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pakrasi H B, Goldenberg A, Sherman L A. Plant Physiol. 1985;79:290–295. doi: 10.1104/pp.79.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debus R J. Met Ions Biol Syst. 2000;37:657–711. [PubMed] [Google Scholar]
- 17.Bricker T M, Ghanotakis D F. In: Oxygenic Photosynthesis: The Light Reactions. Ort D R, Yocum C F, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 113–126. [Google Scholar]
- 18.Barber J. Biochim Biophys Acta. 1998;1365:269–277. doi: 10.1016/s0005-2728(98)00079-6. [DOI] [PubMed] [Google Scholar]
- 19.Nixon P J, Trost J T, Diner B A. Biochemistry. 1992;31:10859–10871. doi: 10.1021/bi00159a029. [DOI] [PubMed] [Google Scholar]
- 20.Anbudurai P R, Mor T S, Ohad I, Shestakov S V, Pakrasi H B. Proc Natl Acad Sci USA. 1994;91:8082–8086. doi: 10.1073/pnas.91.17.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao D I, Qian J, Chisholm D A, Jordan D B, Diner B A. Nat Struct Biol. 2000;7:749–753. doi: 10.1038/78973. [DOI] [PubMed] [Google Scholar]
- 22.Diner B A, Ries D F, Cohen B N, Metz J G. J Biol Chem. 1988;263:8972–8980. [PubMed] [Google Scholar]
- 23.Satoh K. In: Oxygenic Photosynthesis: The Light Reactions. Ort D R, Yocum C F, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 193–211. [Google Scholar]
- 24.Cramer W A, Whitmarsh J. Annu Rev Plant Physiol. 1977;28:133–172. [Google Scholar]
- 25.Whitmarsh J, Pakrasi H B. In: Oxygenic Photosynthesis: The Light Reactions. Ort D R, Yocum C F, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 249–264. [Google Scholar]
- 26.Golbeck J H, Bryant D A. In: Current Topics in Bioenergetics. Lee C P, editor. New York: Academic; 1991. pp. 83–177. [Google Scholar]
- 27.Chitnis P R. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:593–626. doi: 10.1146/annurev.arplant.52.1.593. [DOI] [PubMed] [Google Scholar]
- 28.Chitnis P R. Plant Physiol. 1996;111:661–669. doi: 10.1104/pp.111.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zak E, Norling B, Andersson B, Pakrasi H B. Eur J Biochem. 1999;261:311–316. doi: 10.1046/j.1432-1327.1999.00281.x. [DOI] [PubMed] [Google Scholar]
- 30.Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix J D. EMBO J. 1997;16:6095–6104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilde A, Hartel H, Hubschmann T, Hoffmann P, Shestakov S V, Borner T. Plant Cell. 1995;7:649–658. doi: 10.1105/tpc.7.5.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita F, Murakami A, Aizawa K, Ohki K. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 677–692. [Google Scholar]
- 33.Takahashi Y, Koike H, Katoh S. Arch Biochem Biophys. 1982;219:209–218. doi: 10.1016/0003-9861(82)90151-5. [DOI] [PubMed] [Google Scholar]
- 34.Seibert M, Picorel R, Rubin A B, Connolly J S. Plant Physiol. 1988;87:303–306. doi: 10.1104/pp.87.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metz J G, Pakrasi H B, Seibert M, Arntzen C J. FEBS Lett. 1986;205:269–274. [Google Scholar]
- 36.Trost J T, Chisholm D A, Jordan D B, Diner B A. J Biol Chem. 1997;272:20348–20356. doi: 10.1074/jbc.272.33.20348. [DOI] [PubMed] [Google Scholar]
- 37.Mannan R M, Pakrasi H B. Plant Physiol. 1993;103:971–977. doi: 10.1104/pp.103.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht U C, Soll J, Westhoff P. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westphal S, Heins L, Soll J, Vothknecht U C. Proc Natl Acad Sci USA. 2001;98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelissen B, Van de Peer Y, Wilmotte A, De Wachter R. Mol Biol Evol. 1995;12:1166–1173. doi: 10.1093/oxfordjournals.molbev.a040289. [DOI] [PubMed] [Google Scholar]
- 41.Rippka R, Waterbury J B, Cohen-Bazire G. Arch Microbiol. 1974;100:419–436. [Google Scholar]