Abstract
The new generation of post-genomic targets, such as protein-protein interactions (PPIs), often require new chemotypes not well represented in current compound libraries. This is one reason for why traditional high throughput screening (HTS) approaches are not more successful in delivering medicinal chemistry starting points for PPIs. In silico screening methods of an expanded chemical space are then potential alternatives for developing novel chemical probes to modulate PPIs. In this review, we report on the state-of-the-art pipelines for virtual screening, emphasizing prospectively validated methods capable of addressing the challenge of drugging difficult targets in the human interactome. Collectively, we show that optimal strategies for structure based virtual screening vary depending on receptor structure and degree of flexibility.
Introduction
Small molecules remain an available and increasingly diverse source for new and repurposed drug compounds. As computational resources and algorithm quality have increased, Computer-Aided Drug Design (CADD) has become an integral part of the drug discovery process. With massive compound libraries available[1–3] and the ever increasing quantity and quality of receptor-ligand structures[4] and other biological data, more efficient algorithms and novel techniques will become increasingly necessary to take advantage of new data. In this review, we will discuss advances in computational drug discovery, including increased chemical diversity and virtual screening technologies.
Current libraries of compounds used for screening are mostly derived from historical medicinal-chemistry efforts by pharmaceutical companies. Thus, chemical phenotypes, or “chemotypes”, are dominated by past drug-discovery research into kinases, G-protein-coupled receptors, enzymes and other targets traditionally considered druggable[2,5]. New targets, such as protein-protein interactions, often require new chemotypes that are poorly sampled in chemical libraries[6]. Thus, expanding the diversity of compound libraries is essential in order to identify new chemical probes that could address the chemotypes required for new targets[7].
Virtual small-molecule libraries provide access to an arbitrarily large and potentially more diverse chemical space. However, in order to be useful, these libraries must not only be available or readily synthesizable but also searchable for compounds likely to bind to the target. Many valuable technologies both commercial and open access exist to perform structure-based virtual screening of commercially available compounds[3,7,8]. Of note, the Dömling and Camacho labs have recently developed breakthrough technologies that allow for drug discovery collaboration efforts to be performed in real time by screening millions of compound in seconds[7]. These open access tools are not only capable of performing pharmacophore-based virtual screening of commercially available compounds[3], but can also screen chemical libraries specially designed to disrupt protein-protein interactions (PPIs)[7]. The latter are a target class that has proven to be especially difficult to drug using traditional libraries. These anchor-biased libraries consist of multicomponent reactions (MCR)-derived compounds. MCR chemistry (“one step, one-pot”)[9] is much faster than traditional multistep sequential synthesis, allowing for the timely experimental verification or falsification of virtual compounds[7].
Critical in virtual screening is the prediction of accurate poses and the enrichment of active compounds. When evaluating ranking performance of new virtual screening methods, high correlation values between the predicted ranking of compounds by affinity and the actual rankings are commonly seen when evaluating on known targets[10]. However, these results don't stack up when methods are tested on prospective data sets, even when ample structural information is available[11,12]. In this review we discuss recent advances in both the software and strategies used for CADD. Much of these improvements has more to do with tuning the screening strategy to the type of receptor structure, flexibility, and cofactors than the specific software platform or scoring function.
Recent advances in virtual screening strategies
Pose Prediction
Poses are usually predicted based on a two-step approach: (a) ligand conformer generation followed by (b) docking and scoring to the target. There are several efficient software tools used for conformer generation that can be described as deterministic or stochastic[13]. Although generally accurate, sampling of ring structures is still challenging and can sometimes impact the outcome. Docking programs combine conformer generation with pose scoring[14]. There are many docking programs both commercially and freely available, such as AutoDock Vina[15], Smina[16], Glide[17], and Gold[18]. Smina, for example, is a fork of AutoDock, which is not only faster but also facilitates the development of new scoring functions[16].
Scoring functions often fall into one of three categories: force-field-based, knowledge-based, or empirical[14]. Force-field-based scoring functions use actual representations of forces between the receptor and ligand molecules. These are often based on existing molecular dynamics force field parameters such as the AMBER force field[19,20]. Knowledge-based scoring functions use simplified representations of atomic interactions in order to attempt to reproduce experimental structural data. Empirical scoring functions are generated by fitting parameters to experimental structural and affinity data. There have been continued improvements in scoring functions for docking applications, notably the development of the OPLS3 force field[21]. This force field fit new parameters based on a data set consisting of small molecule and protein-ligand pairs which leads to better parameterization for analysis of protein-ligand interactions. Another recent development has been the use of convolutional neural nets (CNNs)[22,23] which can be used for scoring. CNNs are a type of neural net architecture where connections between layers are spatially restricted, allowing each neuron to learn about nearby features. While neural nets have been used for receptor-ligand scoring previously[24], their use is pushing the boundaries of deep learning techniques by increasing the ability to learn from spatial interactions from known 3D co-crystal structures[22,25,26].
Receptor Flexibility
Another important characteristic of docking programs is how they treat receptor flexibility. While it is not computationally feasible to simulate full protein flexibility when screening large numbers of ligands, various strategies have been developed to approximate receptor flexibility. For example, a common strategy is the application of ensemble docking[27–29], where docking is performed against multiple available receptor structures. Additionally, partial receptor flexibility has been modeled in a variety of ways, such as rotamer libraries[30], side chain flexibility[31], and full backbone flexibility near the binding site[32]. Because of these advances it is becoming increasingly feasible to account for protein flexibility in virtual screening. Recently the use of metadynamics[33] has been applied to protein-ligand binding[34]. Metadynamics is a method of enhanced sampling which introduces an extra variable into the system which is used to steer the simulation away from areas which have been previously sampled[33]. This method has allowed researchers to combine ideas from induced fit in docking.
Lessons from prospective virtual screening predictions
Because the aforementioned developments are generally trained and tested retrospectively, it is difficult to fairly compare different methods. To that end, analysis of prospective community-wide experiments provides a unique opportunity to evaluate methods and identify problems with different approaches. The Drug Design Data Resource (D3R) project was started as a joint project between the NIH and UCSD with the goal of providing blinded datasets for prospective evaluation of drug discovery pipelines[11,12].
Pose Prediction
Given compounds as SMILES strings[35], predictions for targets for which there are one or more publicly available co-crystal structures (Protein Data Bank (PDB)[4]), are generally performed using three major approaches: alignment-based[36–40], standard docking as discussed above[36–39,41–43], or simulation-based[37,41,44]. Alignment- and docking- based methods have been more consistent in prospective tests[11,12]s. In the former, conformers of each compound are generated[45] and aligned to the ligand of an available co-crystal structure. Alignment metrics can involve chemical similarity measured by Tanimoto similarity[36–38], 3D shape similarity[40], and hybrid 3D shape/pharmacophore feature similarity method[39]. Poses are then minimized and ranked. As expected, higher quality poses were generally correlated with number and similarity of available co-crystal ligands[38] (Table 1).
Table 1.
Best prospective pose prediction median RMSD from D3R Grand Challenges.
Selecting the optimal receptor structure
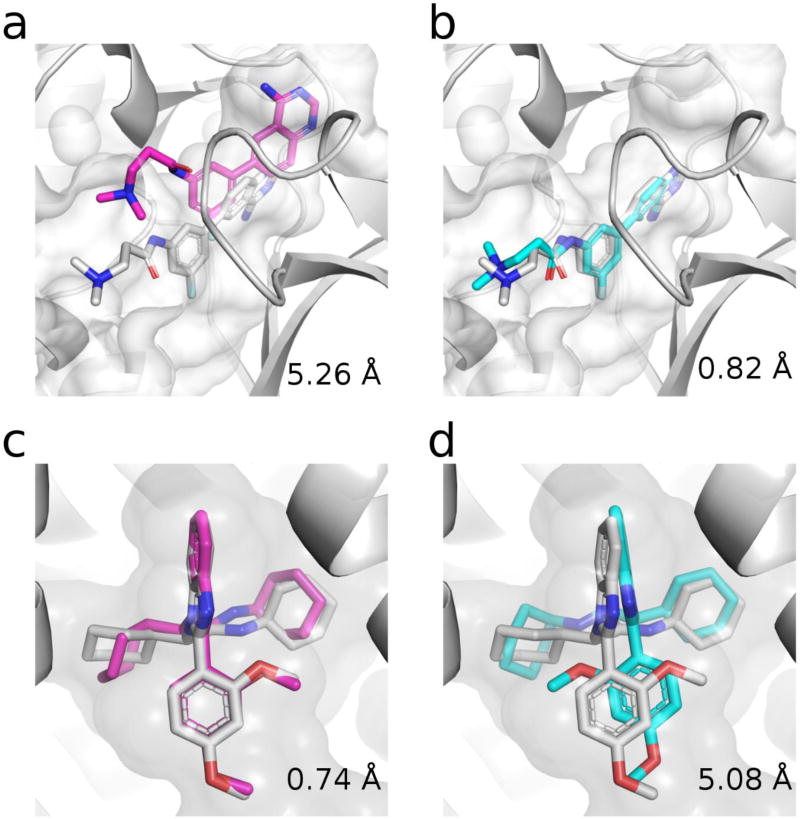
[16,38] is critical for the outcome of pose prediction. Thus, so-called “close” methods that use as receptor the co-crystal structure that had the most similar known ligand perform the best. For example, in the D3R competitions[38] we show that “alignclose” (alignment and minimization) or “dock-close” (docking to closest receptor) methods lead to top-of-the-line predictions, with median predicted poses under 2 Å RMSD[36,38,46], a standard metric for measuring success of pose predictions. We found that the quality of docking vs alignment-based methods depends on both the type of binding pocket being targeted (Figure 1) as well as the similarity level of known ligands[36,38] (Figure 2). For more open or flexible binding pockets (e.g. MAP4K4 [mitogen-activated protein kinase kinase kinase kinase 4]) alignment-based methods performed better than docking methods, whereas for a buried flexible pocket (e.g. FXR [farnesoid X receptor]) minimization is not effective and docking performs better (Table 1). More resource-intensive molecular-dynamics-based methods that used induced fit docking ideas and metadynamics[33,34] were also able to predict similar quality poses[37].
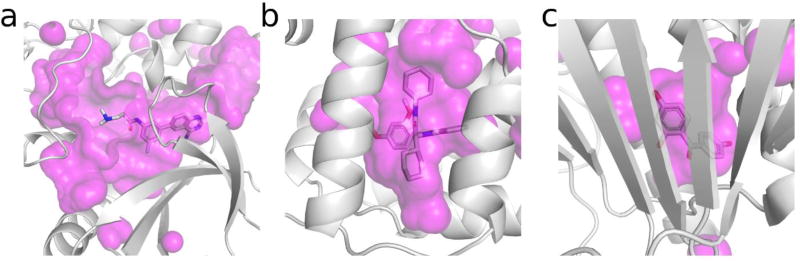
Figure 1. Receptor binding modes of prospectively validated targets.
Surface representations of binding pockets of D3R Grand Challenge 1 and 2 targets: a) MAP4K4, b) FXR, and c) HSP90. Receptor structure shown as cartoons, co-crystal ligand shown as sticks, and volumes shown as magenta surface.
Figure 2. Free docking versus alignment strategies for pose prediction.
Examples of differences in pose predictions for docking (magenta) and alignment-based (cyan) methods for MAP4K4 (a and b) and FXR (c and d). Co-crystal ligand shown as white sticks. Receptor shown as white cartoon and binding pocket as white surface.
Affinity Ranking
A number of techniques have been applied for affinity ranking, from novel scoring functions[25,39,42,43,47], to ranking with free energy prediction methods[48–50]. Prospective analyses have shown that top of the line Spearman ρ/Kendall’s τ correlations for ranking compounds on targets with known co-crystal structures is close to 0.5/0.4 (Table 2). However, again the best ranking methods have been shown to depend on the type of target and the selection of an optimal receptor. We have shown that docking to a single receptor can sometimes lead to better results than methods that use multiple receptor structures[46]. The rationale for this is that the energetics of different receptor structures is difficult to estimate computationally. However, selecting the optimal receptor for docking is not always clear. When there is sufficient data with similar congenerics, rankings are robust[36,38,46], the high degree of compound similarity causing the receptor to have similar binding pocket conformation. Though it is still possible to choose a receptor that doesn’t generalize well to your test set, which can lead to essentially random rankings[38]. For these cases (such as MAP4K4 and p38-α in D3R challenges) methods that take ligand similarity into account have performed better (Table 2).
Table 2.
Best prospective affinity ranking from D3R Grand Challenges.
| RECEPTOR | # Test compounds |
# PDB structures |
Binding Site Description |
Best Ranking [Kendall’s τ] |
Prospective Best method |
|---|---|---|---|---|---|
| HSP90 | 180 | >200 | Four binding modes | 0.32a | Dock close |
| FXR | 102 | 27 | Buried binding modes | 0.46b | Dock close |
| Cathepsin S | 136 | 25 | Narrow surface pocket | 0.39c | Dock close |
| MAP4K4 | 18 | 8 | Large flexible | 0.48d | Min cross |
| p38-α | 72 | 185 | Large flexible | 0.41c | Dock close |
| VEGFR2 | 85 | 4 | Large flexible | 0.45c | Dock cross |
| JAK2 SC2 | 89 | 59 | Large flexible | 0.47c | - |
Other techniques for ranking are being explored, which aim to take advantage of increasing availability of high-quality structural data as well as improving hardware and software resources. For example, CNN-derived scoring functions were used in both the 2015 and 2016 Grand Challenges showing promising results[25]. Interestingly the addition of new co-crystals of more similar compounds did not appear to have any effect on the quality of rankings[11,12], establishing the limitation of current scoring and force fields used.
Discussion
Different methods apply to different targets
The prospective virtual screening discussed here involve targets with publicly available receptor-ligand co-crystal structures. Of note, these targets are generally easier than screening apo structures. The above notwithstanding, these targets present different challenges. As shown in Figure 1, these receptors had distinct binding modes; from relatively exposed ligands (e.g. Cathepsin S) to deeply buried ones (e.g. FXR). Targets also included different types of flexibility: from discrete binding modes such as the HSP90 (heat shock protein 90) catalytic site to large pockets with flexible loops such as in kinases, e.g., MAP4K4. The L2 loop of HSP90 shows a number of conformations induced by binding different ligands[38]. Perhaps the most interesting conclusion is that the receptor structure and pipeline strategy appear to have more effect on screening outcome than any specific software tool[11,12].
Optimal strategies for pose and affinity prediction are generally different. Not surprisingly, pose prediction benefits greatly from known co-crystal information. For these targets, aligning and minimizing to the closest known inhibitor (“align-close” methods) consistently lead to the best poses. However, if the known ligands are not representative of the screening set then docking in the corresponding receptor pocket (“dock-close”) performs better (see Table 1).
For affinity ranking, docking tends to be the best approach. However, “dock-close” (docking each ligand to closest co-crystal receptor) outperforms “dock-cross” (docking ligands to one receptor) methods in constrained pockets with different binding modes. “Align” methods generally do not perform well in these targets because minimization on a constrained environment where clashes are very likely often leads to random poses and scores, whereas docking avoids those clashes and final poses tend to retain the chemotypes of the related co-crystal structure.
Interestingly, prospective predictions for four kinases shows the progress and limitations of virtual screening. Quality of rankings is generally measured by either Spearman’s ρ or Kendall’s τ, both of which are measures of similarity of two rank-orderings and are values between -1 (perfect opposite order) and 1 (perfectly in order). A value of 0 for both would be a random correlation. Namely, with a Spearman’s ρ of around 0.5 or Kendall’s τ around 0.45, structure based virtual screening can produce a significant enrichment of likely binders, yet an orthogonal assessment is still needed to limit the potential number of false positives in real-world applications. Overall, top scoring methods do not incorporate receptor flexibility other than known receptor structures, nor very sophisticated free energy calculations [11,12]. The latter have consistently shown error bars on the order of 0.75 kcal/mol [11,12], which is too large to make a dent on enrichment for large sets of compounds.
Despite progress in the area of CADD, there are still obvious areas of improvements. New force fields and scoring functions are promising[21], yet we have shown that in prospective evaluations with blind data sets simpler virtual screening methods can outperform more complex ones[36,38]. In all likelihood, pose prediction and affinity ranking might have exhausted the benefits of rigid receptors and implicit solvent models. New avenues are being tested but they have yet to be proven in blind tests. Alternatively, new sources of data should be incorporated in the pipeline in order to provide orthogonal validation of the predictions.
Acknowledgments
The work was funded by U.S. National Institutes of Health grant numbers GM097082 and T32EB009403.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koes D, Khoury K, Huang Y, Wang W, Bista M, Popowicz GM, Wolf S, Holak TA, Dömling A, Camacho CJ, et al. Enabling large-scale design, synthesis and validation of small molecule protein-protein antagonists. PLoS One. 2012;7:e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irwin JJ, Shoichet BK. ZINC - A free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koes DR, Camacho CJ. ZINCPharmer: pharmacophore search of the ZINC database. Nucleic Acids Res. 2012;40:W409–W414. doi: 10.1093/nar/gks378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macarron R. Critical review of the role of HTS in drug discovery. Drug Discov Today. 2006;11:277–279. doi: 10.1016/j.drudis.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Sperandio O, Reynès CH, Camproux A-C, Villoutreix BO. Rationalizing the chemical space of protein–protein interaction inhibitors. Drug Discov Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 7•.Koes DR, Dömling A, Camacho CJ. AnchorQuery: Rapid online virtual screening for small-molecule protein-protein interaction inhibitors. Protein Sci. 2018;27:229–232. doi: 10.1002/pro.3303. The authors developed a web server for targeting protein-protein interactions with small molecules with customizable chemistries based on multicomponent reactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunseri J, Koes DR. Pharmit : interactive exploration of chemical space. 2016 doi: 10.1093/nar/gkw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domling A, Wang W, Wang K. Chemistry and Biology Of Multicomponent Reactions. Chem Rev. 2012;112:3083–3135. doi: 10.1021/cr100233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang WJ, Huang Q, Zou J, Li LL, Yang SY. TS-Chemscore, a Target-Specific Scoring Function, Significantly Improves the Performance of Scoring in Virtual Screening. Chem Biol Drug Des. 2015;86:1–8. doi: 10.1111/cbdd.12470. [DOI] [PubMed] [Google Scholar]
- 11•.Gathiaka S, Liu S, Chiu M, Yang H, Stuckey JA, Kang YN, Delproposto J, Kubish G, Dunbar JB, Carlson HA, et al. D3R grand challenge 2015: Evaluation of protein-ligand pose and affinity predictions. J Comput Aided Mol Des. 2016;30:651–668. doi: 10.1007/s10822-016-9946-8. Overview of first D3R Grand Challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Gaieb Z, Liu S, Gathiaka S, Chiu M, Yang H, Shao C, Feher VA, Walters WP, Kuhn B, Rudolph MG, et al. D3R Grand Challenge 2: blind prediction of protein–ligand poses, affinity rankings, and relative binding free energies. J Comput Aided Mol Des. 2018;32:1–20. doi: 10.1007/s10822-017-0088-4. Overview of second D3R Grand Challenge. In-depth discussion of methods used by participants and their effects on prediction outcomes. Looks at progress made through multiple prospective challenges and good discussion on areas for improvement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebejer JP, Morris GM, Deane CM. Freely available conformer generation methods: How good are they? J Chem Inf Model. 2012;52:1146–1158. doi: 10.1021/ci2004658. [DOI] [PubMed] [Google Scholar]
- 14.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 15.Trott O, Olson AJ. Software News and Update AutoDock Vina : Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem. 2009;31 doi: 10.1002/jcc.21334. NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein - Ligand Complexes. 2006 doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 18.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking 1 1. In: Cohen FE, editor. J Mol Biol. Vol. 267. 1997. pp. 727–748. [DOI] [PubMed] [Google Scholar]
- 19.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An all atom force field for simulations of proteins and nucleic acids. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 21•.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J Chem Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. Developed force field specifically designed for small molecule-protein binding. Since been used in numerous succesful virtual screening applications. [DOI] [PubMed] [Google Scholar]
- 22.Ragoza M, Hochuli J, Idrobo E, Sunseri J, Koes DR. Protein-Ligand Scoring with Convolutional Neural Networks. J Chem Inf Model. 2017;57:942–957. doi: 10.1021/acs.jcim.6b00740. Used convolutional neural network (CNN) trained on 3D receptor-ligand structures to predict ligand binding affinities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durrant JD, Carlson KE, Martin TA, Offutt TL, Mayne CG, Katzenellenbogen JA, Amaro RE. Neural-Network Scoring Functions Identify Structurally Novel Estrogen-Receptor Ligands. J Chem Inf Model. 2015;55:1953–1961. doi: 10.1021/acs.jcim.5b00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durrant JD, McCammon JA. NNScore: A neural-network-based scoring function for the characterization of protein-ligand complexes. J Chem Inf Model. 2010;50:1865–1871. doi: 10.1021/ci100244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunseri J, Ragoza M, Collins J, Koes DR. A D3R prospective evaluation of machine learning for protein-ligand scoring. J Comput Aided Mol Des. 2016;30:761–771. doi: 10.1007/s10822-016-9960-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cang Z, Mu L, Wei GW. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selwa E, Martiny VY, Iorga BI. Molecular docking performance evaluated on the D3R Grand Challenge 2015 drug-like ligand datasets. J Comput Aided Mol Des. 2016;30:829–839. doi: 10.1007/s10822-016-9983-3. [DOI] [PubMed] [Google Scholar]
- 28.Feixas F, Lindert S, Sinko W, McCammon JA. Exploring the role of receptor flexibility in structure-based drug discovery. Biophys Chem. 2014;186:31–45. doi: 10.1016/j.bpc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prasad JC, Goldstone JV, Camacho CJ, Vajda S, Stegeman JJ. Ensemble modeling of substrate binding to cytochromes P450: Analysis of catalytic differences between CYP1A orthologs. Biochemistry. 2007;46:2640–2654. doi: 10.1021/bi062320m. [DOI] [PubMed] [Google Scholar]
- 30.Leach AR. Ligand docking to proteins with discrete side-chain flexibility. J Mol Biol. 1994;235:345–356. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 31.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis IW, Baker D. RosettaLigand Docking with Full Ligand and Receptor Flexibility. J Mol Biol. 2009;385:381–392. doi: 10.1016/j.jmb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 33.Laio A, Gervasio FL. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Reports Prog Phys. 2008;71 [Google Scholar]
- 34.Clark AJ, Tiwary P, Borrelli K, Feng S, Miller EB, Abel R, Friesner RA, Berne BJ. Prediction of Protein-Ligand Binding Poses via a Combination of Induced Fit Docking and Metadynamics Simulations. J Chem Theory Comput. 2016;12:2990–2998. doi: 10.1021/acs.jctc.6b00201. [DOI] [PubMed] [Google Scholar]
- 35.Weininger D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J Chem Inf Comput Sci Proc Edinburgh Math Soc J Comput Chem Z Naturforsch Bull Soc Chim MATCH Rev Res Fac Sci-Univ Novi Sad, Math Ser Comput Enumer Substituted Polyhexes Comput Chem. 1988;281413:31–36. [Google Scholar]
- 36••.Wingert BM, Oerlemans R, Camacho CJ. Optimal affinity ranking for automated virtual screening validated in prospective D3R grand challenges. J Comput Aided Mol Des. 2017;0:1–11. doi: 10.1007/s10822-017-0065-y. Application of align/dock and close/cross methods for second D3R Grand Challenge. Analysis of effects of receptor binding pocket on ranking outcomes and retrospective analysis of how to select optimal receptor structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumgartner MP, Evans DA. Lessons learned in induced fit docking and metadynamics in the Drug Design Data Resource Grand Challenge 2. J Comput Aided Mol Des. 2017;32:45–58. doi: 10.1007/s10822-017-0081-y. [DOI] [PubMed] [Google Scholar]
- 38.Ye Z, Baumgartner MP, Wingert BM, Camacho CJ. Optimal strategies for virtual screening of induced-fit and flexible target in the 2015 D3R Grand Challenge. J Comput Aided Mol Des. 2016;30:695–706. doi: 10.1007/s10822-016-9941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan R, Xu X, Zou X. Lessons learned from participating in D3R 2016 Grand Challenge 2: compounds targeting the farnesoid X receptor. J Comput Aided Mol Des. 2017;0:1–9. doi: 10.1007/s10822-017-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Zhang KYJ. Prospective evaluation of shape similarity based pose prediction method in D3R Grand Challenge 2015. J Comput Aided Mol Des. 2016;30:685–693. doi: 10.1007/s10822-016-9931-2. [DOI] [PubMed] [Google Scholar]
- 41.Salmaso V, Sturlese M, Cuzzolin A, Moro S. Combining self- and cross-docking as benchmark tools: the performance of DockBench in the D3R Grand Challenge 2. J Comput Aided Mol Des. 2017;0:1–14. doi: 10.1007/s10822-017-0051-4. [DOI] [PubMed] [Google Scholar]
- 42.Kadukova M, Grudinin S. Docking of small molecules to farnesoid X receptors using AutoDock Vina with the Convex-PL potential: lessons learned from D3R Grand Challenge 2. J Comput Aided Mol Des. 2017;0:1–12. doi: 10.1007/s10822-017-0062-1. [DOI] [PubMed] [Google Scholar]
- 43.Hogues H, Sulea T, Gaudreault F, Corbeil CR, Purisima EO. Binding pose and affinity prediction in the 2016 D3R Grand Challenge 2 using the Wilma-SIE method. J Comput Aided Mol Des. 2017;0:1–8. doi: 10.1007/s10822-017-0071-0. [DOI] [PubMed] [Google Scholar]
- 44.Ding X, Hayes RL, Vilseck JZ, Charles MK, Brooks CL. CDOCKER and λ-dynamics for prospective prediction in D3R Grand Challenge 2. J Comput Aided Mol Des. 2017;0:1–14. doi: 10.1007/s10822-017-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hawkins PCD, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J Chem Inf Model. 2010;50:572–84. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Baumgartner MP, Camacho CJ. Choosing the Optimal Rigid Receptor for Docking and Scoring in the CSAR 2013/2014 Experiment. J Chem Inf Model. 2016;56:1004–1012. doi: 10.1021/acs.jcim.5b00338. Introduction of align/dock and close/cross methods for use in prospective virtual screening challenge. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Réau M, Langenfeld F, Zagury JF, Montes M. Predicting the affinity of Farnesoid X Receptor ligands through a hierarchical ranking protocol: a D3R Grand Challenge 2 case study. J Comput Aided Mol Des. 2017;0:1–8. doi: 10.1007/s10822-017-0063-0. [DOI] [PubMed] [Google Scholar]
- 48.Schindler C, Rippmann F, Kuhn D. Relative binding affinity prediction of farnesoid X receptor in the D3R Grand Challenge 2 using FEP+ J Comput Aided Mol Des. 2017;0:1–8. doi: 10.1007/s10822-017-0064-z. [DOI] [PubMed] [Google Scholar]
- 49.Misini Ignjatovic M, Caldararu O, Dong G, Munoz-Gutierrez C, Adasme-Carreno F, Ryde U. Binding-affinity predictions of HSP90 in the D3R Grand Challenge 2015 with docking, MM/GBSA, QM/MM, and free-energy simulations. J Comput Aided Mol Des. 2016;30:707–730. doi: 10.1007/s10822-016-9942-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng N, Flynn WF, Xia J, Vijayan RSK, Zhang B, He P, Mentes A, Gallicchio E, Levy RM. Large scale free energy calculations for blind predictions of protein-ligand binding: the D3R Grand Challenge 2015. J Comput Aided Mol Des. 2016;30:743–751. doi: 10.1007/s10822-016-9952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]