Abstract
Mitochondria are central organelles to cellular metabolism. Their function relies largely on nuclear-encoded proteins that must be imported from the cytosol, and thus the protein import pathways are important for the maintenance of mitochondrial proteostasis. Mitochondrial HSP70 (mtHsp70) is a key component in facilitating the translocation of proteins through the inner membrane into the mitochondrial matrix. Its protein folding cycle is regulated by the nucleotide-exchange factor GrpE, which triggers the release of folded proteins by ATP rebinding. Vertebrates have two mitochondrial GrpE paralogs, GRPEL1 and 2, but without clearly defined roles. Using BioID proximity labeling to identify potential binding partners of the GRPELs in the mitochondrial matrix, we obtained results supporting a model where both GRPELs regulate mtHsp70 as homodimers. We show that GRPEL2 is not essential in human cultured cells, and its absence does not prevent mitochondrial protein import. Instead we find that GRPEL2 is redox regulated in oxidative stress. In the presence of hydrogen peroxide, GRPEL2 forms dimers through intermolecular disulfide bonds in which Cys87 is the thiol switch. We propose that the dimerization of GRPEL2 may activate the folding machinery responsible for protein import into mitochondrial matrix or enhance the chaperone activity of mtHSP70, thus protecting mitochondrial proteostasis in oxidative stress.
Keywords: GRPEL2, Oxidative stress, mtHSP70, Redox regulation, Mitochondrial protein import, Protein folding
1. Introduction
Redox homeostasis is critical for cell survival and functionality. Reversible formation of disulfide bonds by ROS oxidation of redox-sensitive cysteines is an important mechanism for regulating protein activity directly in response to the redox conditions in a given environment. Endoplasmic reticulum (ER) and the mitochondrial intermembrane space (IMS) are well known oxidative compartments in eukaryotic cells, where protein folding is accompanied by disulfide bond formation [25]. Critically regulating the proteostasis in those compartments, the redox-sensitive ER chaperone protein disulfide isomerase (PDI) acts as a sensor and processes oxidized proteins preventing their misfolding and aggregation [39], whereas the protein import into IMS is controlled by a disulfide relay formed by the Mia40 and Erv1 system [20]. However, such redox regulated switches have not been linked to the protein import and folding machinery of mitochondrial matrix proteins.
Most mitochondrial proteins are nuclear-encoded and must be imported into mitochondria. In the process of mitochondrial matrix protein import, preproteins with a presequence first enter through the translocase complexes of the outer and inner membrane (TOM and TIM, respectively). Mitochondrial HSP70 chaperone (mtHSP70) has a special role as the core of the presequence translocase-associated motor (PAM) in the matrix side, which drives the completion of protein transport into the matrix [30], [41], [4]. The exact mechanism of the import motor is not known, but it may function by a combination of trapping and pulling of the preprotein into the matrix [42]. After preprotein release, the presequence is removed by the mitochondrial processing peptidase, followed by folding of the imported proteins to their active conformation by molecular chaperone Hsp60 in cooperation with Hsp10. MtHSP70 also has a chaperoning role of the imported proteins to prevent misfolding and aggregation [11], [9].
MtHSP70 belongs to the family of 70 kDa heat shock proteins (HSP70) that assist in folding of newly synthesized, misfolded and aggregated proteins [34], [38]. The activity of HSP70s depends on their ability to circulate between ATP- and ADP-bound states. ADP-bound HSP70 has high affinity to the substrate protein, whereas the release of folded substrate requires ADP dissociation and ATP rebinding. The ATPase cycle is regulated by J-domain proteins, which target HSP70s to their substrates by stimulating ATP hydrolysis, and by nucleotide exchange factors, which determine the lifetime of the HSP70-substrate complex by facilitating ADP/ATP exchange and substrate release. In E.coli, the nucleotide exchange factor for DnaK (bacterial Hsp70) is GrpE [15], and its mammalian homologs are mitochondrial GRPEL1 and GRPEL2 [23], whereas the cytosolic HSP70 uses BAG1 as the nucleotide exchange factor [35]. The functional significance of having two GrpE proteins in mammalian mitochondria is not well understood.
We report here that the human mtHSP70 co-chaperone GRPEL2 is not essential for mitochondrial protein import in cultured cells. Instead we show that it forms dimers through redox regulated disulfide bond formation, thus potentially contributing to the regulation of mtHSP70 protein folding cycle in oxidative stress.
2. Results
2.1. GRPELs are ubiquitously expressed mitochondrial proteins
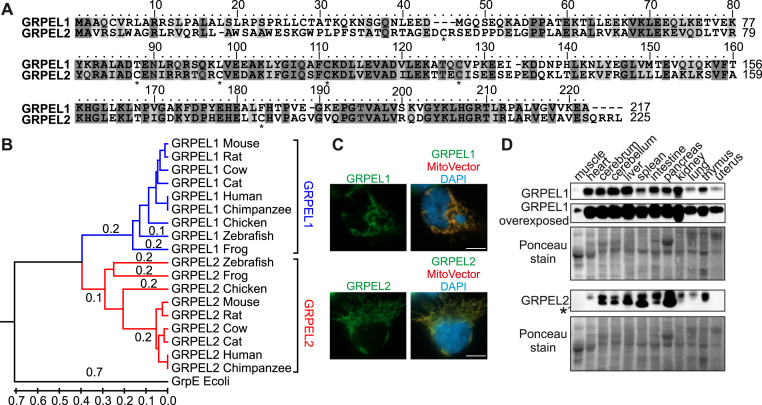
The human GRPEL1 and GRPEL2 are 217 and 225 amino acid proteins, respectively, being 42% identical and 64% similar in amino acid composition (Fig. 1A). Phylogenetic analysis shows that the GRPEL homologs co-exist in vertebrates, including fish and amphibians (Fig. 1B). The endogenous GRPELs localize strictly to mitochondria in human cells (Fig. 1C), as they contain an N-terminal mitochondrial targeting sequence. In all studied mouse tissues both GRPEL proteins were ubiquitously present, albeit at varying levels (Fig. 1D). At equal protein loading, GRPEL1 was particularly abundant in the mouse heart, brain, liver and kidney, while GRPEL2 protein levels were the highest in pancreas, cerebrum, spleen, intestine and thymus. The expression patterns may suggest tissue-specific functions of GRPEL paralogs in mammals.
Fig. 1.
Phylogenetic tree, protein expression profiles and alignment of GRPELs A. Protein sequence alignment of human GRPEL1 and GRPEL2. Identical residues are highlighted in dark gray, similar residues are in light gray. Cysteines in GRPEL2 are marked with asterisks. B. Phylogenetic tree of GRPELs in vertebrates generated from protein sequence alignments. The scale represents evolutionary distances, which were calculated as the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA7 [13]. C. Mitochondrial localization of GRPELs as shown by immunocytochemistry in 143B cells. Transfection of MitoVector was used for labeling mitochondria, DAPI shows nuclei. Scale bar, 10 µm. E. Protein expression profiles of GRPELs in mouse tissues by Western blotting of tissue lysates. Ponceau staining was used as a loading control. Asterisk marks a non-specific protein band.
2.2. GRPEL2 is not essential in cultured human cells
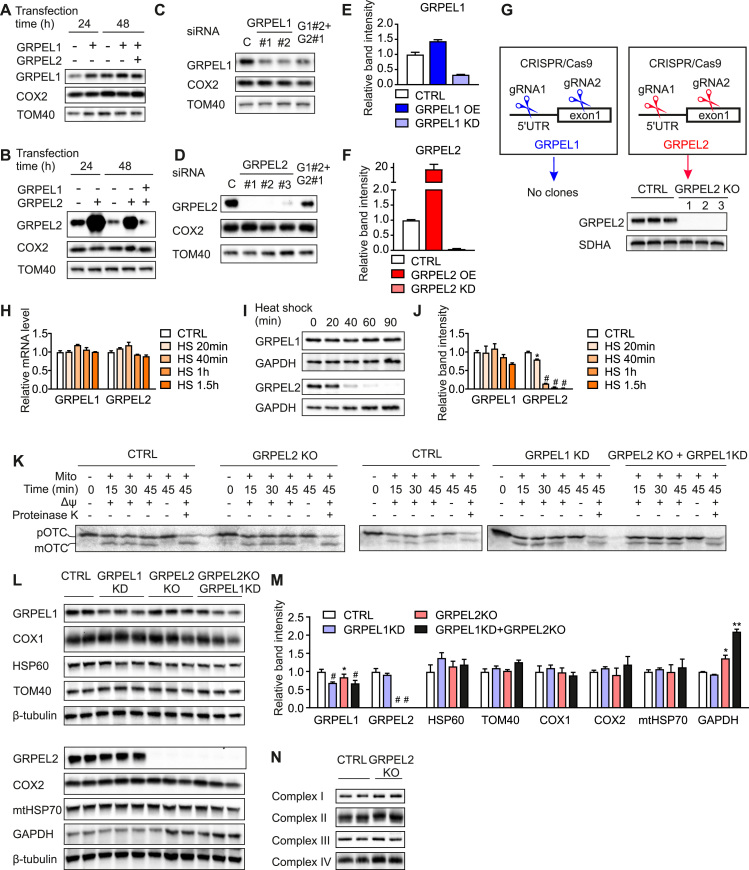
To investigate the importance of GRPELs in human cells, we tested overexpression, knockdown and knockout systems. Using the same transiently transfected overexpression plasmid (pBabe) in 143B osteosarcoma cells separately for GRPEL1 or GRPEL2 resulted in about 20-fold increase in GRPEL2 protein level, while GRPEL1 was only 1.5-fold increased (Fig. 2A,B,E,F). Similarly, siRNA-based knockdown reduced GRPEL1 protein level only about 70%, whereas GRPEL2 was nearly completely lost (Fig. 2C-F). We then tested CRISPR/Cas9-based editing on GRPEL genes in HEK293 cells, where two guide-RNAs were targeted simultaneously to the 5′UTR and exon 1 of each gene, in attempt to generate a deletion disrupting the transcription/translation of the gene. We were able to generate several independent clones lacking GRPEL2. However, no GRPEL1 knockout clones were obtained (Fig. 2G). These results indicate that GRPEL2 is a non-essential protein in cultured human cells, whereas GRPEL1 protein levels cannot be easily modulated. Interestingly, these results are supported by the human variation data in the ExAc database [14], which suggests that GRPEL1 is a haploinsufficient gene being intolerant to loss-of-function variants, unlike GRPEL2 (Table 1).
Fig. 2.
GRPEL2 is not essential for mitochondrial protein import in cultured cells A, B. Western blot analysis of 143B cells overexpressing GRPEL1, GRPEL2 or both. C, D. Western blot analysis of 143B cells depleted by siRNA for GRPEL1, GRPEL2 or both. G1, GRPEL1, G2, GRPEL2. E, F. Quantification of western blots showing the efficiency of GRPEL1 or GRPEL2 protein overexpression (OE) or knock down (KD) in 143B cells. The knock down or overexpression effect was analyzed after 48 h of transfection in comparison to TOM 40. G. CRISPR/Cas9 approach with guideRNAs (gRNA) was successful in generating several GRPEL2 knockout clones in HEK293 cells, but no GRPEL1 knockout clones were obtained. Western blot analysis shows full knockout of GRPEL2. SDHA was used as a loading control. H. mRNA levels of GRPELs in HEK293 cells exposed to heat shock as determined by qPCR assay (n = 4). I. Western blot analysis of HEK293 cells exposed to heat shock for indicated time. GAPDH was used as a loading control. J. Quantification of protein levels of GRPELs in HEK293 cells exposed to heat shock (n = 3). + 45 °C heat shock was used in all experiments. K. Import of [35S] preornithine transcarbamylase (pOTC) into isolated mitochondria of HEK293 cells. [35S] pOTC was incubated with mitochondria in the absence or presence of membrane potential (ΔΨ) for indicated time. As a control, after incubation with [35S] pOTC one sample was treated with proteinase K. Radiolabeled proteins were detected by phosphorimage analysis. Precursor (p) and mature matrix-processed (m) forms of OTC are indicated. L. Western blot analysis of HEK293 cells depleted for GRPEL1, GRPEL2 or for both. M. Quantification of protein levels determined by Western blots. β-tubulin was used as a loading control (n = 3). N. Blue native analysis of OXPHOS complexes in GRPEL2 KO cells. In all graphs data are presented as mean ± SD. CTRL, non-treated cells, *P < 0.05, #P < 0.0005 as compared to untreated cells (unpaired t-tests).
Table 1.
The number of GRPEL1 and GRPEL2 variants in the ExAC database.
|
GRPEL1 |
GRPEL2 |
|||||
|---|---|---|---|---|---|---|
| Constraint from ExAC |
Number of variants |
Metric |
Number of variants |
Metric | ||
| Expected | Observed | Expected | Observed | |||
| Synonymous | 36.0 | 31 | z = 0.51 | 31.4 | 32 | z = − 0.06 |
| Missense | 75.2 | 70 | z = 0.29 | 74.9 | 70 | z = 0.28 |
| Loss of function | 7.1 | 0 | pLI = 0.90 | 9.8 | 5 | pLI = 0.01 |
| Copy number variants | 4.6 | 9 | z = − 0.59 | 3.5 | 0 | z = 0.79 |
Z indicates quantified deviation of observed number of variants from expectation. pLI shows the probability of being loss-of-function intolerant and separates genes into loss-of-function intolerant (pLI ≥ 0.9) or loss-of-function tolerant (pLI ≤ 0.1) categories.
2.3. GRPEL2 protein is highly sensitive to heat stress
The E.coli GrpE is part of the heat shock regulon [16], but the promoters of mammalian GRPEL genes lack heat shock elements [10], [22]. Indeed, we could not detect any changes in the mRNA levels of GRPEL1 and GRPEL2 after heat treatment in HEK293 cells (Fig. 2H). Both the bacterial GrpE and the yeast mitochondrial GrpE (Mge1) were described to be thermosensors [21], [6]. Accordingly, the level of GRPEL2 protein was dramatically reduced already after 40 min of heat stress (45 °C) (Figs. 2I,J and S1). On the contrary, GRPEL1 protein levels were not affected by the similar heat treatment. Thus, despite their high similarity on amino acid sequence, GRPEL1 and GRPEL2 proteins have major differences in their properties including thermostability in vivo.
2.4. GRPEL2 knockout cells import mitochondrial proteins normally
Next we characterized the mitochondrial protein import in GRPEL2 knockout (KO) cells. Standard mitochondrial protein import assays of radiolabeled ornithine carbamoyltransferase (OTC) showed that GRPEL2 KO cells were able to fully import the OTC preprotein into mitochondria and process it into mature OTC (Fig. 2K). No differences in import efficiency were seen in GRPEL1 knockdown cells in which the GRPEL1 knockdown efficiency was limited as described above. Steady state levels of the tested mitochondrial proteins were not changed in cells lacking GRPEL2 or in cells partially depleted for GRPEL1 (Fig. 2L,M). Also, the levels of mitochondrial respiratory chain complexes were unchanged (Fig. 2N). These findings suggest that GRPEL2 is not critical for mitochondrial protein import. We, however, noted that the protein level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key enzyme in glycolysis, was increased in GRPEL2 KO cells.
2.5. Identification of proteins proximal to human GRPELs
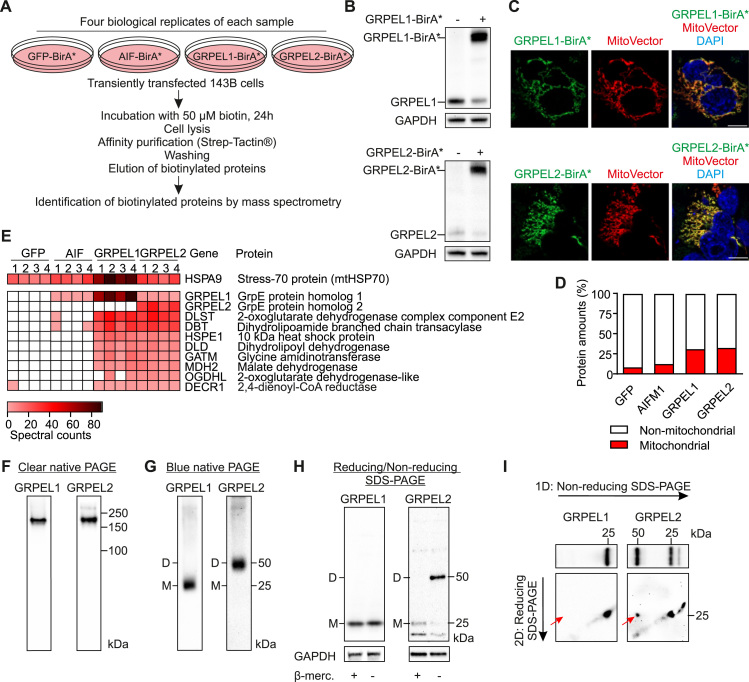
To reveal possible specific functions of GRPEL1 and GRPEL2 in human cells we sought to identify proteins that are in the vicinity of GRPEL1 or GRPEL2 using the proximity-dependent biotin identification (BioID) approach [26]. Promiscuous biotin ligase BirA* was fused to the C-terminus of either GRPEL1 or GRPEL2 (Fig. 3A), and the expression of the fusion constructs in human 143B cells was confirmed by western blotting (Fig. 3B). Immunocytochemistry showed that the GRPEL fusion proteins were localized correctly to mitochondria (Fig. 3C). Notably, overexpression of the fused constructs decreased the endogenous levels of the respective protein, suggesting proper intra-mitochondrial localization and functionality of the GRPEL1-BirA* and GRPEL2-BirA* fusion proteins (Fig. 3B). For the exclusion of non-specifically labeled proteins, we used BirA* fused to green fluorescent protein (GFP-BirA*) or to the apoptosis inducing factor (AIF-BirA*), an IMS protein. Biotinylated proteins from four biological replicates for each four BirA* fusion proteins were extracted using streptavidin beads and analyzed by mass spectrometry (Table S1).
Fig. 3.
Human GRPELs are homodimers with nearly identical proximal proteins A. Design of proteomic experiment using BioID assay. 143B cells were transiently transfected with indicated constructs. Biotin was added to the culture media for 24 h. The cells were lysed and biotinylated proteins were enriched by Strep-Tactin®. The samples were analyzed by mass spectrometry. B. Western blot analysis of GRPEL1-BirA* or GRPEL2-Bir* expression. GAPDH was used as a loading control. C. Intracellular localization of GRPEL1-BirA* or GRPEL2-Bir* determined by immunocytochemistry. MitoVector was used to label mitochondria, DAPI shows nuclei. Scale bar, 10 µm. D. Mitochondrial specificity of GRPEL1 or GRPEL2 proximal candidates before filtering. E. Heat map of the unique proximal proteins of GRPEL1 or/and GRPEL2. The spectral counts for mtHSP70 (HSPA9) are also shown. F. Clear native gel electrophoresis analysis of GRPELs in HEK293 cells G. Blue native gel electrophoresis analysis of GRPELs in HEK293 cells. H. Reducing and non-reducing western blotting of GRPELs. GAPDH was used as a loading control. I. Two-dimensional non-reducing/reducing SDS-PAGE of GRPELs. Red arrows point to the monomer in second dimension.
For the analysis of proximal partners, proteins that had two or more peptide spectrum matches in at least three replicates were included. Of the matches for each BirA* fusion protein, 31% of the proteins for GRPEL1, 32% for GRPEL2, 12% for AIF and 8% for GFP were mitochondrial (Fig. 3D). First, to characterize the high confidence interactors of GRPEL1 and GRPEL2, we excluded the proteins that were found in GFP or AIF control samples (Fig S2, Table S1). This list still contained many proteins that were identified as common mitochondrial matrix BioID matches in our recent study where several matrix proteins were investigated [17]. Thus, to identify unique matrix interactors of GRPEL1 and/or GRPEL2 we excluded matches that had an average fold change of two or more as compared to BioID matches from our previous study (Table S2, Fig S3). This resulted in a total of 10 proteins, which were potential binding partners of GRPEL1 and/or GRPEL2 (Fig. 3E, Table S2).
MtHSP70, the known functional interactor of GRPELs [18], [23], [33], was biotinylated by both GRPELs, as well as by AIF and GFP (Fig. 3E). However, GRPEL1 showed much higher spectral counts for proximity with mtHSP70 than GRPEL2 or the control proteins. GRPEL1 also showed strong proximity to itself, but not at all to GRPEL2. Likewise, GRPEL2 showed marked proximity with itself, but only a few hits with GRPEL1. These findings suggested that GRPELs form homodimers, and that GRPEL1 is the preferred interactor of mtHSP70 in normal culture conditions.
Otherwise the proximal proteomes of GRPEL1 and GRPEL2 were alike (Fig. 3E). Consistent with this, the biotinylation patterns of whole-cell lysates were highly similar for GRPEL1-BirA* and GRPEL2-BirA*(Fig S4). Most of the potential proteins interacting with both GRPELs (7 out of 10) were metabolic enzymes, in particularly dehydrogenases of the TCA cycle and dehydrogenases providing TCA cycle with acetyl-CoA.
2.6. Human GRPELs are homodimers
The crystal structure of bacterial GrpE shows that it is a dimer when interacting with DnaK [43], [7]. The ability of human GRPELs to dimerize in vitro was shown previously [24]. Recent study suggested that human GRPEL1 and GRPEL2 associate with mtHsp70 as a hetero-oligomeric subcomplex [33]. However, our BioID analysis indicated homodimerization of human GRPELs. To further dissect the dimerization of GRPELs we used clear native PAGE, which showed that both GRPELs are part of a high molecular weight complex (150–250 kDa) (Fig. 3F). By using blue native PAGE we could separate GRPEL2 from the large complex and detected a band around 50 kDa, which corresponds to dimeric GRPEL2 (Fig. 3G). Using non-reducing PAGE, we noticed that in the presence of β-mercaptoethanol, the GRPEL2 dimers disappeared, while in the absence of the reducing agent GRPEL2 dimers were preserved (Fig. 3H), suggesting that GRPEL2 formed dimers through a disulfide bond. Notably, monomeric GRPEL2 was depleted in non-reducing conditions, confirming the shift of GRPEL2 monomers toward dimers (Fig. 3H).
In the same conditions using blue native or non-reducing PAGE, we were not able to detect GRPEL1 dimers (Fig. 3G,H). We noted that the level of GRPEL1 monomer in non-reducing conditions was not changed, indicating that even if GRPEL1 dimers were present but not detected, they did not form by disulfide bonds. As it was possible that our GRPEL1 antibody did not recognize GRPEL1 dimers, we applied two-dimensional PAGE to test if GRPEL2 formed homodimers and not heterodimers with GRPEL1. Two-dimensional non-reducing/reducing SDS-PAGE demonstrated that the disulfide bond complex at 50 kDa contained only GRPEL2 and not GRPEL1 (Fig. 3I). Altogether these findings showed that human GRPEL2 is a homodimer formed by disulfide bonds. These results were confirmed on several human cell lines (Fig S5).
2.7. Redox regulation of GRPEL2 dimerization
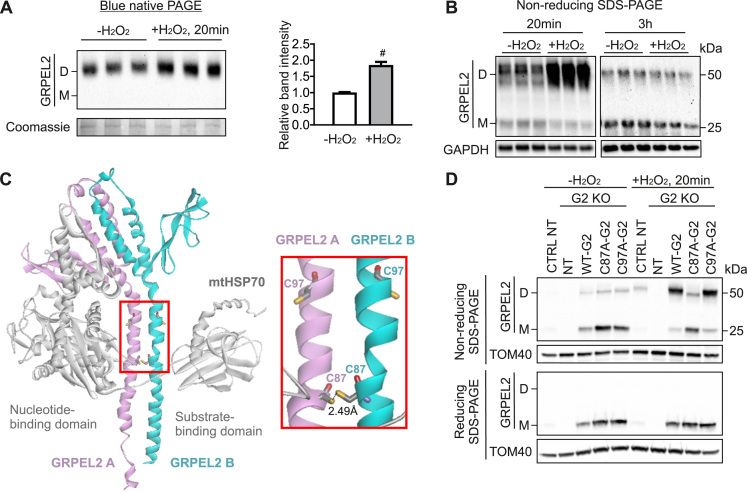
Previously the yeast homolog of GRPELs, Mge1, was suggested to act as an oxidative sensor shifting from an active dimer to inactive monomers by hydrogen peroxide [18], which was proposed to stall the transport of proteins into the mitochondrial matrix to prevent the accumulation of unfolded proteins [18]. To investigate whether human GRPEL2 is responsive to oxidative stress by changing oligomerization status we treated HEK293 cells with hydrogen peroxide and tested GRPEL2 oligomerization by blue native PAGE and non-reducing western blotting. We found that 20 min of H2O2 treatment significantly increased GRPEL2 dimers (Fig. 4A). This effect was transient as after 3 h of H2O2 treatment the dimer amount had returned to normal (Fig. 4B). Reducing western blot of the same samples showed that the overall abundance of GRPEL2 protein was not affected by H2O2 treatment (data not shown).
Fig. 4.
Oxidative stress induces GRPEL2 dimerization by disulfide bond formation A. Blue native analysis of HEK293 cells treated with H2O2 (left panel) and quantification of GRPEL2 dimers (right panel, n = 3). B. GRPEL2 dimerization in HEK293 cells treated with H2O2 for 20 min or for 3 h. C. In the left panel, the structural model of GRPEL2 dimer (magenta, GRPEL2 A; cyan, GRPEL2 B) and mtHSP70 (in gray) is shown. GRPEL2 A and B indicate the two monomers and their intermolecular interaction between cysteine 87 residues is shown in the right panel. Cysteine 97 residues are also indicated. For simplicity, the structure of only one mtHSP70 molecule is displayed. D. Dimerization of wild type (WT) and mutant GRPEL2 in response to H2O2 treatment. GRPEL2 KO HEK293 cells were transiently transfected with WT or mutant GRPEL2 for 24 h. Data are presented as mean ± SD. #P < 0.0005 as compared to not treated cells (unpaired t-tests). CTRL, control, parental cells.
To investigate which cysteines might be responsible for the increased disulfide bond formation of GRPEL2 in oxidative stress, we modelled the GRPEL2 structure based on the crystal structure of GrpE of Geobacillus kaustophilus [43] (Fig. 4C). This bacterial GrpE is a homodimer crystallized with a nearly full length G. kaustophilus DnaK. The GrpE dimer interface comprises two N-terminal α-helices and a C-terminal four-helix bundle. The long N-terminal α-helices interact with the substrate binding and linker domains of DnaK, whereas the C-termini interact with the ATPase domain of DnaK. GRPEL2 contains six cysteine residues (Fig. 1B). Of those, based on the structural model, Cys87 was the likely candidate to be involved in disulfide bond formation between N-terminal α-helixes (Fig. 4C), whereas the close Cys97 is placed outside of GrpE dimer interface. We generated overexpression constructs where the respective cysteine residues were mutated to alanine. We transiently expressed wild type GRPEL2, GRPEL2-C87A or GRPEL2-C97A in GRPEL2 KO cells, and treated the cells with H2O2 for 20 min. The results clearly showed that under oxidative stress the dimerization of wild type GRPEL2 and GRPEL2-C97A was induced, but that GRPEL2-C87A did not respond to H2O2 treatment (Fig. 4D). Cys87 in GRPEL2 is thus a redox regulated residue, which is oxidized under H2O2 and forms an intramolecular disulfide bond between two GRPEL2 molecules. Based on our structural modeling there are no cysteines in GRPEL1, which could potentially make intermolecular disulfide bonds with another GRPEL1 molecule or with GRPEL2. This suggests that redox regulation is a specific feature of GRPEL2.
3. Discussion
We show here that human GRPEL2, a nucleotide exchange factor for mtHSP70, is a redox-sensitive protein forming dimers in oxidative stress. This finding may link redox regulation to import and folding of mitochondrial matrix proteins. Overall our results suggest that GRPEL1 is the housekeeping import co-chaperone, required to maintain protein translocation by the PAM complex, whereas GRPEL2 is not essential in cultured cells but may have developed to fine-tune the protein import and folding in response to altered redox state. This division of labor between GRPEL1 and GRPEL2 was also supported by human variation data, showing that loss of function variants are tolerated in GRPEL2 but not in GRPEL1. In line with our findings, human GRPEL1 was shown to complement the lethality of ∆Mge1 yeast strain [10], while yeast cells transformed with human GRPEL2 were not viable [33]. Furthermore, our BioID analysis suggested that GRPEL1 is found more in close proximity with mtHSP70 than is GRPEL2, indicating the role of GRPEL1 as the main nucleotide-exchange factor in normal culture conditions. Earlier in vitro study also showed that GRPEL2 had 5-fold lower affinity for mtHSP70 as compared to GRPEL1 [33]. Both GRPELs were, however, present in high molecular weight complexes of approximately the same size, and both had many of the same proximal partners in our BioID analysis, suggesting that they may be physically in the same large complexes, although form only or mainly homodimers. Using blue native electrophoresis, which dissociates labile supramolecular assemblies, we could separate the GRPEL2 dimer, while GRPEL1 appeared as a monomer, indicating that the stability of GRPEL1 homodimer may require interaction with mtHSP70.
The yeast Mge1 was previously suggested to switch from active dimers to inactive monomers in response to oxidative stress to stall transport of proteins. This action was dependent on Met155 [18]. In contrast we show that the human GRPEL2 forms dimers in oxidative stress, and uses Cys87 as the thiol switch. While human GRPEL2 has no methionines, the yeast Mge1 does not contain any cysteine residues that would enable the formation of disulfide bridges between Mge1 molecules. Clearly, such regulation of Mge1 and GRPEL2 dimerization to opposite directions has evolved for different purposes. The oxidation of GRPEL2 ensures a rapid effect on mtHSP70 function, before any major changes take place on transcriptional and/or translational levels. We thus propose that the redox regulated GRPEL2 acts as a sensor of oxidative stress enhancing the activity of mtHSP70, in order to prevent misfolding of imported proteins. Further studies under physiological conditions are, however, needed to show whether the disulfide formation in GRPEL2 depends on cellular redox levels. Furthermore, investigation of how the redox regulated disulfide bond actually affects mitochondrial protein import and folding will be of interest. For example, cysteine oxidation of BiP, the Hsp70 chaperone in the ER, enhances its chaperone activity during suboptimal folding conditions [40]. Recent study in yeast suggested that mitochondria have a role in maintaining cytosolic proteostasis by importing cytosolic misfolding proteins into mitochondria for degradation [27]. Should such a quality control process take place in mammalian mitochondria, GRPEL2 could be a sensor for a redox state where proteins aggregate and require enhanced mitochondrial protein import.
The ability of GRPEL2 to form disulfide bonds by hydrogen peroxide using the mechanism described in our study is restricted to primates and rodents as Cys87 is not conserved in other species (Fig S6). Recently it was proposed that such redox regulated mechanisms may be particularly relevant to longer-lived species with a high metabolic rate to counteract the effect of age-associated oxidative stress [3]. Cysteine oxidation of GRPEL2 may thus play a role in the metabolic adaptation to redox stress, which could be important in for example high fat feeding–induced oxidative stress. This remains to be studied in a relevant rodent model.
Recent studies have applied BioID to identify the proximal partners of mitochondrial matrix proteins of interest [32], [5]. We note that many of the studies have reported a nearly identical list of proximal proteins that we observed in our BioID analysis before filtering our data against the common mitochondrial matrix BioID hits from our recent large-scale study where five different mitochondrial matrix proteins were used as baits [17]. The common matrix contaminants included for example Complex I and mitoribosome proteins, CLLP and CLPX, LRPPRC and PRDX3 (Fig S3). It seems that these hits should be excluded as background signal for most biotin-ligated mitochondrial matrix proteins. After filtering out these contaminants, we were left with mostly metabolic enzymes, many of which were dehydrogenases. Further studies are thus required to investigate if there is a direct functional connection between the redox altering potential of GRPEL2 and the dehydrogenase activities.
Disulfide bond formation is the most common posttranslational protein modification in response to potentially damaging redox imbalance [12], and it has an important role in maintaining cellular proteostasis. Mitochondrial proteins can act as sensors of redox state and subsequently fine-tune metabolic and other functions [1], [31], [36]. Thiol-based redox modifications are likely to be common in many more mitochondrial proteins, and it will be interesting to reveal their role in mediating mitochondrial redox state and metabolic function in response to changes in ROS levels. Our study provides a potential link between redox regulation and matrix protein import and folding in human mitochondria.
4. Materials and methods
4.1. Phylogenetic analysis and protein sequence alignment
Protein sequences of GRPEL homologs were obtained from UniProt database (https://www.uniprot.org/). The sequences were aligned using PROMALS3D. The aligned sequences were then used to generate a phylogeny tree in MEGA7 by UPGMA (Unweighted Pair Group Method with Arithmetic Mean) method [13]. The evolutionary distances were computed using the Poisson correction method.
4.2. Cell culture and treatments
Human osteosarcoma (143B) or human embryonic kidney (HEK293) cells were grown in DMEM medium (Lonza, Belgium) with 10% fetal bovine serum, l-glutamine and penicillin/streptomycin. SH-SY5Y cells were cultured in mixture of EMEM/F12 medium (1:1) supplemented with 10% fetal bovine serum, non-essential amino acids, penicillin/streptomycin, sodium pyruvate, D-glucose, uridine and GlutaMAX. Cells were treated with 500 μM H2O2 (Sigma) for indicated time. For heat shock treatments, cells were incubated at 45 °C or 42 °C for indicated times and collected immediately after heat shock. To transfect cells with plasmid DNA or with siRNA JetPrime reagent (Polyplus) was used. For silencing experiments we used sets of three stealth siRNAs for GRPEL1 (Life Technologies, HSS129560, HSS129561, HSS188417) or for GRPEL2 (Life Technologies, HSS134522, HSS134523, HSS175076). After transfection cells were incubated for 24 or 48 h.
4.3. Plasmid construction
Human GRPEL1 or GRPEL2 were cloned into pBabe-puro expression vector using EcoRI and SalI restriction sites. Point mutations for amino acid changes C87A or C97A in human GRPEL2 were introduced by PCR overlap extension using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). To fuse BirA* to C-terminus of GRPEL1 or GRPEL2 we used PCR overlap extension. pHA-BirA* and pBabe-GRPEL1 or pBabe GRPEL2 were used as a templates. The fused constructs were inserted into pBabe-puro vector using EcoRI and SalI restriction sites. GFP-BirA* and AIF-BirA* plasmids [17].
4.4. Immunocytochemistry
143B cells were transiently transfected with pDsRed2-Mito Vector (Clontech, 632421) to label mitochondria. In the experiments with BirA* fused proteins, cells were also co-transfected with GRPEL1-BirA* or GRPEL2-BirA*. After 24 h cells were fixed with 4% paraformaldehyde for 10 min at RT and washed with PBS. Then cells were permeabilized with Triton X-100 for 15 min at RT, washed and blocked with 5% BSA for 2 h at RT. Cells were then incubated with corresponding primary antibody against GRPEL1 (Novus Biological, NBP1–83557) or GRPEL2 (Novus Biological, NBP1–85099) in blocking buffer over night at + 4 °C. After washing cells were incubated with secondary antibody for 1 h at RT (Alexa Fluor 488 goat anti-rabbit, Invitrogen, R37116). Finally, cells were washed, mounted and imaged with Axio Observer Z1 (Zeiss).
4.5. Mouse tissues
Proteins were extracted from tissue samples of C57BL/6JOlaHsd mice. Tissues were homogenized in PBS (100 μl/10 mg of tissue) with protease inhibitor (Halt, Thermo Fisher Scientific). 5xRIPA (Cell signaling technology) was added to homogenates. After 20 min incubation on ice the samples were centrifugated at 14,000 g for 10 min (+ 4 °C).
4.6. Native and SDS–PAGE
For clear native PAGE and non-reducing/reducing SDS-PAGE proteins were extracted from cells using NP-40 lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl (pH 8.0), 10% glycerol, 150 mM NaCl, 5 mM NaF, 5 mM ZnCl, 1 mM Na3VO4, 10 mM EGTA) containing protease inhibitors (Halt, Thermo Fisher Scientific). Lysates were incubated on ice for 10 min before centrifugation at 12,000 g for 20 min at + 4 °C. Protein samples were separated on 10% Mini-PROTEAN TGX Precast Gels (Bio-Rad). For clear native PAGE samples buffer was: 62.5 mM Tris-HCl, pH 6.8, 25% (v/v) glycerol, 0.01% (w/v) bromophenol blue) and running buffer was: 25 mM Tris, 192 mM glycine. For reducing/non-reducing SDS-PAGE sample buffer for non-reducing (250 mM Tris-HCl (pH 6.8), 10% SDS, 30% glycerol, 0.02% bromophenol blue) and reducing (Laemmli sample buffer (Bio-Rad) including 10% β-mercaptoethanol) conditions were used respectively. Protein extracts for reducing SDS-PAGE analysis were prepared by lysing cells in RIPA buffer containing protease inhibitors (Halt, Thermo Fisher Scientific). Mitochondrial protein samples purified from digitonin treated cells were prepared for BN-PAGE analysis as previously described [29]. Samples were run on 6–15% Bis-Tris Native PAGE gels. For 2D analysis samples were separated in the first dimension under non-reducing conditions. Then the gel lanes were cut out and either directly immunoblotted or incubated in reducing Laemmli sample buffer (Bio-Rad) containing 10% β-mercaptoethanol for 10 min at RT and subjected to the second dimension PAGE.
4.7. Immunoblotting
After clear native-, blue native- or SDS-PAGE, proteins were transferred onto PVDF membrane by Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked in 5% milk in TBS–Tween 20 (0.1%). Proteins were immunoblotted with the indicated primary antibodies and corresponding secondary antibodies. The following antibodies were used for protein detection: GRPEL1 (Novus Biological, NBP1–83557), GRPEL2 (Novus Biological, NBP1–85099), Tom40 (Santa Cruz, sc-11414), Complex I subunit NDUFA9 (Mitosciences, MS111), Complex II subunit SDHA (Mitosciences, MS204), Complex III subunit UQCRC2 (Abcam, ab14745), Complex IV subunit 1 (Mitosciences, MS404), Complex IV subunit 2 (GeneTex, GTX62145), mtHSP60 (Santa Cruz, sc-1052), mtHSP70 (Abcam, ab53098) and GAPDH (Cell Signaling, 14C10). Enhanced chemiluminescent substrate and ChemiDoc imaging station (Bio-Rad) were used for signal detection. Quantification of the bands was performed by Image Lab Software (Bio-Rad).
4.8. Generation of gene knockouts
CRISPR/Cas9 was used to generate knockout HEK293 cells. Cells were co-transfected with two gRNA transcriptional cassettes prepared by PCR and CAG-Cas9-T2A-EGFP plasmid (Addgene #7831) as described elsewhere [28]. One of the guide RNAs was targeted to 5′UTR region and the other to exon 1 of the target gene. After 24 h of transfection GFP-positive cells were sorted by FACS and single cell clones generated. The sequences of gRNAs are available on request.
4.9. Real-time PCR assay
Total RNA was extracted using a Mini spin kit (Macherey-Nagel). 1000 ng of RNA was reverse transcribed using Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Fisher scientific). Real-time PCR analysis was done by DyNAmo Flash SYBR Green qPCR Kit (Thermo Fisher scientific) using CFX96™ Real-Time PCR Detection System (Bio-Rad). PCR program started with 95 °C for 7 min followed by 40 cycles at 95 °C for 10 s and 60 °C for 30 s. Beta-2-microglobulin (B2M) was used as a reference gene for normalization and mRNA expression level was calculated using comparative Ct (threshold cycle) method. All primer sequences are available on request.
4.10. Mitochondrial protein import assay
To analyze mitochondrial protein import we synthesized 35S-labled precursor of ornithine transcarbamylase (OTC) by TnT Quick Coupled Transcription/Translation Systems (Promega) using human OTC TnT plasmid (a gift from Prof. Mike Ryan, Monash University, Melbourne, Australia). Mitochondria were isolated from HEK293 cells using trehalose isolation buffer (300 mM trehalose, 10 mM HEPES, 10 mM KCl, 1 mM EGTA, 1 mM EDTA and 2 mM PMSF). Import reaction was performed at 30 °C using import buffer (250 mM sucrose, 80 mM potassium acetate, 5 mM magnesium acetate, 5 mM methionine, 10 mM sodium succinate, 20 mM HEPES/KOH pH 7.4, 0.2 M ATP and 2.5 mg/mL BSA). Samples were run on 12.5% SDS-PAGE. Translocation of 35S-labled OTC to mitochondria was assessed using radioactive detection by storage phosphor screen.
4.11. Biotinylation of the proximal proteins
For each replicate 143B cells were grown on five 15 cm dishes. The cells were transfected with 15 μg of BirA* fusion construct using jetPRIME reagent according to the manufacture's manual. Following 24 h of transfection, 50 μM biotin was added to each plate and allowed to biotinylate the proximal proteins for the next 24 h. The plates were washed with PBS, and cells scraped, pelleted and frozen at − 80 °C. For each sample four biological replicates were analyzed.
4.12. Protein complex purification and mass spectrometry
For BioID purification, cell pellet was thawed in 3 mL ice cold lysis buffer (0.5% IGEPAL, 50 mM Hepes, pH 8.0, 150 mM NaCl, 50 mM NaF, 1.5 mM NaVO3, 5 mM EDTA, 0.1% SDS, supplemented with 0.5 mM PMSF and protease inhibitors; Sigma). Lysates were sonicated, treated with benzonase and following loaded on spin columns (Bio-Rad) containing 200 μl Strep-Tactin beads (IBA, GmbH). Detailed protein complex purification steps were explained in [8]. Liquid chromatography–mass spectrometry (LC-MS) analysis was performed on an Orbitrap Elite ETD mass spectrometer (Thermo Scientific) using the Xcalibur version 2.7.1 coupled to a Thermo Scientific nLCII nanoflow system. LC-MS parameters using for the performance were as described previously [37]. Thermo.RAW files were searched with Proteome Discoverer 1.4 (Thermo Scientific) against SEQUEST search engine. The search parameters were taken from [44]. All reported data were based on high confidence peptides assigned in Proteome Discoverer with a 0.01% FDR by Percolator. Control samples (GFP and AIF) and Contaminant Repository for Affinity Purification (CRAPome, http://www.crapome.org/) database [19] were preliminary used to identify high confidence interactors of GRPELs based on the spectral counts.
4.13. Structure analysis
The modeling of GRPEL and mtHSP70 structures was done by using the crystal structure of bacterial GrpE and DnaK protein complex (PDB entry 4ANI) as template [43]. Multiple sequence alignment of template G. kaustophilus (213 aa) and human GRPEL2 (225 aa) along with a number of GrpE homologs was done using PROMALS3D server. The obtained alignment was used for homology modeling by SWISS-MODEL server [2]. Images of the molecular structure were produced using BIOVIA Discovery Studio Visualizer v17.2.0 software (Dassault Systèmes BIOVIA).
Acknowledgements
Riitta Lehtinen, Maria Shcherbii and Laura Mäenpää are thanked for assistance. Prof. Mike Ryan and Prof. Agnieszka Chacińska are acknowledged for providing their protocols and plasmids for mitochondrial import experiments. Dr. Brendan Battersby and Taina Turunen are acknowledged for sharing expertise on preparing constructs for BioID. Genome Biology Unit (GBU) and FACS Core Facility at Biomedicum of Helsinki are acknowledged for their services. CSC-IT Center for Science and Kimmo Mattila are acknowledged for the help with using BIOVIA Discovery Studio Visualizer v17.2.0 software. This work was supported by European Research Council (Grant no. 637458), Academy of Finland Center of Excellence in Stem Cell Metabolism (312438), University of Helsinki and Sigrid Juselius Foundation.
Acknowledgments
Author contributions
SK designed and executed most experiments with the help of NN, TH, and YY. SB performed the BioID experiment with the expertise support of XL and MV. PM carried out mutagenesis experiments. DB contributed to generating CRISPR/Cas9 knockouts. MT designed and executed the mitochondrial protein import assay experiments. HT and SK supervised the study and wrote the manuscript. All authors reviewed and edited manuscript.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.07.024.
Contributor Information
Svetlana Konovalova, Email: svetlana.konovalova@helsinki.fi.
Henna Tyynismaa, Email: henna.tyynismaa@helsinki.fi.
Appendix A. Supplementary material
Supplementary material
Fig. S1. Heat stress induces GRPEL2 protein degradation and does not change GRPEL2 oligomerization Non-reducing and reducing western blotting of HEK293 cells exposed to heat shock at + 45 °C (left panel) or + 42 °C (right panel) for indicated time. GAPDH was used as a loading control. Fig. S2. Heat map of the identified high confidence interactors of GRPEL1 or/and GRPEL2. The BioID hits shown here for GRPEL1/2 were filtered against the hits for AIF and GFP but not against common mitochondrial matrix hits [16]. Fig. S3. BioID hits of GRPEL interactome before and after filtering against common mitochondrial matrix hits. Classification of the proteins potentially interacting with GRPEL1 or GRPEL2 without (upper panel, high confidence interactors) and with (lower panel, unique interactors) filtering against common mitochondrial matrix hits [16]. The proteins found as common mitochondrial matrix hits are in light gray in the lower panel. Fig. S4. GRPEL1-BirA* and GRPEL2-BirA* have similar biotinylation patterns Western blot analysis of total cell lysates from 143B cells transiently expressing indicated BirA* fusion constructs with or without biotin treatment. Left: stain-free gel image. Right: streptavidin blot. Black arrows show endogenously biotinylated proteins. Red arrows show proteins specifically biotinylated by GRPEL1-BirA* and GRPEL2-BirA*. Fig. S5. GRPEL2 forms disulfide bond dimers in cultured human cells Non-reducing and reducing western blotting of osteosarcoma (143B) and neuroblastoma (SH-SY5Y) cell lysates. Fig. S6. Cysteine 87 is conserved in primates and rodents. Protein sequence alignment showing amino acid residues corresponding to human GRPEL2 cysteine 87. Cysteines are marked with red. Identical residues are highlighted in dark gray, similar residues are in light gray.
References
- 1.Bak D.W., Weerapana E. Cysteine-mediated redox signalling in the mitochondria. Mol. Biosyst. 2015;11:678–697. doi: 10.1039/c4mb00571f. [DOI] [PubMed] [Google Scholar]
- 2.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll B., Otten E.G., Manni D., Stefanatos R., Menzies F.M., Smith G.R., Jurk D., Kenneth N., Wilkinson S., Passos J.F., Attems J., Veal E.A., Teyssou E., Seilhean D., Millecamps S., Eskelinen E.L., Bronowska A.K., Rubinsztein D.C., Sanz A., Korolchuk V.I. Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat. Commun. 2018;9 doi: 10.1038/s41467-017-02746-z. (256-017-02746-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demishtein-Zohary K., Azem A. The TIM23 mitochondrial protein import complex: function and dysfunction. Cell Tissue Res. 2017;367:33–41. doi: 10.1007/s00441-016-2486-7. [DOI] [PubMed] [Google Scholar]
- 5.Fischer F., Langer J.D., Osiewacz H.D. Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism. Sci. Rep. 2015;5:18375. doi: 10.1038/srep18375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimshaw J.P., Jelesarov I., Schonfeld H.J., Christen P. Reversible thermal transition in GrpE, the nucleotide exchange factor of the DnaK heat-shock system. J. Biol. Chem. 2001;276 doi: 10.1074/jbc.M009290200. (6098-6104) [DOI] [PubMed] [Google Scholar]
- 7.Harrison C.J., Hayer-Hartl M., Di Liberto M., Hartl F., Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 8.Heikkinen T., Kampjarvi K., Keskitalo S., von Nandelstadh P., Liu X., Rantanen V., Pitkanen E., Kinnunen M., Kuusanmaki H., Kontro M., Turunen M., Makinen N., Taipale J., Heckman C., Lehti K., Mustjoki S., Varjosalo M., Vahteristo P. Somatic MED12 nonsense mutation escapes mRNA decay and reveals a motif required for nuclear entry. Hum. Mutat. 2017;38:269–274. doi: 10.1002/humu.23157. [DOI] [PubMed] [Google Scholar]
- 9.Horst M., Oppliger W., Rospert S., Schonfeld H.J., Schatz G., Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda E., Yoshida S., Mitsuzawa H., Uno I., Toh-e A. YGE1 is a yeast homologue of Escherichia coli grpE and is required for maintenance of mitochondrial functions. FEBS Lett. 1994;339:265–268. doi: 10.1016/0014-5793(94)80428-1. [DOI] [PubMed] [Google Scholar]
- 11.Kang P.J., Ostermann J., Shilling J., Neupert W., Craig E.A., Pfanner N. Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature. 1990;348:137–143. doi: 10.1038/348137a0. [DOI] [PubMed] [Google Scholar]
- 12.Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Tukiainen T., Birnbaum D.P., Kosmicki J.A., Duncan L.E., Estrada K., Zhao F., Zou J., Pierce-Hoffman E., Berghout J., Cooper D.N. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipinska B., King J., Ang D., Georgopoulos C. Sequence analysis and transcriptional regulation of the Escherichia coli grpE gene, encoding a heat shock protein. Nucleic Acids Res. 1988;16:7545–7562. doi: 10.1093/nar/16.15.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Ohman T., Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-03523-2. (1188-018-03523-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marada A., Allu P.K., Murari A., PullaReddy B., Tammineni P., Thiriveedi V.R., Danduprolu J., Sepuri N.B. Mge1, a nucleotide exchange factor of Hsp70, acts as an oxidative sensor to regulate mitochondrial Hsp70 function. Mol. Biol. Cell. 2013;24:692–703. doi: 10.1091/mbc.E12-10-0719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., Halim V.A., Bagshaw R.D., Hubner N.C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W.H., Goudreault M., Lin Z.Y. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J.M. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Moro F., Muga A. Thermal adaptation of the yeast mitochondrial Hsp70 system is regulated by the reversible unfolding of its nucleotide exchange factor. J. Mol. Biol. 2006;358:1367–1377. doi: 10.1016/j.jmb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 22.Naylor D.J., Hoogenraad N.J., Hoj P.B. Isolation and characterisation of a cDNA encoding rat mitochondrial GrpE, a stress-inducible nucleotide-exchange factor of ubiquitous appearance in mammalian organs. FEBS Lett. 1996;396:181–188. doi: 10.1016/0014-5793(96)01100-3. [DOI] [PubMed] [Google Scholar]
- 23.Naylor D.J., Stines A.P., Hoogenraad N.J., Hoj P.B. Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the Co-chaperones of specific Hsp70 members. J. Biol. Chem. 1998;273:21169–21177. doi: 10.1074/jbc.273.33.21169. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira C.L., Borges J.C., Torriani I.L., Ramos C.H. Low resolution structure and stability studies of human GrpE#2, a mitochondrial nucleotide exchange factor. Arch. Biochem. Biophys. 2006;449:77–86. doi: 10.1016/j.abb.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Riemer J., Bulleid N., Herrmann J.M. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 26.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan L., Zhou C., Jin E., Kucharavy A., Zhang Y., Wen Z., Florens L., Li R. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017;543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saarimaki-Vire J., Balboa D., Russell M.A., Saarikettu J., Kinnunen M., Keskitalo S., Malhi A., Valensisi C., Andrus C., Eurola S., Grym H., Ustinov J., Wartiovaara K., Hawkins R.D., Silvennoinen O., Varjosalo M., Morgan N.G., Otonkoski T. An activating STAT3 mutation causes neonatal diabetes through premature induction of pancreatic differentiation. Cell Rep. 2017;19:281–294. doi: 10.1016/j.celrep.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 29.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 30.Schulz C., Schendzielorz A., Rehling P. Unlocking the presequence import pathway. Trends Cell Biol. 2015;25:265–275. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Shutt T., Geoffrion M., Milne R., McBride H.M. The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 2012;13:909–915. doi: 10.1038/embor.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva J., Aivio S., Knobel P.A., Bailey L.J., Casali A., Vinaixa M., Garcia-Cao I., Coyaud E., Jourdain A.A., Perez-Ferreros P., Rojas A.M., Antolin-Fontes A., Samino-Gene S., Raught B., Gonzalez-Reyes A., Ribas de Pouplana L., Doherty A.J., Yanes O., Stracker T.H. EXD2 governs germ stem cell homeostasis and lifespan by promoting mitoribosome integrity and translation. Nat. Cell Biol. 2018;20:162–174. doi: 10.1038/s41556-017-0016-9. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava S., Savanur M.A., Sinha D., Birje A., R.V., Saha P.P., D'Silva P. Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J. Biol. Chem. 2017;292:18075–18090. doi: 10.1074/jbc.M117.788463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strub A., Lim J.H., Pfanner N., Voos W. The mitochondrial protein import motor. Biol. Chem. 2000;381:943–949. doi: 10.1515/BC.2000.115. [DOI] [PubMed] [Google Scholar]
- 35.Takayama S., Bimston D.N., Matsuzawa S., Freeman B.C., Aime-Sempe C., Xie Z., Morimoto R.I., Reed J.C. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thaher O., Wolf C., Dey P.N., Pouya A., Wullner V., Tenzer S., Methner A. The thiol switch C684 in Mitofusin-2 mediates redox-induced alterations of mitochondrial shape and respiration. Neurochem. Int. 2017 doi: 10.1016/j.neuint.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S., Aebersold R., Bennett K.L., Colinge J., Gstaiger M., Superti-Furga G. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat. Methods. 2013;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 38.Voos W., Rottgers K. Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1592:51–62. doi: 10.1016/s0167-4889(02)00264-1. [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Yu J., Huo L., Wang L., Feng W., Wang C.C. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a′. J. Biol. Chem. 2012;287:1139–1149. doi: 10.1074/jbc.M111.303149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Pareja K.A., Kaiser C.A., Sevier C.S. Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress. eLife. 2014;3:e03496. doi: 10.7554/eLife.03496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiedemann N., Frazier A.E., Pfanner N. The protein import machinery of mitochondria. J. Biol. Chem. 2004;279:14473–14476. doi: 10.1074/jbc.R400003200. [DOI] [PubMed] [Google Scholar]
- 42.Wiedemann N., Pfanner N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 43.Wu C.C., Naveen V., Chien C.H., Chang Y.W., Hsiao C.D. Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system: insight into intermolecular communication. J. Biol. Chem. 2012;287:21461–21470. doi: 10.1074/jbc.M112.344358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yadav L., Tamene F., Goos H., van Drogen A., Katainen R., Aebersold R., Gstaiger M., Varjosalo M. Systematic Analysis of Human Protein Phosphatase Interactions and Dynamics. Cell Syst. 2017;4:430–444. doi: 10.1016/j.cels.2017.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Fig. S1. Heat stress induces GRPEL2 protein degradation and does not change GRPEL2 oligomerization Non-reducing and reducing western blotting of HEK293 cells exposed to heat shock at + 45 °C (left panel) or + 42 °C (right panel) for indicated time. GAPDH was used as a loading control. Fig. S2. Heat map of the identified high confidence interactors of GRPEL1 or/and GRPEL2. The BioID hits shown here for GRPEL1/2 were filtered against the hits for AIF and GFP but not against common mitochondrial matrix hits [16]. Fig. S3. BioID hits of GRPEL interactome before and after filtering against common mitochondrial matrix hits. Classification of the proteins potentially interacting with GRPEL1 or GRPEL2 without (upper panel, high confidence interactors) and with (lower panel, unique interactors) filtering against common mitochondrial matrix hits [16]. The proteins found as common mitochondrial matrix hits are in light gray in the lower panel. Fig. S4. GRPEL1-BirA* and GRPEL2-BirA* have similar biotinylation patterns Western blot analysis of total cell lysates from 143B cells transiently expressing indicated BirA* fusion constructs with or without biotin treatment. Left: stain-free gel image. Right: streptavidin blot. Black arrows show endogenously biotinylated proteins. Red arrows show proteins specifically biotinylated by GRPEL1-BirA* and GRPEL2-BirA*. Fig. S5. GRPEL2 forms disulfide bond dimers in cultured human cells Non-reducing and reducing western blotting of osteosarcoma (143B) and neuroblastoma (SH-SY5Y) cell lysates. Fig. S6. Cysteine 87 is conserved in primates and rodents. Protein sequence alignment showing amino acid residues corresponding to human GRPEL2 cysteine 87. Cysteines are marked with red. Identical residues are highlighted in dark gray, similar residues are in light gray.